Abstract
Centrosomes are the major sites for microtubule nucleation in mammalian cells, although both chromatin- and kinetochore-mediated microtubule nucleation have been observed during spindle assembly. As yet, it is still unclear whether these pathways are coregulated, and the molecular requirements for microtubule nucleation at kinetochore are not fully understood. This work demonstrates that kinetochores are initial sites for microtubule nucleation during spindle reassembly after nocodazole. This process requires local RanGTP accumulation concomitant with delocalization from kinetochores of the hydrolysis factor RanGAP1. Kinetochore-driven microtubule nucleation is also activated after cold-induced microtubule disassembly when centrosome nucleation is impaired, e.g., after Polo-like kinase 1 depletion, indicating that dominant centrosome activity normally masks the kinetochore-driven pathway. In cells with unperturbed centrosome nucleation, defective RanGAP1 recruitment at kinetochores after treatment with the Crm1 inhibitor leptomycin B activates kinetochore microtubule nucleation after cold. Finally, nascent microtubules associate with the RanGTP-regulated microtubule-stabilizing protein HURP in both cold- and nocodazole-treated cells. These data support a model for spindle assembly in which RanGTP-dependent abundance of nucleation/stabilization factors at centrosomes and kinetochores orchestrates the contribution of the two spindle assembly pathways in mammalian cells. The complex of RanGTP, the export receptor Crm1, and nuclear export signal-bearing proteins regulates microtubule nucleation at kinetochores.
INTRODUCTION
It is now clear that mitotic spindle assembly can occur via different pathways (for reviews, see Gadde and Heald 2004; Wadsworth and Khodjakov, 2004; Rieder, 2005, O'Connel and Khodjakov, 2007). In most somatic cell types, centrosomes nucleate microtubules (MTs) to build a mitotic spindle. Functional spindles also form in cells lacking centrosomes, e.g., in higher plants and some animal oocytes. In Xenopus eggs, spindles assemble through MT nucleation from chromatin in a process requiring the guanosine triphosphatase (GTPase) Ran (Carazo-Salas et al., 1999; Kalab et al., 1999) which, in its GTP-bound form, is generated near chromatin under the localized action of the nucleotide exchange factor RCC1 (Moore et al., 2002; Li et al., 2003; Chen et al., 2007). In the Xenopus system, RanGTP releases spindle assembly factors containing nuclear localization signals (NLSs), such as NuMa and TPX2 (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001), from the inhibitory binding to importin α/β, in the immediate vicinity of chromatin, thus promoting nucleation of MTs therein (reviewed by Weis, 2003; Ciciarello et al., 2007). This has led to propose that spindle formation in acentrosomal cells depends on a gradient of RanGTP, concentrating around chromosomes and diluting away from them (Caudron et al., 2005; Kalab et al., 2006). Local activity of RanGTP and importin α/β also contributes to spindle assembly in centrosome-driven cells (Nachury et al., 2001; Ciciarello et al., 2004, Kalab et al., 2006). In these cells, chromatin-associated MT nucleation is normally suppressed by the dominant centrosome activity, yet there remains an intrinsic potential that can be resumed under certain circumstances, as a functional spindle still forms when centrosomes are removed by laser ablation (Khodjakov et al., 2000), or centrosome activity is abolished by specific mutations (Bonaccorsi et al., 1998).
Kinetochores were also reported to direct MT nucleation in mammalian cells after spindle-disrupting drugs, but this idea was subsequently questioned, based on concerns for possible artifactual effects of the drugs (Rieder, 2005, and references therein). More recently, in vivo imaging of mitosis has shown that unattached kinetochores facing away from centrosomes can promote the formation of MTs during mitotic spindle assembly in both mammalian (Khodjakov et al., 2003) and Drosophila cells (Maiato et al., 2004). In both cases, chromosome arms shielded the distal kinetochore from interaction with microtubules generated at centrosomes, possibly providing time for kinetochore nucleation to occur. However, the importance of this kinetochore-driven MT nucleation for spindle assembly in centrosome-containing cells is not clearly defined, as only a few examples were recorded in the live cell studies (Khodjakov et al., 2003; Maiato et al., 2004). Furthermore, the molecular requirements for MT formation at kinetochore are not fully understood. Recently, Tulu et al. (2006) observed microtubule formation at Bub1-positive sites after release from nocodazole (NOC)-induced spindle disassembly in live cell imaging experiments (Tulu et al., 2006). Formation of MT foci in the chromatin region was severely inhibited when the activity of the Ran-dependent spindle assembly factor TPX2 was suppressed by either microinjection of excess importin β, or by direct TPX2 depletion (Tulu et al., 2006). These results suggest that the dissociation of import complexes entrapping NLS-bearing spindle assembly factors contributes to the genesis of MT foci. However, the limited resolution of current live cell imaging methods did not allow discriminating whether the inhibition of extracentrosomal MT foci reflected the suppression of RanGTP-dependent MT nucleation specifically at kinetochores, or, rather a disruption of the overall RanGTP gradient, which is generated along the entire chromosome region by the persisting binding of RCC1 to mitotic chromatin. Furthermore, it remains to be determined what localizes extracentrosomal MT nucleation to the kinetochore area, given that the exchange factor for Ran is distributed along the entire length of the chromosome. We have readdressed these questions by assessing mitotic spindle reassembly after microtubule depolymerization by nocodazole or after cold-induced spindle disassembly in cells with impaired centrosome-nucleating activity. We now report that the localized accumulation of RanGTP at kinetochores is responsible for MT nucleation therein. The complex between RanGTP, the export receptor Crm1 and NES-bearing proteins modulates MT nucleation at kinetochores.
MATERIALS AND METHODS
Cell Cultures and Treatments
Human lung diploid fibroblasts (MRC-5 cells) and human osteosarcoma U2OS cells were obtained from the American Type Culture Collection (Manassas, VA), and they were maintained in minimal essential medium supplemented with 10% fetal calf serum, antibiotics, l-glutamine, nonessential amino acids, and HEPES. Cells were grown at 37°C, in a humidified atmosphere, with 5% CO2. MRC5 or U2OS cultures were treated with 1.5 μM NOC for 16 h to completely disassembly MTs. tsBN2 cells (a gift from M. Dasso, NICHD/NIH, Bethesda, MD) were grown at 32°C, and they received 4 μM NOC for 3 h either at 32 or at 39°C. At the end of the NOC incubation, cells were washed three times in prewarmed medium, reincubated in fresh medium to release the mitotic block, and fixed at the indicated times after NOC release. For MT regrowth after ice-induced depolymerization, U2OS cells were incubated for 60 min on ice, prewarmed media was then added, and cells were incubated at 37°C for the indicated times. When needed, 20 nM leptomycin B was added to culture medium 2 h before incubation on ice.
RNA Interference Experiments
Small interfering RNA (siRNA) oligonucleotides targeting Polo-like kinase 1 (Plk1) or firefly luciferase gene sequences (GL2) were from QIAGEN (Hilden, Germany). siRNAs were mostly used at the final concentration of 80 nM using Oligofectamine (Invitrogen, Carlsbad, CA) as transfection reagent. siRNA sequences and conditions of interference were described previously (De Luca et al., 2006). All analyses were performed 40 h from transfection.
Immunoblotting
An equal number of cells from tsBN2 cultures grown to 85% confluence at 32°C, or shifted to 39°C during the last 4 h of culture, was pelleted by centrifugation for 10 min at 1500 rpm, and then they were washed in phosphate-buffered saline (PBS). Cells were extracted with radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1%, NP-40, 1 mM EGTA, and 0.25% sodium deoxycholate) supplemented with 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 1 mM NaVO4 and 1 mM NaF. Proteins were resolved by electrophoresis on a 10% SDS-polyacrylamide gel, and then they were transferred to a nitrocellulose membrane (Protran; Whatman Schleicher and Schuell, Keene, NH) by using a semidry blotting system (Bio-Rad, Hercules, CA) in transfer buffer (48 mM Tris, 39 mM glycine, 0037% SDS, and 20% methanol). Sixty micrograms of extract per lane was loaded. Blocking and antibody incubations were in 5% low-fat dry milk in Tris-buffered saline (10 mM Tris-HCl, pH 8.0, and 150 mM NaCl) containing 0.1% Tween 20. Membranes were blocked for 30 min, primary antibodies were incubated for 1 h and secondary antibodies for 30 min at room temperature. Primary antibodies were anti-RCC1 (0.5 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA) and anti-RanBP1 (0.5 μg/ml; Santa Cruz Biotechnology). U2OS cells were lysed, resolved, and transferred under similar conditions. Forty micrograms of extract per lane was loaded. Primary antibodies were mouse anti-α-tubulin (ascite fluid, clone B-5-1-2, 1:2000; Sigma-Aldrich, St. Louis, MO) and mouse anti-Plk1 (ab 14210, 1 μg/ml; Abcam, Cambridge, United Kingdom).
Immunostaining
Cells were rinsed in PHEM buffer [60 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 25 mM HEPES, pH 6.9, 10 mM EGTA, and 4 mM MgSO4], and then they were lysed for 45 s in 0.1% Triton-X in PHEM buffer (MRC-5 and U2OS) or for 5 min in 0.5% Triton-X (tsBN2). Cells were fixed for 10 min in 3.7% formaldehyde in PHEM, and then they were rinsed in PBS and postfixed in ice-cold MetOH for 5 min. After fixation, cells were washed, blocked for 30 min in 20% goat serum at room temperature, and incubated at 37°C for 1 h with primary antibodies diluted in 5% goat serum. Primary antibodies were anti-α-tubulin and anti-γ-tubulin (Sigma-Aldrich) diluted 1:300 and 1:2000, respectively; CREST serum (1:50; Antibodies Incorporated, Davis, CA), anti-NuMA (1:50; Calbiochem, San Diego, CA), anti-p150Glued (1:100; BD Biosciences, San Jose, CA), anti-Plk1(14210 and 14209, ABCAM, 1:100), anti-IAK1/Aurora-A (clone 4, 1:1000; BD Biosciences, San Jose, CA) anti-α-tubulin fluorescein isothiocyanate conjugate (1:100; Sigma-Aldrich), and anti-RanGAP1 (1:200; Santa Cruz Biotechnology). Anti-Kif2a (1:1000), anti-centrin 2 (1:1125), anti-HURP (1:300), and anti-RanGTP (AR12; 1:50) were kindly provided by D. Compton (Dartmouth Medical School, Hanover, NH), J. Salisbury (Mayo Clinic Foundation, Rochester, MN), E. A. Nigg (Max Planck Institute of Biochemistry, Martinsried, Germany), H. Sillje (Max Planck Institute of Biochemistry), and I. Macara (University of Virginia School of Medicine, Charlottesville, VA), respectively. DNA was counterstained in 0.1 μg/ml 4,6-diamidino-2-phenylindole. For RanGTP staining (Tedeschi et al., 2007), cells were pretreated 1 min in 1% Triton-X in PHEM buffer, incubated with AR12 antibody in PHEM buffer containing NOC during the final 30 min of the NOC treatment and then fixed immediately as described above or released in fresh medium before fixation to analyze MT regrowth.
Fluorescence Microscopy and Image Acquisition
All preparations were examined under an Olympus Vanox microscope equipped with a 100× (1.35 numerical aperture) oil immersion objective and a SPOT charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI). Color-encoded images were acquired using ISO 2000 software (Deltasistemi, Rome, Italy), and they were processed with Adobe Photoshop 7 software (Adobe Systems, Mountain View, CA). Three-dimensional deconvolution and reconstruction was performed on 0.3-μm Z-serial optical sections by using AutoDeblur 9.3 (AutoQuant Imaging, Troy, NY) image processing program.
RESULTS
MT Nucleation Restarts at Kinetochores after NOC
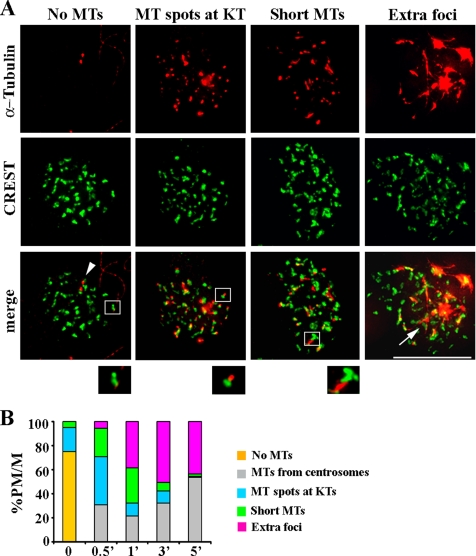
To dissect the relative contribution of chromatin- and kinetochore-mediated MT nucleation during spindle assembly in mammalian cells, we took advantage of the reversibility of NOC-induced MT disassembly and visualized kinetochores at the very early stages of spindle reassembly. Cells were first treated with NOC to completely disassemble MTs. The drug was then carefully removed, and the initial stages of spindle reassembly were followed in a time course analysis. The experiments were performed in human fibroblast MRC-5 cells, but similar results were obtained in other mammalian cell lines, such as osteosarcoma cells (U2OS, Supplemental Figure 1). Combined α-tubulin and kinetochore staining enabled us to unambiguously identify the generation of MTs at kinetochores at very early times after NOC removal, in the form of α-tubulin–positive spots at kinetochores (Figure 1A, MT spots at KTs), short MTs emanating from kinetochores (Figure 1A, Short MTs), or supernumerary tubulin foci at the center of a group of kinetochores (Figure 1A, Extra foci). Thirty seconds after NOC release, ∼70% of cells exhibited evidence of MT nucleation at kinetochores (Figure 1B), demonstrating that MT nucleation at kinetochores is the predominant pathway for MT formation immediately after NOC removal. Over time, the frequency of cells showing kinetochore spots and short kinetochore MTs decreased, with a proportional increase in the frequency of cells with extra foci (Figure 1B), suggesting that kinetochore-nucleated MTs are then organized at their minus ends into pole-like structures. This idea is also supported by the localization to these supernumerary tubulin foci of several noncentrosomal MT minus end-associated proteins with MT-focusing activity (Supplemental Figure 2). The frequency of cells showing only centrosome-based MT arrays increased at later times (Figure 1B), indicating that extracentrosomal foci generated at early times during spindle reassembly are then captured by centrosomal MTs to form a bipolar spindle. Thus, our detailed analysis of MT formation in conjunction with the visualization of the CREST kinetochore marker enabled us to unambiguously identify the generation of MT at kinetochores as the initial moment in MT regrowth in cells recovering from spindle disassembly. This information was still lacking in previous live cell studies (Tulu et al., 2006) that left out the possibility that association of MT to kinetochores was successive to MT nucleation around chromatin. These results, together with those of Tulu et al. (2006), identify kinetochores as the initial sites for MT nucleation after NOC-induced spindle reassembly.
Figure 1.
Chromatin-mediated MT assembly after NOC treatment originates at kinetochores. (A) Kinetochore localization of MT regrowth at early times from NOC release as visualized by CREST (green) and anti-tubulin (red) staining in MRC-5 cells. Representative images of different nucleation patterns are shown. No MTs, no α-tubulin staining at kinetochores, the two α-tubulin spots decorate the centrosomes (arrowhead). MT spots at KTs, kinetochores showing α-tubulin–positive spots. Short MTs, short MTs emanating from kinetochores. Extra foci, supernumerary α-tubulin foci at the center of several kinetochores (arrow). Insets show a twofold magnification of the boxed regions. Bar, 10 μm. (B) Percentages of prometaphase/metaphase cells (PM/M) with different MT nucleation patterns at the indicated minutes from NOC release. Categories are as described in A. One hundred to 250 mitoses in three independent experiments were counted for each time point.
Localized Accumulation of RanGTP at Kinetochores Promotes MT Nucleation
To get some insight into the molecular requirements for MT nucleation at kinetochores, we analyzed several regulators of microtubule nucleation during spindle reassembly after NOC. We initially wondered whether NOC displaced regulators of MT nucleation from centrosomes to chromosomes and whether that in turn promoted kinetochore-dependent MT nucleation. We found that both Aurora-A and Plk1 kinases maintained their localization to centrosomes (Aurora-A, Plk1) and kinetochores (Plk1), with no variation during NOC treatment (Supplemental Figure 3), ruling out the possibility that NOC impaired the association of these crucial regulators with centrosomes and that this in turn activated nucleation from kinetochores.
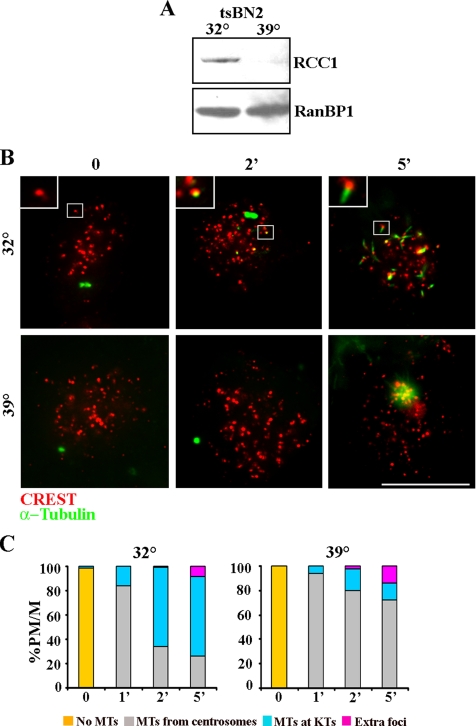
We next investigated Ran, which is clearly required for MT nucleation from chromatin in oocytes and in mammalian cells (reviewed in Ciciarello et al., 2007), and it has been implicated in the kinetochore pathway after NOC (Tulu et al., 2006). To directly investigate the requirement for RanGTP in the kinetochore nucleation pathway, we made use of tsBN2 cells, a baby hamster kidney-derived conditional cell line harboring a mutant version of the chromatin-bound exchange factor RCC1 that is degraded at the nonpermissive temperature (Ohtsubo et al., 1989). Formation of MTs at kinetochores was investigated in tsBN2 cells at the permissive and restrictive temperatures, by combining kinetochore and α-tubulin antibody staining at very early times from NOC release (Figure 2. When NOC release took place at 32°C, when RCC1 is functional (Figure 2A), MT nucleation was activated at kinetochores (Figure 2B, 32°C). The fraction of cells showing MT regrowth at kinetochores increased from 1 to 5 min after NOC release (Figure 2C, 32°C), indicating that MT nucleation from kinetochores greatly contributes to the reformation of the mitotic spindle after NOC release when RanGTP is present. On the contrary, MT nucleation from kinetochores was suppressed at the restrictive temperature for RCC1 activity in the majority of the cells that exhibited MT growing from asters only (Figure 2, B and C, 39°C). This demonstrated that RanGTP production is required for MT nucleation at kinetochores, in addition to its role in nucleation around chromatin.
Figure 2.
MT nucleation at kinetochores depends on the GTPase Ran. (A) tsBN2 cells were grown for 3 h in permissive (32°C) and restrictive (39°C) conditions, and intracellular RCC1 content was analyzed by Western blotting. RCC1 is degraded at the restrictive temperature. RanBP1, loading control. (B) Representative images of MT regrowth in tsBN2 cells incubated with NOC for 3 h at the permissive (32°C) or restrictive (39°C) temperature and then released in NOC-free medium at the two temperatures for the indicated minutes. Nucleation from kinetochores is suppressed in repressive conditions. Insets show a threefold magnification of the boxed regions. Bar, 10 μm. (C) Percentages of prometaphase/metaphase cells (PM/M) with different MT nucleation patterns at the indicated minutes from NOC release. The difference in the frequencies of MTs at kinetochores between the two temperatures was highly significant (t test for 2 and 5 min, p < 0.01). At least 250 cells in three independent experiments were counted for each time point.
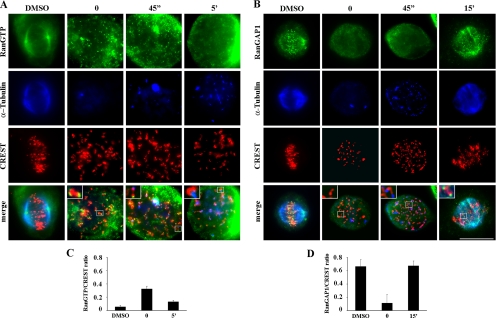
To further substantiate the requirement for RanGTP at kinetochores in MT nucleation, we decided to directly visualize the intracellular localization of RanGTP both during NOC exposure and after drug removal. This was achieved by use of a conformational antibody that specifically reacts with the GTP-bound form of Ran (Richards et al., 1995). That antibody had been previously used to reveal RanGTP at centrosomes and along spindle MTs in mitotic human cells (Keryer et al., 2003; Tedeschi et al., 2007). The specificity of the antibody for the GTP-bound form of Ran in mitosis was further demonstrated by the weak antibody reactivity on the mitotic spindle when the RCC1 exchange factor was inactivated in tsBN2 cells (Supplemental Figure 4). Next, we carried out a triple antibody staining against RanGTP, α-tubulin, and kinetochores in U2OS cells and it did indeed visualize spindle pole- and MT-associated RanGTP in untreated mitotic cells (Figure 3A, dimethyl sulfoxide [DMSO]). When MTs were completely depolymerized in NOC-treated cells, RanGTP did abundantly accumulate on kinetochores, as shown by its localization external to CREST signals (Figure 3A, time 0; see inset). Interestingly, the RanGTP signal was depicted on kinetochores immediately after release from NOC treatment, i.e., before MTs reformed (Figure 3A, time 0). RanGTP localization to kinetochores was still seen at early stages of MT formation at kinetochores, when short MTs appeared from kinetochores (Figure 3A, 45″ release); at later release times, when MTs emanated from most kinetochores, the RanGTP signal still persisted at kinetochores although with a clearly decreased intensity (Figure 3A, 5′ release). Measuring the RanGTP signal intensity and normalizing to the CREST signal confirmed the partial loss of RanGTP from kinetochores at late recovery time (0.32 ± 0.04 vs. 0.13 ± 0.02 at 0 and 5′ recovery, respectively; >40 centromeres from six to eight cells per experimental point; Figure 3C). The temporal pattern of RanGTP at kinetochores suggests that RanGTP accumulation is locally required before the onset of MT nucleation at kinetochores, transiently persists at kinetochores in the earliest stages of MT formation, and it is eventually followed by dissociation from kinetochores and/or hydrolysis therein.
Figure 3.
Enrichment of RanGTP at kinetochores mediates kinetochore MT nucleation. (A) Analysis of RanGTP localization (RanGTP) relative to MTs (α-tubulin) and kinetochores (CREST) in control metaphase cells (DMSO) and in MT-depleted U2OS prometaphases at the end of the NOC treatment (0) or at different times from NOC washout (45″ and 5′). RanGTP accumulates at kinetochores before MTs form and its concentration decreases successively to MT regrowth. (B) Analysis of RanGAP1 localization (RanGAP1) relative to kinetochores (CREST) and MTs (α-tubulin) in control metaphase cells (DMSO) and in MT-depleted prometaphases at the end of the NOC treatment (0) or at different times from NOC washout (45″ and 15′). RanGAP1 is absent from kinetochores after NOC and relocalizes to kinetochores when centrosome-based MTs reach kinetochores. Insets show a threefold magnification of the boxed regions. Bar, 10 μm. (C) Quantification of RanGTP staining intensity at kinetochores normalized for CREST staining (mean ± SE; n > 40 centromeres from 4 to 8 cells per experimental point). (D) Quantification of RanGAP1 staining intensity at kinetochores normalized for CREST staining (mean ± SE; n > 40 centromeres from 4 to 5 cells per experimental point).
RanGTP levels are regulated by the counteracting activities of the chromatin-associated nucleotide exchange factor RCC1 and the cytoplasmic GTP-hydrolyzing factor RanGAP1. During mitosis, RanGAP1 is targeted at kinetochores in a SUMO-conjugated form together with the Ran-binding protein RanBP2, which has SUMO-ligase activity (Joseph et al., 2002). RanGAP1/RanBP2 recruitment at kinetochores is stabilized by its association in a complex containing RanGTP and Crm1, the export receptor for NES-bearing cargoes (Arnaoutov et al., 2005).
In light of the ability of RanGAP1 to activate GTP hydrolysis on Ran and to localize at kinetochores in mitosis, we reasoned that the kinetochore accumulation of RanGTP after NOC might have been associated with a decrease in RanGAP1 concentration therein. In metaphase cells from control cultures, RanGAP1clearly localized at kinetochores (Figure 3B, DMSO), as reported previously (Joseph et al., 2002). In contrast with this defined localization pattern, no preferential RanGAP1 accumulation was observed during NOC exposure, such that the GTP-hydrolysis factor was absent from kinetochores both after MT depolymerization (Figure 3B, time 0, see inset; quantification in Figure 3D) and when MTs began to reform at kinetochores immediately after NOC release (Figure 3B, 45″). RanGAP1 was again observed at kinetochores after several minutes of NOC release, when plus ends of MTs anchored at the centrosome reached kinetochores in prometaphase/metaphase cells (Figure 3B, 15′ and Figure 3D). On the whole, these findings identify the accumulation of the GTP-bound form of Ran as a prerequisite for MT nucleation at kinetochores and demonstrate that NOC-dependent delocalization of RanGAP1 and accumulation of RanGTP is responsible for this localized nucleation.
Centrosome Impairment Stimulates Kinetochore Nucleation
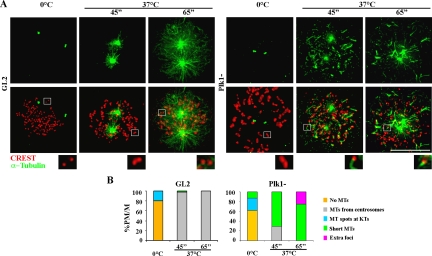
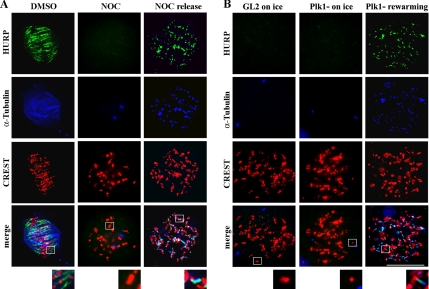
We next asked whether mammalian cells can activate MT nucleation from kinetochores independently of MT-targeting drug treatments. Plk1 is a major centrosomal kinase with key roles in centrosome maturation and spindle assembly (Barr et al., 2004); its inactivation impairs γ-tubulin recruitment at centrosomes (Sumara et al., 2004; van Vugt et al., 2004), producing defective MT regrowth after cold treatment in U2OS cells (De Luca et al., 2006). We therefore used Plk1-specific siRNA to down-regulate the nucleating capacity of centrosomes and to assess whether the kinetochore-associated MT nucleation pathway would be favored. We interfered U2OS cells with control GL2 or Plk1-specific siRNAs, and we verified Plk1 depletion by Western blotting (Supplemental Figure 5). Then, we examined MT regrowth after cold-induced depolymerization in Plk1- and in GL2-interfered U2OS cells shortly after rewarming at 37°C. In GL2-interfered cells, MTs polymerized exclusively from centrosomes upon rewarming (Figure 4, A and B; GL2). On the contrary, in Plk1-depleted cells at 37°C, some MT nucleation was observed around centrosomes, but short MTs also appeared at and between kinetochores (Figure 4A, Plk1− 45″); these MTs further increased in length with time (Figure 4A, Plk1− 65″). MT nucleation was efficiently activated in Plk1-depleted cells, because MTs were observed at all kinetochores and all Plk1-depleted cells exhibited both centrosome-dependent and -independent MT nucleation at 65″ rewarming (Figure 4B); thus, impairing the MT-nucleation ability of centrosomes in Plk1-depleted cells strongly stimulates MT formation at kinetochores. Accordingly, the capacity of centrosomes to develop a nucleating aster was decreased in Plk1-silenced compared with GL2 cells (Figure 4A, 65″, compare GL2 vs. Plk1−). However, kinetochore-nucleated MTs do not elongate much because their minus ends are captured by MTs nucleated by neighbor kinetochores (Figure 4A, 65″, compare microtubule length in centrosome-nucleated MTs in GL2 cells and kinetochore-nucleated MTs in Plk1− cells). These results suggest that nucleation from centrosomes and kinetochores are negatively interrelated, providing new ground for the hypothesis that the two pathways compete for a common supply of tubulin dimers. During a normal mitosis, the high rate of microtubule growth at nucleating asters may depress the capacity of kinetochores to efficiently form MTs.
Figure 4.
MT regrowth from kinetochores after ice-induced MT disassembly is stimulated in centrosome impaired cells. (A) MT depolymerization on ice (0°C) and MT regrowth at different times (45″ and 65″) after rewarming at 37°C in control (GL2) and Plk1-silenced (Plk1−) U2OS cells. Only centrosomes associate residual tubulin as visualized by anti-tubulin staining (green) at 0°C in both GL2 and Plk1− cells. During MT regrowth centrosomes develop large nucleating asters in GL2 cells, whereas tubulin (green) is observed at kinetochores (red) in PLK1-depleted cells. In these cells, the nucleating capacity of centrosomes is defective. Insets show a threefold magnification of the boxed regions. Bar, 10 μm (B) Percentages of prometaphase/metaphase cells (PM/M) with different MT nucleation patterns at the indicated times from NOC release in GL2- or Plk1-silenced cells. The different categories are described in Figure 1A. At least 100 mitoses were counted for each time point.
Crm1/RanGTP/RanGAP1 Complex Regulates MT Nucleation at Kinetochores
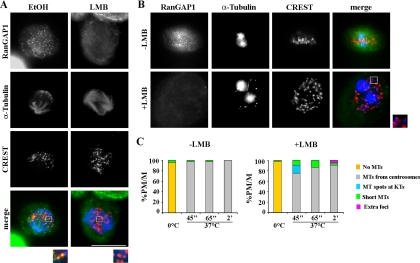
Mitotic RanGTP acts through its interaction with two major effectors: importin β, which regulates NLS-containing spindle assembly factors, and the export receptor for NES proteins, Crm1. In association with Crm1, RanGTP is required to assemble a ternary complex with NES factors (Fornerod et al., 1997), including RanGAP1-RanBP2 (Arnaoutov et al., 2005). In mitotic cells, the Crm1/RanGTP/RanGAP1-RanBP2 complex localizes at kinetochores, where it contributes to regulate the spindle checkpoint activity (Arnaoutov and Dasso, 2003) and K-fiber stability (Arnaoutov et al., 2005). Given that NOC-induced nucleation from kinetochores delocalized RanGAP1, we wondered whether the kinetochore-associated Crm1/RanGTP/RanGAP1-RanBP2 ternary complex also regulates MT nucleation from kinetochores. To address this question, we performed cold-induced depolymerization and MT regrowth assays in the presence of leptomycin B (LMB), a Crm1-specific drug that has been shown to inhibit the formation of the ternary complex among Crm1, RanGTP, and NES proteins (Fornerod et al., 1997; Petosa et al., 2004). We found that RanGAP1 was displaced from kinetochores of U2OS cells after 3 h of LMB treatment (Figure 5A), consistent with a requirement of CRM1 and RanGTP for stable recruitment of RanGAP1 to kinetochores (Arnaoutov et al., 2005). When U2OS cells were exposed to cold and then rewarmed to allow MT depolymerization and regrowth, MTs appeared at centrosomes (Figure 5, B and C, −LMB), consistent with the centrosomal pathway predominantly activated after ice. When cultures were treated with LMB to inhibit RanGAP1 localization at kinetochores before ice incubation, and then MTs were depolymerized by cold treatment, a fraction of cells activated MT nucleation from kinetochores as early as 45″ after rewarming, and subsequently they formed extracentrosomal α-tubulin foci (Figure 5, B and C, +LMB). Accordingly, in these cells RanGAP1 was undetectable on kinetochores during MT regrowth (Figure 5B, +LMB). Interestingly, kinetochore nucleation was achieved in the presence of an intact MT assembly activity of the centrosomes, which developed large nucleating asters upon rewarming (Figure 5B, +LMB). This set of data demonstrates that the Crm1/RanGTP/RanGAP1 complex at kinetochores suppresses kinetochore MT nucleation during spindle assembly in somatic cells and that inhibiting the formation of this complex can reactivate this spindle assembly pathway.
Figure 5.
Disruption of the Crm1/RanGTP/RanGAP1-RanBP2 complex by LMB promotes MT nucleation at kinetochores. (A) RanGAP1(RanGAP, green), MT (α-tubulin, blue) and kinetochore (CREST, red) localization in a metaphase U2OS cell from an asynchronously growing culture incubated for 3 h in the LMB solvent (ethanol [EtOH]) or in a cell incubated for 3 h in 20 nM LMB. Bar, 10 μm. (B) MT regrowth after rewarming U2OS cells from cold-induced MT disassembly (−LMB) or after rewarming LMB pretreated U2OS cells (+LMB). Note the absence of RanGAP1 on kinetochores in LMB-treated cells. MTs (blue) are observed at and between kinetochores (red). Insets show a threefold magnification of the boxed regions in the merge. (C) Percentages of prometaphase/metaphase cells (PM/M) with different MT nucleation patterns in EtOH− and LMB-treated cells after cold induced MT disassembly (0°C) or after the indicated times at 37°C.
HURP Is Required for MT Stabilization at Kinetochores
Having demonstrated that a localized accumulation of RanGTP is required to activate MT nucleation from kinetochores, we then moved to search RanGTP-regulated factors that may act as downstream effectors of RanGTP function in kinetochore nucleation. Most RanGTP regulated MT-nucleating factors, and notably TPX2, are transported by minus-end–directed motors toward centrosomes, where they are activated by RanGTP association with importin β and consequent release of nucleating factors from importin β/α inhibitory activity. HURP (hepatoma up-regulated protein) is a Ran-regulated factor that localizes along kinetochore microtubules in the vicinity of chromosomes (Koffa et al., 2006; Wong and Fang, 2006) in a RanGTP-dependent manner (Sillje et al., 2006). In accordance with its potential role as downstream effector for RanGTP in the vicinity of chromatin, HURP has been shown to stabilize kinetochore MTs and contribute to MT stabilization of both centrosome- and chromatin-nucleated MTs (Koffa et al., 2006; Sillje et al., 2006; Wong and Fang, 2006). This prompted us to investigate whether HURP was also involved in regulating kinetochore MT nucleation during MT regrowth both after NOC and after cold-induced MT disassembly. When MTs were depolymerized by NOC, HURP was diffuse in the mitotic nucleocytoplasm (Figure 6A, NOC), consistent with this protein being a MT-binding protein (Sillje et al., 2006). HURP, however, clearly colocalized with tubulin at kinetochores after NOC release, both on kinetochore tubulin spots and on short MTs in between kinetochores (Figure 6A, NOC release; see inset). Similarly, kinetochores were devoid of tubulin and HURP after cold-induced MT depolymerization in both GL2- and Plk1-depleted cells (Figure 6B, GL2 on ice, Plk1-on ice), but HURP concentrated along short MTs protruding from kinetochores in Plk1-silenced cells during rewarming, with a clear accumulation at the kinetochore–MT interface (Figure 6B, Plk1−, rewarming, inset). These results demonstrate that nucleation of MTs from kinetochores is promptly followed by association of HURP to the nascent MTs. They further suggest that MT plus ends are localized at kinetochores and that they are stabilized by HURP association.
Figure 6.
The Ran-regulated MT-binding protein HURP stabilizes kinetochore-nucleated MTs. (A) HURP (HURP) localization relative to MTs (α-tubulin) and kinetochores (CREST) in a control metaphase cell (DMSO) and in MT-depleted U2OS prometaphases at the end of the NOC treatment (0) or at different times from NOC washout (45″ and 5′). (B) HURP localization relative to MTs (α-tubulin) and kinetochores (CREST) in a GL2-interfered prometaphase cell after ice-induced MT depolymerization (GL2 on ice) or in Plk1-interfered prometaphase cells after ice (Plk1− on ice) or during MT regrowth at 37°C (Plk1− rewarming). Bar, 10 μm.
DISCUSSION
The present work demonstrates that the Ran GTPase network regulates the formation of MTs from kinetochores in mammalian cells. Our study uncovers a specific mechanism that regulates RanGTP production at kinetochores and that is responsible for the formation of MTs from kinetochores. Analysis of MT regrowth in tsBN2 cells demonstrated that RanGTP production specifically at kinetochores, in addition to its formation around chromatin, is required for extracentrosomal MT nucleation in mammalian cells. In addition, the use of a RanGTP-specific antibody after MT disassembly identified the accumulation of the GTP-bound form of Ran as a prerequisite for MT nucleation at kinetochores. Thereafter, nucleation at kinetochores was followed by the association to the kinetochore-nucleated MTs of the Ran-regulated protein HURP, which stabilized their growth.
Recent work has demonstrated that a fraction of Ran localizes during mitosis to kinetochores in a complex with the export receptor Crm1 and the RanGAP1/RanBP2 subcomplex; this complex is proposed to work in an autoregulatory loop in which Crm1, in association with RanGTP, is required to localize the RanGAP1-RanBP2 subcomplex, which in turn promotes RanGTP hydrolysis and therefore catalyzes its own release from the kinetochores (reviewed by Arnaoutov and Dasso, 2005). The effects of disrupting this autoregulatory loop, by either RCC1 inactivation, or by LMB treatment, indicate that the complex functions in maintaining the spindle assembly checkpoint active (Arnaoutov and Dasso, 2003) and in the formation of mature kinetochore fibers in metaphase (Arnaoutov et al., 2005). The present data extend the range of events controlled by the kinetochore-associated Ran network. We have detected GTP-bound Ran at kinetochores before the onset of MT nucleation from these sites in NOC-treated cells. The absence of MTs inhibits RanGAP1 accumulation at kinetochores (Joseph et al., 2002; this study), providing a mechanism whereby the self-regulatory loop that normally regulates nucleotide turnover on Ran (Arnaoutov and Dasso, 2005) is disrupted, and RanGTP is allowed to accumulate at kinetochores. Inhibition of Crm1 by LMB did also result in the failure to localize RanGAP1 to kinetochores and in the activation of MT nucleation in conditions that otherwise would not allow nucleation from kinetochores, i.e., after cold. These results demonstrate that the RanGTP/Crm1/RanGAP1 complex controls MT formation from kinetochores: an unbalance within the self-regulatory loop toward RanGTP accumulation, either by NOC treatment or through LMB exposure, promotes kinetochore nucleation. Finally, we also show that GTP hydrolysis on Ran does not take place during kinetochore-driven MT nucleation, because RanGAP1, the hydrolysis factor for RanGTP, relocalizes to kinetochores only after K fibers are present. The time course analysis with conformational antibody reported here shows that RanGTP is present at kinetochores during the initial stages of MT regrowth, when kinetochore-mediated MT nucleation is highly active, but then its concentration gradually decreases, suggesting that it possibly dissociates from the kinetochores after activation of MT nucleation and becomes hydrolyzed.
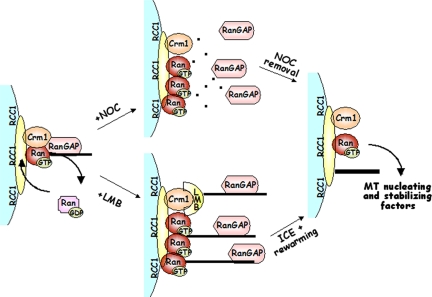
On the basis of our findings, we propose the following model for the role of the Ran network in MT nucleation at kinetochores (Figure 7). Both Crm1 and Ran localize at kinetochores, where the latter is loaded with GTP by chromatin-bound RCC1; this establishes the potential for forming ternary complexes with NES-containing kinetochore proteins. When, in a normal mitosis, MTs capture kinetochores, RanGAP1 is loaded on kinetochores, and therein it can become part of the complex with RanGTP and Crm1: when this occurs, RanGTP hydrolysis is triggered in the complex, and RanGDP is released from the kinetochores (Figure 7, left). A similar autoregulatory Ran “loop,” based on RanGAP1 recruitment at kinetochores and subsequent RanGTP hydrolysis, regulates kinetochore fiber structure and spindle checkpoint timing at the meta- to anaphase transition (Arnaoutov and Dasso, 2005). In the absence of MTs, such as during NOC treatment, RanGAP1 fails to reach kinetochores, with a consequent continuous RanGTP accumulation therein, resulting in the activation of kinetochore MT nucleation (Figure 7, top middle). Crm1 inhibition also activates nucleation from kinetochores by preventing the formation of a stable complex with RanGTP and RanGAP1, thereby also yielding RanGTP accumulation at kinetochores (Figure 7, bottom middle). We propose that accumulated RanGTP at kinetochores promotes the localized release of nucleating and stabilizing factors (Figure 7, right). More than one pathway may be envisaged for activation of MT nucleation from kinetochores when spindle MTs are disrupted or centrosomal nucleation is weakened. First, high concentrations of RanGTP at kinetochores may release NLS-containing MT-regulatory factors from the “sequestering” effect of classical importin α/β complexes, as suggested in TPX2 (Tulu et al.,2006). Furthermore, kinetochore-associated RanGTP may override the inhibitory function exerted by importin β on HURP and/or other stabilizing factors, such as NuSAP, another MT-stabilizing and -bundling protein that is enriched at the central part of the spindle (Ribbeck et al., 2007); such factors would then contribute to MT formation/stabilization at kinetochores. Finally, the kinetochore-associated Crm1 fraction, in concert with RanGTP, may recruit NES-containing proteins therein; some of these NES proteins may have nucleating activity and contribute to kinetochore-activated MT nucleation. The relative importance of these different mechanisms remains to be clarified.
Figure 7.
RanGTP accumulation promotes MT nucleation at kinetochores. Left, in a regular mitosis, RanGTP levels are controlled by the Crm1/RanGTP/RanGAP1-RanBP2 complex that localizes RANGAP1 to kinetochores, where the hydrolysis factor promotes RanGDP formation. Middle, RanGTP accumulates at kinetochores when RanGAP1 does not localize to kinetochores due to NOC-induced MT depolymerization or LMB-mediated inhibition of Crm1. Right, high RanGTP concentrations at kinetochores release spindle assembly and MT-stabilizing factors (e.g., HURP) there. See text for details.
The present work also provides the first evidence for activation of the pathway of kinetochore-driven MT nucleation in response to impaired centrosomal function, such as after Plk1 depletion. This result suggests that during a normal mitosis, the high rate of MT growth at nucleating asters may depress the capacity of kinetochores to efficiently form MTs. Centrosome-nucleating activity may subtract tubulin from the cellular pool, or sequester nucleating/stabilizing factors around the asters. The partial activation of the kinetochore pathway after LMB treatment underscores the dynamic nature of the activation process at the level of single kinetochores. The activation of MT growth is likely to be modulated both by a crucial level of RanGTP accumulation at the kinetochore, and by the intrinsic competition with the centrosome-mediated nucleation activity. Our findings support the idea that kinetochore-mediated MT growth contributes to a great extent to spindle formation when centrosomes are destroyed or inactive as well as in cells with acentrosomal spindles. In addition, kinetochore MT nucleation may be favored in cells with defective centrosome activity or imbalance in the RanGDP/RanGTP cycle.
In conclusion, our data indicate that in normal conditions, a continuous cycle of GTP exchange and hydrolysis takes place on the kinetochore-associated fraction of Ran, which limits MT nucleation and/or does not support the stabilization of nucleated MTs from kinetochores; in contrast, conditions in which the kinetochore RanGTP pool is stabilized activate kinetochore nucleation. We therefore suggest that localized RanGTP abundance orchestrates the relative contribution of MT assembly pathways operating in different subcellular localizations, i.e., centrosomes, chromatin, and kinetochores in mammalian cells. Our study adds a new function for the versatile protein Ran at kinetochores, by showing its role in fine-tuning the multiple paths of spindle formation.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Compton, M. Dasso, I. Macara, E. A. Nigg, J. Salisbury, and H. Sillje for the gift of antibodies and cell lines. This work was supported in part by grants from Associazione Italiana per la Ricerca sul Cancro (to P.L.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1050) on February 20, 2008.
REFERENCES
- Arnaoutov A., Dasso M. The Ran GTPase regulates kinetochore function. Dev. Cell. 2003;5:99–111. doi: 10.1016/s1534-5807(03)00194-1. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Dasso M. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle. 2005;4:1161–1165. doi: 10.4161/cc.4.9.1979. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Azuma Y., Ribbeck K., Joseph J., Boyarchuk Y., Karpova T., McNally J., Dasso M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 2005;7:626–632. doi: 10.1038/ncb1263. [DOI] [PubMed] [Google Scholar]
- Barr F. A., Silljé H., Nigg E. A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti M. G., Gatti M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J. Cell Biol. 1998;142:751–761. doi: 10.1083/jcb.142.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., Mattaj I. W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Caudron M., Bunt G., Bastiaens P., Karsenti E. Spatial coordination of spindle assembly by chromosome mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Ciciarello M., Mangiacasale R., Thibier C., Guarguaglini G., Marchetti E., Di Fiore B., Lavia P. Importin beta is transported to spindle poles during mitosis and regulates Ran-dependent spindle assembly factors in mammalian cells. J. Cell Sci. 2004;117:6511–6522. doi: 10.1242/jcs.01569. [DOI] [PubMed] [Google Scholar]
- Ciciarello M., Mangiacasale R., Lavia P. Spatial control of mitosis by the GTPase Ran. Cell Mol. Life Sci. 2007;64:1891–1914. doi: 10.1007/s00018-007-6568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Muratore T. L., Schaner-Tooley C. E., Shabanowitz J., Hunt D. F., Macara I. G. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M., Lavia P., Guarguaglini G. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle. 2006;5:296–303. doi: 10.4161/cc.5.3.2392. [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I. W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Gadde S., Heald R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Joseph J., Tan S. H., Karpova T. S., McNally J. G., Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pu R. T., Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kalab P., Pralle A., Isacoff E. Y., Heald R., Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Keryer G., Di Fiore B., Celati C., Lechtreck K. F., Mogensen M., Delouvée A., Lavia P., Bornens M., Tassin A. M. Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule organizing activity. Mol. Biol. Cell. 2003;14:4260–4271. doi: 10.1091/mbc.E02-11-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Cole R. W., Oakley B. R., Rieder C. L. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Copenagle L., Gordon M. B., Compton D. A., Kapoor T. M. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 2003;160:671–683. doi: 10.1083/jcb.200208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa M. D., Casanova C. M., Santarella R., Kocher T., Wilm M., Mattaj I. W. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Li H. Y., Wirtz D., Zheng Y. A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 2003;160:635–644. doi: 10.1083/jcb.200211004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Rieder C. L., Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W. J., Zhang C., Clarke P. R. Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr. Biol. 2002;12:1442–1447. doi: 10.1016/s0960-9822(02)01076-x. [DOI] [PubMed] [Google Scholar]
- Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- O'Connel C. B., Khodjakov A. L. Cooperative mechanisms of mitotic spindle formation. J. Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Okazaki H., Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petosa C., Schoehn G., Askjaer P., Bauer U., Moulin M., Steuerwald U., Soler-López M., Baudin F., Mattaj I. W., Müller C. W. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Raemaeckers T., Carmeliet G., Mattaj I. W. A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol. 2007;17:230–236. doi: 10.1016/j.cub.2006.11.050. [DOI] [PubMed] [Google Scholar]
- Richards S. A., Lounsbury K. M., Macara I. G. The C-terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. J. Biol. Chem. 1995;270:14405–14411. doi: 10.1074/jbc.270.24.14405. [DOI] [PubMed] [Google Scholar]
- Rieder C. L. Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to a draw. Chromosoma. 2005;114:310–318. doi: 10.1007/s00412-005-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje H. H., Nagel S., Korner R., Nigg E. A. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Sumara I., Giménez-Abian F. J., Gerlich D., Hirota T., Kraft C., de la Torre C., Ellenberg J., Peters J. M. Roles of Polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 2004;14:1712–1722. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Tedeschi A., Ciciarello M., Mangiacasale R., Roscioli E., Rensen W. M., Lavia P. RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells J. Cell Sci. 2007;120:3748–3761. doi: 10.1242/jcs.009308. [DOI] [PubMed] [Google Scholar]
- Tulu U. S., Fagerstrom C., Ferenz N. P., Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt M. A., van de Weerdt C.M.B., Vader G., Janssen H., Calafat J., Klompmaker R., Wolthuis M.F.R., Medema R. H. Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J. Biol. Chem. 2004;279:36841–36854. doi: 10.1074/jbc.M313681200. [DOI] [PubMed] [Google Scholar]
- Wadsworth P., Khodjakov A. E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol. 2004;14:413–419. doi: 10.1016/j.tcb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M. S., Adam S. A., Merdes A., Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wong J., Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 2006;173:879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.