Abstract
Rapidly cycling proteins of the early secretory pathway can operate as cargo receptors. Known cargo receptors are abundant proteins, but it remains mysterious why their inactivation leads to rather limited secretion phenotypes. Studies of Surf4, the human orthologue of the yeast cargo receptor Erv29p, now reveal a novel function of cargo receptors. Surf4 was found to interact with endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53 and p24 proteins. Silencing Surf4 together with ERGIC-53 or silencing the p24 family member p25 induced an identical phenotype characterized by a reduced number of ERGIC clusters and fragmentation of the Golgi apparatus without effect on anterograde transport. Live imaging showed decreased stability of ERGIC clusters after knockdown of p25. Silencing of Surf4/ERGIC-53 or p25 resulted in partial redistribution of coat protein (COP) I but not Golgi matrix proteins to the cytosol and partial resistance of the cis-Golgi to brefeldin A. These findings imply that cargo receptors are essential for maintaining the architecture of ERGIC and Golgi by controlling COP I recruitment.
INTRODUCTION
The secretory pathway of higher eukaryotic cells is composed of the three membrane organelles endoplasmic reticulum (ER), ER-Golgi intermediate compartment (ERGIC), and Golgi (Bonifacino and Glick, 2004; Appenzeller-Herzog and Hauri, 2006). Maintenance of these organelles requires a balance of anterograde (secretory) and retrograde vesicular traffic. Anterograde traffic from ER to ERGIC is mediated by coat protein (COP) II vesicles that form at ER exit sites (Aridor et al., 1995; Zeuschner et al., 2006) and fuse with the ERGIC that consists of a few hundred tubulovesicular membrane clusters in vicinity of ER exit sites (Appenzeller-Herzog and Hauri, 2006). Transport from ERGIC to Golgi is mediated by pleomorphic vesicles (Ben-Tekaya et al., 2005) that carry COP I (Presley et al., 1997; Scales et al., 1997), although the mechanism of their formation remains unknown. Retrograde traffic mediated by COP I vesicles can occur from ERGIC or Golgi and recycles membrane proteins that possess either dilysine signals, including ERGIC-53 and KDEL-receptor, or diphenylalanine signals, such as members of the 24 protein family. This rapid COP I-dependent recycling is distinct from the slow Golgi-to-ER recycling of Golgi resident proteins that is COP I independent and can be either constitutive or induced (Storrie, 2005).
Major constituents of anterograde and retrograde transport vesicles are transmembrane cargo receptors that mediate protein sorting by linking soluble cargo on the luminal side and coat assembly on the cytoplasmic side. To date, only few cargo receptors have been studied in detail. The polytopic transmembrane protein Erv29p is known to cycle between ER and Golgi in yeast and to operate as a cargo receptor (Belden and Barlowe, 2001). Erv29p is required for efficient packaging of the glycosylated α-factor pheromone precursor into COP II vesicles departing from the ER. Maturation of carboxypeptidase Y and proteinase A, but not other secretory proteins such as invertase, also depends on Erv29p (Caldwell et al., 2001). In support of the cargo receptor concept, a hydrophobic sorting signal was identified in α-factor that is required for its interaction with Erv29p and efficient transport (Belden and Barlowe, 2001; Otte and Barlowe, 2004). Erv29p is conserved among eukaryotes and the mammalian orthologue has been designated Surf4 (Reeves and Fried, 1995). Although its function is unknown, it is possible that Surf4 has a similar role in ER-to-Golgi transport in mammalian cells given the extent of homology with Erv29p that includes a dilysine retrieval motif.
The best characterized cargo receptor in mammalian cells is the mannose-specific leguminous type lectin ERGIC-53 (Hauri et al., 2000; Appenzeller-Herzog and Hauri, 2006). ERGIC-53 is a hexameric type I membrane protein in complex with the luminal EF-hand protein MCFD2 (Zhang et al., 2003; Nyfeler et al., 2006). This cargo receptor complex cycles between ER and ERGIC (Klumperman et al., 1998; Nyfeler et al., 2006), and it facilitates ER-to-ERGIC transport of the lysosomal enzymes glycoproteins cathepsin C (Vollenweider et al., 1998; Nyfeler et al., 2005), cathepsin Z (Appenzeller et al., 1999), and the blood coagulation factors V and VIII (Nichols et al., 1998; Zhang et al., 2003). MCFD2 is dispensable for the transport of the lysosomal enzymes, but it required for the transport of factors V and VIII (Nyfeler et al., 2006). In the ER, high-mannose cathepsin Z binds to ERGIC-53 by a combined glycan/β-hairpin signal, and it is subsequently released from ERGIC-53 in the ERGIC (Appenzeller-Herzog et al., 2005).
Yet another major cargo receptor is Emp24p in yeast. Emp24p is the founding member of the p24 protein family (Kaiser, 2000), and it is required for efficient ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins (Schimmoller et al., 1995; Muniz et al., 2000). It is conceivable that mammalian p24 proteins also operate as cargo receptors although no cargo protein has been identified. Mammalian p24 proteins are localized in the early secretory pathway and rapidly cycle between the ER and Golgi. To achieve their correct targeting within the early secretory pathway they are in a dynamic equilibrium to form homo- and heterodimers with each other (Emery et al., 2000; Jenne et al., 2002). All p24 family members are type I membrane proteins and share a common structure, with a short cytoplasmic tail containing binding signals for COP I and COP II coat complexes and a luminal domain with potential secretory cargo binding capabilities (Fiedler et al., 1996; Sohn et al., 1996; Dominguez et al., 1998; Muniz et al., 2000). Proteomics analysis revealed that p24 family members are major constituents of COP I-coated vesicles (Stamnes et al., 1995). Their involvement in COP I vesicle formation was identified in vitro by using liposomes with Golgi-like lipid composition. Liposomes incubated with the cytoplasmic components Arf1, coatomer, and guanosine triphosphate alone are unable to induce vesicle formation unless cytoplasmic domains of p24 family proteins are present (Bremser et al., 1999). P24 proteins seem to have some morphogenetic potential. p23 of the p24 family is an essential gene in mammals, and a heterozygous deletion reduces the levels of this protein and other family members, resulting in dilation of Golgi cisternae (Denzel et al., 2000). In cell cultures overexpression of p23 leads to its mislocalization to the ER, which causes expansion and clustering of smooth ER membranes. Mislocalization of p23 to the ER also leads to depletion of endogenous p23 from the Golgi, resulting in dispersion of this organelle (Rojo et al., 2000).
In the present study, we have characterized human Surf4, and we found it to localize to and cycle in the early secretory pathway similar to ERGIC-53. Surf4 forms multiprotein complexes with ERGIC-53 and p24 family members. Unexpectedly, silencing of Surf4 together with ERGIC-53 or silencing p25 of the p24 protein family disrupted the Golgi apparatus and led to instability of the ERGIC in conjunction with partial dissociation of COP I.
MATERIALS AND METHODS
Antibodies
The following mouse monoclonal antibodies were used: G1/93 against ERGIC-53 (Schweizer et al., 1988) (ALX-804-602; Alexis, Lausen, Switzerland), A1/182/5 against BAP31 (Klumperman et al., 1998) (ALX-804-601; Alexis), G1/133 against giantin (Linstedt and Hauri, 1993) (ALX-804-600-C100; Alexis), anti-β-COP (kind gift from Thomas Kreis, University of Geneva, Geneva, Switzerland), GM130 (BD Transduction Laboratories, Lexington, KY), and 12CA5 against the hemagglutinin (HA) epitope. Rabbit polyclonal antibodies against the following proteins were used: KDEL-receptor (Majoul et al., 1998; kind gift from H.-D. Söling, Max-Planck-Institut für Biophysikalische Chemie, Göttingen, Germany); Sec31 (Shugrue et al., 1999; (kind gift from F. Gorelick, Yale University, New Haven, CT); p23, p24, and p25 (Jenne et al., 2002; kind gifts of F. Wieland, University of Heidelberg, Germany); p115 and GM130 (Barroso et al., 1995; Nelson et al., 1998; kind gift from D. S. Nelson, University of Alabama Medical School, Birmingham, AL); GRASP65 (Sutterlin et al., 2002; kind gift from V. Malhotra, Division of Biology University of California, San Diego, CA). Alexa 488-, Alexa 568- (Molecular Probes Europe, Leiden, The Netherlands); and horseradish peroxidase-coupled antibodies (The Jackson ImmunoResearch Laboratories, West Grove, PA) were used as secondary antibodies. Polyclonal antibodies against human Surf4 were produced by immunizing rabbits with a recombinant peptide encompassing amino acids 1–60 of Surf4 fused to glutathione transferase (GST). The N-terminal sequence of Surf4 was amplified by polymerase chain reaction (PCR) by using the primers 5′-CAGGATCCCCGGCCAGAACGACCTGATG-3′ and 5′-CGAATTCTTATTACATGTACTGTTTGGGGGAGCTCTC-3′ and cloned into pGEX-5X2 vector via BamHI and EcoRI. The recombinant hybrid protein was expressed in Escherichia coli BL21 and purified by glutathione-Sepharose 4B column chromatography according to the manufacturer's instruction (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The antiserum was affinity-purified by sequential adsorption to Affigel 15 (Bio-Rad, Hercules, CA)-immobilized GST and GST-Surf4 followed by acid elution.
Cell Culture
HeLa cells and HeLa cells stably expressing green fluorescent protein (GFP)-ERGIC-53 (Ben-Tekaya et al., 2005) were grown in DMEM, supplemented with 10% fetal bovine serum and 1× nonessential amino acids. HepG2 cells and HepG2 cells stably expressing HA-Surf4 (Breuza et al., 2004) were grown in minimal essential medium, supplemented with 10% fetal bovine serum. For metabolic labeling and immunoblotting cells were grown in six-well plates. For immunofluorescence microscopy, cells were grown on coverslips in 12-well plates.
Purification of ERGIC Membranes and Blue Native-Polyacrylamide Gel Electrophoresis (PAGE)
Five days after confluence, HepG2 cells were treated with 10 μg/ml brefeldin A (BFA; Epicenter, Madison, WI) for 90 min, and ERGIC membranes were isolated by subcellular fractionation by using Nycodenz gradients (Breuza et al., 2004). Fractions of the Nycodenz gradient enriched in the ERGIC marker ERGIC-53 were pooled and diluted five times with phosphate-buffered saline (PBS). The membranes were centrifuged at 100,000 × g for 1 h, followed by solubilization in 25 mM bis-Tris-HCl, pH 7.0, 2% digitonin, and 500 mM 6-amino-caproic acid. The lysates were cleared at 100,000 × g for 1 h, and then they were separated by Blue Native-PAGE (Hunte et al., 2003).
Mass Spectrometry
Protein complexes separated by Blue Native-PAGE were separated in a second dimension by SDS-PAGE, and proteins were visualized by Colloidal Blue (Invitrogen, Basel, Switzerland). Gel pieces were excised and washed five times for 1 min with 50 μl of 40% n-propanol followed by five washes (1 min each) with 30 μl of 0.2 M NH4HCO3 containing 50% acetonitrile. The gel pieces were dried in a SpeedVac concentrator (Savant, Farmingdale, NY), and they were reswollen in 10 μl of 100 mM NH4HCO3 containing 0.5 μg of modified trypsin (Promega, Madison, WI). Trypsin digestion was performed at 37°C for 18 h. The supernatants were collected, and the gel pieces were extracted with 15 μl of 0.1% formic acid for 5 min, followed by 15 μl of acetonitrile for 1 min. Extraction was repeated twice, and all supernatants were pooled and dried by SpeedVac. For desalting, the peptides were dissolved in 0.1% trifluoroacetic acid (TFA) and adsorbed on C18 ZipTips (P10 size; Millipore, Billerica, MA). The peptides adsorbed on the tips were washed with 0.1% TFA and eluted with 1.5 μl of 80% AcCN, 0.1%TFA, containing 1 μg/μl α-cyano-4-hydroxycinnamic acid (CHCA; Aldrich Chemical, Milwaukee, IL). The eluate (500 nl) was deposited onto anchor spots of a Scout 400-μ m/36 sample support (Bruker Daltonik, Bremen, Germany), and the droplet was left to dry at room temperature. Mass spectra were recorded on a Bruker Scout 26 Reflex III instrument (Bruker Daltonik). The instrument was calibrated with angiotensin II, substance P, bombesin, and ACTH. The peptides were analyzed in reflector mode using delayed ion extraction with a total acceleration voltage of 23 kV. Fifty to 100 single-shot spectra were acquired to improve the signal-to-noise ratio. Spectrum calibration and peak assignment was carried out with the XMASS 5.0 software package provided by the manufacturer. The Mascot search software (http://www.matrixscience.com) was used for protein identification.
Immunoprecipitation
HepG2 cells were solubilized in 50 mM Tris-HCl, pH 7.4, 1% digitonin, 150 mM NaCl, 2 mM CaCl2, and protease inhibitors for 1 h at 4°C, followed by centrifugation at 100,000 × g for 1 h. Supernatants were incubated with anti-ERGIC-53 and anti-HA antibodies covalently coupled via dimethyl pimelimidate to protein A-Sepharose beads (Harlow and Lane, 1999) or with anti-p23 and anti-p24 antibodies bound to protein A-Sepharose. Beads were washed four times in 50 mM Tris-HCl, pH 7.4, 0.1% digitonin, 150 mM NaCl, and 2 mM CaCl2. Proteins were separated by SDS-PAGE, and then they were transferred to nitrocellulose membranes for immunoblotting.
Small Interfering RNA Transfection
siRNA oligos were purchased from Eurogentec (Seraing, Belgium) and QIAGEN (Venlo, The Netherlands). Three siRNA oligonucleotides (oligos) were designed against Surf4 and two against p25. siRNA oligos for ERGIC-53 knockdown were described previously (Nyfeler et al., 2006). The most efficient siRNA oligo was determined by immunoblotting, and it was chosen for all further experiments. Surf4 was knocked down using 5′-CGUAUAUUUCAACGCCUUCdTdT-3′ as sense and 5′-GAAGGCGUUGAAAUAUACGdTdT-3′ as antisense oligo. P25 was knocked down using 5′-CCUCAGAAUCACAGUGUUAdTdT-3′ as sense and 5′-UAACACUGUGAUUCUGAGGdTdG-3′ as antisense oligo. Nonsilencing control siRNA was purchased from QIAGEN (Basel, Switzerland). The siRNA was used at a final concentration of 5 nM for transfection directly after cell plating using Hiperfect (QIAGEN) according to the manufacturer's instructions. All knockdown experiments were performed 72 h after transfection.
Transmission Electron Microscopy
HeLa cells treated with control siRNA, p25 siRNA, or Surf4/ERGIC-53 siRNA were fixed with 3% paraformaldehyde. 0.5% glutaraldehyde in 10 mM PBS, pH 7.4. After washing in PBS, the cells were postfixed in 1% osmium tetroxide. Fixed samples were dehydrated and embedded in Epon 812 resin (Fluka, Buchs, Switzerland). Sections were stained with 6% uranyl acetate and lead acetate, and then they were analyzed with an EM912 Omega EFTEM electron microscope (LEO Electron Microscopy, Oberkochen, Germany).
Immunofluorescence Microscopy and Quantification
Cells were fixed in 3% paraformaldehyde, and then they were permeabilized for 5 min in PBS containing 0.2% Triton X-100, 3% bovine serum albumin (BSA), and 20 mM glycine. For the staining with anti-Surf4 0.5% SDS was included in the permeabilization buffer. For the staining with β-COP antibodies, cells were fixed in methanol:acetone. Primary antibodies were added for 30 min in PBS containing 3% BSA and 20 mM glycine. After washing, secondary antibodies were added for 30 min in PBS containing 3% BSA. Cells were embedded in Mowiol, and then they were analyzed by laser scanning confocal microscopy (TCS NT; Leica Microsystems, Wetzlar, Germany). For the quantification of ERGIC clusters and ER exit sites, cells were stained for KDEL-receptor and Sec31, respectively, and costained for giantin. Knockdown cells were chosen based on a dispersed Golgi pattern indicated by giantin. The Golgi area was subtracted from the KDEL-receptor-stained image, and the ERGIC clusters were counted using Image-Pro Plus software (Media Cybernetics, Bethesda, MD). For the quantification of ER exit sites, spots positive for Sec31 were counted. For quantification of β-COP staining, cells were imaged with a charge-coupled device (CCD) camera (SensiCam; PCO Computer Optics, Kelheim, Germany) connected to a Polyvar microscope (VWR, West Chester, PA). The Golgi area was chosen according to giantin staining. The intensity of β-COP and giantin staining in the Golgi area was determined using Image-Pro Plus software.
Live Cell Imaging
HeLa cells expressing GFP-ERGIC-53 were treated with sodium butyrate overnight and plated at a density of 4.5 × 104 cells/ml on 18-mm round coverslips, followed by transfection with siRNA. Eighty hours after transfection, living cells were imaged (Ben-Tekaya et al., 2005). Images were acquired with a CCD camera (Orca-3CCD; Hamamtsu Photonics, Hamamatsu City, Japan) by using a Lambda DG4 (Sutter Instrument, Novato, CA) for high-speed filter switching. Image-Pro Plus software was used for image recording and processing. Additionally an Edge filter was used to decrease the background signal. Life spans of individual ERGIC clusters were assessed in eight cells for each condition using the automatic tracking tool of Image-Pro Plus. Diameter and intensity filters were used to exclude the Golgi area and to monitor only prominent ERGIC structures. The life span corresponds to the average life span of all ERGIC structures counted per cell. It was measured for no longer than 5 min with image acquisition every 2 s because with these conditions movement of spots could be tracked pixel by pixel. Increasing the analysis time (and imaging interval) resulted in tedious manual tracking, and it gave similar results. Statistical significance (p ≤ 0.05) of the life span between control and knockdown conditions was probed by Student's t test.
Immunoblotting
Cells were lysed for 1 h at 4°C in PBS containing 1% digitonin, supplemented with protease inhibitors. Lysates were centrifuged at 20,000 × g for 30 min at 4°C. Forty micrograms of protein per lane were separated by SDS-PAGE, transferred to nitrocellulose membranes, immunoblotted sequentially with primary and secondary antibodies, and visualized by enhanced chemiluminescence (GE Healthcare).
Metabolic Labeling
HeLa cells were deprived of l-methionine for 20 min, pulsed for 10 min with 100 μCi of [35S]methionine (PerkinElmer Life and Analytical Sciences, Boston, MA), and chased for the indicated times in HeLa culture medium containing 10 mM l-methionine. At the end of the chase, the medium was collected and centrifuged for 10 min at 10,000 × g to remove cell debris. For the 0-min chase time, cells of parallel cultures were homogenized in PBS by passing them 10 times through a 25-gauge needle. Five-microliter aliquots of homogenate and media were trichloroacetic acid (TCA)-precipitated, and radioactivity was measured by scintillation counting. Total protein secretion into the medium was normalized to the total counts in cell homogenates at 0-min chase.
Osmotic Stress Treatment
HeLa cells grown on 18-mm glass coverslips were incubated in 37°C hypotonic medium (60 mM NaCl, 20 mM HEPES, pH 7.4, and 2.5 mM MgOAc) for 5 min at 37°C. The cells were washed two times in ice-cold PBS, and then they were fixed on ice using 3% paraformaldehyde and processed for immunofluorescence microscopy.
RESULTS
Human Surf4 Localizes to the ERGIC and Cycles in the Early Secretory Pathway
Although discovered quite some time ago, mammalian Surf4 remains largely uncharacterized. Even its subcellular localization is uncertain. N-terminally tagged Surf4 localizes to the ER, whereas C-terminally tagged Surf4 localizes to the Golgi in transfected cells (Reeves and Fried, 1995). In contrast, endogenous Surf4 was identified by mass spectrometry in an ERGIC fraction isolated BFA-treated HepG2 cells (Breuza et al., 2004). To characterize endogenous Surf4, we prepared polyclonal antibodies to the N-terminal 60 amino acids of Surf4 fused to GST, and we purified them by affinity chromatography. Affinity-purified anti-Surf4 recognized a protein of ∼22 kDa on Western blots in reasonable agreement with the Mr of Surf4 deduced from conceptual translation (Supplemental Figure S1A). The specificity of the antibody was confirmed by silencing Surf4 in HeLa cells by using siRNA (Supplemental Figure S3). In control cells, anti-Surf4 gave an immunofluorescence pattern similar to that of the ERGIC marker ERGIC-53, whereas the staining disappeared after siRNA-mediated knockdown of Surf4 without affecting the distribution of ERGIC-53 (Supplemental Figure S1B). These results indicate specificity of our antibodies against Surf4.
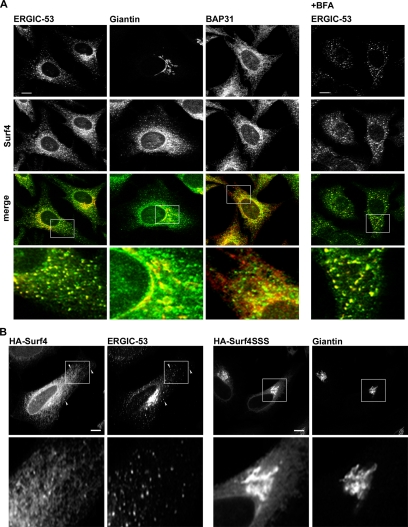
To more precisely establish the localization of Surf4, colocalization studies with various organelle markers were performed by immunofluorescence microscopy. Surf4 prominently stained peripheral ERGIC clusters positive for ERGIC-53 (Figure 1A; also see Supplemental Figure S1B) and partially colocalized with the ER marker BAP31 and the Golgi marker giantin (Figure 1A). The predominant localization of endogenous Surf4 in the ERGIC suggests that Surf4 might be a cycling protein of the early secretory pathway. To test this, the distribution of endogenous Surf4 was studied in HeLa cells treated with BFA that is known to accumulate rapidly cycling proteins in the ERGIC (Lippincott-Schwartz et al., 1990). Indeed, this treatment concentrated Surf4 in ERGIC-53–positive structures (Figure 1A) supporting the notion that Surf4 is a cycling protein.
Figure 1.
Surf4 localizes mainly to the ERGIC. (A) Localization of endogenous Surf4 in HeLa cells by confocal immunofluorescence microscopy using affinity-purified antibodies against Surf4 in combination with antibodies against ERGIC-53, giantin, and BAP31. Right, HeLa cells were treated with 10 μg/ml BFA for 90 min (+BFA) and labeled with antibodies against Surf4 and ERGIC-53. (B) HeLa cells were transfected with HA-Surf4 or HA-Surf4SSS. The tagged versions of Surf4 were stained with anti-HA and costained with anti-ERGIC-53 and anti-giantin antibodies. Arrowheads indicate colocalization of HA-Surf4 with ERGIC-53. Bars, 10 μm.
Surf4 is a multispanning membrane protein with its C terminus predicted to face the cytosol. The cytosolic tail carries lysine residues in positions −3, −4, and −5 from the C terminus. Two lysines in positions −3 and −4 are known to function as ER targeting signal mediating retrieval (Teasdale and Jackson, 1996) or retention (Andersson et al., 1999), depending on amino acids in position −1 and −2. We tested the functionality of the lysine motif by mutating the three lysines to serines in hemagglutinin-tagged Surf4 (HA-Surf4SSS). HA-Surf4 localized to ER and ERGIC very much in contrast to HA-Surf4SSS that localized to the Golgi region (Figure 1B). This result is consistent with and explains the Golgi localization of C-terminally tagged Surf4 (Reeves and Fried, 1995). In that study, the C-terminal tagging obviously inactivated the dilysine signal, which is known to be position dependent (Teasdale and Jackson, 1996). Collectively, our results indicate that Surf4 cycles early in the secretory pathway in a lysine signal-dependent way, similarly to ERGIC-53.
Surf4 Interacts with Members of the p24 Protein Family and ERGIC-53
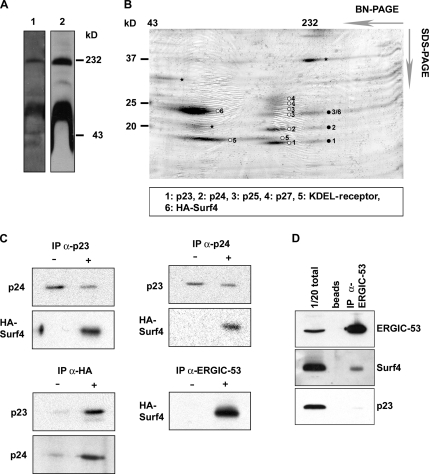
In search of the function of Surf4, we attempted to identify interacting proteins. To maximize such interactions, Surf4 was accumulated in the ERGIC by treating HepG2 cells with BFA. Both nontransfected and HA-Surf4–transfected cells were analyzed. ERGIC membranes were isolated by Nycodenz gradient centrifugation (Breuza et al., 2004), and the gradient fractions were analyzed for organelle markers by Western blotting. Surf4 largely codistributed with the ERGIC marker ERGIC-53 (Supplemental Figure S2). The ERGIC fractions were collected, and the membranes were subjected to Blue Native-PAGE. Western blotting revealed that both Surf4 and HA-tagged Surf4 formed protein complexes of ∼60 and 232 kDa (Figure 2A). Because HA-Surf4 behaved like endogenous Surf4 on Blue Native gels (Figure 2A), and it was more abundant, some of the further experiments were performed with HA-Surf4. Separation of the protein complexes by SDS-PAGE in a second dimension showed distinct protein spots of 15–37 kDa, which were identified by mass spectrometry as Surf4 and members of the p24 protein family (Figure 2B). This approach also identified the previously described protein complex of p23, p24, p25, and p27 (Fullekrug et al., 1999), demonstrating the accuracy of the method (Figure 2B). The 60-kDa complex seems to contain Surf4 and KDEL-receptor, but the possibility of an interaction of the two proteins has not been investigated in the current study.
Figure 2.
Surf4 forms protein complexes with p24 family members and ERGIC-53. (A) ERGIC membranes were isolated from parent HepG2 cells (lane 1) and HepG2 cells stably expressing HA-Surf4 (lane 2) (see Supplemental Figure 1). Isolated membranes were separated by Blue Native-PAGE followed by Western blotting with antibodies against Surf4 and the HA epitope. (B) ERGIC membranes of HepG2 cells stably expressing HA-Surf4 were separated by Blue Native-PAGE as described in A, followed by SDS-PAGE in a second dimension. Proteins were stained with Coomassie Blue and excised for mass spectrometry analysis. Black circles indicate the protein complex containing HA-Surf4 and the p24 family members p23, p24, and p25. Open circles indicate complexes of identified proteins that were not further analyzed. Asterisks, proteins not identified by mass spectrometry. (C) Coimmunoprecipitation experiments: ERGIC membranes of parent HepG2 cells (−) and HepG2 cells stably expressing HA-Surf4 (+) were isolated, lysed, and subjected to immunoprecipitation with anti-HA, anti-ERGIC-53, anti-p23, and anti-p24 followed by Western blotting by using antibodies against the HA-epitope, p23, p24, and p25. (D) HeLa cell lysates were immunoprecipitated with anti-ERGIC-53 antibodies coupled to beads or with beads alone. The total lysate (1/20) was loaded as indicator for protein amount in the cell. Proteins were visualized by Western blotting with antibodies to ERGIC-53 or p23.
To confirm the interaction between Surf4 and p24 family members, coimmunoprecipitation experiments were performed. Because the antibody against Surf4 did not immunoprecipitate endogenous Surf4, the HA-tagged protein was studied in transfected HepG2 cells. Figure 2C shows that both anti-p23 and anti-p24 pulled down HA-Surf4. Inversely, anti-HA pulled down both p23 and p24. Surprisingly, a highly specific monoclonal antibody against ERGIC-53, used as (presumed) negative control, also pulled down HA-Surf4 (Figure 2C). This unexpected result was confirmed for endogenous Surf4 in HeLa cells. Anti-ERGIC-53 pulled down Surf4 but not p23 (Figure 2D). We conclude that Surf4 forms hetero-oligomeric complexes with members of the p24 family and in addition interacts with ERGIC-53.
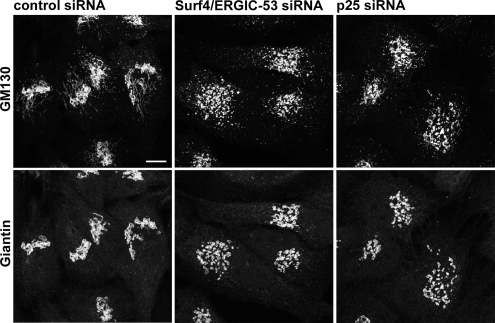
Silencing of Surf4 and ERGIC-53 or p25 Disrupts the Golgi
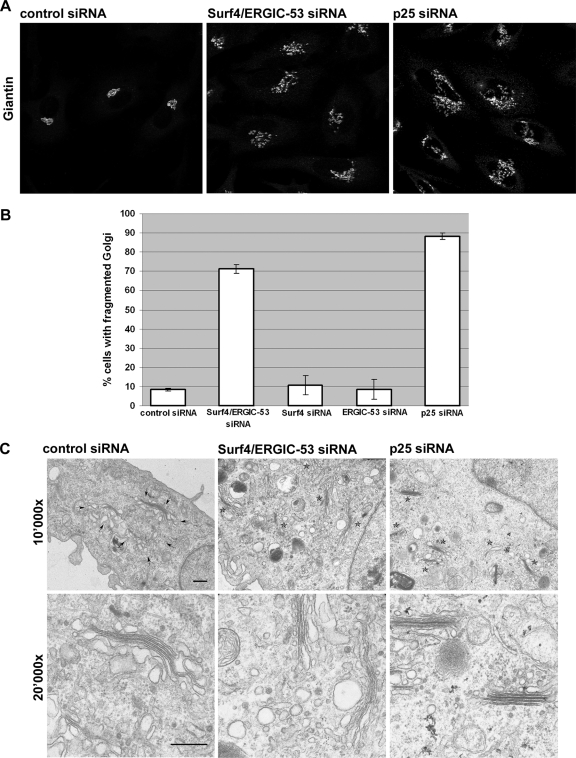
To obtain more insight into the function of Surf4, we took a silencing approach using siRNA (Supplemental Figure S3A and S3B). A knockdown of Surf4 down to 10% in HeLa cells, had no effect on the distribution of organelle markers for ER (unpublished data), ERGIC, and Golgi (Figure 3B and Supplemental Figures S1B and S4A), nor was total secretion of [35S]methonine-labeled proteins impaired 3 d after siRNA transfection (Supplemental Figure S4B). The serendipitous finding of coimmunoprecipation of ERGIC-53 and Surf4 mentioned above led us to test the combined requirement of Surf4 and ERGIC-53. Strikingly, a double knockdown of Surf4 and ERGIC-53 by siRNA disrupted the Golgi apparatus as visualized by staining for giantin (Figure 3A). Quantification showed that 70% of the cells had a dispersed Golgi (Figure 3B). In contrast, a single knockdown of ERGIC-53 down to 10% (Supplemental Figure S3A and S3B) had no effect on Golgi morphology (Figure 3B and Supplemental Figure S4A), consistent with previous knockdown data (Nyfeler et al., 2006) and the observation that mislocalization of ERGIC-53 to the ER did not induce changes of the early secretory pathway (Vollenweider et al., 1998). Because ERGIC-53 is known to form a complex with the soluble protein MCFD2 and a knockdown of ERGIC-53 leads to secretion of MCFD2 (Nyfeler et al., 2006), we wondered whether the Golgi change was due to the lack of MCFD2 rather than ERGIC-53. However, a double knockdown of Surf4 and MCFD2 had no effect on Golgi morphology (unpublished data), strongly suggesting the specific involvement of ERGIC-53 in maintaining normal Golgi structure together with Surf4.
Figure 3.
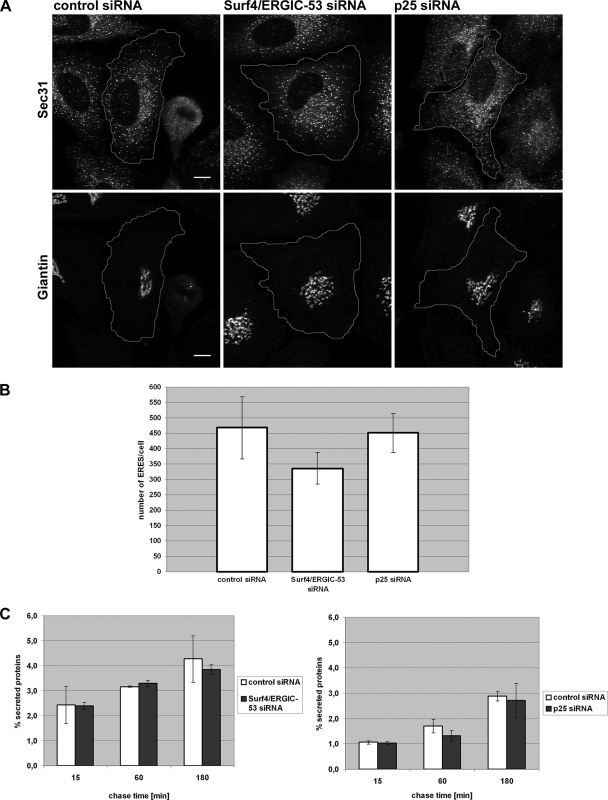
Double knockdown of Surf4/ERGIC-53 or single knockdown of p25 leads to Golgi dispersal. (A) HeLa cells were transfected with control siRNA, Surf4/ERGIC-53 siRNA or p25 siRNA. The Golgi was visualized by immunofluorescence microscopy using anti-giantin. (B) Quantitative analysis of the Golgi phenotype in Surf4/ERGIC-53 and p25 knockdowns. More than 100 cells of three independent experiments were counted for each condition, and the percentage of cells with fragmented Golgi plotted. Results are means ± SD. Bar, 10 μm. (C) Cells treated with control, Surf4/ERGIC-53, or p25 siRNA were processed for electron microscopy and sections of 10,000× and 20,000× magnifications are shown. Arrows, Golgi ribbon. Asterisks, dispersed Golgi stacks. Bars, 0.5 μm.
Next, we asked whether the silencing of p24 proteins would also affect Golgi structure. Besides their proposed role as cargo receptors, p24 family members are thought to function as morphogens in the early secretory pathway. Such a function has mainly been derived from overexpression studies (Blum et al., 1999; Rojo et al., 2000). In addition, the inactivation of one allele of p23 in mice induces structural changes in the Golgi apparatus, and it reduces the levels of p23, p24, and p25 (Denzel et al., 2000). p25 is the only p24 family member containing a canonical dilysine signal. Similar to Surf4 and ERGIC-53, inactivation of the dilysine signal in p25 leads to its mislocalization due to inefficient retrieval back to the ER (Emery et al., 2003). Although knockdowns of p23 were reported previously (Vetrivel et al., 2007), no knockdown experiments have been performed for p25. The known hetero-oligomerization and interdependence of p24 family members complicates such an analysis. Accordingly, we found that a knockdown of p24 reduced p23 levels and vice versa (unpublished data). Depletion of p25 down to 25% (Supplemental Figure S3A and S3B), however, did not affect the protein levels of p24 or p23, which led us to focus on p25 (unpublished data). Strikingly, the knockdown of p25 in HeLa cells induced a change in Golgi morphology that was indistinguishable from that obtained by the Surf4/ERGIC-53 double knockdown (Figure 3A). Ninety percent of the transfected cells showed fragmentation of the Golgi as visualized by immunofluorescence microscopy (Figure 3B). Importantly, the knockdown of p25 did not change the protein levels of Surf4 or ERGIC-53 and vice versa (Supplemental Figure S3C).
Are the changed Golgi structures identical in Surf4/ ERGIC-53 and p25 knockdowns? As a test, we analyzed the silenced cells by transmission electron microscopy. This analysis indicated that under both knockdown conditions the Golgi ribbon was converted to mini-stacks that otherwise looked unchanged. In particular, the cisternae were not swollen and cisternal stacking was intact, suggesting normal cis-trans topology (Figure 3C). Thus, the changes in Golgi morphology induced by a knockdown of Surf4 and ERGIC-53 or p25 are indistinguishable by both light and electron microscopy.
Cargo Receptor Silencing Destabilizes the ERGIC without Affecting ER Exit Sites or Protein Secretion
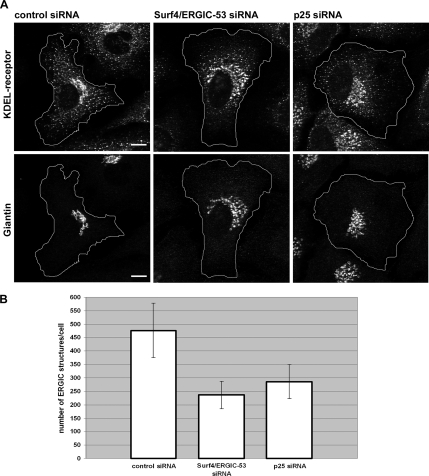
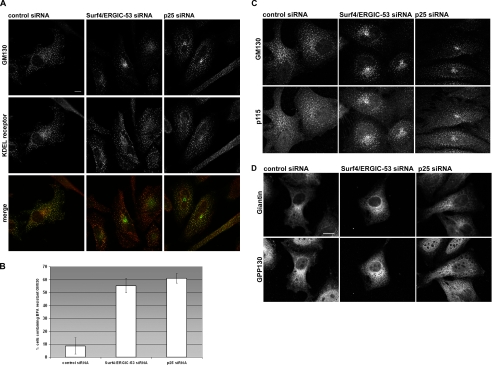
The finding that a double knockdown of Surf4 and ERGIC-53 and a single knockdown of p25 induced a Golgi phenotype was unexpected because all three proteins are mainly associated with the ERGIC, although they also cycle through the Golgi to some extent (Schweizer et al., 1988; Dominguez et al., 1998; Klumperman et al., 1998). Furthermore, a study on the reconstitution of the secretory pathway in a cell-free assay suggests that p25 plays a role in the de novo formation of the ERGIC (Lavoie et al., 1999). Based on these findings, we considered the possibility that the knockdowns might also induce changes at the level of the ERGIC. To detect such changes, HeLa cells depleted of Surf4/ERGIC-53 or p25 were double labeled for the ERGIC/cis-Golgi marker KDEL-receptor and the Golgi marker giantin. Peripheral ERGIC structures were quantified in the knockdown cells, which could readily be identified by a dispersed giantin pattern (Figure 4A). Quantification showed that control cells exhibited 490 KDEL-receptor-positive ERGIC structures on average, whereas cells depleted of Surf4/ERGIC-53 had only 230 and cells depleted of p25 only 300 ERGIC structures per cell (Figure 4, A and B). The reduction of KDEL-receptor-positive ERGIC structures was not due to reduced levels of KDEL-receptor (Supplemental Figure S3C). Clearly, both the Surf4/ERGIC-53 knockdown and the p25 knockdown reduced the number of peripheral ERGIC clusters.
Figure 4.
ERGIC structures are reduced in cells depleted of Surf4/ERGIC-53 or p25. (A) HeLa cells transfected with control, Surf4/ERGIC-53, or p25 siRNA were immunostained with antibodies against KDEL-receptor and giantin, and then they were analyzed by confocal microscopy. The giantin staining was used as indication for efficient knockdown of Surf4/ERGIC-53 and p25. The cell borders are outlined in white. Bars, 10 μm. (B) Quantitative analysis of the ERGIC structures. More than 18 cells per condition of three independent experiments were analyzed. KDEL-receptor–positive ERGIC structures were counted after removal of the Golgi area defined by giantin staining (see Materials and Methods). Results are means ± SD.
ER export activity is known to be modulated by the cargo load (Aridor et al., 1999; Guo and Linstedt, 2006). Accordingly, the depletion of cargo receptors may impair ER export that would explain the reduction in ERGIC cluster numbers. If true, one would expect that the number of ER exit sites is reduced in parallel. Therefore, we determined the number of ER exit sites labeled by antibodies against the COP II coat protein Sec31 (Figure 5A). P25 knockdown cells showed 400 ER exit sites on average, which was comparable with the 420 ER exit sites counted in control cells (Figure 5B). Surf4/ERGIC-53 knockdowns exhibited a slightly reduced number of 320 ER exit sites (Figure 5B). These numbers show that the reduction of ERGIC clusters is not paralleled by a similar reduction of ER exit sites.
Figure 5.
ER exit site formation and anterograde transport are not affected in Surf4/ERGIC-53 or p25 knockdown cells. (A) HeLa cells transfected with control, Surf4/ERGIC-53 or p25 siRNAs were processed for immunofluorescence microscopy using antibodies against Sec31 and giantin. The giantin staining was used as indication for efficient knockdown of Surf4/ERGIC-53 and p25. The cell borders are outlined in white. Bars, 10 μm. (B) Quantitative analysis of ER exit sites. More than 25 cells per condition of three independent experiments were analyzed. ER exit sites were counted according to the Sec31 staining (see Materials and Methods). Results are means ± SD. Bars, 10 μm. (C) HeLa cells were transfected with control, Surf4/ERGIC-53 and p25 siRNA and subjected to pulse-chase analysis using [35S]methionine. Media from cells were collected and assayed for incorporated radioactivity. Results are means ± SD of at least three independent experiments.
Do the structural changes of ERGIC Golgi impair total protein secretion? We used a pulse-chase approach to address this question. HeLa cells in which p25 or Surf4 together with ERGIC-53 had been silenced, were pulse-labeled with [35S]methionine and the radioactive proteins secreted into the medium during chase were quantified by scintillation counting after TCA precipitation. Figure 5C shows that neither silencing Surf4/ERGIC-53 nor p25 significantly affected total protein secretion after 3 d, although after 4 d the Surf4/ERGIC-53 reduced secretion (Supplemental Figure 4B), implying that the secretion assay is sensitive enough to detect inefficient anterograde protein transport. Because maximal protein silencing was reached already after 3 d of transfection, we conclude that secretion is initially unaffected by the two knockdown conditions.
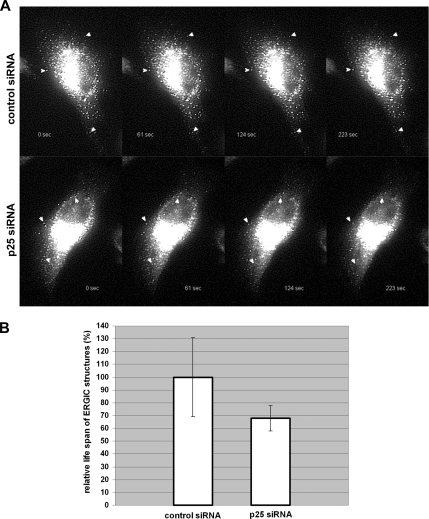
To study the dynamics of the ERGIC, we used live cell imaging of HeLa cells stably expressing GFP-ERGIC-53. In this cell line GFP-ERGIC-53 behaves like endogenous ERGIC-53 (Ben-Tekaya et al., 2005). Strikingly after p25 knockdown, stationary ERGIC structures hovered about in place more actively and disappeared faster than in control cells (Figure 6 and Supplemental Movie 1). Tracking peripheral ERGIC structures revealed that their relative life span was reduced by 35% (Supplemental Figure S5). Thus, the reduction of ERGIC clusters in p25 knockdown cells can, at least in part, be attributed to a shorter half-life. The ERGIC structures in Surf4/ERGIC-53 knockdown cells could not be analyzed in living cells because no acceptable GFP-tagged marker was available to identify the ERGIC-53 in the absence of ERGIC-53. We speculate, however, that the ERGIC structures in Surf4/ERGIC-53 depleted cells would behave similarly.
Figure 6.
Live imaging of GFP-ERGIC-53 reveals a shorter life span of ERGIC structures in p25 knockdown cells. (A) Time series from Supplemental Movies 1 and 2. Cells were transfected with control or p25 siRNA and imaged with an interval of ∼2 s. Representative frames from a control cell show stationary ERGIC structures that hardly move throughout the imaging period (top, arrowheads). In p25 knockdown cells the stationary ERGIC structures do not move either, but they disappear with time (bottom, arrowheads). (B) Life span of ERGIC structures in p25-depleted cells. Quantification of the relative life span of GFP-ERGIC-53 structures presented in Figure 6. The average life span is plotted in percentage. Note that in p25 knockdown cells the life span of ERGIC structures is reduced by ∼35%. This difference is statistically significant (Student's t test, p ≤ 0.05). Results are means ± SD (n = 8).
Collectively, the morphological, biochemical, and live cell imaging results indicate that cargo receptor silencing destabilizes the ERGIC without initial impairment of overall protein secretion.
Golgi Matrix Proteins Remain Associated with the Dispersed Golgi
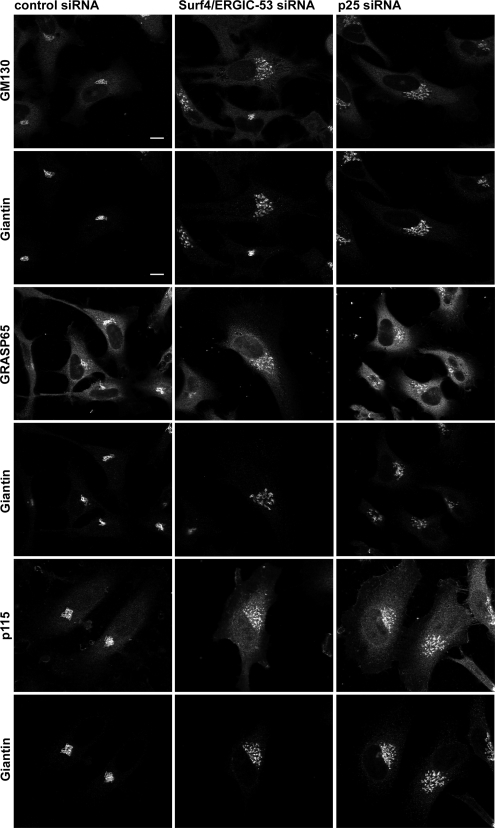
P24 family members are known to form complexes with the Golgi matrix proteins GM130, GRASP65 and GRASP55 (Barr et al., 2001). These matrix proteins are required for normal Golgi morphology. GM130 is a cis-Golgi localized coiled-coil protein targeted to membranes via the peripheral membrane protein GRASP65 (Barr et al., 1997, 1998). It also binds the vesicle tethering factor p115 (Nakamura et al., 1997; Nelson et al., 1998). GM130 and GRASP65 are key determinants for maintaining Golgi morphology as their knockdown transforms the Golgi ribbon to mini-stacks (Sohda et al., 2005; Puthenveedu et al., 2006). The knockdowns of p25 and Surf4/ERGIC-53 produced a Golgi phenotype reminiscent of that observed after a knockdown of GM130 and GRASP65 (Sohda et al., 2005; Puthenveedu et al., 2006). This led us to study the distribution of Golgi matrix proteins in p25- and Surf4/ERGIC-53–depleted cells. Figure 7 clearly shows that GM130, GRASP65 and p115 remained associated with the dispersed Golgi in both p25 and Surf4/ERGIC-53 knockdown cells. We conclude that the morphological changes of the Golgi are unlikely to be due to impaired binding of matrix proteins to Golgi membranes.
Figure 7.
Golgi matrix proteins remain associated with the dispersed Golgi. HeLa cells in which Surf4/ERGIC-53 or p25 was silenced by siRNA were processed for immunofluorescence microscopy by using anti-GM130, anti-GRASP65, and anti-p115 and colabeled with anti-giantin antibodies. The giantin staining was used as indication for efficient knockdown of Surf4/ERGIC-53 and p25. Bars, 10 μm.
Silencing of Surf4 and ERGIC-53 or p25 Dissociates COP I
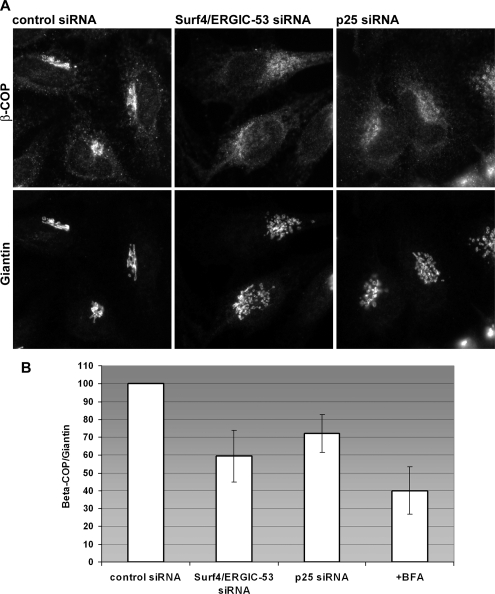
Apart from cycling, a common feature of Surf4, ERGIC-53, and p25 is a dilysine ER retention/recycling signal. Because dilysine signals mediate COP I binding and tails of p24 family members are essential for COP I vesicle formation in vitro (Bremser et al., 1999) we wondered whether the depletion of the three cycling proteins would affect COP I recruitment. To this end, Surf4 and ERGIC-53 or p25 were silenced in HeLa cells, and the COP I coat subunit β-COP was localized by immunofluorescence microscopy. Strikingly, the overall signal for β-COP was reduced in both knockdown conditions (Figure 8A), which was not due to lower protein levels (Supplemental Figure S3C). The Golgi region identified by giantin showed less prominent staining for β-COP compared with cells treated with control siRNA (Figure 8, A and B). Quantification of the Golgi area revealed that the β-COP signal was reduced by 40% in Surf4/ERGIC-53 and 30% in p25 knockdowns compared with control cells, whereas the signal for giantin remained unchanged (Figure 8, A and B). After 5-min BFA treatment of control cells, 60% of β-COP staining was lost from the Golgi region, indicating β-COP redistribution from the Golgi to the cytosol (Figure 8B). The results indicate that Surf4/ERGIC-53 and p25 are required for COP I recruitment to membranes of the early secretory pathway.
Figure 8.
β-COP is dispersed in Surf4/ERGIC-53 and p25 knockdown cells. (A) HeLa cell treated with Surf4/ERGIC-53 or p25 siRNA were immunostained for β-COP and giantin. Shown are representative images of three independent experiments. (B) Quantification of β-COP and giantin intensities in the Golgi region. The Golgi region was defined by giantin staining. Shown are intensity ratios of β-COP and giantin normalized to 100%. +BFA indicates control siRNA-transfected HeLa cells treated with 10 μg/ml BFA for 5 min. Results are means ± SD (n = 3). Bars, 10 μm.
A loss of COP I from Golgi membranes is known to change the structure of this organelle to the extent that it rapidly tubulates and fuses with the ER. Such an outcome is well known for cells treated with the fungal metabolite BFA. The Golgi changes induced by silencing Surf4 and ERGIC-53 or p25 are clearly different from those induced by BFA. We wondered whether knockdown cells would respond normally to BFA. Figure 9D shows that a 30-min BFA treatment of control cells induced an almost complete disappearance of the Golgi. As expected, giantin showed an ER-like pattern and GM130 redistributed to the ERGIC (Figure 9, A and D). In contrast, after Surf4/ERGIC-53 or p25 silencing, GM130 and p115 were largely resistant to BFA, and they remained in the juxtanuclear area (Figure 9, A–C), very much in contrast to KDEL-receptor that redistributed to the ERGIC (Figure 9A) and the two Golgi markers giantin and GPP130, which redistributed to the ER (Figure 9D).
Figure 9.
Cis-Golgi remains partially resistant to BFA in Surf4/ERGIC-53– and p25-depleted cells. HeLa cells transfected with Surf4/ERGIC-53, p25 siRNA, or control siRNA were treated with 10 μg/ml BFA for 30 min. Cells were processed for immunofluorescence microscopy by using anti-GM130 and anti-KDEL-receptor (A), anti-GM130 and anti-p115 (C), or anti-giantin and anti-GPP130 (D) antibodies. (B) Quantification of cells showing BFA resistant cis-Golgi according to GM130 staining. Results are means ± SD (n = 3). Bars, 10 μm.
Obviously, COP I dissociation induced by cargo receptor silencing does not result in a BFA-like effect. Thus, COP I dissociation cannot explain the absence of tubulation of the cis-Golgi. Together with the partial resistance of the cis-Golgi to BFA after cargo receptor silencing, the lack of tubules implies that cargo receptors are required for efficient tubulation. The role of cargo receptors in promoting tubulation of the cis-Golgi indicated by GM130 was assessed by subjecting cells depleted of cargo receptors to hypotonic stress known to cause tubulation of the Golgi (Lee and Linstedt, 1999). Cells depleted of Surf4/ERGIC-53 or p25 showed no tubulation of the cis-Golgi, whereas in control cells the cis-Golgi was extensively tubulated after hypotonic stress (Figure 10).
Figure 10.
Cargo receptors are required for tubulation of the cis-Golgi. HeLa cells treated with control, Surf4/ERGIC-53, or p25 siRNA were incubated in hypotonic medium for 5 min. Cells were processed for immunofluorescence microscopy by using anti-GM130 and anti-giantin antibodies. Note that the knockdown conditions prevented the tubulation of the cis-Golgi indicated by GM130. Bars, 10 μm.
Collectively, these data indicate that silencing Surf4 together with ERGIC-53 or silencing p25 leads to partial dissociation of COP I. Moreover, the partial resistance of the cis-Golgi to BFA and the lack of tubulation after cargo receptor silencing imply that cargo receptors are required for efficient tubulation of the cis-Golgi.
DISCUSSION
In this study, we characterized human Surf4, and we found it to be associated with the ERGIC and to cycle in the early secretory pathway in a dilysine signal-dependent manner. Erv29p, the yeast orthologue of Surf4, acts as a cargo receptor for glycosylated α-factor in yeast (Belden and Barlowe, 2001; Otte and Barlowe, 2004). Although a knockdown of Surf4 had no effect on total protein secretion, it remains possible that human Surf4 also operates as a cargo receptor for a limited set of proteins that would not be apparent in a global secretion assay. Previous studies have also implicated Erv29p in ER quality control. In yeast cells lacking Erv29p, misfolded soluble proteins are stabilized, and it was proposed that efficient degradation of these misfolded proteins requires transport between ER and Golgi mediated by Erv29p (Caldwell et al., 2001). We found no equivalent function for human Surf4. An efficient knockdown of Surf4 had no effect on the degradation of the Z mutant of α1-antitrypsin a prototype ERAD substrate (data not shown). This observation argues against a general role of Surf4 in ER degradation of misfolded soluble proteins as suggested for Erv29p.
The characterization of Surf4-interacting proteins uncovered a novel role of cargo receptors in maintaining the architecture of ERGIC and Golgi. Surf4 was found to form at least two protein complexes, one complex that has an Mr of 232 kDa and comprises p23, p24, and p25; and another complex of ∼60 kDa, which was not further characterized but may contain KDEL-receptor. The serendipitous finding of a coimmunoprecipitation of Surf4 and ERGIC-53 suggests the existence of a third complex. Because ERGIC-53 forms homohexamers (Schweizer et al., 1988), this complex can be expected to be very large so that it may not have entered the Blue Native gel. It is widely recognized that p24 family proteins form heterooligomeric complexes with one another, which complicates the functional analysis of these proteins (Dominguez et al., 1998). The current study suggests that the situation is even more complex. The major known cargo receptors can form various protein complexes with one another with functional implications for organelle maintenance. Although this was unexpected, an even greater surprise was the observation that a double knockdown of Surf4/ERGIC-53 and a single knockdown of p25 resulted in an identical Golgi and ERGIC phenotype, particularly because the Surf4/ERGIC-53 knockdown did not affect p25 levels and vice versa. There are no indications, however, for a major difference of the phenotypes resulting from the two different knockdowns, neither at the light nor at the ultrastructural level. The phenotype is characterized by a reduced number of ERGIC clusters and fragmentation of the Golgi apparatus whereby the Golgi elements were not randomly distributed in the cytoplasm but largely remained in the original area of the initially compact Golgi.
Numerous situations are known in which the Golgi assumes a fragmented phenotype. How do these phenotypes compare with that observed in the present study? The classical phenotype of dispersed Golgi is due to disruption of microtubules by microtubule-active drugs, such as nocodazole. By contrast, silencing of Surf4/ERGIC-53 or p25 had no effect on microtubules (unpublished data) and the Golgi mini-stacks were not randomly distributed in the cytoplasm as in nocodazole-treated cells. Some other knockdown conditions can lead to Golgi fragmentation similar to that described here, although effects on the ERGIC have not been studied. For example, silencing the SNARE protein syntaxin 5 results in Golgi fragmentation that barely affects anterograde transport of VSV-G, but the underlying mechanism is unknown (Suga et al., 2005). Silencing of KAP3, the nonmotor subunit of kinesin 2, also results in fragmentation of the Golgi (Stauber et al., 2006). Again, anterograde secretory traffic is unaffected, but KDEL-receptor–dependent retrograde transport is abrogated, presumably due to an unexplained redistribution of the KDEL-receptor to the ER. Thus, this phenotype is different. Yet another type of Golgi fragmentation results from silencing golgin-84 (Diao et al., 2003). However, this phenotype is accompanied by changes of the ER, and it has been attributed to a defect in anterograde trafficking. Comparing all the known Golgi fragmentation phenotypes, the Golgi phenotype induced by cargo receptor silencing is strikingly similar to that recently reported for knockdowns of the Golgi matrix proteins GM130 and GRASP65 (Puthenveedu et al., 2006). Either knockdown prevents lateral linking of Golgi stacks resulting in mini-stacks. GM130 mediates stabilization and targeting of GRASP65, and the two proteins are required for Golgi ribbon formation. As a further similarity to the current work, secretory transport is independent of GM130-mediated Golgi ribbon formation (Puthenveedu et al., 2006). Importantly, however, there was no indication of dissociation of GM130 or GRASP65 in cargo receptor knockdowns in the current study, indicating that these two Golgi matrix proteins are not sufficient for Golgi ribbon formation. Moreover, a knockdown of GM130 has no effect on the stability of the ERGIC (our unpublished observations).
Reduced COP I binding for both knockdowns of Surf4/ERIGC-53 and p25 provided a mechanistic explanation for at least some aspects of the phenotype. There are two major different functions of COP I: vesicle formation and stabilization of membranes (Klausner et al., 1992; Rothman, 1994; Storrie, 2005; Bethune et al., 2006a). COP I vesicles mediate membrane traffic within the Golgi, from cis-Golgi to ERGIC, and from ERGIC to ER. Some rapidly cycling transmembrane proteins are actively recruited to retrograde vesicles by a dilysine signal of their cytosolic tail that directly interacts with COP I subunits (Jackson et al., 1990; Cosson and Letourneur, 1994; Bethune et al., 2006a). Surf4, ERGIC-53, and p25 contain such a dilysine signal that is functional in all three proteins (Itin et al., 1995; Emery et al., 2003; this study). In vitro, the formation of COP I vesicles requires the presence of the cytoplasmic domains of p24 family proteins (Bremser et al., 1999). Thus, COP I dissociation from cis-Golgi and ERGIC observed in the current study renders retrograde traffic less efficient. Because anterograde secretory traffic is unaffected this obviously leads to a shortage of ERGIC membranes, which would explain the reduced number and perhaps also shortened life span of ERGIC clusters. For such an outcome with reduced ERGIC-53 cluster numbers one would have to also postulate that in the knockdown cells ERGIC-to-ER transport, although reduced, is slightly more efficient than cis-Golgi to ERGIC transport. This is plausible in view of the proximity of ERGIC and ER, but it cannot be assessed experimentally with current technology.
A function of COP I in membrane stabilization is known from experiments with BFA. On BFA treatment, COP I dissociates from Golgi membranes, and these membranes rapidly tubulate and fuse with the ER. Obviously, COP I protects membranes from tubulation and thereby guarantees organelle integrity and identity. Importantly, neither silencing Surf4/ERGIC-53 nor p25-induced Golgi tubulation despite considerable dissociation of COP I. Under these knockdown conditions COP I dissociation can be assumed to occur at the level of the ERGIC and cis-Golgi, the recycling sites of these cargo receptors. In contrast, overexpression of p25 containing an inactivated dilysine signal does not affect COP I distribution or induce fragmentation of the Golgi apparatus, although it mislocalizes p24 family members to the cell surface (Emery et al., 2003). Inversely, the depletion of p25 did not lead to mislocalization of endogenous p24 to the cell surface (unpublished data). Obviously, overexpression of mutated p25 does not impair the function of p25 to the same extent as a knockdown of p25.
Clearly, COP I dissociation induced by cargo receptor silencing does not result in a BFA-like effect. Thus, COP I depletion cannot explain the absence of tubulation of the cis-Golgi. Together with the partial resistance of the cis-Golgi to BFA after cargo receptor silencing, the lack of tubules implies that cargo receptors are required for efficient tubulation. A likely scenario is that cargo receptor tails mediate the interaction of cis-Golgi membranes with microtubules. Microtubules are required for BFA-induced tubulation of Golgi membranes after COP I dissociation and their subsequent consumption by the ER (Lippincott-Schwartz et al., 1990). Receptor tails may recruit kinesine-type motor proteins, such as kinesin II (Stauber et al., 2006), in the absence of protective COP I coats. Consistent with such a mechanism, the tubulation of anterograde transport intermediates also depends on cargo receptor tails as microinjection of cytosolic tails of p23 and p24 efficiently inhibits tubule formation (Simpson et al., 2006). Obviously, p24 and presumably other cargo receptor tails have an inherent tubulation potential which needs to be controlled by COP I coats to maintain Golgi integrity.
Is the Golgi fragmentation in Surf4/ERGIC-53 or p25 knockdown cells due to COP I dissociation? The close similarity of phenotypes resulting from matrix or cargo receptor knockdowns raises the question of whether an interaction of the two classes of proteins is required for maintaining the Golgi ribbon. If so, a knockdown of either protein class would cause an identical Golgi mini-stack phenotype. Such a notion is not entirely hypothetical because p23, p24, and p25 have been reported to be in a complex with GRASP65, GRASP55, and GM130 in vivo and purified GRASPs directly bind to cytoplasmic tails of p24s (Barr et al., 2001). In contrast to these observations, we have not seen an interaction of p25, Surf4, or ERGIC-53 with GM130 in immunoprecipitation experiments with antibodies to GM130 (data not shown). Thus, more detailed studies will be required to assess a putative dual interaction of cargo receptors with COP I and matrix proteins. It is worth noting, however, that the ERGIC phenotype induced by cargo receptor silencing is unlikely to be due to impaired matrix/tail interactions, because GM130 is primarily associated with the first Golgi cisterna at steady state (Nakamura et al., 1995; Taguchi et al., 2003) and is not detectable in the ERGIC (Figure 7). An alternative possibility to explain the Golgi phenotype induced by receptor silencing is a disturbed balance of the amount of Golgi membranes and matrix proteins. Reduced retrograde traffic from cis-Golgi to ERGIC may result in an increase in Golgi membranes without a corresponding increase in matrix proteins, which may affect Golgi ribbon maintenance.
Why does a single knockdown of Surf4 or ERGIC-53 not change Golgi morphology, whereas p25 does? Currently, we can only speculate about the underlying mechanism. One possibility is that the individual levels of ERGIC-53 and Surf4 in the cis-Golgi are lower than those of p25; therefore, only a combined knockdown of Surf4 and ERGIC-53 leads to sufficient dissociation of COP I from the cis-Golgi. Although no information for Surf4 is available, the levels of ERGIC-53 in the cis-Golgi are indeed low, because the recycling of ERGIC-53 between ERGIC and ER largely bypasses the cis-Golgi (Klumperman et al., 1998; Ben-Tekaya et al., 2005). Alternatively, p25 may not act in isolation because it forms complexes with other p24 proteins that are known to interact with COP I coats via a diphenylalanine rather than a dilysine signal (Bethune et al., 2006a,b). By indirectly affecting other p24 family members, silencing of p25 may have a greater impact.
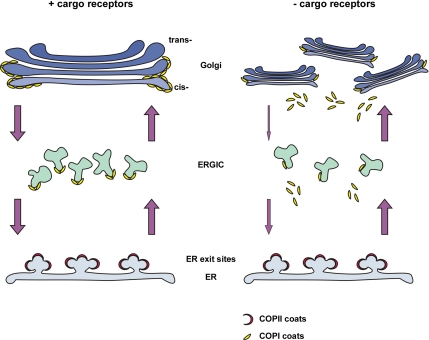
In conclusion, we propose the following model for the changes of the early secretory pathway induced by the depletion of Surf4/ERGIC-53 or p25 (Figure 11). The reduction of cargo receptor tails reduces COP I binding to cis-Golgi and ERGIC and impairs retrograde vesicular traffic. Because anterograde traffic is unchanged this defect results in fewer ERGIC clusters. The reduction of cargo receptors in the cis-Golgi also leads to Golgi mini-stacks either due to insufficient cross-linking of cargo receptor tails with Golgi matrix or due to an imbalance of Golgi membranes and Golgi matrix. According to the maturation model, mini-stack formation would start at the cis-Golgi and gradually be completed as the first cis-Golgi cisterna moves and matures in cis-to-trans direction. Whatever the precise mechanism, the current study shows that networks of established and putative cargo receptors are required to maintain the architecture of ERGIC and Golgi. Thus, cargo receptors of the early secretory pathway can have multiple functions by operating both individually and in concert with one another. This striking dual mode of operation will have to be taken into consideration in future attempts to understand the organization and function of the secretory pathway.
Figure 11.
Model depicting the effect of silencing Surf4/ERGIC-53 or p25 on the early secretory pathway. In the presence of cargo receptors (+cargo receptors), the architecture of the organelles is guaranteed by balanced anterograde and retrograde trafficking indicated by arrows. Depletion of cargo receptors such as Surf4/ERGIC-53 or p25 (−cargo receptors) dissociate COP I coats from cis-Golgi and ERGIC membranes, impairing retrograde transport from cis-Golgi to ERGIC and ERGIC to ER. The sum of this reaction results in dispersal of the Golgi apparatus and reduction of ERGIC structures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anne Spang for helpful suggestion; Käthy Bucher for technical assistance; Paul Jenö for mass spectrometry analysis; Ursula Sauder for electron microscopy analysis; Adam Linstedt for anti-giantin; David S. Nelson for anti-p115 and anti-GM130; Felix Wieland for antibodies to p23, p24, and p25; Fred Gorelick for anti-Sec31; and Vivek Malhotra for anti-GRASP65. This work was supported by the Swiss National Science Foundation and the University of Basel.
Abbreviations used:
- BFA
brefeldin A
- COP
coat protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-10-0989) on February 20, 2008.
REFERENCES
- Andersson H., Kappeler F., Hauri H. P. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J. Biol. Chem. 1999;274:15080–15084. doi: 10.1074/jbc.274.21.15080. [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Hauri H. P. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell Sci. 2006;119:2173–2183. doi: 10.1242/jcs.03019. [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Nyfeler B., Burkhard P., Santamaria I., Lopez-Otin C., Hauri H. P. Carbohydrate- and conformation-dependent cargo capture for ER-exit. Mol. Biol. Cell. 2005;16:1258–1267. doi: 10.1091/mbc.E04-08-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller C., Andersson H., Kappeler F., Hauri H. P. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1999;1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- Aridor M., Bannykh S. I., Rowe T., Balch W. E. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J. Cell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M., Bannykh S. I., Rowe T., Balch W. E. Cargo can modulate COPII vesicle formation from the endoplasmic reticulum. J. Biol. Chem. 1999;274:4389–4399. doi: 10.1074/jbc.274.7.4389. [DOI] [PubMed] [Google Scholar]
- Barr F. A., Nakamura N., Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F. A., Preisinger C., Kopajtich R., Korner R. Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. J. Cell Biol. 2001;155:885–891. doi: 10.1083/jcb.200108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F. A., Puype M., Vandekerckhove J., Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- Barroso M., Nelson D. S., Sztul E. Transcytosis-associated protein (TAP)/p115 is a general fusion factor required for binding of vesicles to acceptor membranes. Proc. Natl. Acad. Sci. USA. 1995;92:527–531. doi: 10.1073/pnas.92.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden W. J., Barlowe C. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 2001;294:1528–1531. doi: 10.1126/science.1065224. [DOI] [PubMed] [Google Scholar]
- Ben-Tekaya H., Miura K., Pepperkok R., Hauri H. P. Live imaging of bidirectional traffic from the ERGIC. J. Cell Sci. 2005;118:357–367. doi: 10.1242/jcs.01615. [DOI] [PubMed] [Google Scholar]
- Bethune J., Kol M., Hoffmann J., Reckmann I., Brugger B., Wieland F. Coatomer, the coat protein of COPI transport vesicles, discriminates endoplasmic reticulum residents from p24 proteins. Mol. Cell. Biol. 2006a;26:8011–8021. doi: 10.1128/MCB.01055-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune J., Wieland F., Moelleken J. COPI-mediated transport. J. Membr. Biol. 2006b;211:65–79. doi: 10.1007/s00232-006-0859-7. [DOI] [PubMed] [Google Scholar]
- Blum R., Pfeiffer F., Feick P., Nastainczyk W., Kohler B., Schafer K. H., Schulz I. Intracellular localization and in vivo trafficking of p24A and p23. J. Cell Sci. 1999;112:537–548. doi: 10.1242/jcs.112.4.537. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bremser M., Nickel W., Schweikert M., Ravazzola M., Amherdt M., Hughes C. A., Sollner T. H., Rothman J. E., Wieland F. T. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Breuza L., Halbeisen R., Jeno P., Otte S., Barlowe C., Hong W., Hauri H. P. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 2004;279:47242–47253. doi: 10.1074/jbc.M406644200. [DOI] [PubMed] [Google Scholar]
- Caldwell S. R., Hill K. J., Cooper A. A. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- Cosson P., Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- Denzel A., Otto F., Girod A., Pepperkok R., Watson R., Rosewell I., Bergeron J. J., Solari R. C., Owen M. J. The p24 family member p23 is required for early embryonic development. Curr. Biol. 2000;10:55–58. doi: 10.1016/s0960-9822(99)00266-3. [DOI] [PubMed] [Google Scholar]
- Diao A., Rahman D., Pappin D. J., Lucocq J., Lowe M. The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 2003;160:201–212. doi: 10.1083/jcb.200207045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M., Dejgaard K., Fullekrug J., Dahan S., Fazel A., Paccaud J. P., Thomas D. Y., Bergeron J. J., Nilsson T. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J. Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G., Parton R. G., Rojo M., Gruenberg J. The trans-membrane protein p25 forms highly specialized domains that regulate membrane composition and dynamics. J. Cell Sci. 2003;116:4821–4832. doi: 10.1242/jcs.00802. [DOI] [PubMed] [Google Scholar]
- Emery G., Rojo M., Gruenberg J. Coupled transport of p24 family members. J. Cell Sci. 2000;113:2507–2516. doi: 10.1242/jcs.113.13.2507. [DOI] [PubMed] [Google Scholar]
- Fiedler K., Veit M., Stamnes M. A., Rothman J. E. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1396–1399. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- Fullekrug J., Suganuma T., Tang B. L., Hong W., Storrie B., Nilsson T. Localization and recycling of gp27 (hp24gamma3): complex formation with other p24 family members. Mol. Biol. Cell. 1999;10:1939–1955. doi: 10.1091/mbc.10.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Linstedt A. D. COPII-Golgi protein interactions regulate COPII coat assembly and Golgi size. J. Cell Biol. 2006;174:53–63. doi: 10.1083/jcb.200604058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Hauri H. P., Kappeler F., Andersson H., Appenzeller C. ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 2000;113:587–596. doi: 10.1242/jcs.113.4.587. [DOI] [PubMed] [Google Scholar]
- Hunte C., von Jagow G., Schägger H. Membrane Protein Purification and Crystallization. San Diego, CA: Elsevier Science; 2003. [Google Scholar]
- Itin C., Schindler R., Hauri H. P. Targeting of protein ERGIC-53 to the ER/ERGIC/cis-Golgi recycling pathway. J. Cell Biol. 1995;131:57–67. doi: 10.1083/jcb.131.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. R., Nilsson T., Peterson P. A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne N., Frey K., Brugger B., Wieland F. T. Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. J. Biol. Chem. 2002;277:46504–46511. doi: 10.1074/jbc.M206989200. [DOI] [PubMed] [Google Scholar]
- Kaiser C. Thinking about p24 proteins and how transport vesicles select their cargo. Proc. Natl. Acad. Sci. USA. 2000;97:3783–3785. doi: 10.1073/pnas.97.8.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Schweizer A., Clausen H., Tang B. L., Hong W., Oorschot V., Hauri H. P. The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J Cell Sci. 1998;111:3411–3425. doi: 10.1242/jcs.111.22.3411. [DOI] [PubMed] [Google Scholar]
- Lavoie C., Paiement J., Dominguez M., Roy L., Dahan S., Gushue J. N., Bergeron J. J. Roles for alpha(2)p24 and COPI in endoplasmic reticulum cargo exit site formation. J. Cell Biol. 1999;146:285–299. doi: 10.1083/jcb.146.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Linstedt A. D. Osmotically induced cell volume changes alter anterograde and retrograde transport, Golgi structure, and COPI dissociation. Mol. Biol. Cell. 1999;10:1445–1462. doi: 10.1091/mbc.10.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D., Hauri H. P. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Donaldson J. G., Schweizer A., Berger E. G., Hauri H. P., Yuan L. C., Klausner R. D. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- Majoul I., Sohn K., Wieland F. T., Pepperkok R., Pizza M., Hillemann J., Soling H. D. KDEL receptor (Erd2p)-mediated retrograde transport of the cholera toxin A subunit from the Golgi involves COPI, p23, and the COOH terminus of Erd2p. J. Cell Biol. 1998;143:601–612. doi: 10.1083/jcb.143.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz M., Nuoffer C., Hauri H. P., Riezman H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Lowe M., Levine T. P., Rabouille C., Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T. E., Warren G. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. S., Alvarez C., Gao Y. S., Garcia-Mata R., Fialkowski E., Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J. Cell Biol. 1998;143:319–331. doi: 10.1083/jcb.143.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols W. C., et al. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell. 1998;93:61–70. doi: 10.1016/s0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- Nyfeler B., Michnick S. W., Hauri H. P. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. USA. 2005;102:6350–6355. doi: 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler B., Zhang B., Ginsburg D., Kaufman R. J., Hauri H. P. Cargo selectivity of the ERGIC-53/MCFD2 transport receptor complex. Traffic. 2006;7:1473–1481. doi: 10.1111/j.1600-0854.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- Otte S., Barlowe C. Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat. Cell Biol. 2004;6:1189–1194. doi: 10.1038/ncb1195. [DOI] [PubMed] [Google Scholar]
- Presley J. F., Cole N. B., Schroer T. A., Hirschberg K., Zaal K. J., Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Reeves J. E., Fried M. The surf-4 gene encodes a novel 30 kDa integral membrane protein. Mol. Membr. Biol. 1995;12:201–208. doi: 10.3109/09687689509027508. [DOI] [PubMed] [Google Scholar]
- Rojo M., Emery G., Marjomaki V., McDowall A. W., Parton R. G., Gruenberg J. The transmembrane protein p23 contributes to the organization of the Golgi apparatus. J. Cell Science. 2000;113:1043–1057. doi: 10.1242/jcs.113.6.1043. [DOI] [PubMed] [Google Scholar]
- Rothman J. E. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Scales S. J., Pepperkok R., Kreis T. E. Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Schimmoller F., Singer-Kruger B., Schroder S., Kruger U., Barlowe C., Riezman H. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A., Fransen J. A., Bachi T., Ginsel L., Hauri H. P. Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J. Cell Biol. 1988;107:1643–1653. doi: 10.1083/jcb.107.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugrue C. A., Kolen E. R., Peters H., Czernik A., Kaiser C., Matovcik L., Hubbard A. L., Gorelick F. Identification of the putative mammalian orthologue of Sec31P, a component of the COPII coat. J. Cell Sci. 1999;112:4547–4556. doi: 10.1242/jcs.112.24.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J. C., Nilsson T., Pepperkok R. Biogenesis of tubular ER-to-Golgi transport intermediates. Mol. Biol. Cell. 2006;17:723–737. doi: 10.1091/mbc.E05-06-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohda M., Misumi Y., Yoshimura S., Nakamura N., Fusano T., Sakisaka S., Ogata S., Fujimoto J., Kiyokawa N., Ikehara Y. Depletion of vesicle-tethering factor p115 causes mini-stacked Golgi fragments with delayed protein transport. Biochem. Biophys. Res. Commun. 2005;338:1268–1274. doi: 10.1016/j.bbrc.2005.10.084. [DOI] [PubMed] [Google Scholar]
- Sohn K., Orci L., Ravazzola M., Amherdt M., Bremser M., Lottspeich F., Fiedler K., Helms J. B., Wieland F. T. A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. J. Cell Biol. 1996;135:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes M. A., Craighead M. W., Hoe M. H., Lampen N., Geromanos S., Tempst P., Rothman J. E. An integral membrane component of coatomer-coated transport vesicles defines a family of proteins involved in budding. Proc. Natl. Acad. Sci. USA. 1995;92:8011–8015. doi: 10.1073/pnas.92.17.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber T., Simpson J. C., Pepperkok R., Vernos I. A role for kinesin-2 in COPI-dependent recycling between the ER and the Golgi complex. Curr. Biol. 2006;16:2245–2251. doi: 10.1016/j.cub.2006.09.060. [DOI] [PubMed] [Google Scholar]
- Storrie B. Maintenance of Golgi apparatus structure in the face of continuous protein recycling to the endoplasmic reticulum: making ends meet. Int. Rev. Cytol. 2005;244:69–94. doi: 10.1016/S0074-7696(05)44002-4. [DOI] [PubMed] [Google Scholar]
- Suga K., Hattori H., Saito A., Akagawa K. RNA interference-mediated silencing of the syntaxin 5 gene induces Golgi fragmentation but capable of transporting vesicles. FEBS Lett. 2005;579:4226–4234. doi: 10.1016/j.febslet.2005.06.053. [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Hsu P., Mallabiabarrena A., Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- Taguchi T., Pypaert M., Warren G. Biochemical sub-fractionation of the mammalian Golgi apparatus. Traffic. 2003;4:344–352. doi: 10.1034/j.1600-0854.2003.00091.x. [DOI] [PubMed] [Google Scholar]
- Teasdale R. D., Jackson M. R. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- Vetrivel K. S., et al. Dual roles of the transmembrane protein p23/TMP21 in the modulation of amyloid precursor protein metabolism. Mol. Neurodegener. 2007;2:4. doi: 10.1186/1750-1326-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollenweider F., Kappeler F., Itin C., Hauri H. P. Mistargeting of the lectin ERGIC-53 to the endoplasmic reticulum of HeLa cells impairs the secretion of a lysosomal enzyme. J. Cell Biol. 1998;142:377–389. doi: 10.1083/jcb.142.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuschner D., Geerts W. J., van Donselaar E., Humbel B. M., Slot J. W., Koster A. J., Klumperman J. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat. Cell Biol. 2006;8:377–383. doi: 10.1038/ncb1371. [DOI] [PubMed] [Google Scholar]
- Zhang B., et al. Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nature Genet. 2003;34:220–225. doi: 10.1038/ng1153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.