Abstract
The AP-3 adaptor complex targets selected transmembrane proteins to lysosomes and lysosome-related organelles. We reconstituted its preferred interaction with liposomes containing the ADP ribosylation factor (ARF)-1 guanosine triphosphatase (GTPase), specific cargo tails, and phosphatidylinositol-3 phosphate, and then we performed a proteomic screen to identify new proteins supporting its sorting function. We identified ≈30 proteins belonging to three networks regulating either AP-3 coat assembly or septin polymerization or Rab7-dependent lysosomal transport. RNA interference shows that, among these proteins, the ARF-1 exchange factor brefeldin A-inhibited exchange factor 1, the ARF-1 GTPase-activating protein 1, the Cdc42-interacting Cdc42 effector protein 4, an effector of septin-polymerizing GTPases, and the phosphatidylinositol-3 kinase IIIC3 are key components regulating the targeting of lysosomal membrane proteins to lysosomes in vivo. This analysis reveals that these proteins, together with AP-3, play an essential role in protein sorting at early endosomes, thereby regulating the integrity of these organelles.
INTRODUCTION
In eukaryotic cells, the identity of organelles is maintained by mechanisms controlling their protein and lipid contents. Thus, protein and lipid sorting into transport intermediates shuttling between the Golgi and the endosomal/lysosomal system contributes to maintain the integrity of both systems (Munro, 2005). This process requires coat proteins that concentrate selected transmembrane proteins into specific transport intermediates by interacting directly or indirectly with sorting signals contained in their cytoplasmic domains (Bonifacino and Glick, 2004).
Many studies in yeast, Drosophila, and mammalian cells have illustrated that the heterotetrameric AP-3 adaptor complex, which belongs to a family composed of AP-1, AP-2, and AP-4, functions in the targeting of selected transmembrane proteins to lysosomes and lysosome-related organelles (Owen et al., 2004; Robinson, 2004). Thus, AP-3 dysfunction in mammalian cells leads to a missorting of lysosomal proteins such as lysosomal-associated membrane protein (LAMP)-1, lysosomal integral membrane protein (LIMP)-II, or CD63 toward the plasma membrane (Le Borgne et al., 1998; Dell'Angelica et al., 1999; Feng et al., 1999). The sorting function of AP-3, which localizes to the trans-Golgi network (TGN) and to early endosomes (Dell'Angelica et al., 1997; Simpson et al., 1997; Peden et al., 2004), relies on interactions of its subunits with tyrosine- or dileucine-based sorting signals contained in cytoplasmic domains of lysosomal membrane proteins (Bonifacino and Traub, 2003). These cargo proteins are believed to follow a direct intracellular pathway from the biosynthetic to the endosomal/lysosomal system (Harter and Mellman, 1992). However, recent studies have raised the possibility that they could also follow an indirect route via the plasma membrane for subsequent delivery to lysosomes (Janvier and Bonifacino, 2005).
Thus far, only a few molecules of the AP-3 sorting machinery have been identified, making it difficult to fully understand where AP-3 functions and how it contributes to maintaining organelle identity. The ADP ribosylation factor (ARF)-1) guanosine triphosphatase (GTPase) regulates AP-3 binding onto membranes (Ooi et al., 1998; Drake et al., 2000) as it does for other coat components, such as AP-1 or Golgi-localized, γ-ear-containing, ARF-binding proteins that function in the sorting of mannose 6-phosphate receptors (MPRs), or other proteins shuttling between the biosynthetic and the endocytic pathway (D'Souza-Schorey and Chavrier, 2006). Phosphatidylinositides (PIPs) are known to confer, at least in part, identities to membrane domains, by regulating the activities of ARF effectors and thus local ARF activation necessary for subsequent coat binding (De Matteis and Godi, 2004; Di Paolo and De Camilli, 2006). Thus, the ARF-1 GTPase-activating protein (AGAP)1, whose activity is modulated by specific phosphatidylinositols (PIs), plays a critical role in AP-3-dependent sorting (Nie et al., 2003). PIPs also provide additional binding sites for APs. For example, phosphatidylinositol 4-phosphate (PI-4P) is required for AP-1 binding (Heldwein et al., 2004; Baust et al., 2006), whereas phosphatidylinositol 4,5-bisphosphate is required for AP-2–dependent sorting (Honing et al., 2005). PIPs are synthesized by specific kinases (Di Paolo and De Camilli, 2006). For example, the PI-4 kinase (PI-4K) IIα has been implicated in AP-1 binding in vivo (Wang et al., 2003). This kinase is also present on purified AP-3–coated membranes (Salazar et al., 2005), thereby suggesting that AP-3, like AP-1 (Wang et al., 2003), could sort membrane proteins in PI-4P–rich membrane domains, i.e., the TGN.
We have shown previously that proteomic screens applied to in vitro systems fully recapitulating AP-1 coat assembly could be used to identify protein networks supporting its sorting function (Baust et al., 2006). Following a similar strategy here, we first set out to reconstitute AP-3 recruitment onto synthetic membranes. We then identified the recruited proteins by mass spectrometry, and we investigated the potential implication of several key components in the AP-3–dependent pathway by using RNA interference.
MATERIALS AND METHODS
Chemicals, Antibodies, and Plasmids
Chemicals were purchased from Sigma-Aldrich (Munich, Germany), lipids were from Avanti Polar Lipids (Alabaster, AL), and phosphoinositides were from Echelon Biosciences (Salt Lake City, UT). The antibodies used were as follows: polyclonal antibodies against the AP-3 s-subunit (Le Borgne et al., 1998), Rab5 (United States Biological, Swampscott, MA), Myc (Upstate Biotechnology, Charlottesville, VA), septin2 (a gift from Dr. W. S. Trimble, University of Toronto, Toronto, Canada), septin7 (a gift from Dr. I. G. Macara, University of Virginia School of Medicine, Charlottesville, VA), and p60 anti-Rab7 (a gift from Dr. A. Wandinger-Ness, University of New Mexico, Albuquerque, NM); and monoclonal antibodies against ARF-1 (1D9; Dianova, Hamburg, Germany); COP-I β-subunit (maD), AP-1 γ-subunit (100/3), AP-2α-subunit (100/2), and β-tubulin (Sigma-Aldrich); clathrin heavy chain, AP-3 δ-subunit, β-PIX, and early endosomal antigen 1 (EEA1) (BD Biosciences, Heidelberg, Germany), Myc (9E10; MPI-CBG Antibody Facility, Dresden, Germany); green fluorescent protein (GFP; Roche Applied Science, Mannheim, Germany); and LAMP-1 and LAMP-2 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). Secondary antibodies were peroxidase-conjugated goat anti-mouse immunoglobulin (Ig)G and goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Suffolk, United Kingdom), Alexa Fluor 546-conjugated goat anti-mouse IgG, and Alexa Fluor 488 goat anti-mouse IgG (Invitrogen, Karlsruhe, Germany). The cDNA for human Cdc42 effector protein 4 (Borg4) (RZPD, Berlin, Germany) was cloned in a pCMV-Tag3B vector (Stratagene, La Jolla, CA).
Liposomes and Peptides
Liposomes were formed and sized to 400 nm from a mixture of PC:PE:PS:cholesterol:anchor (4:3:1:1:1 M ratio) in chloroform/methanol as described previously (Baust et al., 2006). Phosphoinositides in chloroform/methanol were added as 1% molar ratio. In standard assays, 10 μl of liposomes were used corresponding to ≈10 nmol of lipid and 0.5 nmol of peptide per reaction. The peptides used in this study were chemically modified at their N termini as described previously (Baust et al., 2006), and they had the following amino acid sequences: LAMP-1 wt (GGRKRSHAGYQTI), LAMP-1 Y10A (GGRKRSHAGAQTI), LIMP-II wt (GRGQGSTDEGTADERAPLIRT), LIMP-II L18/I19A (GRGQGSTDEGTADERAPAART), gpI wt (GGKRMRVKAYRVDKSPYNQSMYYAGLPVDDFEDSESTDTEE), and gpI Y10, 23A DAC (GGKRMRVKAARVDKSPYNQSMYAAGLPVDDF).
Coat Recruitment Assay
Assays were performed in a total volume of 200 μl of recruitment buffer (25 mM HEPES-KOH, pH 7.2, 125 mM potassium acetate, 2.5 mM magnesium acetate, and 1 mM dithiothreitol), containing brain cytosol (10 mg/ml), liposomes (0.5 nmol of peptide per reaction) and guanosine 5′-O-(3-thio)triphosphate (GTPγS) (0.1 mM), and binding reactions were performed as described previously (Baust et al., 2006). ARF-depleted cytosol was generated by gel filtration on Sephadex G50 and tested by Western blotting by using anti-ARF-1 antibodies. For mass spectrometric analyses, LIMP-II/PI-3P–containing liposomes were incubated with mouse brain cytosol and GTPγS, purified by flotation on sucrose density gradients, and concentrated by centrifugation. Liposome-bound proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE).
Electron Microscopy
AP-3–coated liposomes recovered by centrifugation were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, and then they were postfixed with 1% osmium tetroxide. Membranes were then embedded in Epon. Thin sections were contrasted with uranyl acetate and lead citrate.
Protein Identification by Mass Spectrometry
Sample preparation, in-gel digestion, peptide extraction, and peptide separation were performed as described previously (Czupalla et al., 2005). Liquid chromatography/tandem mass spectrometry analysis was performed on a Micromass CapLC liquid chromatography system and a quadrupole orthogonal acceleration time-of-flight mass spectrometer Q-TOF Ultima (Micromass, Manchester, United Kingdom) equipped with a Z-spray nanoelectrospray source. Protein identification by fragment ion analysis was performed using MASCOT (Matrix Science, London, United Kingdom). Search criteria were as follows: database, SwissProt (version 271205, 204086 sequences); taxonomy, mouse; mass accuracy, 0.5 Da for fragment analysis; and modifications, carbamidomethylation and methionine oxidation, maximum one missed cleavage site.
Small Interfering RNA (siRNA) Treatment and Antibody Uptake Assay
siRNAs targeting human proteins were used, with the following sequences (5′→3′): PI-4K IIα (GGAUCAUUCCUGUCUUCAAtt, GGUAGUAUACCUGGCCAGUtt, ACCUAUAUGAACUCUUCTT), phosphatidylinositol 3-kinase (PI-3K) IIIC3 (GGAACAACGGUUUCGCUCUtt, GCUGUUAUCCUCUCAUUACtt, GCAUUGUUGAAGGGUGAUAtt), Borg4 (GGAAUAGGUUUUCCUCUGUtt, CCCUUGAUUCAGACCAUGGtt), ARAP1 (GGAAGGUGUGGACAGAAACtt, GGUAGUGAAAAUGAAAUGCtt), AP-1μ (GCCAGAUCAGUAUUGACCCtt, GCUCCCAGGUACCAAAAAGtt), AP-1γ (GGAAGUUAUGUUCGUGAUGtt, GGCCAAUGAUUUAUUGGAUtt), and AP-3μ (GGAAUUACAGUGACAGUUCtt, GGUAGUAAAAGGGAACAUUtt). These siRNAs and a nontargeting control (siNon2) were purchased from Ambion (Foster City, CA). siRNAs against human brefeldin A-inhibited exchange factor 1 (Big1) (5′-UGAAUCACCUCAACUUAGAuu-3′) (Shen et al., 2006) were purchased from Dharmacon Research (Chicago, IL), and siRNAs against β-PIX (CCUUCAUGCGCCUGGAUAA, GCAGACCAGUGAGAAGUUA) were purchased from Invitrogen. The siRNAs used for knockdowns in the antibody uptake experiments are shown in italics.
HeLa cells stably expressing a GFP-tagged MPR (Waguri et al., 2003) were transferred to fresh medium, and then they were incubated with either 100 nM siRNAs and Oligofectamine transfection reagent (Invitrogen) or with 20 nM siRNAs and Interferin transfection reagent (Polyplus, Illkirch, France), as recommended. Similar knockdown efficiencies have been observed for the two transfection conditions, as evaluated by Western blot when antibodies were available, or by quantitative polymerase chain reaction (QPCR) performed using the Brilliant SYBR Green QPCR Master Mix and a Mx400 QPCR System (Stratagene). The primers used for PCR were as follows: glyceraldehyde-3-phosphate dehydrogenase (5′-TCACCACCATGGAGAAGGC-3′, 5′-GCTAAGCAGTTGGTGGTGCA-3′), BIG1 (5′-ACCGTCTCCAGTAAGCCA-3′, 5′-TCAGGCTCTGTGTCATCC-3′), PI-3K IIIC3 (5′-CAGTCAGTTCCTGTGGCTG-3′, 5′-TATCCAGGTGCCTGTCTC-3′ and 5′-CTACATCCATCGGCTTCC-3′, 5′-GGACCTTCTGACCACGAT-3′), Borg4 (5′-CCAATCCTCAAGCAACTGG-3′, 5′-CTTCCTGGACAGGAGACT-3′ and 5′-GGCCAACAGAGGAAAACCT-3′, 5′-CAGGACCCTTGATTCAGAC-3′), and ARAP1 (5′-ACTTGCCCTCACGGTATG-3′, 5′-ATGGACAGGAAGGTGTGG-3′ and 5′-AGACGTTGCGCAGAAGAGA-3′, 5′-GCTGTGACACACAGATGGA-3′).
For antibody uptake, the siRNA-treated cells were incubated with antibodies diluted in DMEM with 1% bovine serum albumin and 25 nM HEPES, pH 7.4. HeLa cells were incubated either for 4 h with antibodies against LAMP-1 (0.8 μg/ml) or LAMP-2 (0.7 μg/ml) to monitor the integrity of the AP-3 pathway, or for 4 h with anti-GFP antibodies (1 μg/ml) to assess the integrity of the AP-1/MPR pathway. Cells were further washed with phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde for 15 min at room temperature, permeabilized with 0.1% Triton X-100 for 5 min at room temperature, blocked with 3% bovine serum albumin for 30 min at room temperature, and then incubated with Alexa Fluor-labeled secondary antibodies for 20 min at room temperature. Finally, they were washed with PBS and mounted on microscope slides by using Mowiol (Calbiochem, Nottingham, United Kingdom). Samples were analyzed by confocal fluorescence microscopy, using a confocal laser scanning microscope LSM510 Meta and C-Apochromat 63×/1.20W and 40×/1.20W objectives (Carl Zeiss MicroImaging, Göttingen, Germany). For the analyses of the localizations of AP-3δ, AP-1γ, and of anti-GFP (4-h uptake), we used a Leica SP5 inverse confocal microscope, with HCX plan apochromat lambda blue 63×/1.4–0.60 or HCX plan apochromat 40×/1.25–0.75 objectives (Leica Microsystems, Mannheim, Germany). Cell fluorescence intensities per area unit were measured and quantified using Adobe Photoshop 7 (Adobe Systems, Mountain View, CA) and ImageJ software (http://rsb.info.nih.gov/ij/). We observed similar knockdown efficiencies and similar effects on antibody uptakes for all indicated siRNAs that targeted PI-3K IIIC3, ARAP1, and AP-1γ, and the results shown represent an average of the oligonucleotides used for one protein. For Borg4 and β-PIX, we used only one siRNA for the antibody uptake experiments because the others lead to only ∼40% reduction in mRNA levels (data not shown). Such assays gave similar results as fluorescence-activated cell sorting analyses to monitor the amount of lysosomal membrane proteins or MPR present at the cell surface in siRNA-treated cells (data not shown).
RESULTS
We reconstituted AP-3 coat formation on synthetic membranes by using a strategy that we developed previously (Baust et al., 2006). Briefly, synthetic peptides corresponding to the cytoplasmic domains of the lysosomal membrane proteins LAMP-1 and LIMP-II, harboring a reactive group at their N termini, were first covalently coupled to a reactive lipid anchor incorporated into liposomes. These proteoliposomes were then incubated with a pig brain cytosol in the presence of the slowly hydrolysable GTPγS to stabilize GTPases in an active conformation. The proteoliposomes were purified, and the bound proteins were analyzed by Western blotting and identified by mass spectrometry.
AP-3 Binding Requires a Mosaic of Membrane Components: Intact Signals in Cargo Tails, ARF-1 and PI-3P
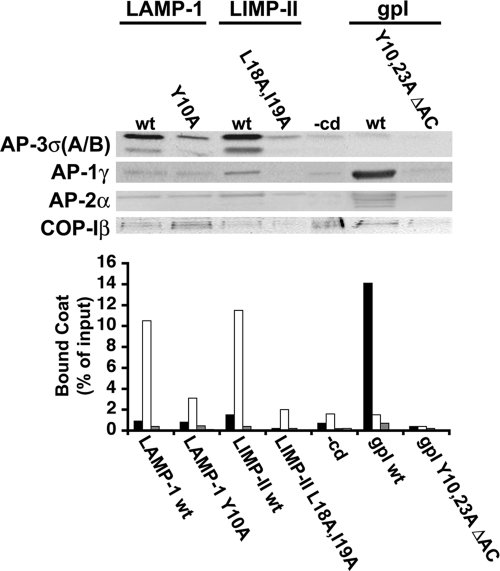
Figure 1 shows that AP-3, like AP-1, AP-2, and COP-I coats, bound to protein-free liposomes, as shown previously (Donaldson et al., 1991; Zhu et al., 1999; Drake et al., 2000; Haucke et al., 2000). However, AP-3 binding became more efficient and specific when LAMP-1 or LIMP-II cytoplasmic domains were incorporated onto liposomes. In contrast, AP-1, AP-2, and COP-I binding remained unaffected. Furthermore, AP-3 was not recruited onto liposomes containing LAMP-1 or LIMP-II tails with mutations in their tyrosine- or dileucine-based sorting signals, or onto liposomes containing the cytoplasmic tail of the varicella zoster virus glycoprotein I (gpI) that permits an efficient and specific recruitment of AP-1A (Baust et al., 2006).
Figure 1.
AP-3 recruitment requires intact sorting signals. Liposomes with cytoplasmic domains (wild type or mutants) of LAMP-1 or LIMP-II, or gpI, or no cytoplasmic domain (−cd) were incubated with cytosol, and GTPγS. AP-1, AP-3, AP-2, and COP-I bound to liposomes were detected and quantified after SDS-PAGE and Western blotting by using specific antibodies against AP-1γ (black bars), AP-3σA/B (white bars), AP-2α (dark gray bars), and COP-Iβ (light gray bars).
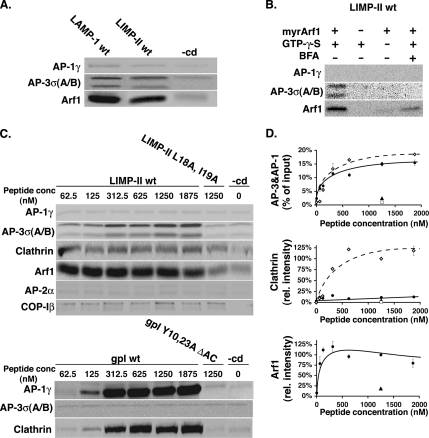
Interestingly, more ARF-1 was recovered onto liposomes when intact cytoplasmic domains of LIMP-II or LAMP-1 were present, probably due to the stabilization of AP-3 coats on liposomes (Figure 2A). The implication of ARF-1 in AP-3 recruitment was confirmed by using ARF-depleted cytosol complemented with myristoylated, recombinant ARF-1, supporting AP-3 recruitment in a brefeldin A-sensitive manner (Figure 2B). The amount of ARF-1 recruited onto liposomes, as that of AP-3, was influenced by the amount of LIMP-II coupled onto liposomes, reaching a 10-fold increase at maximal concentrations of cargo tail (Figure 2, C and D). This effect was not observed in the absence of an intact dileucine sorting signal or for other coats. Although ARF-1 and AP-3 recruitments were sensitive to the presence of LIMP-II tail, that of clathrin remained mostly unaffected. However, clathrin recruitment was readily detected when liposomes exposed at their surface the gpI cytoplasmic domain that also recruited AP-1A and ARF-1, as described previously (Baust et al., 2006), and relied on the presence of intact tyrosine-based sorting signals and acidic clusters in the gpI tail.
Figure 2.
AP-3 recruitment is ARF-1 dependent. (A) Liposomes with LAMP-1, LIMP-II, and no cytoplasmic domain (−cd) were incubated with cytosol and GTPγS. AP-1γ, AP-3σA/B, and ARF-1 bound to liposomes were detected after SDS-PAGE and Western blotting by using specific antibodies. (B) Liposomes with LIMP-II cytoplasmic domains were incubated with ARF-depleted cytosol (3 mg/ml) and supplemented or not with recombinant, myristoylated ARF-1 (30 μg/ml) with or without GTPγS and with or without brefeldin A (100 μg/ml). AP-1γ, AP-3σA/B, and ARF-1 bound to liposomes were detected after SDS-PAGE and Western blotting. (C) Top, liposomes with increasing amounts of LIMP-II wt (♦), LIMP-II L18A, I19A (■), or no cytoplasmic domain (−cd) (♦) were incubated with cytosol and GTPγS. Bottom, liposomes with increasing amounts of gpI (♦), gpI Y10, 23A, DAC (■), or no cytoplasmic domain (−cd) (♦) were incubated with cytosol in the presence of GTPγS. After incubation, AP-1γ, AP-2α, AP-3σA/B, clathrin, and COP-Iβ bound to liposomes were detected after SDS-PAGE and Western blotting. (D) Quantification of C. The clathrin signal was quantified and normalized to the values obtained with gpI-containing liposomes.
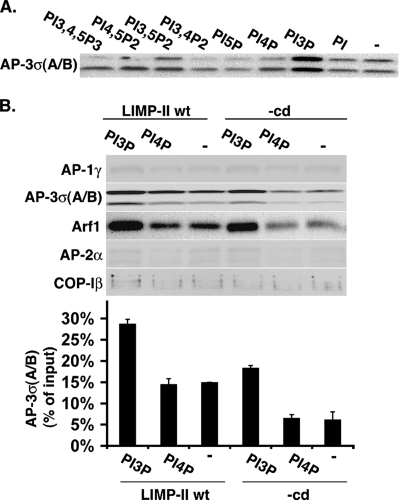
To determine which PIP regulates AP-3 binding, liposomes containing LIMP-II tails and various PIPs were incubated with GTPγS and limiting concentrations of cytosol (3 mg/ml). Figure 3A shows that AP-3 recruitment increased in the presence of PI-3P, whereas the other PIPs tested had no effect. Under optimal conditions, >25% of the cytosolic AP-3 was recruited onto liposomes containing LIMP-II and PI-3P. In contrast, the recruitments of AP-1, AP-2, and COP-I remained unaffected (Figure 3B). Interestingly, PI-3P also increased ARF-1 recruitment, even onto liposomes devoid of LIMP-II tail.
Figure 3.
PI-3P increases AP-3 recruitment. (A) Liposomes with LIMP-II tails containing different or no PIPs were incubated with cytosol (3 mg/ml), GTPγS, and phosphatase inhibitors. AP-3σA/B bound to liposomes was detected after SDS-PAGE and Western blotting. (B) Liposomes with or without LIMP-II tails and either PI-3P or PI-4P or no PIP were incubated with cytosol (3 mg/ml), GTPγS, and phosphatase inhibitors. AP-1γ, AP-2α, AP-3σA/B, COP-Iβ, and ARF-1 bound to liposomes were detected after SDS-PAGE and Western blotting. The AP-3σA/B signal was quantified.
Together, these results demonstrate that optimal AP-3 binding to synthetic membranes requires a mosaic of membrane components, minimally composed of intact sorting signals in specific cargoes, ARF-1 and PI-3P. In addition, AP-3 seems to be a poor clathrin interactor. Accordingly, electron microscopy detected only a few clathrin-coated structures, probably due to a residual, nonspecific AP-1 binding (Supplemental Figure S1).
Proteomic Analysis of AP-3–coated Liposomes
The cytosolic proteins bound onto LIMP-II– and PI-3P–containing liposomes were identified by mass spectrometry after fractionation by SDS-PAGE (Supplemental Figure S2). Our first analyses revealed that these proteoliposomes contained AP-3 as expected, but they also contained a significant amount of AP-1 (up to 30% of total mass), although our biochemical analysis showed that such liposomes did not recruit AP-1 in a specific manner. This observation probably reflects the low abundance of cytosolic AP-3 compared with AP-1. Thus, Table 1 only presents the ≈30 proteins specifically recruited together with AP-3 onto LIMP-II- and PI-3P–containing liposomes and it excludes those previously found on AP-1A–coated liposomes (Baust et al., 2006). Several groups of proteins were distinguished. The first group comprises coat components, i.e., the AP-3 subunits, ARF-1 effectors such as Big1 (Shen et al., 2006), the AP-3–associated AGAP1 (Nie et al., 2003), as well as ARAP1, an ARF-1 GTPase-activating protein that also contains a Rho GTPase-activating domain (Miura et al., 2002). Second, we found a Cdc42-dependent septin nucleation machinery, consisting of the Cdc42 effector protein 4 (Borg4) (Joberty et al., 2001) and seven members of the septin family (Kinoshita, 2006). This analysis also identified the Rho GTPase-activating protein 1 (p50RhoGAP), a potential linker between Rho and Rab GTPases (Sirokmany et al., 2006). The third group comprises Rab GTPases, i.e., Rab5C, Rab7, and Rab3C, as well as the Rab GTPase-activating protein TBC1 domain family member 10. The fourth group contains Rab5 interactors, such as the Rab5 effectors EEA1, Rabaptin-5, Rabex-5, the orthologue of the yeast Vps45 (Zerial and McBride, 2001), and huntingtin (Pal et al., 2006). The fifth group represents kinases and kinase effectors, such as the Cdc42-binding protein kinase α, protein kinase Cβ, and protein kinase Cα-binding protein (PICK1). Finally, several other proteins were also identified such as subunits of the vacuolar ATP synthase; GRIP1-associated protein 1 (GRASP-1), an endosomal protein with Ras GTPase nucleotide exchange activity (Stinton et al., 2005); the N-ethylmaleimide sensitive factor (NSF), or the syntaxin-binding protein 1.
Table 1.
Proteins bound to LIPM-II- and PI-3P-containing liposomes
| Identified proteins | NCBI gene identifier | Predicted Mr (Da) | Total MS/MS score | No. of sequenced peptides | Sequence coverage (%) |
|---|---|---|---|---|---|
| Coat components | |||||
| AP-3 d1 | 81882150 | 135,081 | 655 | 11 | 11 |
| AP-3 b1 | 18203657 | 122,798 | 548 | 10 | 11 |
| AP-3 b2 | 61219108 | 119,118 | 2009 | 33 | 42 |
| AP-3 m1 | 20531985 | 46,906 | 501 | 8 | 32 |
| AP-3 m2 | 66774020 | 46,886 | 654 | 10 | 43 |
| AP-3 s1 | 33112223 | 21,718 | 241 | 4 | 23 |
| AP-3 s2 | 33112221 | 22,003 | 185 | 4 | 23 |
| BIG1 | 82795382 | 184,075 | 526 | 10 | 7 |
| AGAP1 | 51338821 | 45,260 | 331 | 7 | 19 |
| ARAP1 | 68299130 | 162,234 | 134 | 3 | 2 |
| CDC42 effectors and septin proteins | |||||
| BORG4 | 21362404 | 37,846 | 600 | 8 | 37 |
| Neuronal-specific septin 3 | 13124538 | 52,792 | 186 | 3 | 8 |
| Septin-4 | 114978 | 54,901 | 298 | 5 | 12 |
| Septin-5 | 83305642 | 42,721 | 264 | 5 | 16 |
| Septin-6 | 20178348 | 49,457 | 59 | 1a | 5 |
| Septin-7 | 9789726 | 50,518 | 280 | 5 | 14 |
| Septin-8 | 45477305 | 49,781 | 249 | 5b | 19 |
| Septin-11 | 50401563 | 49,532 | 208 | 4b | 14 |
| Rho-GTPase-activating protein 1 (p50RhoGAP) | 3024550c | 50,404 | 191 | 4 | 12 |
| Rabs and effectors | |||||
| Rab5C | 38258917 | 23,398 | 722 | 10 | 62 |
| Rab7 | 46397834 | 23,475 | 677 | 11 | 63 |
| Rab3C | 51338605 | 25,856 | 484 | 7 | 39 |
| TBC1 domain family member 10 | 20454885 | 56,167 | 309 | 6 | 11 |
| EEA1 | 76363511 | 160,915 | 1491 | 26 | 22 |
| Rabaptin-5 | 47605961 | 99,490 | 574 | 10 | 14 |
| Rabex-5 | 56405101 | 56,833 | 278 | 4 | 10 |
| Vacuolar protein sorting-associated protein 45 | 23396903 | 65,012 | 568 | 10 | 20 |
| Huntingtin | 1708161 | 344,471 | 322 | 5 | 2 |
| Kinases and effectors | |||||
| CDC42 binding protein kinase alpha | 81174934d | 196,940 | 293 | 6 | 4 |
| Protein kinase C, β type | 55977813 | 76,844 | 275 | 6 | 10 |
| PICK1 | 22095978 | 46,517 | 489 | 8 | 21 |
| 14-3-3 ζ/δ (protein kinase C inhibitor protein 1) | 52000885 | 27,754 | 337 | 6 | 30 |
| A kinase anchor protein 10 | 71153491 | 73,586 | 559 | 9 | 20 |
| Others | |||||
| Vacuolar ATP synthase catalytic subunit A | 1718086 | 68,225 | 172 | 4 | 7 |
| Vacuolar ATP synthase catalytic subunit E | 1718091 | 26,571 | 120 | 2 | 8 |
| GRASP-1 | 76363168 | 92,715 | 412 | 6 | 8 |
| Syntaxin binding protein 1 | 48429206 | 67,526 | 235 | 4 | 6 |
| Vesicular-fusion protein NSF | 1171774 | 82,513 | 778 | 13 | 19 |
LIMP-II and PI-3P–containing liposomes were incubated with a mouse brain cytosol and GTPγS, and then they were purified on flotation gradient and analyzed by mass spectrometry as indicated in Materials and Methods.
a More sequenced peptides match to SEPT3/6/8/11.
b More sequenced peptides match to SEPT6/8/11.
c Human protein; no SwissProt entry available for an according mouse protein. All sequenced peptides also match with the mouse protein gi 13879250.
d Rat protein; no SwissProt entry available for an according mouse protein. All sequenced peptides also match with the mouse protein gi 94364571.
AP-3, Septins, and Rab7 Are Stabilized on the Same Membrane Domains
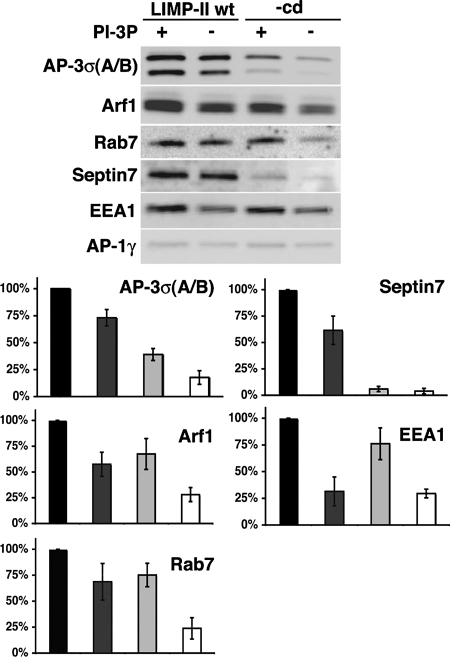
We then asked whether membrane domains containing LIMP-II tails and PI-3P could also promote the recruitment of several key proteins identified in our proteomic screen, as they do for AP-3. Figure 4 shows that the presence of LIMP-II tails not only stabilized AP-3 and ARF-1 but also septin7, known to form a complex with septin11, and Rab7. PI-3P could, to some degree, stabilize Rab7 on synthetic membranes, and efficient recruitment of Rab7 was also observed in the presence of LIMP-II tails alone. The most efficient binding of Rab7 required both LIMP-II tails and PI-3P. In contrast, Rab5 (data not shown) and its effector EEA1 were mostly stabilized on synthetic membranes by PI-3P. Thus, LIMP-II not only stabilized ARF-1 and AP-3, a process enhanced by PI-3P but also contributed to stabilize other components required for septin polymerization and subsequent membrane fusion.
Figure 4.
Requirements for stabilization of AP-3, septins, and Rab7 on liposomes. Liposomes with or without LIMP-II and with or without PI-3P were incubated with cytosol and GTPγS. After incubation, AP-3σA/B, AP-1γ, ARF-1, Rab7, and Septin 7 were analyzed by Western blotting by using specific antibodies. The recruitment of these markers was quantified (black bar, LIMP-II and PI-3P; dark gray bar, LIMP-II no PI-3P; light gray bar, PI-3P and no LIMP-II; and white bar, no LIMP-II and no PI-3P).
siRNA-based Functional Analyses
In AP-3–deficient mammalian cells, lysosomal membrane proteins are missorted toward the cell surface (Le Borgne et al., 1998; Dell'Angelica et al., 1999; Feng et al., 1999). This phenomenon can be quantified by measuring the uptake of exogenously added antibodies directed against the luminal domain of these cargoes (e.g., LAMP-1 or LAMP-2). This assay coupled to RNA interference was used to evaluate the potential importance of proteins in AP-3–dependent transport (Supplemental Figures S3 and S4). We also monitored possible alterations in MPR traffic. MPRs are in dynamic equilibrium among the TGN, endosomes, and the plasma membrane (Duncan and Kornfeld, 1988). Thus, we measured the uptake of anti-GFP antibodies exogenously added to siRNA-treated HeLa cells stably expressing a GFP-tagged MPR (Supplemental Figure S5). The intracellular distributions of AP-3, AP-1, and GFP-MPR were also monitored (Figure 6 and Supplemental S5 and S6).
Figure 6.
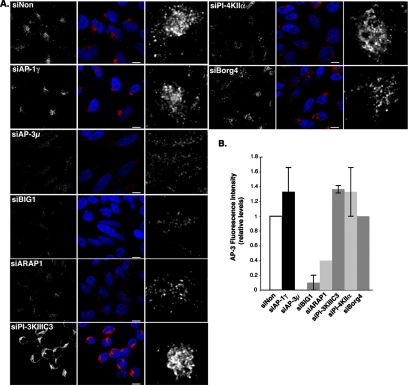
AP-3 localization in siRNA-treated cells. (A) Cells transfected for 72 h with siRNAs against the indicated molecules or with control nontargeting siRNAs (siNon) were then fixed, permeabilized, and labeled with antibodies against AP-3δ (red). The cells were analyzed by fluorescence confocal microscopy. Right, AP-3δ signal at higher magnifications. Bars, 10 μm. (B) Total cellular intensity of the AP-3δ signal was quantified for each condition, and the averages were plotted relative to control (value = 1) and to the siAP-3 (value = 0). n = 2 independent experiments performed in triplicates with 100 cells/condition.
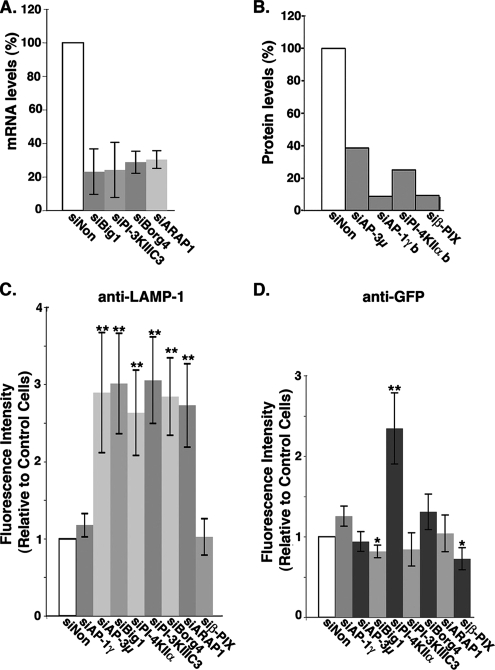
A first protein network identified in our proteomic screen comprises AP-3 subunits and proteins potentially regulating its ARF-1–dependent binding. A 75% reduction in the expression the ARF-1 guanine nucleotide exchange factor (GEF) Big1, a 70% reduction in the expression of ARF-1 GTPase activation protein (GAP) ARAP1 (Figure 5A), or a 60% reduction of AP-3 expression (Figure 5B and Supplemental Figure S3) resulted in a similar LAMP-1 missorting compared with control cells (Figure 5C). Accordingly, the amount of AP-3 bound to intracellular membranes was significantly reduced (Figure 6). The same knockdowns only slightly reduced the uptake of anti-GFP antibodies (Figure 5D and Supplemental Figure S5), although AP-3 and Big1 knockdowns resulted in the scattering of GFP-MPR–containing structures coated with AP-1 (Supplemental Figure S6). Alternately, AP-1γ knockdown resulted in a slightly higher accessibility of the GFP-MPR to the cell surface, whereas that of β-PIX, a Rho exchange factor previously found associated with AP-1 (Baust et al., 2006), resulted in a slight decrease (Figure 5D), without affecting LAMP-1 trafficking (Figure 5C). Thus, the ARF-1 GEF Big1 and the ARF-1 GAP ARAP1 seem to be key regulators of the AP-3–dependent sorting of LAMP-1.
Figure 5.
LAMP-1 lysosomal targeting in siRNA-treated HeLa cells. Cells were transfected for 72 h with siRNAs against the indicated molecules or with control nontargeting siRNAs (siNon). (A) mRNA levels after siRNA-mediated interference with siBig1, siBorg4, siARAP1, and siPI-3KIII C3 were evaluated by QPCR, and data were plotted relative to those in control siNon-treated cells. (B) siRNA-mediated knockdowns of AP-1, AP-3, PI-4K IIα, and β-PIX were evaluated by Western blots (also see Supplemental Figure S4) and plotted relative to control siNon-treated cells. (C and D) The uptakes of anti LAMP-1 (C) or anti-GFP antibodies (D) after the knockdown of the indicated molecules were analyzed by fluorescence confocal microscopy. Fluorescence levels were normalized, and data were plotted relative to those in control siNon-treated cells. Bars indicate average values ± SD (n = 4 independent experiments). **p < 0.005 and *p < 0.02 (analysis of variance single factor analysis).
Although our proteomic screen did not identify any PI-3 kinase, we explored the potential implication of the PI-3 kinase IIIC3, the mammalian orthologue of the yeast Vps34, whose activity is spatially controlled by the Rab5 GTPase (Shin et al., 2005). The knockdown of the PI-3 kinase IIIC3 (80% reduction) resulted in clear phenotypes. It induced a drastic accessibility of LAMP-1 to the cell surface (Figure 5), without affecting that of GFP-MPR (Figure 5 and Supplemental Figure S5), and it did not change either the intracellular distribution of GFP-MPR or that of AP-1 (Supplemental Figure 6). Interestingly, PI-3 kinase IIIC3 knockdown resulted in the loss of AP-3 binding onto peripheral structures, but it did not affect AP-3 binding onto perinuclear compartments (Figure 6). We also investigated the functional importance of the PI-4 kinase IIα, previously found to not only regulate AP-1 binding (Wang et al., 2003) but also recovered on purified AP-3–coated membranes (Salazar et al., 2005). The knockdown of the PI-4 kinase IIα (70% reduction) (Figure 5 and Supplemental Figure S3) resulted in phenotypes that were more difficult to interpret. First, it resulted in higher accessibilities of both LAMP-1 and GFP-MPR to the cell surface (Figure 5 and Supplemental S4 and S5). Second, GFP-MPR exhibited a more scattered distribution, being detected in structures only partially coated with AP-1 (Supplemental Figure S6). These phenotypes were not detected in the AP-1γ or β-PIX knockdowns (Figure 5 and Supplemental S5 and S6). Thus, the PI-3 kinase IIIC3 is a key regulator of the AP-3–dependent sorting of LAMP-1. However, it would seem that the PI-4 kinase IIα has a more pleiotropic effect, regulating both LAMP-1 and MPR trafficking in vivo.
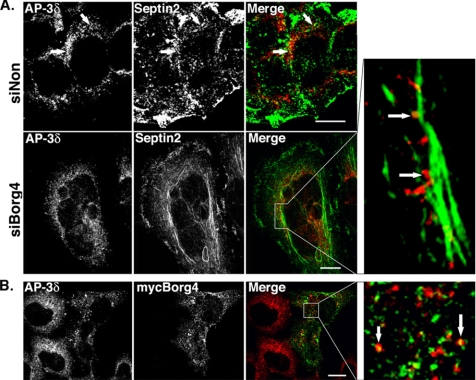
Another protein network identified in our proteomic screen is made of septin GTPases and Borg4, a Cdc42 effector protein that regulates their polymerization (Joberty et al., 2001). A 70% reduction in Borg4 expression (Figure 5A) or a 60% reduction in AP-3 expression (Figure 5B and Supplemental Figure S3) resulted in a similar LAMP-1 missorting (Figure 5C). Borg4 knockdown induced a slight scattering of AP-3–coated structures (Figure 6), and it resulted in the formation of longer septin2-positive filaments, sometimes forming bundles with actin (data not shown), along which AP-3–positive structures could be detected (Figure 7A). Borg4 knockdown resulted in a slight increase in anti-GFP antibody uptake (Figure 5 and Supplemental Figure S5) and in a dispersion of intracellular, GFP-MPR–containing structures still partially coated with AP-1 (Supplemental Figures S5 and S6). When a myc-tagged Borg4 was overexpressed in HeLa cells, the number of AP-3–coated structures decreased drastically. Nevertheless, the few remaining AP-3–coated structures partly localized with myc-tagged Borg4 (Figure 7B). Thus, the Cdc42-effector protein Borg4, and probably septin assembly, is an important regulator of AP-3–dependent sorting of LAMP-1 to lysosomes.
Figure 7.
Localization of AP-3δ in siBorg4 and myc-Borg4-treated cells. (A) HeLa cells were transfected with either control siNon or siBorg4, double labeled with antibodies against AP-3δ (red) and septin2 (green). (B) HeLa cells expressing myc-Borg4 were fixed, permeabilized, and labeled with antibodies against AP-3δ (red) and myc (green). Cells were then fixed and analyzed by fluorescence confocal microscopy. The arrows indicate AP-3δ–positive structures that overlap with septin2 (A) or myc-Borg4 (Figure 7B) are aligned along septin2-positive filaments. Bars, 10 μm.
DISCUSSION
Our study demonstrates that AP-3 binding to synthetic membranes requires a mosaic of membrane components, minimally composed of ARF-1, PI-3P, and sorting signals present in selected transmembrane proteins. These defined membrane domains bind other proteins and cellular machines, in particular, septin7 and Rab7, a GTPase controlling transport from early to late endosomes. Among the proteins, Big1, ARAP1, Borg4, and the PI-3 kinase IIIC3 were found to be essential for AP-3–dependent lysosomal targeting in vivo. Thus, our findings strongly suggest that AP-3 and the other machineries contribute to maintain the identity of early endosomes by sorting specific cargoes toward late endosomes.
A Mosaic of Membrane Components Stabilizes AP-3 on Synthetic Membranes
AP-3 binding requires several membrane components. First, the ARF-1 GTPase is a key regulator. If this was expected from previous studies (Drake et al., 2000, and references therein), our data show that specific ARF-1 effectors, i.e., Big1 and ARAP1, regulate AP-3–dependent lysosomal targeting, most likely by modulating a local ARF-1 activation and subsequent AP-3 binding. Our analysis also identified AGAP1, another ARF-1 GAP shown to regulate AP-3 function in vivo (Nie et al., 2003). Second, sorting signals present in specific cargo transmembrane proteins play a key role by increasing and providing specificity to AP-3 binding, an observation that fully supports our previous findings (Le Borgne et al., 1998). Interestingly, the presence of cargoes also enhances ARF-1 recruitment on synthetic membranes, as we observed previously for other cargoes on ARF-1 and AP-1A recruitment (Baust et al., 2006). This suggests that cargoes, probably by stabilizing APs on membranes, can trigger amplification loops to expand specialized membrane domains able to sort incoming cargo molecules. Finally, PI-3P is the most efficient phosphatidylinositide for stabilizing AP-3 on membranes. Accordingly, the PI-3 kinase IIIC3, the orthologue of the yeast Vps34p whose activity is spatially controlled by the Rab5 GTPase (Zerial and McBride, 2001; Shin et al., 2005), confers a selective LAMP-1 targeting. In contrast, the PI-4 kinase IIα, a kinase regulating AP-1 binding onto TGN membranes (Wang et al., 2003) and recovered on purified AP-3–coated structures (Salazar et al., 2005), is involved not only in MPR trafficking but also in LAMP-1 trafficking. This finding was surprising because our biochemical study did not detect any requirement of PI-4P for AP-3 binding. Clearly, further studies are needed to elucidate the mechanisms by which this kinase regulates protein trafficking and organelle homeostasis.
Collectively, our present and former in vitro studies (Baust et al., 2006) indicate that the two related AP-3 and AP-1 adaptor complexes, both functioning in lysosomal targeting, follow similar principles of binding to synthetic membranes. It is the combinatorial use of membrane components that determines their stable interactions. Their selective interactions however require different but related components: specific PIPs and lipid kinases; specific ARF GEFs and GAPs whose compartmentalization could mediate a local ARF-1 activation; and finally, selected cargoes with intact sorting signals exhibiting preferred interactions for either AP-3 or AP-1.
Protein Networks Detected on LIMP-II– and PI-3P–containing Liposomes
The identified proteins belong to three major protein networks involved in either AP-3 binding or septin dynamics or early sorting endosome dynamics. In addition, we identified several kinases and interacting proteins that could regulate the recruitment of these components onto membranes. Interestingly, several of these proteins, including Rab5C, Rab7, and Rab3C, the early endosomal marker EEA1, the catalytic subunits of the vacuolar ATP synthase, and PICK1, were found associated with AP-3–coated organelles isolated from cells (Salazar et al., 2005). The recruitment of Rab5 and its effectors, such as EEA1, is only determined by the presence of PI-3P on synthetic membranes, in agreement with previous studies (Zerial and McBride, 2001; Shin et al., 2005). In contrast, the recruitments of AP-3, the septin GTPases, and Rab7 need the presence of cargoes (septin7), or both cargoes and PI-3P (AP-3, Rab7). It is difficult to understand how cargo tails, which bind to AP coats, could stabilize septins and Rab7 on membranes. The simplest explanation is that AP-3 binds first to membranes in a cargo-dependent manner, and then it stabilizes the other machineries by virtue of protein–protein interactions.
We showed earlier that well-defined synthetic membrane domains not only recruit AP-1A coats, including ARF-1 and its specific effectors, but also a machinery regulating actin nucleation (Rac-1, its effectors, and the Wave/Scar complex) and the Rab11 and Rab14 GTPases regulating membrane dynamics between the TGN and endosomes (Baust et al., 2006). Collectively, our studies identifying >80 proteins, strongly suggesting that distinct, defined membrane domains stabilize either the AP-3 or the AP-1A coat, together with well-defined components: septin- or actin-based polymerization devices, respectively, as well as distinct Rab GTPases functioning along the two distinct steps of membrane traffic. Although several proteins identified in our screens contain domains that could molecularly connect coat assembly, cytoskeleton assembly and Rab-dependent membrane fusion, a future challenge will be to establish in terms of protein–protein interactions how these different protein networks are connected and how they precisely function in transport intermediate formation.
AP-3 Functions on Early Sorting Endosomes
Our analysis showing that PI-3P and the PI-3 kinase IIIC3 modulate AP-3 binding and LAMP-1 targeting indicates that AP-3 functions most efficiently in sorting on early sorting endosomes. This interpretation would agree with other morphological studies showing that AP-3 localizes to early endosomal exit sites together with LAMP-1 (Peden et al., 2004). The high affinity of AP-3 for specific cargoes would therefore prevent their access to the cell surface and permit their selective, intracellular transport to lysosomes where they reside. Disturbing early endosome homeostasis would result in their missorting to the cell surface and most likely perturb the recycling of other endocytosed cargoes recycling back to the TGN, such as MPRs, as also observed in wortmannin-treated cells (Kundra and Kornfeld, 1998). That AP-3 could contribute to stabilize Rab7 on membranes is interesting because this Rab GTPase controls protein transport from early to late endosomes (Press et al., 1998), a maturation process in which Rab5-rich membrane domains are converted into Rab7-rich membrane domains (Rink et al., 2005). In this regard, some proteins identified in our proteomic screen, such as the TBC1 domain family member 10 containing a Rab GAP domain, could potentially regulate this transition.
Borg4, Septins, and AP-3–dependent Protein Sorting
A surprising finding is the implication of Borg4 in AP-3–dependent lysosomal targeting. Borgs control septin organization, and they are negatively regulated by Cdc42 (Joberty et al., 2001). Interestingly, several septin (SEPT) family members, in particular septin11 and septin7, which form complexes, were identified in our proteomic screen. Septins are GTP-binding proteins involved in various cellular processes, including cytoskeleton organization, cytokinesis, and diffusion of molecules between different cellular domains (Barral et al., 2000; Kinoshita, 2006; Spiliotis and Nelson, 2006). In mammalian cells, septins regulate actin organization (Kremer at al., 2007) and also bind to microtubules (Surka et al., 2002; Nagata et al., 2003; Spiliotis et al., 2005), Very recently, tubulin-associated septin2 was shown to facilitate vesicle transport from the Golgi to the plasma membrane by maintaining polyglutamylated microtubule tracks and impeding tubulin binding of microtubule-associated protein 4, a regulatory step required for polarized, columnar-shaped epithelia biogenesis (Spiliotis et al., 2008). This process could potentially involve IQGAP1, a Cdc42 target, which associates with septin2–exocyst complexes and regulates insulin secretion by pancreatic β cells (Rittmeyer et al., 2008). How Borg4 and septins control AP-3–dependent targeting remains unknown. Protein transport from early to late endosomes includes several critical steps of protein sorting. During multivesicular body formation for example, ubiquitinated transmembrane proteins destined for degradation must be segregated away from other transmembrane proteins, such as LAMPs or LIMPs, which remain in the outer membrane. Alternately, cargo transport form early to late endosomes is facilitated by cytoskeleton elements, in particular, microtubules (Aniento et al., 1993). If our study highlights the functional importance of Borg4, and probably specific septins in AP-3–dependent transport, additional studies will be needed to elucidate their precise function in coordinating membrane traffic with cytoskeleton organization.
Supplementary Material
ACKNOWLEDGMENTS
We thank the laboratory members for advice and helpful discussions and Drs K. Simons and M. Zerial for the critical reading of the manuscript. We are grateful to M. Wilsch-Breuninger for performing the electron microscopy analysis. We are also thankful to Drs A. Wandinger-Ness, W. S. Trimble, and I. Macara for generous gifts of reagents. This work was supported in part by grants from TU-Dresden (HWP-1207), SMWK-EFRE 1203, Bundesministerium für Bildung und Forschung (0313815B), and Deutsche Forschungsgemeinschaft (TRR 13/2-08, HO 2584/2-1, and HO 2584/1-1).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0110) on February 20, 2008.
REFERENCES
- Aniento F., Emans N., Griffiths G., Gruenberg J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J. Cell Biol. 1993;123:1373–1387. doi: 10.1083/jcb.123.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y., Mermall V., Mooseker M. S., Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Baust T., Czupalla C., Krause E., Bourel-Bonnet L., Hoflack B. Proteomic analysis of adaptor protein 1A coats selectively assembled on liposomes. Proc. Natl. Acad. Sci. USA. 2006;103:3159–3164. doi: 10.1073/pnas.0511062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Traub L. M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Czupalla C., Mansukoski H., Pursche T., Krause E., Hoflack B. Comparative study of protein and mRNA expression during osteoclastogenesis. Proteomics. 2005;5:3868–3875. doi: 10.1002/pmic.200402059. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- De Matteis M. A., Godi A. PI-loting membrane traffic. Nat. Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Ohno H., Ooi C. E., Rabinovich E., Roche K. W., Bonifacino J. S. AP-3, an adaptor-like protein complex with ubiquitous expression. EMBO J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica E. C., Shotelersuk V., Aguilar R. C., Gahl W. A., Bonifacino J. S. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol. Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Di Fiore P. P., Polo S., Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat. Rev. Mol. Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G., Kahn R. A., Lippincott-Schwartz J., Klausner R. D. Binding of ARF and beta-COP to Golgi membranes: possible regulation by a trimeric G protein. Science. 1991;254:1197–1199. doi: 10.1126/science.1957170. [DOI] [PubMed] [Google Scholar]
- Drake M. T., Zhu Y., Kornfeld S. The assembly of AP-3 adaptor complex-containing clathrin-coated vesicles on synthetic liposomes. Mol. Biol. Cell. 2000;11:3723–3736. doi: 10.1091/mbc.11.11.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan J. R., Kornfeld S. Intracellular movement of two mannose 6-phosphate receptors: return to the Golgi apparatus. J. Cell Biol. 1988;106:617–628. doi: 10.1083/jcb.106.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., et al. The beta3A subunit gene (Ap3b1) of the AP-3 adaptor complex is altered in the mouse hypopigmentation mutant pearl, a model for Hermansky-Pudlak syndrome and night blindness. Hum. Mol. Genet. 1999;8:323–330. doi: 10.1093/hmg/8.2.323. [DOI] [PubMed] [Google Scholar]
- Harter C., Mellman I. Transport of the lysosomal membrane glycoprotein lgp120 (lgp-A) to lysosomes does not require appearance on the plasma membrane. J. Cell Biol. 1992;117:311–325. doi: 10.1083/jcb.117.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haucke V., Wenk M. R., Chapman E. R., Farsad K., De Camilli P. Dual interaction of synaptotagmin with mu2- and alpha-adaptin facilitates clathrin-coated pit nucleation. EMBO J. 2000;19:6011–6019. doi: 10.1093/emboj/19.22.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein E. E., Macia E., Wang J., Yin H. L., Kirchhausen T., Harrison S. C. Crystal structure of the clathrin adaptor protein 1 core. Proc. Natl. Acad. Sci. USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honing S., Ricotta D., Krauss M., Spate K., Spolaore B., Motley A., Robinson M., Robinson C., Haucke V., Owen D. J. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol. Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Janvier K., Bonifacino J. S. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell. 2005;16:4231–4242. doi: 10.1091/mbc.E05-03-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G., Perlungher R. R., Sheffield P. J., Kinoshita M., Noda M., Haystead T., Macara I. G. Borg proteins control septin organization and are negatively regulated by Cdc42. Nat. Cell Biol. 2001;3:861–866. doi: 10.1038/ncb1001-861. [DOI] [PubMed] [Google Scholar]
- Kinoshita M. Diversity of septin scaffolds. Curr. Opin. Cell Biol. 2006;18:54–60. doi: 10.1016/j.ceb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Kremer B. E., Adang L. A., Macara I. G. Septins regulate actin organization and cell-cycle arrest through nuclear accumulation of NCK mediated by SOCS7. Cell. 2007;130:837–850. doi: 10.1016/j.cell.2007.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundra R., Kornfeld S. Wortmannin retards the movement of the mannose 6-phosphate/insulin-like growth factor II receptor and its ligand out of endosomes. J. Biol. Chem. 1998;273:3848–3853. doi: 10.1074/jbc.273.7.3848. [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Alconada A., Bauer U., Hoflack B. The mammalian AP-3 adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J. Biol. Chem. 1998;273:29451–29461. doi: 10.1074/jbc.273.45.29451. [DOI] [PubMed] [Google Scholar]
- Miura K., Jacques K. M., Stauffer S., Kubosaki A., Zhu K., Hirsch D. S., Resau J., Zheng Y., Randazzo P. A. ARAP 1, a point of convergence for Arf and Rho signaling. Mol. Cell. 2002;9:109–119. doi: 10.1016/s1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- Munro S. The Golgi apparatus: defining the identity of Golgi membranes. Curr. Opin. Cell Biol. 2005;17:395–401. doi: 10.1016/j.ceb.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Nagata K., et al. Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules. J. Biol. Chem. 2003;278:18538–18543. doi: 10.1074/jbc.M205246200. [DOI] [PubMed] [Google Scholar]
- Nie Z., Boehm M., Boja E. S., Vass W. C., Bonifacino J. S., Fales H. M., Randazzo P. A. Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev. Cell. 2003;5:513–521. doi: 10.1016/s1534-5807(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Ooi C. E., Dell'Angelica E. C., Bonifacino J. S. ADP-Ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J. Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Pal A., Severin F., Lommer B., Shevchenko A., Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington's disease. J. Cell Biol. 2006;172:605–618. doi: 10.1083/jcb.200509091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden A. A., Oorschot V., Hesser B. A., Austin C. D., Scheller R. H., Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press B., Feng Y., Hoflack B., Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Rittmeyer E. N., Daniel S., Hsu S. C., Osman M. A. A dual role for IQGAP1 in regulating exocytosis. J. Cell Sci. 2008;121:391–403. doi: 10.1242/jcs.016881. [DOI] [PubMed] [Google Scholar]
- Robinson M. S. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Shen X., Xu K. F., Fan Q., Pacheco-Rodriguez G., Moss J., Vaughan M. Association of brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) with recycling endosomes during transferrin uptake. Proc. Natl. Acad. Sci. USA. 2006;103:2635–2640. doi: 10.1073/pnas.0510599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H. W., et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F., Peden A. A., Christopoulou L., Robinson M. S. Characterization of the adaptor-related protein complex, AP-3. J. Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirokmany G., Szidonya L., Kaldi K., Gaborik Z., Ligeti E., Geiszt M. Sec14 homology domain targets p50RhoGAP to endosomes and provides a link between Rab and Rho GTPases. J. Biol. Chem. 2006;281:6096–6105. doi: 10.1074/jbc.M510619200. [DOI] [PubMed] [Google Scholar]
- Spiliotis E. T., Kinoshita M., Nelson W. J. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science. 2005;307:1781–1785. doi: 10.1126/science.1106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis E. T., Nelson W. J. Here come the septins: novel polymers that coordinate intracellular functions and organization. J. Cell Sci. 2006;119:4–10. doi: 10.1242/jcs.02746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis E. T., Hunt S. J., Hu Q., Kinoshita M., Nelson W. J. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J. Cell Biol. 2008;180:295–303. doi: 10.1083/jcb.200710039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinton L. M., Selak S., Fritzler M. J. Identification of GRASP-1 as a novel 97 kDa autoantigen localized to endosomes. Clin. Immunol. 2005;116:108–117. doi: 10.1016/j.clim.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Surka M. C., Tsang C. W., Trimble W. S. The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol. Biol. Cell. 2002;13:3532–3545. doi: 10.1091/mbc.E02-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waguri S., Dewitte F., Le Borgne R., Rouille Y., Uchiyama Y., Dubremetz J. F., Hoflack B. Visualization of TGN to endosome trafficking through fluorescently labeled MPR and AP-1 in living cells. Mol. Biol. Cell. 2003;14:142–155. doi: 10.1091/mbc.E02-06-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. J., Wang J., Sun H. Q., Martinez M., Sun Y. X., Macia E., Kirchhausen T., Albanesi J. P., Roth M. G., Yin H. L. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Drake M. T., Kornfeld S. ADP-ribosylation factor 1 dependent clathrin-coat assembly on synthetic liposomes. Proc. Natl. Acad. Sci. USA. 1999;96:5013–5018. doi: 10.1073/pnas.96.9.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.