Abstract
Throughout the cell cycle, the histones remain associated with DNA, but the repertoire of proteins associated with the chromatin fiber continuously changes. The chromatin interaction of HMGNs, a family of nucleosome binding proteins that modulates the structure and activity of chromatin, during the cell cycle is controversial. Immunofluorescence studies demonstrated that HMGNs are not associated with chromatin, whereas live cell imaging indicated that they are present in mitotic chromosomes.
To resolve this controversy, we examined the organization of wild-type and mutated HMGN1 and HMGN2 proteins in the cell nucleus by using immunofluorescence studies, live cell imaging, gel mobility shift assays, and bimolecular fluorescence complementation (BiFC). We find that during interphase, HMGNs bind specifically to nucleosomes and form homodimeric complexes that yield distinct BiFC signals. In metaphase, the nucleosomal binding domain of the protein is inactivated, and the proteins associate with chromatin with low affinity as monomers, and they do not form specific complexes. Our studies demonstrate that the mode of binding of HMGNs to chromatin is cell cycle dependent.
INTRODUCTION
Proper progression through the cell cycle is crucial for cell survival and for the correct transmission of genetic information to newly formed progeny cells. Passage through the various stages of the cell cycle involves major rearrangement in the structure and activity of the chromatin fiber. As the cell enters the mitotic phase, the chromatin becomes highly condensed and most of the transcriptional activity is inhibited (Gottesfeld and Forbes, 1997; Li et al., 1998; Belmont, 2006). These processes are associated with changes in the posttranslational modifications in the amino terminal tails of the core histones (Cheung et al., 2000; Kouskouti and Talianidis, 2005; Bonenfant et al., 2007), altered nucleosomal organization in the promoter of certain genes (Komura and Ono, 2005), and alterations in the intracellular organization of chromatin binding proteins and transcription factors (Martinez-Balbas et al., 1995; Hock et al., 1998; He and Davie, 2006). Whereas proteins necessary for chromatin condensation and mitotic chromosome formation localize to the mitotic chromosome, some but not all transcription factors and chromatin regulatory proteins are displaced from mitotic chromatin and are dispersed throughout the mitotic cell (Isackson et al., 1980; Hock et al., 1998; Zaidi et al., 2003; Belmont, 2006). Although the relocation of chromatin binding protein is an integral part of the mitotic condensation, this process has not been studied in detail. It seems that the displacement of chromatin binding protein from mitotic chromatin is not due to changes in the chromatin structure because the condensed mitotic chromatin remains fully accessible to both transcription factors and structural proteins (Chen et al., 2005). More likely alterations in the proteins themselves play a major role in their displacement from mitotic chromatin.
The location of the high mobility group (HMG) proteins during mitosis has been the subject of several studies (Isackson et al., 1980; Falciola et al., 1997; Hock et al., 1998; Prymakowska-Bosak et al., 2001; Pallier et al., 2003; Harrer et al., 2004; Disney et al., 1989; Saitoh and Laemmli, 1994); however, to date their intranuclear organization during the various stages of the cell cycle remains controversial (Pallier et al., 2003; Dyson et al., 2005). HMG proteins are a superfamily of abundant and ubiquitous nuclear proteins that bind to chromatin without any known DNA sequence specificity, and they induce structural and functional changes in their binding sites (Bustin, 1999; Reeves, 2001; Sgarra et al., 2004; Bianchi and Agresti, 2005). Although their exact cellular function is still not fully understood, several types of experiments indicate that altered expression of these proteins leads to developmental abnormalities and that it is associated with the etiology of several diseases (Hock et al., 2007). The interaction of all HMGs with chromatin is highly dynamic: the proteins move constantly throughout the nucleus and sample the nucleosomes for potential binding sites in a “stop and go” manner (Scaffidi et al., 2002; Catez et al., 2004; Harrer et al., 2004; Phair et al., 2004). Thus, at the level of the single nucleosome, there is a continuous turnover of HMGs. However, because the “stop” step is longer than the “go” step, at the global level most of the HMGs are associated with chromatin most of the time.
The HMG superfamily is composed of three families, named HMGA, HMGB, and HMGN (Bustin, 1999; Bianchi and Agresti, 2005). HMGA proteins remain associated with chromatin throughout the cell cycle, and they are located in the scaffold of the metaphase chromosome (Disney et al., 1989; Saitoh and Laemmli, 1994; Harrer et al., 2004). Early immunofluorescence studies indicated that both HMGB and HMGN are displaced from mitotic chromatin (Isackson et al., 1980; Falciola et al., 1997; Hock et al., 1998; Prymakowska-Bosak et al., 2001); however, more recent studies with fluorescently labeled proteins suggested that in living cells both HMGBs and HMGNs remain associated with the mitotic chromosome (Pallier et al., 2003). For HMGNs this finding is especially puzzling because biochemical experiments demonstrated that during mitosis a large fraction of cellular HMGN is phosphorylated, a modification that abolishes the specific binding of these proteins to nucleosomes and chromatin (Prymakowska-Bosak et al., 2001).
Because HMGN have been shown to affect chromatin structure and function, it is important to determine unequivocally their fate during mitotic condensation. Here, we examined the organization of HMGN proteins in the cell nucleus by using immunofluorescence studies, live cell imaging, gel mobility shift assays (EMSAs), and bimolecular fluorescence complementation and also by comparing the chromatin binding properties of wild-type and HMGN mutant proteins. We find that indeed, HMGNs can associate with mitotic chromatin; however, this type of interaction is markedly different from their specific binding to nucleosomes in interphase chromatin. The binding of HMGN to mitotic chromatin is not dependent on a functional HMGN nucleosomal binding domain, and it is weaker than the binding to interphase nucleosomes in which HMGNs form specific complexes with nucleosomes. We conclude that the interaction of HMGNs with chromatin is cell cycle dependent.
MATERIALS AND METHODS
Cell Culture and Synchronization
HeLa ccl2 cells (American Type Culture Collection, Manassas, VA) were grown in DMEM (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 2 mM l-glutamine, and 1% penicillin-streptomycin at 37°C in a 5% CO2 incubator. Logarithmically growing cells were enriched for mitotic phase by an 18.5-h treatment with 0.4 μg/ml nocodazole (Knehr et al., 1995).
Transient and Stable Expression of HMGN-Fusion Proteins
For examination of HMGN localization of transiently expressed HMGN1-yellow fluorescent protein (YFP) and HMGN-YFP, HeLa ccl2 cells were transfected with plasmids expressing either fluorescent wild-type or mutant HMGN-green fluorescent protein (GFP) fusion proteins by using FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN). The localization of HMGN1 was also analyzed in stably transformed rat choriocarcinoma (Rcho-1) and mouse embryonic stem (ES) cells stably expressing HMGN1-GFP. Rcho-1 cells (gift from Dr. M. J. Soares, University of Kansas Medical Center, Kansas City, KS) were transfected with hHMGN1-GFP plasmid by using Lipofectamine 2000, selected for 2 wk on 250 μg/ml Geneticin (G418; Invitrogen), fluorescence-activated cell sorted into 96-well plate (30 cells/well), and propagated as clones in the presence of antibiotic. After 2 wk of growth, clones were up scaled in medium containing 0.1 μg/ml G418. ES cells (gift from Dr. A. Nagy, Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Canada) were transfected with hHMGN1-GFP plasmid by using Dotap kit (Roche Applied Science), selected for 2 wk on 400 μg/ml G418 (Invitrogen), and HMGN-GFP expressing clones were identified by direct observation under epifluorescence microscope. Selected single ES clones were propagated on gamma-irradiated feeder layers, in mouse embryonic fibroblasts in DMEM with 15% fetal calf serum (ES tested), 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, and 1000 U/ml leukemia inhibitory factor.
For live cell imaging, cells were seeded on Mat-Tek dishes (MatTek Corporation, Ashland, MA) and labeled with Hoechst 33342 for 15 min just before imaging. Images were collected using a confocal microscope (Carl Zeiss, Thornwood, NY) at 63× objective with zoom 2.
Preparation of Nucleosomes and Proteins
Nucleosome core particles were prepared from chicken red blood cells as described previously (Ausio et al., 1989). Wild-type and mutant HMGN proteins were expressed in and purified from Escherichia coli cells as described previously (Lim et al., 2004).
Electrophoretic Mobility Shift Assay
Core particles and core particle DNA were incubated with increasing concentrations of HMGN1 or HMGN1-S2024EE proteins in 2× Tris borate-EDTA (TBE; 180 mM Tris, 180 mM boric acid, and 2 mM EDTA, pH 8.3) containing 1% (wt/vol) Ficoll 400 on ice for 15 min. The mixtures were electrophoresed on 5% native polyacrylamide gel electrophoresis in 2× TBE at 4°C. After electrophoresis, gels were stained with ethidium bromide and then processed for photography.
Confocal Microscopy
Cells were grown on coverslips in DMEM supplemented with 10% FBS at 37°C in a 5% CO2 incubator. Cells were washed with phosphate-buffered saline (PBS), fixed with 4% formaldehyde in PBS for 10 min at room temperature, washed with PBS, and then permeabilized with 1% Triton X-100 for 5 min at room temperature. The cells were then incubated in blocking buffer (PBS containing 3% bovine serum albumin, 0.05% Tween 20, and 0.1% Triton X-100) for 20 min, followed by incubation with the primary antibody in blocking buffer for 2 h at room temperature. The cells were washed with PBS and incubated with secondary antibody conjugated to fluorescein isothiocyanate (Invitrogen) for 1 h at room temperature. After the final wash steps, cover slips were mounted onto glass slides using Vectashield mounting solution containing 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Confocal laser scanning microscopy was performed using a Zeiss LSM 510 laser scanning confocal microscope using the 354-nm line of a UV laser and the 488- and 543-nm lines of argon and helium-neon lasers.
For analysis of living cells, DNA was stained with the cell-permeant Hoechst 33342 and the images were processed using Zeiss software (Carl Zeiss).
Salt Extraction of HMGN Proteins and Western Blotting
Logarithmically growing and metaphase HeLa cells were collected and washed with PBS. The cells were incubated in HEPES buffer containing 0.1% Triton X-100 and various concentrations of NaCl ranging between 0.1 and 0.4 M for 10 min at room temperature. Cells were centrifuged at 10,000 rpm for 10 min, and the supernatant and the pellet were separated and resuspended in 2× Laemmli sample buffer (Bio-Rad, Hercules, CA). Proteins were separated using polyacrylamide gels (Bio-Rad), and they were transferred onto a polyvinylidene difluoride membrane (Immobilon P; Millipore, Billerica, MA) by semidry transfer. After transfer, the membranes were blocked with 1× casein/Tris-buffered saline (TBS) blocker (Bio-Rad) for 1 h at room temperature and incubated overnight at 4°C with anti-HMGN2 antibodies. The membrane was washed with TBS containing 0.1% Tween 20, and then they were incubated with goat anti-rabbit IgG secondary antibody coupled to peroxidase (Pierce Chemical, Rockford, IL). The HMGN antibodies were detected by enhanced chemical luminescence (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Bimolecular Fluorescence Complementation Assay (BiFC)
HMGN1 and HMGN2 cDNAs were subcloned into pBiFC-YC155 and pBiFC-YN155 expression vectors (kindly provided by Tom Kerppola, University of Michigan School of Medicine, Ann Arbor, MI), which produce either hemagglutinin (HA)-tagged-YC or Flag-tagged-YN fusion proteins. HepG2 were transiently transfected with FuGENE HD according to the provider's instructions (Roche Applied Science). For cotransfections, each construct was used at 1 μg. Fluorescence complementation in living cells was inspected ∼18–24 h after transfection by using a Leica TCS-SP2/AOBS and the 514-nm laser line of an argon laser. Interference contrast was used to control viability and integrity of cells. Parallel immunolocalizations were used to control transfection, coexpression, and localization of HMGN fusion proteins on chromosomes. For immunolocalization, cells grown on coverslips were washed in PBS, fixed in 2% formaldehyde/PBS for 15 min, washed again, and permeabilized for 5 min with ice cold 0.5% Triton/PBS. Expression of YC-fusion proteins was controlled using monoclonal rat-antibodies directed against HA-tag (1:50; gift from Aloys Schepers, GSF-National Research Center for Environment and Health, Munich, Germany); expression of YN-fusion proteins was controlled using mouse antibodies directed against Flag-tags (1:500; Sigma-Aldrich, St. Louis, MO). Secondary antibodies were anti-rat cyanine (Cy)5 (1:100; Dianova, Hamburg, Germany) and anti-mouse Texas Red (Tx-Red, 1:100; Dianova). Antibodies were diluted in PBS, pH 7.4, and incubated subsequently for 45 min in a humidified chamber at room temperature. To stain DNA, 10 μl of 5 μg/ml Hoechst/PBS was added and incubated for a further 10 min. After two final wash steps, coverslips were mounted in Mowiol. Immunofluorescences were analyzed using a HCX Pl APO lbd.Bl. 63× 1.4 oil immersion objective by sequential scanning by using the 405-nm diode for Hoechst staining, the 514-nm laser line of an argon laser for YFP, the 561-nm DPSS-Laser for Tx-Red, and a HeNe-Laser at 633 nm for Cy5.
RESULTS
HMGNs Localize to Mitotic Chromosomes in Live but Not Fixed Cells
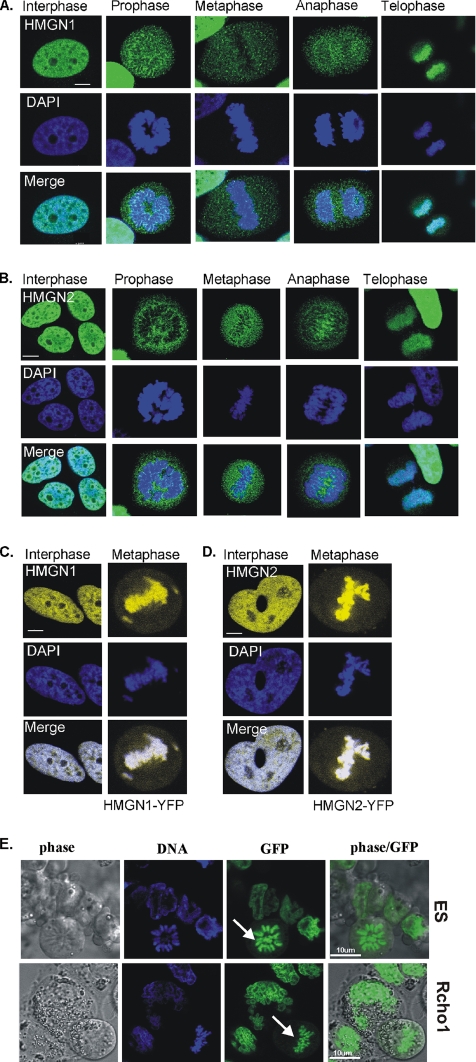
Because of the conflicting results regarding the association of HMGNs with mitotic chromatin, we first reinvestigated the intracellular distribution of the HMGN1 and HMGN2 proteins in HeLa cells during different stages of the cell cycle by using both immunofluorescence and live cell imaging with GFP-tagged HMGN proteins. Confocal immunofluorescence microscopy by using affinity-purified antibodies specific for HMGN1 and HMGN2 revealed that in interphase nuclei, both HMGN1 and HMGN2 proteins were dispersed throughout the entire nucleus, but they are excluded from nucleoli (Figure 1, A and B, left). During the immunofluorescence studies, we noted that exclusion of HMGN protein from nuclei serves as a measure that the fixation procedure did not alter the intranuclear organization of the proteins. Indeed, mutants that do not bind to chromatin accumulate in nucleoli (Prymakowska-Bosak et al., 2002; also see Figure 4). Therefore, we avoided cell fixation conditions that lead to nucleolar localization of wild-type HMGNs.
Figure 1.
Visualization of HMGNs in fixed and living cells. (A and B) Immunolocalization of HMGN1 and HMGN2 during the cell cycle. Immunostaining of HMGNs is presented in green (top), corresponding DAPI staining is shown in blue (middle), and the merged figures are shown in bottom panels. The cell cycle stages are indicated above the figures. Note in interphase, HMGN1 and HMGN2 proteins are distributed all over the nucleus, excluding the nucleoli. In prophase, metaphase, and anaphase cells, HMGN proteins are undetectable on chromatin. In telophase, HMGN proteins reassociate with chromatin. (C and D) Fluorescent images of HMGN fusion proteins in transiently transfected live cell. In living cells, transiently expressed HMGN1-YFP or HMGN2-YFP fusion proteins associate with chromatin in both interphase and mitotic cells. (E) Fluorescent images of HMGN1-GFP fusion proteins in stably transfected, live, mouse embryonic (top row) and Rcho-1 (bottom row) cells. In living cells stably expressed HMGN1-GFP, the fusion proteins associate with chromatin in both interphase and mitotic cells. Arrow points to mitotic cells. HeLa cells (A–D), ES cells (E, top), and Rcho-1 cells (E, bottom). All pictures shown are optical sections made with a confocal microscope.
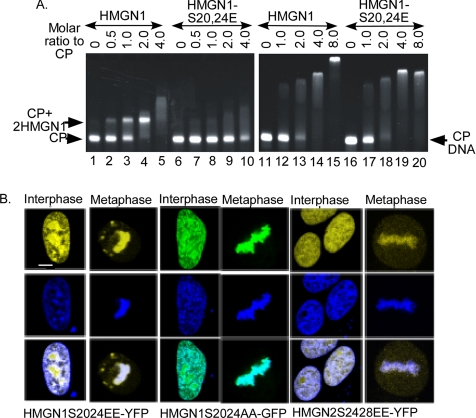
Figure 4.
Chromatin interactions of HMGN proteins bearing mutations in their NBD. (A) EMSA of wild-type and HMGN1S20,24E mutants with core particle (CP) and purified DNA prepared from core particles (CP-DNA). CPs (left) or CP-DNA (right) were incubated with increasing concentrations of either HMGN1 or HMGN1S2024E proteins, and then the complexes were separated on 5% native page in 2× TBE. Arrows indicate nucleosome-HMGN complexes (CP-2HMGN1), nucleosome core particles (CP), and naked DNA (CP-DNA). Note that high concentrations of HMGN1 S20,24E mutant bind to CPs and that the wild-type and mutant HMGNs bind with the same affinity to CP-DNA. (B) HMGN mutants bind to mitotic chromosome, in living cells. HMGN1-S20,24EE-YFP, HMGN1-S20,24AA-GFP, or HMGN2S24,28EE-YFP fusion proteins were expressed in HeLa cells. Confocal microscopy of live cell images show that all the mutant fusion proteins associate with both interphase and mitotic chromatin. Top row, protein fluorescence; middle row, DNA; and bottom, merge images.
As the cells enter mitosis, both proteins seem to dissociate from chromatin, and in prophase, metaphase, and anaphase cells (Figure 1, A and B, middle three panels) the HMGN signal associated with mitotic chromatin is much weaker. Most of the protein is dispersed throughout the entire cell, and it does not colocalize with the DNA. As the nuclear envelope reforms upon entering telophase, both HMGN1 and HMGN2 proteins seem to reassociate with the chromatin, and they colocalized with DNA (Figure 1, A and B, right). These results are fully consistent with most previous reports that in fixed cells HMGN proteins are highly depleted from mitotic chromatin (Hock et al., 1998; Prymakowska-Bosak et al., 2001).
In contrast, confocal analyses of live HeLa cells transiently expressing HMGN1-YFP and HMGN2-YFP proteins reveal that a large portion of these fusion proteins, which are highly expressed (Supplemental Figure 1), remain associated with mitotic chromatin (Figure 1, C and D), a finding that also is in agreement with previous results (Pallier et al., 2003). Likewise, in both live mouse embryonic stem cells and rat Cho-1 cells that stably express HMGN1-GFP, the green fusion protein is clearly associated with mitotic chromosomes (Figure 1E). Thus, indeed, the various experimental approaches yield conflicting results, raising the question whether HMGNs and similar chromatin binding proteins are associated with mitotic chromatin.
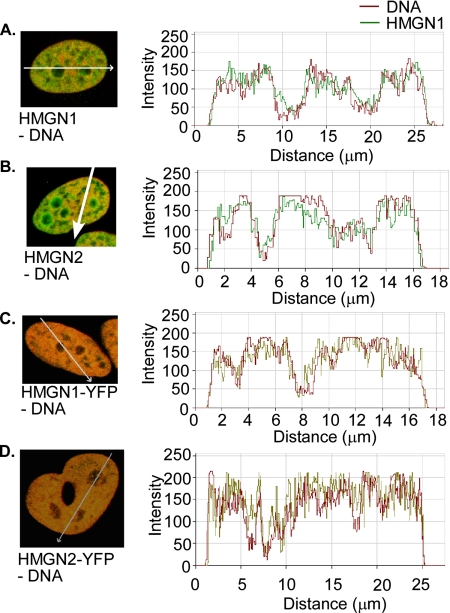
It has been suggested that the discrepancy between the results obtained by immunofluorescence and those obtained by imaging live cells are due to the fixation procedure, and that paraformaldehyde fixation “releases” HMGN from mitotic chromosomes (Pallier et al., 2003). This explanation is puzzling because the chemical composition of the mitotic chromatin fiber is similar to that of the interphase chromatin fibers. Antibodies to histones stain intensely the core histones in both fixed interphase chromatin and in fixed condensed mitotic chromosomes, an indication of similar accessibility of antigenic determinants. Indeed, the condensed mitotic chromatin is fully accessible to structural proteins (Chen et al., 2005). We therefore reasoned that if the organization of HMGN in mitotic chromatin is similar to that of interphase chromatin, paraformaldehyde fixation should also disrupt the binding of HMGN to interphase chromatin and examined in detail whether in fixed interphase chromatin HMGN remains associated with DNA. Profiling of the fixed cells clearly demonstrates that in fixed interphase cells, HMGN remain associated with the nucleosomal DNA and the intranuclear distribution of the wild-type HMGN1 in interphase fixed cells (Figure 2, A and B) is indistinguishable from that HMGN-GFP in live cells (Figure 2, C and D). These findings are fully compatible with previous observations that wild-type HMGN compete with HMGN-GFP for nucleosome binding sites (Catez et al., 2003). Our finding that HMGN is associated with paraformaldehyde-treated interphase chromatin, but not with similarly treated mitotic chromatin, suggests that the fixation “prevents” their binding to, rather than “releasing” them from, mitotic chromosomes, a significant conceptual difference that is relevant to the understanding of the organization of HMGNs in the mitotic chromatin.
Figure 2.
Paraformaldehyde fixation does not remove HMGN from interphase chromatin. In fixed cells (A and B), HMGNs and DNA were detected by immunofluorescence as in Figure 1. In Living cells (C and D), HMGN-YFPs were directly visualized through YFP fluorescence, and DNA was stained with Hoechst 33342. The graphs show relative intensity of each signal along the white arrows and localization profiles of DNA and HMGNs in fixed and live cells. Note that in all cells the DNA and HMGNs colocalize.
Mitotic Phosphorylation of the HMGN Nucleosomal Binding Domain
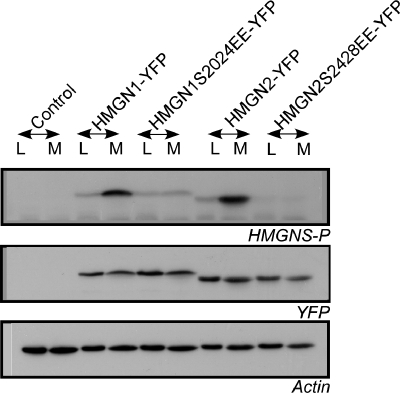
HMGNs bind specifically to nucleosome cores through a highly conserved region known as the nucleosome binding domain (NBD) (Bustin, 1999). During mitosis, two serines located in the NBD are phosphorylated, a modification that abolishes the interaction of HMGNs with chromatin. NBD phosphorylation has been proposed as the main mechanism preventing the binding of HMGN proteins to mitotic chromatin (Prymakowska-Bosak et al., 2001). Conceivably, the HMGN-YFP fusion proteins remain associated with mitotic chromatin, whereas the wild-type HMGN does not, because the YFP fused to the HMGN interferes with the mitotic phosphorylation of the NBD. To test this possibility, we examined the phosphorylation of transfected HMGN1-YFP and HMGN2-YFP in logarithmically growing and mitotic HeLa cells. As controls, HeLa cells were transfected with vectors expressing HMGN-NBD-mutant proteins in which the two serines are replaced by glutamic acid; therefore, they cannot be phosphorylated. Western analyses of whole cell extracts with an affinity purified antibody that specifically recognizes the phosphorylated NBD of HMGNs (Prymakowska-Bosak et al., 2001) demonstrate that during mitosis both HMGN1-YFP and HMGN2-YFP fusion proteins are efficiently phosphorylated in their NBD, just like the endogenous proteins (Figure 3). Thus, although efficiently phosphorylated, HMGN-YFP fusion proteins do associate with mitotic chromatin, suggesting that the binding of HMGs to mitotic chromatin differs from the binding to interphase chromatin.
Figure 3.
Cell cycle-dependent phosphorylation of HMGN-YFP fusion proteins. HeLa cells were transfected with HMGN1-YFP, HMGN2-YFP, or the mutated fusion proteins HMGN1S2024E-YFP or HMGN2S2428E-YFP. Total cell extracts were prepared from log phase (L) or mitotic cells (M), and they were subjected to immunoblot analysis using antibodies recognizing HMGNs phosphorylated at the two serines located in their NBDs (Prymakowska-Bosak et al., 2001). Wild-type HMGN-YFP fusion proteins show mitotic specific serine phosphorylation in the NBD (top). Western analyses with actin demonstrate equal loading, and Western analyses for YFP (middle) demonstrates that all extracts contained comparable amounts of fusion proteins.
NBD-independent Binding of HMGN-YFP to Mitotic Chromatin
Previous studies established that phosphorylation of two highly conserved serines in the HMGN NBD, which in HMGN1 are located in position 20 and 24 and in HMGN2 at position 24 and 28, abolishes the specific binding of HMGN to chromatin, both in vivo and in vitro. The primary binding target of HMGNs in chromatin is the nucleosome core particle. At physiological ionic strength, each nucleosome core particle binds two molecules of HMGN and forms a complex that can be detected by mobility shift assays (Postnikov and Bustin, 1999). Because we now find that although HMGN-GFP fusion proteins are efficiently phosphorylated during mitosis they still bind to mitotic chromosomes, we first retested whether indeed, the HMGN1 S20,24E mutant does not bind to nucleosomes. Direct comparison of mobility shift of equal amount of wild-type and HMGN1S20,24E double mutant clearly demonstrates that the mutant does not bind to nucleosomes and that it does not produce a mobility shift (Figure 4A, left). However, at high concentrations this mutant does interact with the core particle, and it produces a smear (lanes 9 and 10), an indication of nonspecific, or low affinity, binding. Similarly, high concentrations of both wild type HMGN1 and double mutant HMGN1-S20,24E also produce smears with purified, 147-base pair core particle DNA (CP DNA), which is devoid of core histone (Figure 4A, right). Thus, HMGN can bind with low affinity to DNA and even chromatin, independently of a functional NBD. We estimate (Postnikov et al., 1994) that under our experimental conditions, the affinity of HMGN1 for nucleosomes is ∼3 times higher than for purified DNA and that wild-type and HMGN1S20,24E mutant bind with the same affinity to purified DNA.
Given the possibility of an NBD-independent binding of HMGN proteins to DNA and in some cases even chromatin, we investigated the binding of NBD mutants to chromatin in living cells. We transiently transfected HeLa cells with either HMGN1-S20,24AA-GFP or HMGN1-S20,24EE-YFP or HMGN2-S24,28EE-YFP construct, and we examined their distribution in interphase and metaphase chromatin in living cells by using confocal microscopy. In the interphase nuclei, both alanine and glutamic acid mutant proteins were distributed all over the nucleus, including the nucleolus, suggesting that these mutant proteins behave differently than the wild-type proteins, which invariably are excluded from nucleoli (compare with Figure 1). Interestingly, in metaphase cells, the mutant HMGN-fusion proteins localized to condensed chromosomes just like the wild-type fusion proteins (compare Figure 1C with Figure 4B), an unexpected finding because our in vitro mobility shift analyses (Prymakowska-Bosak et al., 2001) and our fluorescence recovery after photobleaching (FRAP) measurements (Catez et al., 2003) clearly indicate that these mutants do not bind to nucleosomes. Together with our finding that mitotic HMGNs are phosphorylated, these results indicate that phosphorylation does not preclude the association of HMGNs with chromatin in living cells; in mitotic cells, the binding of HMGNs to chromatin is independent of a functional NBD and therefore different from their binding to interphase chromatin.
Low-Affinity Binding of HMGN1/2 Proteins to Mitotic Chromosomes
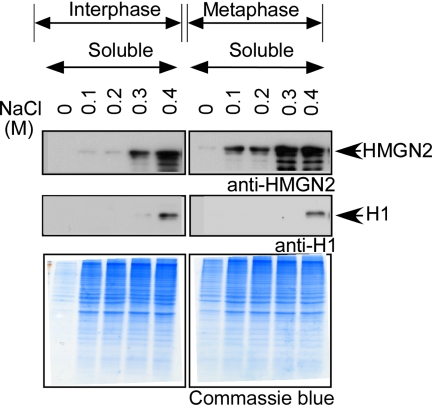
Because mutations in the NBD of HMGN weakens their binding to nucleosomes and because the HMGNs associated with mitotic chromatin could be phosphorylated, we compared the chromatin-binding of endogenous HMGN in interphase and mitotic cells by testing their extractability from chromatin with various NaCl concentrations. Interphase and nocodazole-arrested mitotic HeLa cells were collected and extracted using increasing salt concentration ranging from 0.1 to 0.4 M NaCl. The soluble fractions were collected and subjected to immunoblot analysis (Figure 5). The results reveal that with <0.2 M NaCl, very little HMGN2 was extracted into the supernatant of the interphase cells; the protein was extracted starting 0.3 M NaCl, and significant HMGN accumulated in the extract when interphase cells were extracted with 0.4 M NaCl. In contrast, significant amounts of endogenous HMGN2 could be extracted from mitotic cells with NaCl concentrations as low as 0.1 M and the amount of HMGN2 extracted with 0.3 M NaCl concentrations from mitotic cells was significantly higher than that extracted from interphase cells. These results indicate that the association of HMGN with mitotic chromatin is weaker compared with interphase chromatin.
Figure 5.
Salt extractability of HMGN2 protein from log phase and mitotic HeLa cells. Log phase and mitotic HeLa cells HMGN2 proteins were extracted with increasing concentrations of NaCl. The soluble protein fractions were subjected to immunoblot analysis. As expected, histone H1 is not extracted at the NaCl concentrations used. The Coomassie Blue panels indicate that at each NaCl concentration, equal amount of protein from interphase and metaphase cells were loaded on the gels.
Bimolecular Fluorescence Complementation Analyses Reveal HMGN Homo-Complementation in Interphase, but Not in Mitotic Cells
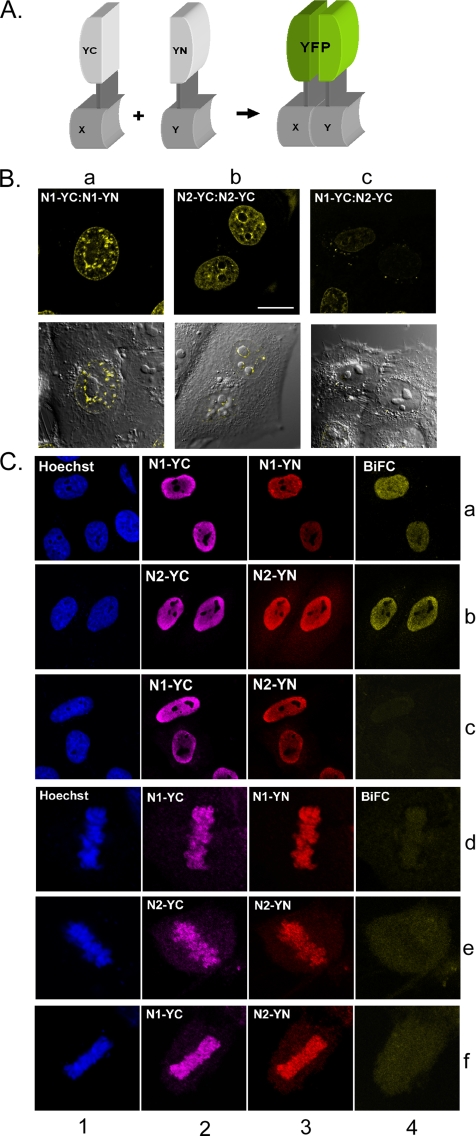
Previous in vitro analyses revealed that HMGNs bind to nucleosomes and form complexes containing two molecules of either HMGN1 or HMGN2. Nucleosomes containing only one HMGN molecule, or nucleosomes containing one molecule of each HMGN1 and HMGN2 are not detected under physiological conditions (Postnikov et al., 1997; Postnikov and Bustin, 1999). We reasoned that if the binding of HMGNs to mitotic chromatin differs from their binding to interphase chromatin the frequency of homodimer formation would also be altered. We therefore used BiFC analysis (Kerppola, 2006) to compare the binding of HMGNs to interphase and mitotic chromatin, in living cells. BiFC enables direct visualization of protein interaction in living cells and is based on the principle that tethering two nonfluorescent constituents of a potentially fluorescent protein to the same macromolecular complex will yield a fluorescent signal (Hu and Kerppola, 2003). The generation of a BiFC is not instantaneous; to generate a signal the interacting partners need to be tethered in proximity for at least several seconds (Kerppola, 2006). For our BiFC analyses, we cotransfected HepG2 cells with the combinations HMGN1-YC:HMGN1-YN, HMGN2-YC:HMGN2-YN, or HMGN1-YC:HMGN2-YN. These expression vectors contained Flag-tags on the YN (YFP N-terminal half) partner and HA-tags on the YC (YFP C-terminal half) partner to allow specific localization of both fusion proteins. Close tethering of an HMGN-YN and HMGN-YC to the same nucleosome for a sufficiently long period, would yield a fluorescent signal, whereas random binding to distant nucleosomes, or colocalization for very short times, would not. The cells were analyzed for the presence of fluorescence complementation in live cells and by immunofluorescence in fixed cells (Figure 6B) 18–24 h after transfection (Figure 6C).
Figure 6.
BiFC assays indicate difference in the organization of HMGNs in interphase and mitotic chromatin. (A) Schematic diagram showing the principle of BiFC. For BiFC, two proteins were fused to the nonfluorescent halves (YC-partner and YN-partner) of YFP. Complementation of functional fluorescent fluorophores is diagnostic of stable interaction of the fusion proteins. Because YC-partners also contain an HA-tag and the YN-partners contain a Flag-tag, expression of both BiFC partners can be visualized by immunofluorescence. (B) BiFC assays of live cells. The fusion proteins expressed in the cells are indicated. Note that cells expressing one type of HMGN fused to the two parts of YFP produced a BiFC signal (a and b), whereas cells transfected with HMGN1 and HMGN2, each fused to a different fragment of the YFP, did not (c). (C) Immunofluorescence analyses of the expression of the fusion proteins. Note that combinations that produce strong BiFC in interphase did not produce a signal in metaphase even though the proteins are associated with chromosomes (compare a, b with d, e; lanes 4). DNA staining is shown in blue; the immunolocalization of YC-partners bearing HA-tags, detected with anti Cy5-labeled secondary antibody, is shown in magenta. The immunolocalization of YN-partners bearing Flag-tag HA-tags, detected with Tx-Red labeled secondary antibody is shown in red. The complemented YFP-fluorescence (BiFC) is shown in yellow. Absence of yellow (in row 4, c and d) indicates lack of BiFC.
Examination of live interphase cells transfected with a mixture of plasmids expressing both HMGN1-YC and HMGN1-YN showed strong fluorescence, an indication that the differentially labeled HMGN1 proteins remained sufficiently close, for a sufficiently long time, to produce a fluorescent signal (Figure 6Ba). Likewise, cells transfected with a mixture of plasmids expressing both HMGN2-YN and HMGN2-YC also produced a prominent fluorescent signal (Figure 6Bb). Live cells transfected with a mixture expressing HMGN1-YC and HMGN2-YN (Figure 6Bc) produce a drastically lower signal an indication that the frequency of heterodimerization, in which HMGN1 and HMGN2 are in proximity for sufficient time, is significantly lower than in cells transfected with only one type of HMGN. Live mitotic cells did not produce a significant BiFC signal in any of the combination tested, suggesting that that most of the HMGNs are not in proximity, or if they are, they remain so for only a very short period.
To further verify that all tested cell do indeed express the various HMGN fusion proteins, we fixed the cells and visualized the expression of the HMGN-YN and HMGN-YC fusion proteins with antibodies to either the HA- or the Flag-tag (Figure 6C). In agreement with the results obtained in live cells, coexpression of either HMGN1-YC or HMGN-YN or coexpression of HMGN2-YC and HMGN2-YN produced a BiFC signal, but coexpression of any combination of HMGN1 and HMGN2 did not. In the fixed mitotic cells, the fusion proteins could be detected associated with chromosomes using the antibodies to the Flag- and HA-tags. Even though significant amount of protein remain associated with these chromosomes, none of the protein combinations produced a significant BiFC signal. The faint signals are due to random, occasional colocalization of molecules bearing complementary fragments of the fluorescent protein. We therefore conclude that the association of these proteins with mitotic chromatin differs from their organization in interphase chromatin, where they form specific homodimeric complexes with nucleosomes.
DISCUSSION
The mechanisms whereby the cell lineage-specific genetic information is accurately transmitted from generation to generation, without being disrupted by the major reorganization of the chromatin fiber that occurs during each mitotic cycle, are not fully understood. Because HMGN, and similar structural proteins, affect chromatin structure and activity, it is important to determine whether they are associated with the mitotic chromatin. Our study demonstrates that the interaction of HMGNs with chromatin is cell cycle dependent and that the binding of these proteins to interphase chromatin differs from their association with the condensed chromosome. In interphase, the binding is dependent on a functional nucleosome binding domain, whereas in mitotic chromatin it is not.
Our conclusions are based on the following experimental observations. First, most immunofluorescence analyses indicate that HMGN proteins are highly depleted from mitotic chromosomes. Immunofluorescence is a widely used technique that has been repeatedly used to visualize proteins in mitotic chromosome. There is no obvious reason why the various procedures used in immunofluorescence should specifically dislodge HMGNs from mitotic chromatin because it does not dislodge them from interphase chromatin (Figure 2). Second, bimolecular fluorescence complementation assays demonstrate that in living interphase cells HMGNs form complexes that yield a signal, whereas in mitotic cells they do not. Previous studies established in HMGN form homodimeric complexes with nucleosomes, two molecules of either HMGN2 or HMGN1 are bound to the same nucleosomes. Heterodimeric complexes in which one molecule of HMGN1 and one molecule of HMGN2 are bound to the same nucleosome are not stably formed under physiological conditions. The BiFC results are fully compatible with the notion that in interphase cells HMGN homodimeric complexes are sufficiently stable to generate a signal, whereas in mitotic chromatin they are not. The slight differences between HMGN1 and HMGN2 in the appearance of the BiFC pattern may be indicative of functional specificity. Third, the NaCl concentration at which HMGN can be extracted from mitotic cells is lower than that required to extract them from interphase cells, an indication that the binding to interphase chromatin is stronger. Fourth, HMGN bearing mutations in their NBD do associate with mitotic chromatin. Numerous in vitro and in vivo experiments demonstrated that mutations of the conserved serines located in the NBD abolish the specific interaction of HMGNs with nucleosomes. Thus, the association of HMGNs with mitotic chromatin does not involve a functional NBD.
HMGN proteins are the only well characterized proteins known to bind specifically to the 147-base pair nucleosome core particle. Their nucleosomal binding is conferred by the highly conserved eight-amino acid motif “RRSARLSA,” which is present in all HMGN proteins. Our EMSA experiments with wild type and HMGN1 mutants demonstrate that the serines 20, 24 are critical amino acid residues for specific nucleosome binding. Mutation of these serines abolished the nucleosome binding of HMGN1 proteins, indicating that these two serines are essential for NBD-dependent nucleosome association. Previous studies revealed that these serine residues are phosphorylated during mitosis (Prymakowska-Bosak et al., 2001) and that phosphorylation prevents specific binding to nucleosomes. In cells treated with phosphatase inhibitors phosphorylated HMGN1/N2 proteins are found in the cytoplasm (Louie et al., 2000). Furthermore, the intranuclear distribution of the HMGN1 S20,24E-YFP mutant in living cells is different from that of the wild-type fusion proteins, an indication of altered binding to chromatin. Likewise, FRAP analyses indicated that the intranuclear mobility of the HMGN1 S20,24 E mutant is faster than that of the wild-type HMGN1, an indication that its chromatin residence time is shorter (Catez et al., 2003). The low salt extractability of the NBD-mutants corroborates that NBD-independent binding affinity is low. Together, the data indicate that the HMGNs associated with mitotic chromosomes do not form specific nucleosome complexes, as they do in interphase chromatin.
Our in vitro EMSA assays showed that at high concentrations HMGN1 mutants lacking a functional NBD do bind to nucleosomes (Figure 4). However, they produce a smear rather than a specific band, an indication that the binding does not form a stable, discrete complex. Likewise, both the HMGN1S20,24E mutant and the wild-type HMGN produce a smear with purified DNA. These results indicate that the proteins can associate with chromatin and DNA even when their NBD is not fully functional. In fact, although loss of NBD abolishes the specific binding of HMGN to nucleosomes their chromatin binding affinity is reduced by only threefold. Thus, it is possible that in some cases HMGNs that lack an intact NBD domain will associate with DNA and chromatin. In these cells, or in cells in which the NDB is not efficiently phosphorylated, under certain fixation conditions, HMGNs will remain associated with mitotic chromosomes. As noted herein, results obtained with fixation conditions that show association of HMGNs with nucleoli should be viewed with caution.
Our findings provide an explanation for the apparent discrepancy between the immunostaining experiments, which demonstrate significant depletion of HMGNs from mitotic chromatin, and live cell imaging, which indicates that fluorescent HMGN fusion proteins are present on mitotic chromatin. In fixed cells the endogenous proteins are depleted from chromosomes and the proteins are immobilized by the fixation procedure. In contrast, in the living cells, the fusion proteins can repeatedly reassociate with DNA or chromatin, creating a steady state in which a significant portion of the protein seems to be permanently bound to the mitotic chromosomes. This effect is most obvious in transiently transfected cells expressing high levels of HMGN-YFP fusion proteins (Supplemental Figure 1) augmenting the visualization of the low-affinity binding to mitotic chromosomes. However, we find HMGN-GFP binding to mitotic chromosomes also in stably transformed, live mouse embryonic stem cells and Rcho-1 cells, which usually express lower amounts of exogenous HMGN protein.
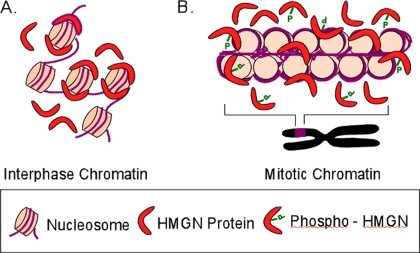
In summary, we find that the mode of HMGN binding to mitotic chromosomes differs from their mode of binding to interphase chromatin. The data suggest that in interphase, chromatin HMGNs bind to nucleosomes specifically and form homodimeric complexes, whereas during mitosis the proteins associate with chromatin weakly and do not form specific complexes with nucleosomes (Figure 7). Our results may also be relevant to the interaction of HMGB proteins with interphase and mitotic chromatin, because immunofluorescence studies indicate that the proteins are not (Isackson et al., 1980; Falciola et al., 1997), whereas live cell imaging suggest that HMGBs are (Pallier et al., 2003), present in chromosomes.
Figure 7.
Model depicting the chromatin binding characteristics of HMGN proteins during cell cycle. In interphase nuclei, two molecules of HMGN1 or two molecules of HMGN2 proteins occupy one nucleosome core particle. These homodimeric complexes are sufficiently stable to yield a BiFC signal. In mitosis, some HMGNs could still be associated with chromatin; however, the binding is not specific, and they do not form homodimers and mitotic cells do not yield a BiFC signal.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Tom Kerppola, University of Michigan School of Medicine, for providing the initial vector backbones for BiFC, Dr. Aloys Schepers for HA-tag antibodies, Dr. A. Nagy, Toronto, Canada, For ES cells, Dr. M. J. Soares, University of Kansan Medical Center, for Rcho-1 and Eyal Rand, NCI protein section, for producing growing and analyzing ES cells stably expressing HMGN-YFP. This work was supported by a grant of the Deutsche Forschungsgemeinschaft to R. Hock, by the Intramural Program of NIH, NCI, and by a Jacob and Lena Joels Foundation Visiting Professorship Award, from the Hebrew University, Israel, to MB.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-1018) on February 20, 2008.
REFERENCES
- Ausio J., Dong F., van Holde K. E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- Belmont A. S. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Bianchi M. E., Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Bonenfant D., Towbin H., Coulot M., Schindler P., Mueller D. R., van Oostrum J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell Proteomics. 2007;6:1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catez F., Lim J. H., Hock R., Postnikov Y. V., Bustin M. HMGN dynamics and chromatin function. Biochem. Cell Biol. 2003;81:113–122. doi: 10.1139/o03-040. [DOI] [PubMed] [Google Scholar]
- Catez F., Yang H., Tracey K. J., Reeves R., Misteli T., Bustin M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 2004;24:4321–4328. doi: 10.1128/MCB.24.10.4321-4328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Dundr M., Wang C., Leung A., Lamond A., Misteli T., Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Allis C. D., Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Disney J. E., Johnson K. R., Magnuson N. S., Sylvester S. R., Reeves R. High-mobility group protein HMG-I localizes to G/Q- and C-bands of human and mouse chromosomes. J. Cell Biol. 1989;109:1975–1982. doi: 10.1083/jcb.109.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson M. H., Thomson S., Mahadevan L. C. Heat shock, histone H3 phosphorylation and the cell cycle. Cell Cycle. 2005;4:13–17. doi: 10.4161/cc.4.1.1362. [DOI] [PubMed] [Google Scholar]
- Falciola L., Spada F., Calogero S., Langst G., Voit R., Grummt I., Bianchi M. E. High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J. Cell Biol. 1997;137:19–26. doi: 10.1083/jcb.137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Forbes D. J. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Harrer M., Luhrs H., Bustin M., Scheer U., Hock R. Dynamic interaction of HMGA1a proteins with chromatin. J. Cell Sci. 2004;117:3459–3471. doi: 10.1242/jcs.01160. [DOI] [PubMed] [Google Scholar]
- He S., Davie J. R. Sp1 and Sp3 foci distribution throughout mitosis. J. Cell Sci. 2006;119:1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- Hock R., Furusawa T., Ueda T., Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock R., Scheer U., Bustin M. Chromosomal proteins HMG-14 and HMG-17 are released from mitotic chromosomes and imported into the nucleus by active transport. J. Cell Biol. 1998;143:1427–1436. doi: 10.1083/jcb.143.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. D., Kerppola T. K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isackson P. J., Bidney D. L., Reeck G. R., Neihart N. K., Bustin M. High mobility group chromosomal proteins isolated from nuclei and cytosol of cultured hepatoma cells are similar. Biochemistry. 1980;19:4466–4471. doi: 10.1021/bi00560a013. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knehr M., Poppe M., Enulescu M., Eickelbaum W., Stoehr M., Schroeter D., Paweletz N. A critical appraisal of synchronization methods applied to achieve maximal enrichment of HeLa cells in specific cell cycle phases. Exp. Cell Res. 1995;217:546–553. doi: 10.1006/excr.1995.1121. [DOI] [PubMed] [Google Scholar]
- Komura J., Ono T. Disappearance of nucleosome positioning in mitotic chromatin in vivo. J. Biol. Chem. 2005;280:14530–14535. doi: 10.1074/jbc.M500637200. [DOI] [PubMed] [Google Scholar]
- Kouskouti A., Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Sudlow G., Belmont A. S. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 1998;140:975–989. doi: 10.1083/jcb.140.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. H., Catez F., Birger Y., Postnikov Y. V., Bustin M. Preparation and functional analysis of HMGN proteins. Methods Enzymol. 2004;375:323–342. doi: 10.1016/s0076-6879(03)75021-6. [DOI] [PubMed] [Google Scholar]
- Louie D. F., Gloor K. K., Galasinski S. C., Resing K. A., Ahn N. G. Phosphorylation and subcellular redistribution of high mobility group proteins 14 and 17, analyzed by mass spectrometry. Protein Sci. 2000;9:170–179. doi: 10.1110/ps.9.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M. A., Dey A., Rabindran S. K., Ozato K., Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Pallier C., Scaffidi P., Chopineau-Proust S., Agresti A., Nordmann P., Bianchi M. E., Marechal V. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol. Biol. Cell. 2003;14:3414–3426. doi: 10.1091/mbc.E02-09-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R. D., Scaffidi P., Elbi C., Vecerova J., Dey A., Ozato K., Brown D. T., Hager G., Bustin M., Misteli T. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikov Y. V., Bustin M. Reconstitution of high mobility group 14/17 proteins into nucleosomes and chromatin. Methods Enzymol. 1999;304:133–155. doi: 10.1016/s0076-6879(99)04010-0. [DOI] [PubMed] [Google Scholar]
- Postnikov Y. V., Herrera J. E., Hock R., Scheer U., Bustin M. Clusters of nucleosomes containing chromosomal protein HMG-17 in chromatin. J. Mol. Biol. 1997;274:454–465. doi: 10.1006/jmbi.1997.1391. [DOI] [PubMed] [Google Scholar]
- Postnikov Y. V., Lehn D. A., Robinson R. C., Friedman F. K., Shiloach J., Bustin M. The cooperative binding of chromosomal protein HMG-14 to nucleosome cores is reduced by single point mutations in the nucleosomal binding domain. Nucleic Acids Res. 1994;22:4520–4526. doi: 10.1093/nar/22.21.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prymakowska-Bosak M., Hock R., Catez F., Lim J. H., Birger Y., Shirakawa H., Lee K., Bustin M. Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14.3.3 proteins. Mol. Cell. Biol. 2002;22:6809–6819. doi: 10.1128/MCB.22.19.6809-6819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prymakowska-Bosak M., Misteli T., Herrera J. E., Shirakawa H., Birger Y., Garfield S., Bustin M. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 2001;21:5169–5178. doi: 10.1128/MCB.21.15.5169-5178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- Saitoh Y., Laemmli U. K. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T., Bianchi M. E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- Sgarra R., Rustighi A., Tessari M. A., Di Bernardo J., Altamura S., Fusco A., Manfioletti G., Giancotti V. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett. 2004;574:1–8. doi: 10.1016/j.febslet.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Zaidi S. K., Young D. W., Pockwinse S. M., Javed A., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc. Natl. Acad. Sci. USA. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.