Abstract
Two ubiquitin-like molecules, Atg12 and LC3/Atg8, are involved in autophagosome biogenesis. Atg12 is conjugated to Atg5 and forms an ∼800-kDa protein complex with Atg16L (referred to as Atg16L complex). LC3/Atg8 is conjugated to phosphatidylethanolamine and is associated with autophagosome formation, perhaps by enabling membrane elongation. Although the Atg16L complex is required for efficient LC3 lipidation, its role is unknown. Here, we show that overexpression of Atg12 or Atg16L inhibits autophagosome formation. Mechanistically, the site of LC3 lipidation is determined by the membrane localization of the Atg16L complex as well as the interaction of Atg12 with Atg3, the E2 enzyme for the LC3 lipidation process. Forced localization of Atg16L to the plasma membrane enabled ectopic LC3 lipidation at that site. We propose that the Atg16L complex is a new type of E3-like enzyme that functions as a scaffold for LC3 lipidation by dynamically localizing to the putative source membranes for autophagosome formation.
INTRODUCTION
Macroautophagy, referred to here as autophagy, is an intracellular process by which double-membrane structures, called autophagosomes, sequester and deliver cytosol and organelles to the lysosome/vacuole for degradation. In addition to its well understood physiological role as a starvation response, there is growing evidence for the participation of autophagy in processes such as cellular differentiation, tissue remodeling, growth control, adaptation to adverse environments, and cellular immunity (Cuervo, 2004; Levine and Klionsky, 2004; Yoshimori, 2004).
The mode of autophagosome formation stands apart from vesicle formation in other membrane trafficking processes, such as endocytosis and the secretory pathway. Transport vesicles in these pathways are generated by budding and scission from the membranes of donor organelles. In contrast, autophagosomes are formed de novo in the cytosol (Noda et al., 2002): a flattened membrane sac, the so-called isolation membrane in mammals, elongates and curves until the ends merge to enclose the cargo and form the autophagosome. This process requires unique mechanisms other than those used in the canonical membrane-trafficking pathways. Yeast genetic studies have revealed a set of genes involved in the autophagic pathway termed ATG, for autophagy-related, and at least 17 ATG genes have been shown to be involved in autophagosome formation to date (Suzuki and Ohsumi, 2007).
A hallmark of the Atg machinery is its two ubiquitin-like conjugation systems (Ohsumi, 2001; Mizushima et al., 2002). The first ubiquitin-like molecule is Atg12. The C-terminal carboxyl base of Atg12 is covalently conjugated to an internal Lys residue of Atg5 through an isopeptide bond by a series of ubiquitination-like reactions that involves an E1 homologue (Atg7) and an E2 analogue (Atg10) (Mizushima et al., 1998). The Atg12-Atg5 conjugate then associates with Atg16 (Atg16L in mammals), and these complexes then homo-oligomerize. As a result, Atg12-Atg5 and Atg16L form an ∼800-kDa protein complex (referred to as Atg16L complex). A fraction of the multimeric complex localizes to isolation membranes, whereas most of it is diffused throughout the cytoplasm (Mizushima et al., 2001, 2003). The complex is released from the membrane just before or after completion of autophagosomes, so it is a good marker for isolation membranes. Although Atg5 is essential for isolation membrane elongation, the function of the complex is unclear. Recently, three independent studies by genome-wide association scans have shown that ATG16L mutation is a risk factor for ileal Crohn's disease (Consortium TWTCC, 2007; Hampe et al., 2007; Rioux et al., 2007), suggesting that Atg16L is a key molecule in preventing the onset of the disease.
The second ubiquitin-like molecule is Atg8 (LC3 in mammals). The C-terminal carboxyl base of LC3/Atg8 is conjugated to the head group amine of phosphatidylethanolamine (PE) through an amide bond by a sequence of ubiquitination-like reactions that involves a protease (Atg4), Atg7, and a different E2 analogue (Atg3) (Ichimura et al., 2000). The lipidated form of LC3/Atg8 is associated with the autophagosomal membrane (Kabeya et al., 2000; Kirisako et al., 2000). The lipidation process is reversible, because the Atg4 protease can also catalyze delipidation of LC3/Atg8 in vitro (Kirisako et al., 2000; Kabeya et al., 2004). In fact, most LC3/Atg8 is liberated from the membrane during the final stage of autophagy, fusion of autophagosomes and lysosomes (Kirisako et al., 1999; Kimura et al., 2007). Similar to Atg5, Atg8 is required for autophagosome formation. Although lipidated LC3/Atg8 might be directly involved in membrane elongation, its actual function is unknown.
Interestingly, in the absence of the Atg12-Atg5 conjugate, the amount of PE-conjugated Atg8/LC3 is severely reduced, and cytosolic Atg8/LC3 is not targeted to the membrane in yeast or mammals (Mizushima et al., 2001; Suzuki et al., 2001). Very recently, it has been reported that the Atg12-Atg5 conjugate facilitates PE conjugation of Atg8 in vitro (Fujioka et al., 2007; Hanada et al., 2007). However, the mechanisms by which the Atg16L complex, and particularly the Atg16L molecule, affects Atg8/LC3 lipidation in vivo is still largely unknown.
One of the most fundamental questions in autophagy is the origin of the membrane that forms the autophagosome (Stromhaug et al., 1998; Juhasz and Neufeld, 2006). A small crescent-like membrane-bound compartment, to which Atg5 localizes, has been identified as an early autophagic structure in mammalian cells (Mizushima et al., 2001); however, little is known about this structure. In yeast, the preautophagosomal structure (PAS) has been proposed as the origin of autophagosomes, as most Atg proteins accumulate there at least transiently (Suzuki et al., 2001). The nature of the PAS, however, is still controversial, and whether it is a membranous structure remains a question. Moreover, a structure corresponding to the PAS has not been identified in mammalian cells. Where, then, does the initial crescent-like membrane structure come from? How are lipids supplied to the isolation membrane for elongation? Elucidation of the molecular mechanisms governing how and where LC3 lipidation takes place would add significant insight.
To address these questions, we took advantage of an inhibitory effect arising upon overexpression, and succeeded in dissecting multiple functions of the Atg16L complex. Overexpression of Atg16L or Atg12 inhibited autophagosome formation at different stages, although these two proteins are in one complex together with Atg5. Based on these results, we discuss how and where LC3 is lipidated. We believe this study provides important insight into the process of autophagosome biogenesis.
MATERIALS AND METHODS
Reagents and Antibodies
Cell culture reagents were purchased from Invitrogen (Carlsbad, CA). The following antibodies were used: rabbit polyclonal anti-rat LC3 (Kabeya et al., 2000); anti-human Atg5 (Mizushima et al., 2001); anti-mouse Atg16L (Mizushima et al., 2003); anti-human Atg12 (Mizushima et al., 2001); anti-monomeric red fluorescent protein, which reacts with mStrawberry (MBL, Nagoya, Japan); mouse monoclonal anti-GFP (clone 7.1 and 13.1; Roche, Indianapolis, IN); anti-Flag M2 (Sigma, St. Louis, MO); anti-c-myc (clone 9E10; Gentaur Molecular Products, Kobe, Japan); anti-α-tubulin (clone B5-1-2; Sigma). Wortmannin (Calbiochem, La Jolla, CA) was prepared as a 100 μM stock in Me2SO. All other reagents were purchased from Sigma-Aldrich.
DNA Construction and Recombinant Adenoviruses
A plasmid encoding monomeric red fluorescent protein (mStrawberry) was a generous gift of Dr. Roger Y. Tsien (University of California, San Diego, CA) (Shaner et al., 2004). Expression vectors for GFP-LC3, Myc-LC3-HA, Myc-LC3G120A-HA, and GFP-Atg5 have previously been described (Kabeya et al., 2000; Mizushima et al., 2001). pCI-neo-Atg12G140A or -Atg12G140A,F108 double mutants (F108A, F108L, or F108D) were generated using the QuikChange Site-Directed mutagenesis system (Stratagene, La Jolla, CA). The cDNAs encoding myc-tagged Atg7WT and -Atg7C572S, 3xFlag-tagged Atg3WT and -Atg3C264S, Atg5, Atg12, mStrawberry-tagged Atg12G140A, mStrawberry-tagged Atg12F108A,G140A, mStrawberry-tagged Atg16L deletions, Flag-tagged Atg16L deletions (see Figure 1A), mStrawberry-tagged Atg16L-N chimera with the C-terminal 17 amino acids (KDGKKKKKKSKTKCVIM) of K-ras (Atg16L-NKras-CAAX), Flag-tagged Atg16L-NKras-CAAX, and mStrawberry were subcloned into the pENTR 1A plasmid (Invitrogen). The cDNA inserts in pENTR-1A were transferred to the pAd/CMV/V5-DEST vector (Invitrogen) by the Gateway system using LR clonase (Invitrogen). Recombinant adenoviruses were prepared with the ViraPower Adenovirus Expression System (Invitrogen) according to the manufacturer's instructions.
Figure 1.
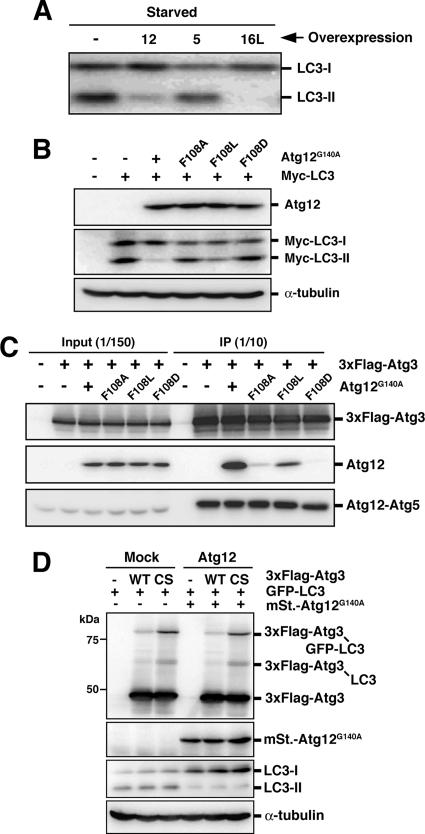
The role of the interaction between Atg12 and Atg3 in LC3 lipidation. (A) PC12 cells were either left untreated or infected with adenovirus bearing Atg12 (12), Atg5 (5), or Atg16L (16L). After 40-h incubation, the cells were cultured in HBSS (Starved) for 2 h and collected. Lysates were examined by Western blotting using anti-LC3 antibody. (B) HEK293A cells were transiently cotransfected with Myc-tagged LC3 and Atg12G140A or Atg12G140A,F108 double mutants (F108A, F108L, or F108D). Lysates were subjected to Western blotting with each antibody. Top panel, anti-Atg12; middle panel, anti-myc; bottom panel, anti-α-tubulin. (C) HEK293A cells were transiently transfected with 3xFlag-tagged Atg3 and Atg12G140A or Atg12G140A,F108 double mutants (F108A, F108L, or F108D). Lysates were subjected to anti-Flag immunoprecipitation, and the immune complexes were examined by Western blotting using each antibody. Top panel, anti-Flag; middle panel, anti-Atg12; bottom panel, anti-Atg5. (D) PC12 cells were coinfected using a combination of recombinant adenoviruses as indicated. After 40-h incubation, the cells were collected. Lysate samples were examined by Western blotting using each antibody. From top panel, anti-Flag, anti-RFP, anti-LC3, and anti-α-tubulin.
Cell Culture, Plasmid Transfections, and Adenovirus Infections
MDCK, PC12, HEK293A, and MCF7 cells were grown in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and appropriate antibiotics in a 5% CO2 incubator at 37°C. For nutrient starvation, cells were cultured in HBSS (Invitrogen) for 2 h. Transient transfection was carried out using LipofectAMINE2000 reagent (Invitrogen) according to the manufacturer's protocol. Stable lines were selected in growth medium supplemented with 500 μg/ml G418. Adenovirus infection was carried out as follows: on the day before infection, ∼2 × 105 cells were plated into six-well plates and incubated at 37°C overnight in a CO2 incubator. The medium was then replaced with 1.5 ml of culture medium containing recombinant adenoviruses. After a 16-h incubation, the medium was replaced with 1.5 ml of culture medium. After an additional 24 h of incubation, the cells were used for experiments.
Western Blotting
Cells were rinsed with ice-cold PBS, scraped, collected by centrifugation at 4°C, and lysed in PBS containing 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and Protease inhibitor cocktail (Roche). Lysates were centrifuged at 15,000 × g for 15 min at 4°C, and supernatants were collected. Samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane. The membranes were blocked with 1% skim milk in 0.1% Tween 20/TBS and incubated with primary antibodies. Immunoreactive bands were detected using horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) and luminol solution (1.25 mM luminol, 65 mM Tris-HCl, pH 8.0, 0.2 mM coumaric acid, 0.01% H2O2).
Fluorescence Microscopy
Cells cultured on coverslips were fixed with 4% paraformaldehyde/PBS. Samples were examined under a fluorescence laser scanning confocal microscope (FV1000; Olympus, Tokyo, Japan) or Olympus IX81 microscope equipped with a mercury lamp and cooled charge-coupled device camera (Roper Cool Snap HQ, Tucson, AZ), under control of SlideBook software (Intelligent Imaging Innovations, Denver, CO). More than 100 cells were examined, and the number of GFP-LC3 or GFP-Atg5 dots within each cell was determined.
Gel Filtration
Gel filtration analysis was performed as previously described (Mizushima et al., 2003). Briefly, PC12 cells were homogenized in homogenization buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and Protease inhibitor cocktail; Roche) by repeated passage (∼15 times) through a 1-ml syringe with a 23-gauge needle. The homogenate was centrifuged at 10,000 × g for 10 min, and the supernatant was further centrifuged at 100,000 × g for 60 min. The resulting supernatants (cytosolic fraction) were separated by size exclusion chromatography on a Superose 6 column (GE Healthcare, Waukesha, WI).
Electron Microscopy
Conventional electron microscopy was performed as described previously (Mizushima et al., 2001).
Statistics
All values shown in figures are represented as means and SDs.
RESULTS
Inhibition of LC3 Lipidation by Atg12 Overexpression Is Dependent on the Interaction between Atg12 and Atg3
Atg12, Atg5, and Atg16L form a stoichiometric complex (referred to as Atg16L complex), and the monomeric forms of each protein are nearly undetectable under steady-state conditions (Mizushima et al., 2003). In an attempt to unbalance the stoichiometry, we used an adenovirus-based system to express each protein at high levels. Overexpression of Atg5 did not affect autophagy; in contrast, overexpression of Atg12 or Atg16L had severe effects on LC3 lipidation and autophagy (Figure 1A and see below).
We first explored the molecular basis of the effect of Atg12 overexpression. To determine whether Atg12 had an inhibitory effect as part of the Atg12-Atg5 conjugate or as the unconjugated form, we replaced the C-terminal glycine (G140) of Atg12 with alanine, which renders the protein incompetent for conjugation to Atg5. The mutant provided a similar inhibitory effect on LC3 lipidation as conjugation-competent Atg12, indicating that unconjugated Atg12 is an inhibitor of LC3 lipidation (Figure 1B). It has been reported that a hydrophobic residue [F154] in yeast Atg12 is important for autophagy. Mutation at F154 has no effect on Atg12-Atg5 conjugation, but the residue is important for Atg8-PE conjugation (Hanada and Ohsumi, 2005). F154 in yeast Atg12 corresponds to F108 in mouse Atg12. We therefore examined how the F108 mutation would affect the inhibition of LC3 lipidation observed upon Atg12 overexpression. Atg12F108A formed the Atg12-Atg5 conjugate to the same extent as wild-type Atg12, and as well as yeast Atg12F154A (data not shown). However, the inhibition of LC3 lipidation was significantly diminished by the F108 mutation (Figure 1B), suggesting that the F108 residue is critical for Atg12 function (and thus for blocking LC3 lipidation when Atg12 is overexpressed).
There is a physical interaction between Atg12 and Atg3 in both yeast and mammals (Uetz et al., 2000; Tanida et al., 2002; Hanada et al., 2007); however, the functional significance of this interaction has not been fully elucidated. We therefore analyzed the interaction between Atg3 and Atg12 using Atg12F108 mutants. Flag-tagged mouse Atg3 was immunoprecipitated from HEK293A cells transiently transfected with mouse Atg12 mutants (Atg12G140A or double mutants with F108A, F108L, or F108D). The interaction of the coexpressed proteins was then examined by Western blotting. As shown in Figure 1C, Atg12G140A was efficiently coimmunoprecipiated with Atg3. In contrast, Atg12F108A,G140A and Atg12F108D,G140A were hardly coimmunoprecipitated, whereas Atg12F108L,G140A was coimmunoprecipitated with Atg3 albeit to a lesser extent than Atg12G140A. These results show that F108 of Atg12 is necessary for interaction with Atg3. Because the strength of the inhibitory effect of the Atg12 mutants on LC3 lipidation correlates with the binding capacity of the Atg12 mutants for Atg3 (Figure 1, B and C), it is conceivable that the Atg3-LC3 intermediate is titrated out by the excessive amount of Atg12 monomer.
To confirm whether an Atg3-LC3 intermediate was formed in the Atg12-overexpressing cells, we used a mutant construct of the E2 (Atg3C264S) enzyme whose active-site cysteine residue is replaced by serine. By using this mutant, high-molecular-mass Atg3C264S-LC3 intermediates are stabilized, because a stable ester bond forms between the enzyme and substrate instead of an unstable thioester bond (Tanida et al., 2002). As shown in Figure 1D, an LC3-Atg3C264S intermediate was formed equally well in either mock cells or mStrawberry-Atg12G140A–overexpressing cells. Moreover, the reaction kinetics was hardly affected by Atg12G140A overexpression (Supplementary Figure 1, A, B, and D). Therefore, it is likely that the transfer of LC3 from Atg3 to PE is blocked by Atg12G140A, rather than the preceding E1 or E2 steps.
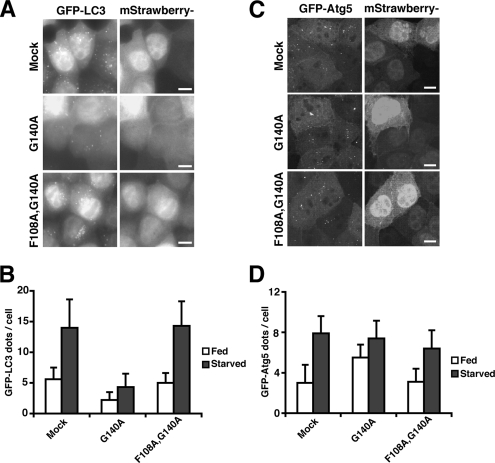
Finally, to determine whether autophagosome formation is inhibited by the overexpression of Atg12 mutants, MDCK cells stably expressing GFP-LC3 were infected with recombinant adenoviruses bearing mStrawberry, mStrawberry-Atg12G140A, or mStrawberry-Atg12F108A,G140A, and the number of GFP-LC3 dots (an autophagosome marker) was counted. mStrawberry-Atg12G140A (but not mStrawberry-Atg12F108A,G140A)-infected cells showed a significant decrease in GFP-LC3 dots under both nutrient-rich and starvation conditions (Figure 2, A and B). Similarly, we examined the effects of overexpression of Atg12 mutants on GFP-Atg5 dot formation. GFP-Atg5, together with Atg16L, is localized to the isolation membrane, a nascent autophagosome, and upon completion, it detaches into the cytosol. In contrast to the effects on GFP-LC3, overexpression of mStrawberry-Atg12G140A had little inhibitory effect on GFP-Atg5 dot formation (Figure 2, C and D). These results suggest that F108 of mouse Atg12 has a role in the targeting and lipidation of LC3, but not the membrane localization of the Atg16L complex. Collectively, we propose that the interaction between Atg12 and Atg3 is important for PE conjugation of LC3 in vivo.
Figure 2.
F108 of mouse Atg12 is important for targeting of LC3. (A and B) MDCK cells stably expressing GFP-LC3 were infected with adenovirus bearing mStrawberry (Mock), mStrawberry-Atg12G140A (G140A), or mStrawberry-Atg12F108A,G140A (F108A, G140A) and incubated for 40 h. The cells were then cultured in HBSS (Starved; A and B) or growth medium (Fed; B) for 2 h, fixed, and observed using fluorescence microscopy (A). Bar, 10 μm. More than 100 mStrawberry-positive cells were examined for the number of GFP-LC3 dots per cell (B). Data are represented as means ± SD. (C and D) MDCK cells stably expressing GFP-Atg5 were infected with adenovirus bearing mStrawberry (Mock), mStrawberry-Atg12G140A (G140A), or mStrawberry-Atg12F108A,G140A (F108A, G140A) and incubated for 40 h. The cells were then cultured in HBSS (Starved; C and D) or growth medium (Fed; D) for 2 h and fixed. Three-dimensional image stacks were obtained from sequential optical sections acquired 0.3 μm apart by confocal laser scanning microscopy (FV1000, Olympus) (C). Bar, 10 μm. More than 100 mStrawberry-positive cells were examined for the number of GFP-Atg5 dots per cell (D). Data are represented as means ± SD.
Overexpression of the Atg16L Coiled-Coil Domain Inhibits Autophagy
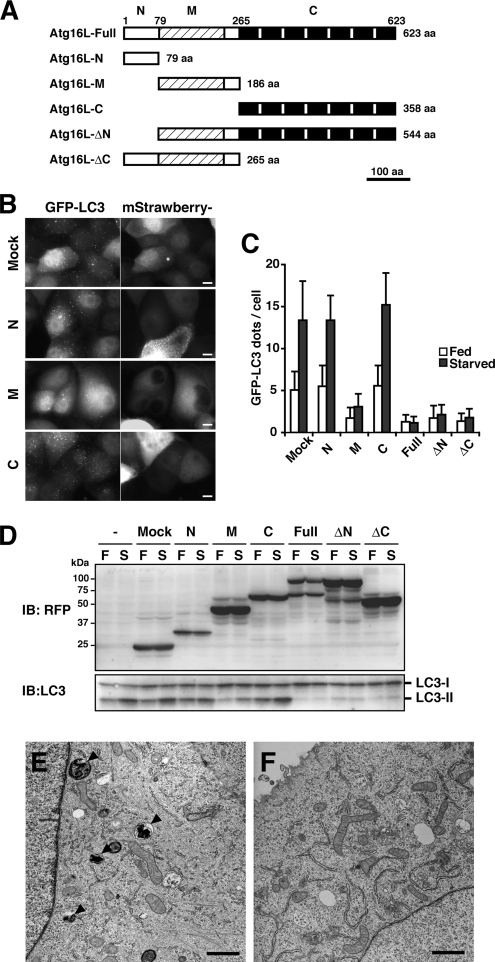
The above results suggest the Atg16L complex plays a role in LC3 lipidation. To further explore the function of the Atg16L complex, we next examined the effects of Atg16L overexpression. We generated a series of adenoviruses bearing various mStrawberry-tagged Atg16L deletion mutants (Figure 3A). The Atg16L protein consists of an N-terminal domain involved in Atg5-binding, a central coiled-coil domain required for self-multimerization, and a C-terminal WD repeat domain whose role is unknown. At least three isoforms, Atg16L-α, -β, and -γ, are generated in mammals by alternative splicing events (Mizushima et al., 2003). MDCK cells stably expressing GFP-LC3 were infected with adenovirus bearing constructs from the Atg16L deletion series, and the number of GFP-LC3 dots was counted. All constructs bearing the coiled-coil region (16L-M, -ΔN, or -ΔC) inhibited the formation of GFP-LC3 dots equivalently to full-length Atg16L (Figure 3, B and C, and Supplementary Figure 2). Next, we assessed whether LC3 lipidation, which is necessary for LC3 dot formation, was affected by the overexpression of the deletion constructs. PC12 cells were infected with the same adenoviruses, and cell lysates were subjected to Western blotting for LC3. As shown in Figure 3D, LC3 lipidation was inhibited by overexpression of Atg16L constructs containing the coiled-coil region. To determine the effects on autophagosome formation, we examined cells by electron microscopy. Autophagosomes were rarely observed in cells overexpressing Atg16L (Figure 3, E and F). Taken together, these results show that overexpression of the Atg16L coiled-coil region inhibits autophagosome formation.
Figure 3.
The effect of overexpression of the Atg16L coiled-coil domain on autophagy. (A) Schematic diagram of mouse Atg16L and the deletion constructs. The coiled-coil region is indicated by shading, and the WD repeats are shown as black boxes. (B and C) MDCK cells stably expressing GFP-LC3 were infected with adenovirus bearing mStrawberry (Mock) or mStrawberry-fused Atg16L deletion constructs and incubated for 40 h. The cells were then cultured in HBSS (Starved; B and C) or growth medium (Fed; C) for 2 h, fixed, and observed using fluorescence microscopy (B). Bar, 10 μm. More than 100 mStrawberry-positive cells were examined for the number of GFP-LC3 dots per the cell (C). Data are represented as means ± SD. (D) PC12 cells were either left untreated or infected with adenovirus bearing mStrawberry (Mock) or mStrawberry-fused Atg16L deletion constructs. After 40-h incubation, the cells were cultured either in growth medium (F; Fed) or HBSS (S; Starved) for 2 h and collected. Lysates were examined by Western blotting using the indicated antibodies. Anti-RFP antibody reacts with mStrawberry. (E and F) MDCK cells stably expressing GFP-LC3 were infected with adenovirus bearing mStrawberry (E) or mStrawberry-fused Atg16L (F) and incubated for 40 h. The cells were then cultured in HBSS for 2 h, fixed, and subjected to conventional electron microscopic analysis. Autophagic structures are indicated (arrowheads). Bar, 1 μm.
Overexpression of Atg16L Disturbs Membrane Localization of the Atg16L Complex and the Transfer of LC3 from Atg3 to PE
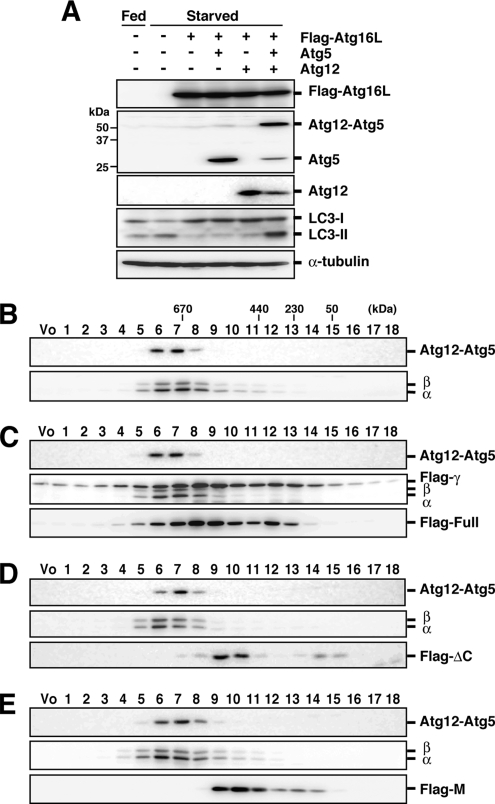
It is possible that Atg16L overexpression causes an indirect effect on autophagy. Alternatively, altering the proper subunit stoichiometry of the Atg16L complex may cause the inhibitory effect on LC3 lipidation. To assess these possibilities, PC12 cells were infected with a combination of recombinant adenoviruses bearing Atg16L, Atg5, and Atg12, and LC3 lipidation was determined. The inhibitory effect of full-length Atg16L overexpression was completely suppressed only by overexpression of Atg12 and Atg5 together, but not Atg12 or Atg5 alone (Figure 4A). It was previously reported that Atg12-Atg5 and Atg16L form an ∼800-kDa complex via self-multimerization through the coiled-coil region of Atg16L (Mizushima et al., 2003). We confirmed that coexpressed Atg12, Atg5, and Flag-tagged Atg16L formed an ∼800-kDa multimeric complex in the cytosol (Supplementary Figure 3). This result demonstrates that inhibition of LC3 lipidation is caused by alteration of subunit stoichiometry rather than by an indirect effect of Atg16L overexpression.
Figure 4.
Organization of the Atg16L complex in the presence of excess Atg16L. (A) PC12 cells were coinfected using a combination of recombinant adenoviruses as indicated. After 40-h incubation, cells were cultured in growth medium (Fed) or HBSS (Starved) for 2 h, and lysates were analyzed by Western blotting were performed with each antibody. From top panel, anti-Flag, anti-Atg5, anti-Atg12, anti-LC3, and anti-α-tubulin. (B–E) Cytosolic fractions of cell homogenates of control and PC12 cells overexpressing Flag-Atg16L constructs were separated by size exclusion chromatography. Each fraction was subjected to Western blotting using each antibody. Top panel, anti-Atg5; middle panel, anti-Atg16L; bottom panel, anti-Flag (except for B). (B) Mock. (C) Flag-Atg16L. (D) Flag-Atg16L-ΔC. (E) Flag-Atg16L-M. The positions of the molecular mass standards are shown. PC12 cells express only the Atg16L-α and -β isoforms. Vo, void fraction.
On the basis of these results, we hypothesized that the ∼800-kDa multimeric complex is a functional complex and that overexpression of the Atg16L coiled-coil region leads to the formation of incomplete and nonfunctional complexes that lack the proper ratio of Atg12-Atg5 subunits. To examine this possibility, we tested the effects of overexpression of Flag-Atg16L or deletion constructs containing the coiled-coil domain on the molecular mass of the Atg16L complex by size exclusion chromatography. Unexpectedly, the molecular mass of the Atg16L complex was not shifted by the overexpression of these constructs (Figure 4, B–E). Moreover, only very low levels of the deletion constructs were found in fractions four to seven, which contained the Atg16L complex (Figure 4, D and E). These results indicate that the Atg16L complex remained intact in the presence of excess Atg16L coiled-coil region. We therefore reasoned that the Atg16L complex is stable, that it does not undergo frequent subunit exchange, and that transient overexpression of Atg16L does not interfere with pre-existing Atg16L complexes.
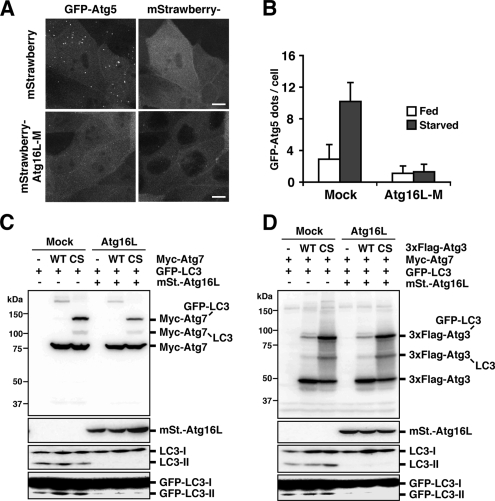
We next observed the localization of the complex to determine whether it was affected by overexpression of Atg16L. Administration of wortmannin, an inhibitor of phosphatidylinositol 3-kinases (PI3Ks), is the sole condition known to inhibit Atg5 localization (Mizushima et al., 2001). MDCK cells stably expressing GFP-Atg5 were infected with recombinant adenoviruses bearing mStrawberry or mStrawberry-Atg16L-M, and the number of GFP-Atg5 dots was counted. In contrast to the effect seen with Atg12 (Figure 2, C and D), cells expressing mStrawberry-Atg16L-M showed a dramatic reduction in GFP-Atg5 dots (Figure 5, A and B). These results indicate that overexpression of Atg16L precludes the ∼800-kDa multimeric complex from localizing to the membrane. A probable explanation for this observation is that a hypothetical Atg16L-interacting factor, which is required for membrane localization of the Atg16L complex, is titrated out by the excess amount of Atg16L. Loss of the correct localization of the complex at the hypothetical source of the isolation membrane leads to inhibition of LC3 lipidation.
Figure 5.
The effect of Atg16L overexpression on membrane localization of the Atg16L complex and the LC3 lipidation reaction. (A and B) MDCK cells stably expressing GFP-Atg5 were infected with adenovirus bearing mStrawberry (Mock) or mStrawberry-Atg16L-M and incubated for 40 h. The cells were then cultured in HBSS (Starved; A and B) or growth medium (Fed; B) for 2 h and fixed. Three-dimensional image stacks were obtained from sequential optical sections acquired 0.3 μm apart by confocal laser scanning microscopy (FV1000, Olympus) (A). Bar, 10 μm. More than 100 mStrawberry-positive cells were examined for the number of GFP-Atg5 dots per cell (B). Data are represented as means ± SD. (C and D) PC12 cells were coinfected using a combination of recombinant adenoviruses as indicated. After 40-h incubation, cells were collected. Lysates were examined by Western blotting using each antibody. (C) From top panel, anti-myc, anti-RFP, anti-LC3, and anti-GFP. (D) From top panel, anti-Flag, anti-RFP, anti-LC3, and anti-GFP.
Finally, we asked which step in the lipidation of LC3 was inhibited by Atg16L overexpression. We used mutant constructs of the E1 (Atg7C567S) and E2 (Atg3C264S) enzymes whose active-site cysteine residues are replaced by serine (Tanida et al., 2001, 2002). As shown in Figure 5C, in PC12 cells overexpressing Atg16L, Atg7-LC3 was generated at levels comparable to the mock control. Similarly, normal amounts of Atg3-LC3 were generated (Figure 5D). The Atg3-LC3 reaction kinetics was also unaffected by Atg16L overexpression (Supplementary Figure 1, A, C, and D). These results show that the excess amount of Atg16L inhibited the step that followed the formation of the Atg3-LC3 intermediate, but had no effect before the E1 and E2 reaction steps.
Ectopic Atg12-Atg5 Directs the LC3-Conjugation Reaction to the Plasma Membrane
Collectively, the above results suggest that the Atg16L complex is directly involved in the LC3 conjugation reaction. One hypothesis is that the Atg16L complex is the direct determinant of the location of the conjugation reaction. If so, LC3 lipidation would take place on membranes where the Atg16L complex is localized. To directly assess this hypothesis, we ectopically targeted the Atg12-Atg5 conjugate to the plasma membrane (PM). We chose the PM as the target in this experiment because PE is abundant in the inner leaflet of this membrane (Emoto and Umeda, 2000).
To target the Atg12-Atg5 conjugate to the PM, the C-terminus of full-length Atg16L or the Atg16L-N terminus involved in Atg5 binding were fused to 17 amino acids(KDGKKKKKKSKTKCVIM) of K-ras (Atg16L-NKras-CAAX), which is sufficient to target the heterologous protein to the PM (Hancock et al., 1991). Both constructs were predominantly localized to the PM, but mStrawberry-Atg16L-NKras-CAAX showed a more clear PM localization; therefore we used this construct for further analysis (Figure 6A and not shown). The Atg16L-NKras-CAAX protein expressed in cells was recovered entirely in the pellet fraction by centrifugation at 100,000 × g for 60 min (data not shown). GFP-Atg5 was also recruited to the PM by mStrawberry-Atg16L-NKras-CAAX expression (Supplementary Figure 4).
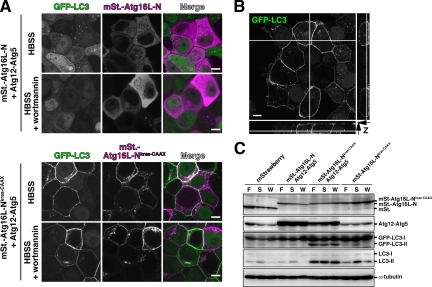
Figure 6.
The effect of ectopic Atg12-Atg5 localized to the plasma membrane on the LC3 lipidation reaction. (A) MCF7 cells stably expressing GFP-LC3 were coinfected using a combination of recombinant adenoviruses. After 40-h incubation, the cells were cultured in HBSS or HBSS with 100 nM wortmannin for 2 h, fixed, and observed by confocal laser-scanning microscopy (FV1000, Olympus). Bar, 10 μm. (B) MCF7 cells stably expressing GFP-LC3 were coinfected with recombinant adenovirus bearing Atg12, Atg5, and Flag-Atg16L-NKras-CAAX. After 40-h incubation, the cells were cultured in HBSS for 2 h, prepermeabilized for 5 min in PEM buffer (80 mM PIPES-KOH, pH 6.8, 5 mM EGTA, 1 mM MgCl2) containing 50 μg/ml digitonin, and fixed. Samples were observed by confocal laser scanning microscopy (FV1000, Olympus). Lateral images reconstituted from Z-sectioning are also shown. Bar, 10 μm. (C) MCF7 cells stably expressing GFP-LC3 were infected using a combination of recombinant adenoviruses as indicated. After 40-h incubation, the cells were grown for 2 h in growth medium (F), HBSS (S), or HBSS with 100 nM wortmannin (W). Lysates were examined by Western blotting using each antibody. From top panel, anti-RFP, anti-Atg5, anti-GFP, anti-LC3, and anti-α-tubulin.
Having established that we can target/mislocalize the Atg16L and Atg5 proteins to the PM, we examined the effect on LC3 lipidation. MCF7 cells stably expressing GFP-LC3 were coinfected with recombinant adenoviruses bearing mStrawberry-Atg16L-NKras-CAAX, Atg12, and Atg5. As shown in Figure 6, A and B, GFP-LC3 signals appeared clearly on the PM, and this was not affected by wortmannin treatment; however, starvation-induced cytosolic LC3 dots representing autophagosomes were greatly reduced. Similar results were obtained in MDCK and PC12 cells. We then examined how LC3 lipidation was affected by the Atg12-Atg5· mStrawberry-Atg16L-NKras-CAAX complex. As shown in Figure 6C, LC3 lipidation was robustly facilitated when mStrawberry-Atg16L-NKras-CAAX, Atg12, and Atg5 were coexpressed. Interestingly, the Atg16LKras-CAAX-dependent LC3 lipidation was phosphatidylinositol 3-phosphate- and starvation signal-independent. Even under nutrient-rich conditions that support only low basal levels of LC3 lipidation, these constructs facilitated LC3 lipidation. LC3 lipidation is usually dependent on PI3K activity, because wortmannin treatment prevents this modification. However, these constructs enable LC3 lipidation in the presence of wortmannin. Under these conditions, the intracellular GFP-LC3 dots representing autophagosomes virtually disappear, but the PM-localized GFP-LC3 remains, indicating that lipidated LC3 must be localized on the PM. Overexpression of the Atg16L coiled-coil region did not inhibit the PM localization of GFP-LC3 (Supplementary Figure 5). This result indicates that the inhibitory effect of Atg16L on autophagy is due to the mistargeting of the Atg16L complex; the overexpressed protein no longer localizes to the site at which the lipidation reaction normally occurs.
DISCUSSION
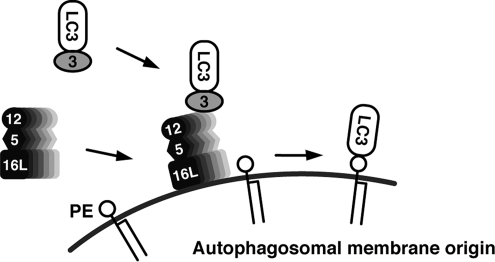
In this study, we have elucidated the function of the Atg16L complex. This complex brings LC3 to the site of lipidation and catalyzes the final step of the reaction. On the basis of these results, we propose a dynamic localization model of LC3 lipidation (Figure 7). On induction of autophagy, the Atg16L complex relocalizes from the cytosol to an undetermined membrane in an Atg16L-dependent manner. The high-energy Atg3-LC3 intermediate, which has been activated by Atg7, is recruited to the membrane via the interaction between Atg3 and Atg12 in the Atg16L complex. This brings LC3 into proximity with PE in the membrane, leading to subsequent lipidation.
Figure 7.
The dynamic localization model of LC3 lipidation. On induction of autophagy, the Atg16L complex in the cytosol localizes on a yet undetermined membrane in an Atg16L-dependent manner. The high-energy Atg3-LC3 intermediate activated by Atg7 is recruited to that membrane via the interaction between Atg3 and Atg12. LC3 is thus brought into proximity with PE in the membrane, leading to lipidation. The Atg16L complex functions as a scaffold on which LC3 is transferred from Atg3 to PE.
In vitro reconstitution experiments show that the minimum components necessary for Atg8/LC3 lipidation are Atg7, Atg3, Atg8/LC3, phospholipid-containing liposomes, and ATP (Ichimura et al., 2004; Sou et al., 2006). In the presence of Atg8/LC3 and PE at much higher concentrations than those seen in vivo, PE-containing micelles/vesicles and Atg8/LC3 will encounter each other at some frequency sufficient to support the lipidation reaction. The Atg12-Atg5 conjugate has been reported to facilitate the formation of Atg8-PE in vitro (Hanada et al., 2007). Atg16 is necessary for efficient Atg8-PE formation in vivo; however, little additive effect of Atg16 on PE conjugation of Atg8 was observed in vitro (Hanada et al., 2007). According to our model, the putative membrane factor that is recognized by Atg16 is probably not present on artificial liposomes. However, we show herein that Atg16L is directly involved in LC3 lipidation by specifying the site of the reaction. Based on both the in vitro and in vivo results, it is reasonable to propose that in vivo, the Atg16L complex functions as a scaffold on which LC3 is transferred from Atg3 to PE. In an analogy to the ubiquitination reaction, the complex may be regarded as an E3-like factor in the LC3 conjugation system in the sense that the E3 component plays a role in associating the substrate with the E2 enzyme (Suzuki et al., 2005; Matsushita et al., 2007). Generally, an E3 specifically recognizes its protein substrates; however, in this case, the recognition process may be attributed to the binding of the complex to the membrane. Thus, the Atg16L complex is a unique and new type of E3, which recognizes specific membranes as substrates. The mechanism by which LC3 is targeted to specific membranes has been mysterious, because PE is an abundant phospholipid that may be present in all membrane organelles, and unlipidated LC3 is a soluble protein. Our results show that membrane targeting of LC3 is defined by the Atg16L complex, and not some intrinsic targeting information within LC3. The in vivo target of LC3 is restricted to PE, but phosphatidylserine (PS) can be a target of LC3 in vitro (Sou et al., 2006). The selectivity in vivo may be related to the membrane localization of the Atg16L complex.
Our results demonstrate that the site of Atg16L complex localization is the site of lipidation. This place is likely critical for autophagosome formation, because Atg16L complex forced to localize to the PM enables LC3 lipidation, but not autophagosome formation. The only known place where Atg5 (or Atg16L) localizes is the outer surface of the isolation membrane, the forming autophagosome in mammalian cells (Mizushima et al., 2001). Therefore, the isolation membrane is the premier candidate for the location of LC3 lipidation. The riddle associated with this possibility is that recruitment of the Atg3-LC3 intermediate to forming autophagosomes does not explain how new lipid, including PE, is supplied. Recently, Nakatogawa et al. (2007) showed that Atg8-PE is potent in causing the hemifusion of vesicles/micelles in vitro, and this property may be related to the membrane elongation step of autophagosome formation. An alternative possibility is that there may be an undetected, and probably transient, association of the Atg16L complex to a membrane structure that enables the lipidation of LC3. In this case, lipidated LC3 would be delivered to the autophagosome after lipidation, possibly together with the Atg16L complex. Such a membrane could be assigned as the source of autophagosomal membrane lipids. In the case of yeast, the PAS is such a candidate. Yeast genetic studies showing a reciprocal dependence of PAS localization of the Atg12 system and Atg8 can be explained by our model (Suzuki et al., 2007). However, a membrane structure has not yet been detected at the PAS. Whether the PAS model is applicable to mammals is a further interesting question.
The PM-localized Atg16L complex bypasses two important factors that are normally critical for LC3 lipidation: starvation signaling and PI3K signaling (Kabeya et al., 2000). This result indicates that the output of these signaling pathways is the targeting of the Atg16L complex to the source membrane. The simplest model is that the autophagy-specific PI3K complex, which includes Beclin-1, is activated by starvation, and the resulting phosphatidylinositol 3-phosphate recruits the Atg16L complex; however, another more complex mechanism may be involved. Most importantly, we showed that targeting of the Atg16L complex to the membrane is sufficient for LC3 lipidation. This indicates that targeting of the Atg16L complex is an important step in terms of the regulation of autophagosome formation.
We showed that overexpresssion of the Atg16L coiled-coil region affects only the localization of the Atg16L complex, but not its function as a lipidation catalyst. The mechanism of inhibition remains unknown, but we speculate that a factor associating with the Atg16L coiled-coil region is titrated out by the excess Atg16L. Such a factor would interact with the Atg16L complex and be necessary for its localization. We are now pursuing the identification of this factor, which must be key for autophagosome formation.
We have uncovered a direct functional linkage between the Atg12 and LC3 systems. The most important finding is that the Atg16L complex may specifically recognize the membrane origin of the autophagosome. Hence, the mechanism of membrane localization of the Atg16L complex is key to understanding membrane dynamics in autophagy. By pursuing the details, we are approaching the core of this longstanding fundamental question in autophagy.
Crohn's disease is a common form of chronic inflammatory bowel disease of unknown etiology (Travis et al., 2006). Recently, an association between Crohn's disease and variant Atg16L has been reported, although the mechanisms by which the variant predisposes to intestinal inflammation is unknown (Consortium TWTCC, 2007; Hampe et al., 2007; Rioux et al., 2007). Our findings may be an important step toward understanding this disease, by providing information about the precise function of Atg16L and the regulation of autophagy via Atg16L.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Shunsuke Kimura (Yoshimori lab) for the gift of MDCK cells stably expressing GFP-LC3 or GFP-Atg5; Dr. Kouichi Matsunaga (Yoshimori lab) for the gift of MCF7 cells stably expressing GFP-LC3; Dr. Roger Y. Tsien for the gift of mStrawberry cDNA; Dr. Noboru Mizushima (Tokyo Medical and Dental University, Japan) for the gift of anti-Atg16L antibody; Dr. Kyouhei Umebayashi (Yoshimori lab) for helpful discussions; and Dr. Daniel Klionsky (University of Michigan) for critical reading of the manuscript. The work described in this report was supported in part by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT).
Abbreviations used:
- HBSS
Hanks' balanced salt solution
- PE
phosphatidylethanolamine
- PI3K
phosphatidylinositol 3-kinase
- PM
plasma membrane.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1257) on March 5, 2008.
REFERENCES
- Consortium TWTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Emoto K., Umeda M. An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 2000;149:1215–1224. doi: 10.1083/jcb.149.6.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Noda N. N., Fujii K., Yoshimoto K., Ohsumi Y., Inagaki F. In vitro reconstitution of plant ATG8 and ATG12 conjugation systems essential for autophagy. J. Biol. Chem. 2007;283:1921–1928. doi: 10.1074/jbc.M706214200. [DOI] [PubMed] [Google Scholar]
- Hampe J., et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn's disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohusmi Y. The ATG12-ATG5 conjugate has a novel e3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hanada T., Ohsumi Y. Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy. 2005;1:110–118. doi: 10.4161/auto.1.2.1858. [DOI] [PubMed] [Google Scholar]
- Hancock J. F., Cadwallader K., Paterson H., Marshall C. J. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Imamura Y., Emoto K., Umeda M., Noda T., Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Juhasz G., Neufeld T. P. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Matsushita M., Suzuki N. N., Obara K., Fujioka Y., Ohsumi Y., Inagaki F. Structure of Atg5.Atg16, a complex essential for autophagy. J. Biol. Chem. 2007;282:6763–6772. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Ohsumi Y., Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Noda T., Suzuki K., Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Rioux J. D., et al. Genome-wide association study identifies new susceptibility loci for Crohn's disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sou Y. S., Tanida I., Komatsu M., Ueno T., Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- Stromhaug P. E., Berg T. O., Fengsrud M., Seglen P. O. Purification and characterization of autophagosomes from rat hepatocytes. Biochem. J. 1998;335(Pt 2):217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Suzuki N. N., Yoshimoto K., Fujioka Y., Ohsumi Y., Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;1:119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- Tanida I., Tanida-Miyake E., Komatsu M., Ueno T., Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Tanida I., Tanida-Miyake E., Ueno T., Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- Travis S. P., et al. European evidence based consensus on the diagnosis and management of Crohn's disease: current management. Gut. 2006;55(Suppl 1):i16–i35. doi: 10.1136/gut.2005.081950b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Yoshimori T. Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 2004;313:453–458. doi: 10.1016/j.bbrc.2003.07.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.