Abstract
The autonomously replicating sequence binding factor 1 (Abf1) was initially identified as an essential DNA replication factor and later shown to be a component of the regulatory network controlling mitotic and meiotic cell cycle progression in budding yeast. The protein is thought to exert its functions via specific interaction with its target site as part of distinct protein complexes, but its roles during mitotic growth and meiotic development are only partially understood. Here, we report a comprehensive approach aiming at the identification of direct Abf1-target genes expressed during fermentation, respiration, and sporulation. Computational prediction of the protein's target sites was integrated with a genome-wide DNA binding assay in growing and sporulating cells. The resulting data were combined with the output of expression profiling studies using wild-type versus temperature-sensitive alleles. This work identified 434 protein-coding loci as being transcriptionally dependent on Abf1. More than 60% of their putative promoter regions contained a computationally predicted Abf1 binding site and/or were bound by Abf1 in vivo, identifying them as direct targets. The present study revealed numerous loci previously unknown to be under Abf1 control, and it yielded evidence for the protein's variable DNA binding pattern during mitotic growth and meiotic development.
INTRODUCTION
Progression through the mitotic and meiotic cell cycles in budding yeast is in part controlled by underlying expression programs that coordinate timing of induction with time of function of many genes essential for these processes (for review, see Futcher, 2002; Schlecht and Primig, 2003). Transcriptional control requires a complex interplay between activators and repressors, basal transcription factors, and enzymes involved in chromatin modification together with general regulatory factors such as Abf1. This protein was initially shown to be required for normal activity of autonomously replicating sequence (ARS) elements by direct interaction with its specific DNA binding motif (Rhode et al., 1992). Subsequent studies revealed its contribution to mitotic and meiotic promoter activities via mutational analysis of its N-terminal DNA binding module, its C-terminal protein interaction domains, and its DNA target site (Kovari and Cooper, 1991; Cho et al., 1995; Gailus-Durner et al., 1996; Ozsarac et al., 1997; Miyake et al., 2002). The precise mechanism of action of Abf1 is not known, but its association with DNA has an effect on nucleosome positioning (Lascaris et al., 2000; Yarragudi et al., 2004). Exchanging Abf1-dependent regulatory elements in promoters mediating early and middle meiotic transcriptional activation suggested a role for the protein in enhancing gene expression to wild-type levels during spore development rather than controlling timing of induction (Pierce et al., 1998).
Abf1 was proposed to be a hub protein that is part of many distinct protein complexes (Luscombe et al., 2004). Direct physical interaction or coprecipitation was demonstrated for proteins likely involved in Abf1 phosphorylation (Cka1/Cka2 and Ckb1/Ckb2), nuclear localization (Pse1), SUMOylation (Smt3), and ARS binding activity (Cdc6). Other studies found the protein to coprecipitate with factors required for chromatin assembly and chromatin remodeling (Hta2, Htb1, Rvb1, Rvb2, and Isw2), transcriptional regulation (Mot1, Sth1, and Taf5), activity of RNA polymerase I (Hmo1 and Rpc40) and III (Rpc34, Rpc53, and Rpo31), mRNA export from the nucleus (Yra1), and nucleotide excision repair (Rad7 and Rad16) (see BioGRID for references; Stark et al., 2006).
Genome-wide DNA binding assays and protein–protein interaction analyses have shown that Abf1 binds to a large number of promoters during mitotic growth and that it controls coregulated protein clusters (also termed “regulonic complexes” (Zhang et al., 2005) involved for example in cytoplasmic transport and histone deacetylation (Tan et al., 2007). Some of these Abf1-dependent clusters, such as motor proteins, seem to be conserved between yeast and fly (Tan et al., 2007). Dynamic network analysis covering multiple conditions revealed that Abf1 controls cell growth and in addition regulates intracellular transport during stress response, indicating that it can activate distinct target genes when cells are exposed to different environmental cues (Luscombe et al., 2004). The correlation between its role as a transcriptional regulator and its DNA binding activity, as assessed by chromatin immunoprecipitation in combination with microarrays (ChIP-Chip) or protein binding microarrays is unclear because many promoters that are bound do not seem to contain a match to the target site. In contrast, a statistical model of the sequence-dependent binding energy of Abf1, which is similar to the position-specific weight matrix model used in this study, is in good agreement with in vitro and in vivo binding data (Kinney et al., 2007, and references therein). Recently, the temperature-sensitive abf1-1 allele was used to identify target genes in mitotically growing haploid cells by using microarrays because the mutant protein fails to interact with its target site at the restrictive temperature due to a point mutation in the DNA binding domain (Rhode et al., 1992; Miyake et al., 2004; Yarragudi et al., 2007). However, these studies lacked wild-type and mutant controls at the permissive temperature, they failed to include growth in the absence of a fermentable carbon source, and they did not cover meiotic development.
In this article, we report the results of a comprehensive approach to studying the role of Abf1 in controlling mitotic growth during fermentation, respiration, and sporulation. We first characterized the growth properties of diploid wild-type versus mutant yeast cells containing one or two abf1-1 alleles at the permissive (25°C), semipermissive (30°C), and restrictive (37°C) temperatures on solid and in liquid growth media containing glucose (YPD) or acetate (YPA). Subsequently, the ability of diploid yeast cells containing one wild-type or abf1-1 temperature-sensitive mutant allele to form spores was monitored using plate and liquid sporulation assays carried out at temperatures that permit (25, 28, and 33°C) or inhibit (37°C) sporulation. To identify mitotic and meiotic genes under direct transcriptional control of Abf1, a combination of computational target site prediction, a genome-wide DNA binding assay of mitotic and meiotic cells, and expression profiling of wild-type versus mutant strains was used. Obtaining results from three complementary methods enabled us to set optimally low filtration thresholds for each data type, to allow for efficient mitotic and meiotic target gene discovery. The microarray expression profiling and binding data are available for downloading via the certified public ArrayExpress repository at the European Bioinformatics Institute (Parkinson et al., 2005). A graphical display in the context of genome annotation is accessible via the Internet through the GermOnline database (http://www.germonline.org/) (Gattiker et al., 2007).
MATERIALS AND METHODS
Yeast Culture Conditions
Diploid W303 wild-type and abf1-1 mutant strains (Table 1) were grown in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose) or YPA (1% yeast extract, 2% bactopeptone, and 1% potassium acetate) at 25°C to a density of 2 × 107 cells/ml. The cultures were split and incubated either at 37 or 25°C first in a water bath and subsequently in a rotatory shaker at 37°C (260 rpm). Cells were harvested after 60 min, washed with sterile water, snap-frozen in liquid nitrogen, and stored at −80°C. Diploid W303 strains containing one wild-type or mutant allele were sporulated in SPII (2% acetate, pH 7.0) at 28°C for 5 and 9 h, respectively, and then kept at 28, 33, or 37°C for 1 h before harvesting, washing, and storing at −80°C. A diploid SK1 strain was grown and sporulated using standard conditions (Hochwagen et al., 2005), and 50-ml samples were taken for a genome-wide DNA binding assay in YPD, YPA, and SPII 4 and 8 h after initiation of meiosis.
Table 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| KM7 | MATa leu2-3 his4-519 ade1-100 ura3-52 GAL-ABF1::URA3 | Gonçalves et al. (1992) |
| JCA30 | W303 MATa trp1 Δ his3Δ200 ura3-52 lys2-801 ade2-1 gal ABF1 HIS3 | Rhode et al. (1992) |
| JCA31 | W303 MATa trp1 Δ his3Δ200 ura3-52 lys2-801 ade2-1 gal abf1-1 HIS3 | Rhode et al. (1992) |
| JCA40 | W303 MATα trp1 Δ his3Δ200 ura3-52 lys2-801 ade2-1 gal ABF1 HIS3 | Rhode et al. (1992) |
| JCA41 | W303 MATα trp1 Δ his3Δ200 ura3-52 lys2-801 ade2-1 gal abf1-1 HIS3 | Rhode et al. (1992) |
| MPY170 | SK1 MATa/α ho::LYS2 ura3 lys2 leu2::hisG arg4-Nsp/arg4-Bgl his4x::LEU2-URA3/his4B::LEU2 | Primig et al. (2000) |
| MPY284 | MATa/α trp1Δ/trp1Δ his3Δ200/his3Δ200 ura3-52/ura3-52 lys2-801/lys2-801 ade2-1/ade2-1 gal/gal ABF1/ABF1 | This study |
| MPY283 | MATa/α trp1Δ/trp1Δ his3Δ200/his3Δ200 ura3-52/ura3-52 lys2-801/lys2-801 ade2-1/ade2-1 gal/gal abf1-1/abf1-1 | This study |
| MPY285 | MPY283 pRS416-ABF1 | This study |
| MPY125 | MATa/α trp1Δ/TRP1 his3Δ200/his4-519 ura3-52/ura3-52 ade2-1/ade1-100 gal ABF1/GAL-ABF1::URA3 | This study |
| MPY128 | MATa/α trp1Δ/TRP1 his3Δ200/his4-519 ura3-52/ura3-52 ade2-1/ade1-100 gal abf1-1/GAL-ABF1::URA3 | This study |
| USY368 | MPY284 CDC3/CDC3-eGFP | This study |
| USY364 | MPY285 CDC3/CDC3-eGFP | This study |
Antibodies
Rabbit antibodies were raised against His6-Abf1 (amino acids 264–513) affinity purified from a bacterial extract by using XL10-GOLD (Stratagene, La Jolla, CA). The final bleed was aliquoted, snap-frozen, and stored at −80°C. The commercial hemagglutinin (HA) antibody 12CA5 (Roche Diagnostics, Mannheim, Germany) was used to detect HA3-tagged Abf1 protein. Commercial Abf1 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Western Blotting
Protein extracts were prepared using acid-washed glass beads and a BeadBeater apparatus (BioSpec, Bartlesville, OK). Three cycles of bead beating were done at maximal intensity for 30 s. Extracts were kept on ice for 2 min between each cycle. The concentration of protein extracts was adjusted to 15 μg/μl. For electrophoretic analysis, 60 μg of protein extract was loaded onto a 10% SDS gel run at 100 V for 2 h in the cold room. Western blotting was performed using standard conditions and nitrocellulose membranes pretreated with 3% milk in 0.1% phosphate-buffered saline (PBS)-Tween and then washed in 0.1% PBS-Tween. The secondary anti-rabbit immunoglobulin G antibody was diluted 1:20,000 (Sigma-Aldrich, Paris, France). The enhanced chemiluminescence kit was used to reveal Western blot signals (Bio-Rad, Hercules, CA).
Immunofluorescence
To visualize nuclei and Cdc3-green fluorescent protein (GFP), cells were fixed for 1 h in 3.7% formaldehyde in growth media, washed once in 1× PBS, and resuspended in a 5 μg/ml solution of Hoechst dye (Invitrogen, Carlsbad, CA). Cells were incubated for 30 min at room temperature, and then they were washed three times in 1× PBS. Cells were analyzed using an Axioplan 2 imaging microscope (Carl Zeiss, Jena, Germany) equipped with a Plan Neofluar 100× Ph3 numerical aperture 1.3 objective. Digital images were produced with a TE/CCD-1000PB camera (Princeton Scientific Instruments, Monmouth Junction, NJ). The excitation intensity was controlled with different neutral density filters (Chroma Technology, Brattleboro, VT).
Electrophoretic Mobility Shift Assay (EMSA)
Probe annealing, labeling, and the binding reaction were done as described previously (Primig et al., 1991, 1992). Antibody supershift assays were carried out using diluted antisera that were added to the probe–protein mix for 5 min before loading on a 6% polyacrylamide gel (0.5× Tris-EDTA) run at room temperature for 120 min at 200 V. Gels were dried and analyzed using a PhosphorImager Storm 860 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Computational Binding Site Prediction
For each intergenic region in Saccharomyces cerevisiae, orthologous intergenic regions from the four other budding yeast species were obtained using the open reading frame (ORF) annotations from Kellis et al. (2003) and Cliften et al. (2003), and all groups of orthologous intergenic regions were aligned with T-Coffee (Notredame et al., 2000). The phylogenetic tree of the species was reconstructed using third positions of fourfold degenerate codons in orthologous genes. A weight matrix was reconstructed by simultaneously clustering and aligning known Abf1 sites from the SCPD collection (Zhu and Zhang, 1999). We then predicted binding sites in all intergenic region alignments by using the MotEvo algorithm (Erb and van Nimwegen, 2006). Briefly, MotEvo finds all intergenic sequence segments in the S. cerevisiae reference genome that match the weight matrix better than a third order Markov model of background intergenic sequences. It then collects the corresponding sequence segments from the other species from the alignment. Each of these segments is first scored under the weight matrix and a species-dependent background model and those orthologous segments that score better under the weight matrix are marked as “under selection.” The probability of the observed sequence alignment is calculated under an evolutionary model that assumes all sequences under selection were constrained to retain their match to the weight matrix, by using the inferred phylogenetic tree. This probability is then compared with the probability of the observed alignment under background evolution, and under evolution that is constrained by a different (unknown) selective pressure, to obtain a posterior probability for the site. The positional profile was generated by summing the posterior probabilities of all binding sites that occur upstream of a single gene (i.e., excluding divergently transcribed regions) for which a transcription start site has been determined in Zhang and Dietrich (2005).
Comparison of the Methods for Genome-Wide Abf1 Target Identification
To determine the log-odds ratios of site occurrence, we combined, for each intergenic region, the posterior probabilities of all sites to calculate the probability p that at least one true site occurs. The log-odds ratio r is then given by r = log(p/(1 − p)). The log-odds ratios for each intergenic region to be bound by Abf1 were determined from the ChIP-Chip data as follows. First, probes were mapped to the intergenic regions: a probe was considered to belong to a region if at least 80% of its sequence was contained in the region. Usually, several probes mapped to one region. Using the log-ratio of binding at each probe we calculated, for each region, the probability that binding occurred at least one probe in the region. The final probability of binding p is given by the taking the maximum of this probability over all experiments. The log-odds ratio r is then given by r = log(p/(1 − p)). For each experiment the noise in the expression changes of both wild-type and mutant was estimated from the variance of the duplicate measurements to calculate z-statistics. For each gene, the z-statistic was given by the difference in the observed expression change in mutant and wild-type, relative to the estimated noise, summed over all experiments. To assign z-statistics to intergenic regions, the maximal z-value of the two downstream genes was taken for divergently transcribed regions. We thus obtained, for each intergenic region, a log-odds ratio for site occurrence, a log-odds ratio for binding, and a z-statistic for expression change. A set of cut-off values for the three variables partitions the intergenic regions into eight groups, varying from “unregulated” according to all three methods to “regulated” by all three methods. The overall agreement of the three methods was calculated by the mutual information in the partition into eight groups. We calculated the fractions f(x,y,z) of regions in each of the eight groups whereby x, y, and z were either 0 (unregulated) or 1 (regulated). The mutual information of the partition is then given by
 |
where fs(1) is the overall fraction of regions with binding site predictions over the cut-off, fb(1) the fraction of regions with binding over the cut-off, and fb(1) the fraction of regions with expression change over the cut-off. We then determined the combination of cut-offs for the three methods that maximized the mutual information I. We also calculated the mutual information (at the optimal cut-offs) for each pair of methods.
Chromatin Immunoprecipitation
Cultures (20 ml) containing ∼4 × 108 cells were treated with 1% formaldehyde for 15 min at room temperature. DNA was processed and analyzed by semiquantitative polymerase chain reaction (PCR) as described previously (Borde et al., 2004).
ChIP-Chip Assay
DNA was eluted from the protein G beads, purified, amplified, and labeled as described previously (Borde et al., 2004). Hybridization was carried out using spotted microarrays containing yeast ORFs and intergenic regions as described in Robine et al. (2007). Spotted probes were assigned to promoters as in (Lee et al., 2002), a reference table is available for downloading from http://staffa.wi.mit.edu/cgi-bin/young_public/navframe.cgi?s = 17&f = downloaddata.
ChIP-Chip Data Production and Analysis
Microarrays were scanned using an Axon 4000B scanner (Axon Instruments, Foster City, CA), and the images were analyzed using GenePix 5.0. Each experiment was done in triplicate. Normalization and data analyses were not only carried out as described but also with a lower statistical cut-off value (Median Percentile Rank of 0.7 instead of 0.9) to decrease the selection stringency of regions showing significant fold-enrichment (Robine et al., 2007). Binding data were obtained with cells in YPD, YPA, and SPII at 4 and 8 h after initiation of meiosis. Cyanine (Cy)3/Cy5 and Cy5/Cy3 dye swap array data sets obtained with YPD cultures only were not averaged to determine the influence of the labeling reaction on the data output.
RNA Isolation and cRNA Target Synthesis
Culture conditions and one-channel microarray hybridization protocols were carried out as published with a few modifications. cRNA target molecules were prepared from 50-ml cultures at 3–5 × 107 cells/ml. Samples were hybridized to yeast S98 GeneChips (Affymetrix, Santa Clara, CA) that contain probes for ∼6400 transcripts (http://www.affymetrix.com/). Fifteen micrograms of yeast total RNA was purified using RNeasy mini-spin columns (QIAGEN, Hilden, Germany) by using standard protocols provided by the manufacturer. The cell pellets were resuspended in RLT buffer (QIAGEN) and lysed by shearing in a 2-ml syringe. Supernatant (600 μl) was mixed with 600 μl of 70% ethanol, loaded onto an RNeasy column, washed, and eluted in 100 μl of double-distilled water. The total RNA quality was monitored using RNA Nano 6000 Chips processed with the 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA). Biotin labeling of RNA was done as outlined in the Expression Analysis Technical Manual with minor modifications. Single-stranded cDNA was synthesized by mixing 13 μg of total RNA with oligo(dT) and incubating the reaction mix with SuperScript II reverse transcriptase (Invitrogen) at 42°C for 1 h. After synthesis of the second cDNA strand using the Superscript double-stranded cDNA Synthesis kit (Invitrogen), the material was extracted with phenol-chloroform-isoamyl alcohol and precipitated with 0.5 volumes of 7.5 M ammonium acetate and 2.5 volumes of ethanol. The cDNA was used for an in vitro transcription reaction by using the BioArray high yield RNA transcript labeling kit T7 (Enzo Diagnostics, New York, NY) to synthesize cRNA in the presence of biotin-conjugated uridine 5′-diphosphate and cytidine 5′-triphosphate analogs. Approximately 50 μg of cRNA was purified over RNeasy Mini-Spin columns and analyzed again on RNA Nano 6000 Chips. The cRNA targets were incubated at 94°C for 35 min, and the resulting fragments of 50–150 nucleotides were again analyzed with RNA Nano 6000 Chips. All synthesis reactions were carried out in a PCR machine (T1 thermocycler; Biometra, Göttingen, Germany) to ensure optimal temperature control.
GeneChip Hybridization and Scanning
Hybridization cocktails (220 μl) containing heat-fragmented and biotin-labeled cRNA at a concentration of 0.05 μg/μl were injected into GeneChips and incubated at 45°C on a rotator in a hybridization oven 640 (Affymetrix) overnight at 60 rpm. The arrays are washed and stained with a streptavidin-phycoerythrin conjugate (Invitrogen). To increase signal strength, a standard antibody amplification protocol was used (EukGE-WS2v4; see Affymetrix Expression Analysis Manual). The S98 GeneChips were processed with a GeneArray Scanner 300 7G (Affymetrix) as described previously (Chalmel et al., 2007).
Microarray Data Processing and Statistical Analysis
Raw data were preprocessed (background adjustment, normalization, and summarization of probe sets) by using the Robust Multiarray Analysis package from BioConductor as described in Schlecht et al. (2004). First, probe sets for which we observed a signal equal or >100 (empirical conservative background threshold) in at least one condition were identified. Among those, differentially expressed probe sets were identified by analysis of variance. Genes were filtered using a p value <0.005 and a SD >0.3.
Cluster Analysis
Among the probe sets we identified those that were clearly associated with predicted genes, thus eliminating cases related to SAGE data and TY-elements for which probes are present on the S98 GeneChip. Genes identified as differentially expressed were then clustered using the k-means algorithm. Note that because of this approach, the sum of all genes in YPD and YPA clusters 1–3 shown in Figure 4D, clusters 4 and 5 shown in Supplemental Figure 5, and in SPII clusters shown in Figure 7B is smaller than the number indicated in the Venn diagram in Figure 4B.
Figure 4.
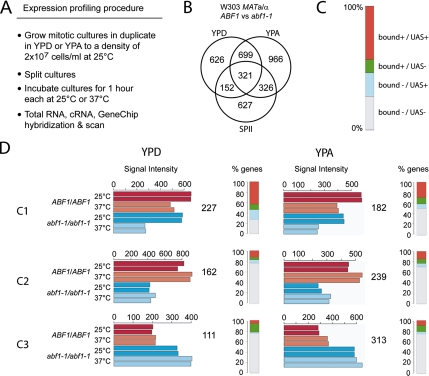
Identification of mitotic Abf1 target genes by using Affymetrix S98 GeneChips. (A) Outline of the approach. (B) Venn diagram of a comparative profiling experiment using diploid wild-type ABF1 versus abf1-1 mutant strains compared at 25 versus 37°C (log-phase growth, mitosis) and at 28°C (6- and 10-h time points, meiosis). Genes detected in three media as indicated are given. (C) Color-coded bar diagram displaying differentially expressed genes whose promoters are bound and contain a UAS motif (red, bound+/UAS+), whose promoters are bound but lack a UAS motif (green, bound+/UAS−), whose promoters are not bound but do contain a UAS motif (blue, bound−/UAS+) and whose promoters are not bound and lack UAS (bound−/UAS−). (D) Overview of three different expression clusters (C1–C3) identified among genes in rich medium with glucose (YPD) or acetate (YPA). Diploid wild-type (ABF1/ABF1, red) and temperature-sensitive mutant strains (abf1-1/abf1-1, blue) are indicated. Median signal intensities are plotted against samples analyzed in duplicate at the temperatures shown. The total number of genes in each cluster is given. The gene content of clusters C1–C3 identified in YPD and YPA overlaps but is not identical. Percentages of genes falling into any of four categories outlined in C are shown.
Figure 7.
Identification of meiotic Abf1 target genes. (A) Outline of the approach. (B) Five expression clusters (C1–C5) identified among genes in sporulation medium at 28°C. Note that although C3-C5 seem rather similar, we empirically determined five clusters to be the number that identifies the largest percentage of genes in C1 confirmed by in vivo binding and presence of a putative target site in the promoter. Strains containing one wild-type (ABF1/abf1, red) and one mutant allele (abf1-1/abf1, blue) and a GAL-ABF1 transgene are indicated. Median signal intensities are plotted against samples analyzed in duplicate at the time points shown. The total number of genes in a cluster is given. Percentages of genes falling into any of four categories outlined in Figure 4C are shown.
Gene Ontology (GO) Analysis
Expression clusters were searched for enrichment of functions using the Annotation, Mapping, Expression and Network (AMEN) analysis tool (see SourceForge at http://sourceforge.net/projects/amen/), a new array data analysis and visualization software package (Chalmel and Primig, 2008). A specific annotation term was considered as enriched in a group of coexpressed genes if the p value was <0.005 and the number of genes in this cluster showing this annotation was >5. The results are output in a color-coded graph that facilitates the interpretation of expression patterns and their functional significance.
MIAME Compliance
CEL feature level data files and transcript level text files corresponding to rich medium (YPD), presporulation medium (YPA), and sporulation medium (SPII) samples are available via the EBI ArrayExpress public data repository at http://www.ebi.ac.uk/arrayexpress/ under accession number E-TABM-291. Genome-wide DNA binding data are available via E-MEXP-1439.
RESULTS
abf1-1 ts Mutant Cells Fail to Undergo Normal Cytokinesis
Haploid abf1-1 cells grow slowly at 30°C, become very large, form elongated buds, and arrest growth at several steps in the cell cycle when shifted to 37°C (Rhode et al., 1992). Homozygous diploid abf1-1 mutant cells show similar temperature-dependent growth both on solid (Figure 1A) and in liquid rich medium (Figure 1C). This effect is complemented in cells containing a single-copy plasmid bearing a wild-type copy of ABF1 controlled by its native promoter (Figure 1A, bottom row). Diploid strains heterozygous for ABF1 grow both at the permissive and restrictive temperatures (Figure 1B, top), which is likely due to Abf1 being an abundant protein (Planta et al., 1995; Ghaemmaghami et al., 2003). However, MATa/α cells containing only one abf1-1 allele show a severe growth phenotype at the semipermissive temperature of 30°C (Figure 1B, top), indicating a dosage effect because diploid cells that contain two abf1-1 alleles display only a mild phenotype (Figure 1A, middle). As expected, diploid strains that contain a wild-type copy of ABF1 under the control of a galactose-inducible promoter and an endogenous wild-type or abf1-1 mutant allele grow on rich medium containing galactose at the semipermissive temperature (Figure 1B, bottom). Note that the galactose-dependent induction of ABF1 only partially complements the Abf1-1 phenotype at 37°C (Figure 1B, bottom). The GAL-Abf1 allele is a priori fully functional because the GAL1-GAL10 intergenic region fused to a full-length ABF1 locus can complement an ABF1 gene deletion in haploid (Goncalves et al., 1992) and diploid cells (our own observation). However, overexpression of ABF1 is toxic under certain conditions and additional heat stress may therefore result in partial growth inhibition (Donovan et al., 1998).
Figure 1.
Growth properties of diploid ABF1 wild-type versus abf1-1 mutant strains. (A) Shown are serial fivefold dilutions of diploid wild-type (ABF1/ABF1; top), mutant (abf1-1/abf1-1; middle), and mutant cells transformed with a plasmid containing ABF1 (abf1-1/abf1-1 p[+]; bottom), grown for 2 d on YPD plates at the temperatures indicated. (B) Dilutions are given of an ABF1/abf1 heterozygous or abf1/abf1 homozygous strain containing an ABF1 transgene fused to a galactose-inducible promoter as indicated, grown on YPD (top) or YPGal (bottom). (C) Growth rate of diploid wild-type (ABF1/ABF1) and mutant (abf1-1/abf1-1) strains in liquid YPD during 9 h at the temperatures indicated. Mean values from three independent experiments are shown. The bars represent the SD. (D) Cell morphology by differential interference contrast (DIC) and nuclear staining (DAPI) of wild-type and mutant strains at 25, 30, and 37°C as indicated. (E) Quantification of a cell separation defect in the abf1-1 background. Cells were incubated at 25, 30, and 37°C during 4 h and the percentage of unbudded cells (white bar), cells with a single bud (light gray), and multicellular bodies (dark gray) was plotted on the y-axis against the wild-type and mutant strains on the x-axis. Mean values from three independent experiments are shown. Bars represent the SD.
A detailed morphological examination revealed no difference between wild-type and mutant cells at the permissive temperature (Figure 1, D and E, left) of 25°C. As opposed to that, mutant cells grown at the semipermissive temperature of 30°C accumulated bodies of more than three unseparated and mostly unbudded enlarged cells (middle). This effect was also observed when mutant cells were cultured at 37°C, albeit at a lesser degree, because cells stop budding at that temperature (right). Together, these data indicate a dose-dependent growth effect of the abf1-1 mutant allele in diploid cells and suggest a novel role for Abf1 in the regulation of cytokinesis.
The abf1-1 ts Mutant Is Sporulation Deficient at the Permissive Temperature
Previous work showed that normal expression of genes specifically transcribed during meiotic development required a functional Abf1 upstream activation sequence (UAS) target site (Prinz et al., 1995; Gailus-Durner et al., 1996; Ozsarac et al., 1997; Pierce et al., 1998). To directly test the role of the Abf1 protein in meiotic gene expression, diploid strains containing one wild-type or mutant abf1-1 allele together with a GAL1-10-ABF1 fusion, respectively, were constructed. This was done for two reasons: first, we sought to outcross the Abf1-1 allele into a different genetic background because the original isogenic W303 wild-type strain sporulated only very poorly, making it difficult to assess the effect of the mutated allele (Schlecht and Primig, unpublished observation). Second, we expected a strain containing only one transcriptionally active abf1-1 temperature-sensitive allele to be suitable for analysis of meiosis at semipermissive conditions (28–30°C) because this process is inhibited by temperatures >34°C (Simchen, 1974).
Using fluorescence-activated cell sorting to measure the DNA content of cells, we determined that the abf1-1 mutant strain was able to undergo premeiotic DNA replication with the same kinetics as the wild-type control at all three temperatures permissive for meiotic development tested (25, 28, and 33°C) (Figure 2A). This is in keeping with the fact that Abf1-1 does not affect transition through the mitotic S phase (Rhode et al., 1992). As expected, both strains failed to initiate premeiotic DNA replication at 37°C (Figure 2A). Next, we cultured the abf1-1 strain at 25 and 28°C to ask whether temperatures still allowing for mitotic growth (permissive/semipermissive conditions) would impair the onset of meiotic M phase. To test this, the accumulation of bi- and tetranucleate cells as well as the formation of asci was monitored in wild-type and mutant cells. In a diploid strain containing one abf1-1 allele, the efficiency of meiotic development was decreased at 25°C, severely reduced at 28°C, and abolished at 33°C, whereas a strain containing one wild-type allele sporulated normally at all permissive temperatures. Neither of the strains was able to enter meiotic M phase at 37°C (Figure 2B). These results show a role for the Abf1 protein in controlling the onset of meiotic M phase and spore development, which is coherent with previous reports on the role of its target sites in meiotic promoters.
Figure 2.
Determining meiotic landmark events in ABF1 versus abf1-1 strains. (A) Fluorescence-activated cell sorting analysis of premeiotic DNA replication in hetero- or homozygous abf1 deletion strains containing a GAL-ABF1 construct at different temperatures and at the time points indicated. The DNA content of G1 and G2 cells is given as 2N and 4N. (B) Sporulation properties of strains containing one wild-type or mutant allele as indicated. The percentage of cells undergoing meiosis I, meiosis II, and ascus formation at the temperature indicated are plotted on the y-axis against time points on the x-axis from 0 to 120 h. Data from wild-type versus mutant alleles are color coded as given in the legend.
The data also demonstrated that the abf1-1 allele is a suitable tool for identifying not only mitotic but also meiotic Abf1 target genes via expression profiling at temperature conditions that impair Abf1-1 activity without inhibiting the meiotic process itself. Therefore, we set out to define genes directly controlled by Abf1 by validating target site predictions with genome-wide in vivo DNA binding assays and microarray profiling of wild-type versus temperature-sensitive mutant abf1-1 strains at permissive, semipermissive, and restrictive conditions.
Computational Prediction of Abf1 Binding Sites Reveals Positional Similarity with Reb1
To identify putative Abf1 binding sites, a position-specific weight matrix was built by applying the PROCSE clustering algorithm (van Nimwegen et al., 2002) to experimentally verified Abf1 binding sites present in the promoters of genes such as HOP1, CAR1, and SPR3 (Kovari and Cooper, 1991; Gailus-Durner et al., 1996; Ozsarac et al., 1997). Subsequently, we searched the multiple alignments of all intergenic regions in five related yeast species (Cliften et al., 2003) for matches to the Abf1 weight matrix, by using the MotEvo algorithm based on a Bayesian probabilistic approach that explicitly models the evolution of functional regulatory sites and nonfunctional intergenic DNA (Erb and van Nimwegen, 2006). We identified 1049 distinct intergenic regions that contain one or more putative Abf1 binding sites with significant probability. As expected, the weight matrix constructed from all predicted binding sites present in the budding yeast genome matches matrices obtained by in vitro selection of Abf1 binding sites (Mukherjee et al., 2004; Beinoraviciute-Kellner et al., 2005) and from high-throughput data (Kinney et al., 2007). Predicted motifs were found to be located preferentially ∼100 base pairs upstream of the transcription start site, which is in keeping with a previous report (Yarragudi et al., 2007). Among >75 other transcription factors, only Reb1 binds to target sites that show a similar positional bias (Erb and van Nimwegen, 2006). Although Reb1 and Abf1 bind very different sequence motifs, both are general regulatory factors that enhance transcription by displacing nucleosomes (Lascaris et al., 2000; Angermayr et al., 2003; Yarragudi et al., 2004; Raisner et al., 2005).
Abf1 Displays Partially Distinct in Vivo DNA Binding Patterns during Mitosis and Meiosis
We next monitored Abf1/DNA interaction during mitotic growth in the presence of glucose (fermentation) and acetate (respiration) and during early and middle stages of meiotic development in diploid SK1, a strain we had previously used for growth and sporulation experiments (Primig et al., 2000). Tagging Abf1 with a C-terminal HA epitope decreased sporulation efficiency and spore viability (our unpublished data). Therefore, we raised a polyclonal anti-Abf1 antibody against a 60-amino acid fragment between the DNA binding domains. The serum was shown to recognize the antigen (Supplemental Figure 1A) and the HA-tagged Abf1 expressed from its native promoter and Abf1 under the control of a galactose-inducible promoter (Supplemental Figure 1B). The result was confirmed by restaining the blot with a monoclonal antibody against the HA epitope (Supplemental Figure 1C). We next validated the Abf1 serum in an electrophoretic mobility antibody supershift assay by using an oligonucleotide probe containing the HOP1 UAS. The free probe migrated as a single band (Supplemental Figure 1D, lane 1). On addition of a protein extract from mitotic cells, a slowly migrating complex was observed (lane 2) and found to be abolished in the presence of a molar excess of unlabeled HOP1UAS and UME6UAS wild-type (lanes 3 and 5) but not UAS mutant probes (lanes 4 and 6). No effect was observed upon addition of an antibody against Ndt80 (lanes 7–9), whereas the anti-Abf1 serum shifted the Abf1/DNA complex in a concentration-dependent manner (lanes 10–12). To test the ability of the Abf1 antibody to recognize the protein in vivo during mitotic growth, we performed qualitative chromatin immunoprecipitation control experiments that confirmed known binding activities to the MCK1 (Harbison et al., 2004) and ZIP1 (Prinz et al., 1995) promoters (Supplemental Figure 1E, top and middle). Note that no in vivo binding activity was observed in the HOP1 promoter whose UAS element is thought to be bound by Abf1 (bottom) (Prinz et al., 1995; Gailus-Durner et al., 1996).
The genome-wide binding experiments using spotted microarrays covering ∼13,000 intergenic sequences and open reading frames were done in triplicate (Figure 3A and Supplemental Figure 2A). Standard controls (dye-swap, mock immunoprecipitation, and input-overinput) were carried out to correct for systematic errors using cells cultured in YPD (Supplemental Figure 2B). A comparison of binding data from mitotic and meiotic cells revealed the upstream sequences of 1169 genes to be bound in three media by using nonstringent selection criteria (see Materials and Methods); a subset of these potential promoter regions such as iMRPS18/iMCK1 and iMSS4/iUME6 also contained at least one predicted binding motif (p value of 5.4 × 10−254) (Figure 3B). Importantly, our data showed a significant overlap (p value of 2.1 × 10−149) with a previous study that identified 458 Abf1 target promoters in haploid cells cultured in rich medium (Harbison et al., 2004). Like in a qualitative chromatin immunoprecipitation assay (Supplemental Figure 1E), we did not observe any interaction of Abf1 with its target site in the HOP1 promoter during mitosis or meiosis (Figure 3B, bottom right), whereas we show that the promoter of ZIP1 is bound in vivo during growth and meiotic development (bottom left). It is noteworthy that binding to 82, 105, and 331 fragments was only detected during fermentation (YPD), respiration (YPA), or sporulation (SPII), respectively, indicating that the DNA binding affinity of Abf1 to some of its targets or their accessibility changes during these processes. Lists of genes whose promoters are detected as constitutively bound or specifically enriched during any of the conditions tested are available (Supplemental Table 1). We applied a less stringent selection procedure than a previous study (Robine et al., 2007; see Materials and Methods) because the binding data are integrated with results of motif prediction and expression profiling methods. Importantly, using the more selective filtration approach also reproducibly yields different binding patterns (Supplemental Figure 3, A and B).
Figure 3.
Genome-wide Abf1 in vivo binding assay. (A) Summary of the approach and Venn diagram of fragments found to be enriched above background in a diploid SK1 strain grown in rich medium with glucose (YPD) or acetate (YPA) or incubated for 4 and 8 h in sporulation medium (SPII). (B) Graphical display of four intergenic regions containing matches to UAS motifs (black vertical bars) and the corresponding genes on the top (red) and bottom (blue) strands. The fold-enrichment is plotted on the y-axis against experiments by using cells cultured in media as indicated. Sporulating cells were harvested 4 h (SPII 4h) and 8 h (SPII 8h) after induction of meiosis. Control experiments with cells cultured in YPD at 30°C were done using reversed fluorophor labeling (dye-swap), an unrelated antibody (mock), and no antibody (input over input). A color code indicates significant enrichment (red, orange) or background signals (blue, yellow).
Many promoter regions bound by Abf1 also seem to interact with other site-specific DNA binding regulators involved in metabolic functions (Gcn4 and Ino4), cell cycle regulation (Swi4 and Fkh1), chromatin remodeling (Cbf1 and Spt10), induction of pseudohyphal growth (Phd1), and the mitotic repression of meiosis-specific genes (Ume6 and Sum1) (Table 2). This is in keeping with the fact that Abf1 was often found to contribute to normal expression via chromatin opening rather than being the sole essential factor activating a promoter (Yarragudi et al., 2004).
Table 2.
Comparison of genome-wide DNA binding assays
| Gene | Target genes (Harbison et al. 2005) (p < 0.005) | Overlap with this study | Significance of overlap |
|---|---|---|---|
| Abf1 | 468 | 387 | 2.06 × 10−149 |
| Ume6 | 244 | 113 | 7.24 × 10−11 |
| Gcn4 | 103 | 45 | 7.24 × 10−4 |
| Fkh1 | 239 | 86 | 0.00016 |
| Swi4 | 218 | 78 | 0.00041 |
| Ino4 | 123 | 46 | 0.00171 |
| Phd1 | 131 | 47 | 0.00414 |
| Spt10 | 8 | 5 | 0.00516 |
| Cbf1 | 73 | 27 | 0.01300 |
| Abt1 | 3 | 2 | 0.01748 |
| Sum1 | 96 | 33 | 0.02454 |
Identification of Mitotic Abf1 Target Genes by Expression Profiling
Diploid wild-type and abf1-1 mutant strains were cultured in rich medium containing glucose (YPD) or acetate (YPA) at the permissive (25°C) temperature until mid-log phase and then shifted to restrictive (37°C) conditions for 1 h before harvesting. Samples were analyzed in duplicate using high-density oligonucleotide microarrays containing probes covering ∼6400 yeast protein coding loci (S98 GeneChips) (Figure 4A). Total RNA and cRNA quality was assessed using the BioAnalyzer and found to be excellent and very homogenous (Supplemental Figure 4A). Differentially expressed genes were filtered using SD and a permutation test as published and clustered using the k-means algorithm (Schlecht et al., 2004). Expression clusters were searched for enriched functional annotation using data from the Gene Ontology consortium (www.geneontology.org/) and AMEN software (Chalmel and Primig, 2008).
In total, 3717 probe sets that map to 3214 unique protein-coding yeast genes representing ∼47% of the transcripts detected by the S98 Affymetrix GeneChip were identified as being differentially expressed between wild-type and mutant strains in at least one condition tested. This fairly large gene output compared with previous studies is likely due to relaxed selection criteria (see Materials and Methods) and inclusion of three growth and developmental conditions (Miyake et al., 2004; Yarragudi et al., 2007). Three hundred twenty-one genes were detected in three media, whereas 626, 966, and 627 were filtered only in YPD, YPA, or SPII medium, respectively (Figure 4B). This does not necessarily reflect growth- or sporulation-specific regulation but rather a media-specific threshold effect (i.e., some genes are differentially expressed in two or all media, but the effect is strong enough to be detected by our filtration procedure only in one condition). We then grouped the genes into coregulated clusters and correlated transcriptional deregulation with the presence of a predicted binding motif and in vivo Abf1 promoter binding (Figure 4C).
The loci detected during fermentation in YPD (1798) and/or during respiration in YPA (2312) were separately organized into five expression clusters that turned out to be highly reproducible between media, including two that are temperature dependent (compare C1–C3 YPD vs. YPA in Figure 4D and C4 and C5 in Supplemental Figure 5). Genes in cluster 1 showed the lowest level of expression in mutant cells incubated at the restrictive temperature (37°C), which is the pattern we expected to find. Importantly, this cluster is highly enriched for loci whose promoters are bound by Abf1 and/or contain a predicted binding motif (73% in YPD and 56% in YPA). Moreover, it contains known Abf1 targets reported in molecular biological work (ARO3; Kunzler et al., 1995) or expression studies (TRP3; Martens and Brandl, 1994). GO term analysis of YPD data shows that this cluster is enriched for genes involved in ribosome biogenesis and assembly (p = 4.5 × 10−4; 23 genes observed, 11 expected) and rRNA processing (p = 1.4 × 10−4; 20/8) (Figure 5A). This confirms previous reports that Abf1 is important for protein translation and amino acid biosynthesis (Mager and Planta, 1991). Furthermore, we find cellular localization (p = 10−4; 43/24), protein targeting (p = 2.3 × 10−5; 24/10) and protein transport (p = 2.2 × 10−4; 34/18) as well as cytokinesis (p = 6.6 × 10−3; 11/5). The latter category includes genes required for budding (BUD9, BUD31, GIC2, PWP2, and RAX2), actin cytoskeletal organization and cell wall morphogenesis (END3, PIK1, and SUN4), and septation (CDC3 and CDC10). YPA cluster 1 data yield for example metal ion transport (p = 5.1 × 10−6x; 12/3) and lipid metabolic process (p = 5.3 × 10−5; 20/8) (Figure 5B). Note that we display only enriched GO terms specific for genes in the YPA cluster to avoid partial redundancy with the YPD cluster.
Figure 5.
Biological Process GO term enrichment in mitotic expression clusters. (A) GO terms identified in five clusters from cells grown in YPD as given on top of the diagram. Rectangles contain bold numbers of enriched genes associated with a specific GO term as observed and expected. The total number of genes in the genome bearing a given GO term is shown. The numbers of genes bearing a GO term for each cluster are indicated at the top. Overrepresentation (red) and underrepresentation (blue) are color coded as shown in the scale bar (top left). (B) Data for gene clusters identified in YPA.
We furthermore identified two clusters showing lower (cluster 2) or higher (cluster 3) expression in the mutant than in the wild-type strain at both the permissive and restrictive temperatures. This reveals an expression phenotype of abf1-1 cells under conditions where normal growth and sporulation assays fail to indicate any pronounced defect. We note that ABF1 fell into cluster 3 (increased transcription in the mutant at permissive and restrictive temperature), which confirms a previous observation that the protein regulates its own expression via a negative feedback loop (Halfter et al., 1989; Miyake et al., 2004). YPD cluster 2 is enriched for genes involved in DNA-mediated transposition (p = 4.5 × 10−64; 75/7), amine transport (p = 1.7 × 10−5; 11/2), and carboxylic acid transport (p = 1.1 × 10−6; 13/3). YPD cluster 3 is enriched for aerobic respiration (p = 7.1 × 10−4; 7/2), coenzyme biosynthetic process (p = 9.5 × 10−3; 5/1), and electron transport (p = 1.4 × 10−4; 8/2) (Figure 5A). YPA cluster 2 is, like its YPD counterpart, enriched in genes (albeit different ones) associated with DNA-mediated transposition (p = 1.1 × 10−8; 29/9) (Figure 5B). Interestingly, YPA cluster 3 is enriched for ribosome biogenesis and assembly (p = 1.1 × 10−26; 63/14), a term that is found in YPD cluster 1 (whose genes are expressed at the lowest level in the Abf1-1 mutant at 37°C). This may reflect different transcriptional regulation of ribosome biogenesis during fermentation and respiration (Schawalder et al., 2004).
The remaining classes identified genes up- (cluster 4) or down-regulated (cluster 5) in both wild-type and mutant strains when cultured at the restrictive temperature (see Supplemental Figure 5). As expected, YPD cluster 4 is enriched in stress response (p = 1.4 × 10−8; 87/49) loci, including heat shock-inducible genes (HSP12, HSP26, HSP30, HSP42, HSP78, HSP104), whereas YPD cluster 5 is enriched for ribosomal genes (DBP2, DBP9, LRP1, NIP7, NOP1, NOP12, RPL7B, RPL18B, and RPL36B) repressed under heat shock conditions (Grigull et al., 2004) (Figure 5, A and B). These results demonstrate that including wild-type and permissive temperature controls is crucial for the analysis and interpretation of expression data obtained with a temperature-sensitive mutant.
Abf1 Core Targets Include Multicomponent Factors Required for Cytokinesis
We identified 434 genes that were down-regulated in the Abf1-1 background during mitotic growth and/or meiotic development (cluster 1 genes). Among those, we found 157 core target genes whose promoters contained a predicted target motif, and they were bound by Abf1 in vivo. This group revealed (among other genes required for budding, actin cytoskeletal organization, and septation) CDC3, a member of the family of bud neck filaments required for growth and spore formation as a potential Abf1 target gene (Fares et al., 1996; for review, see Longtine et al., 1996; www.germonline.org/). Because absence of Cdc3 function causes a defect in cytokinesis and abnormal budding (Kim et al., 1991), we concluded that Abf1 might in part regulate bud formation and daughter cell separation by directly controlling expression of Cdc3. To confirm the chromatin immunoprecipitation experiment and to test whether the predicted CDC3UAS was indeed bound by Abf1, we performed an EMSA by using a radioactive oligonucleotide probe (Figure 6A, lane 1). A slowly migrating binding activity observed in a protein extract from growing wild-type cells (lane 2) was not shifted by the preimmune serum (lane 3), whereas an antibody against Abf1 did induce a supershift (lane 4) that was absent when a control antibody against Ndt80 was added (lane 5). The binding signal was abolished by an excess of unlabeled oligonucleotides containing wild-type CDC3UAS and HOP1UAS elements (lanes 6 and 8) but not by a mutated HOP1UAS (lane 7). These results indicate that Abf1 participates in the regulation of normal budding and cytokinesis by direct UAS-mediated transcriptional control of the critical septin CDC3. Next, we investigated whether formation of the septin ring structure necessary for daughter cell separation would be impaired in cells lacking fully functional Abf1. To this end, we visualized the structure by using Abf1 wild-type and mutant strains containing a Cdc3-green fluorescence protein fusion reporter gene. The data show that only the wild-type (Figure 6B, left) but not the mutant strain (right) is capable of forming the septin ring structure localized to the bud neck when cultured at the restrictive temperature of 37°C, whereas both strains form the septin ring at the permissive temperature of 25°C (Figure 6B as indicated). Thus, expression profiling, genome-wide and biochemical DNA binding assays, and cell biological data together suggest Abf1 to control septation via transcriptional regulation of CDC3 expression. This finding is in keeping with the cytokinesis phenotype observed in abf1-1 cells (Figure 1, D and E). We consider it unlikely, however, that suboptimal expression of Cdc3 alone is responsible for the defect, because other factors, such as Cdc10, also fail to reach normal expression levels in the mutant at the restrictive temperature (see YPD C1 and www.germonline.org/).
Figure 6.
Abf1 binds to CDC3UAS and is required for septin formation. (A) Electrophoretic mobility antibody supershift assay by using a polyclonal anti-Abf1 antibody and a radioactive CDC3UAS probe. The free probe (lane 1) is followed by a binding activity present in an extract from cells cultured in YPD (lane 2). Preimmune serum (lane 3), Abf1 (lane 4, α-Abf1), and Ndt80 (lane 5, α-Ndt80) antibodies were diluted 1:30 and added to the binding reaction. A 100-fold molar excess of unlabeled wild-type (lane 6) and mutated (lane 7) HOP1UAS as well as wild-type (lane 8) CDC3UAS probes was added. Free probe (fp), a nonspecific band (*), and Abf1/DNA (Abf1p) and Abf1/antibody/DNA (Abf1p/ab) complexes are marked with arrows. (B) Fluorescence microscopic analysis of diploid wild-type (ABF1/ABF1) and mutant (abf1-1/abf1-1) cells containing a green fluorescent protein-tagged Cdc3. The strains were incubated in YPD at 25 and 37°C. The cell morphology (DIC, top) and GFP-signals (Cdc3-Gfp, middle) are shown individually and in an overlapping view (Merge, bottom).
Abf1 Controls Genes Involved in Inhibition of Meiosis, Spore Formation and Germination
Diploid strains containing one wild-type or Abf1-1 mutant allele were cultured in sporulation medium containing acetate and standard supplements (SPII) at the permissive temperature (25°C) until 5 and 9 h after induction of meiosis before they were shifted to 28, 33, or 37°C for 1 h (Figure 7A). Samples were analyzed in duplicate as described for the mitotic experiment (Supplemental Figure 4B). Note that only the meiotic samples harvested at 28°C were used for clustering (Figure 7B). Meiotic cluster 1 contained genes that failed to be induced to wild-type levels in the Abf1-1 mutant strain. As anticipated, the promoters of 40% of the genes in this cluster were bound by Abf1 and/or contained a predicted binding motif. The remaining expression clusters contained genes that were either repressed (cluster 2) or induced to various levels (clusters 3–5) during middle stages of sporulation in wild-type and mutant cells. All clusters showing unanticipated patterns contained a certain percentage of confirmed promoters (bound by Abf1 and/or predicted motif) and the level of induction observed in clusters 3–5 was indeed lower in the mutant strain than in the wild type. To rule out that different signal intensities were due to slower meiotic progression (hence, lower gene induction) by the mutant strain per se, we compared the induction profiles of known meiotic genes in Abf1 versus abf1-1 cells at the permissive temperature of 28°C, and we found no substantial difference (Supplemental Figure 6A); this was observed for very early, early, middle, and mid-late genes (Supplemental Figure 6B). Meiotic expression clusters displayed no significantly enriched relevant GO terms; however, we identified genes essential for inhibition of meiosis (APS2, PPH21, SET3, and UBR2), synaptonemal complex (SC) formation (RED1 and ZIP1), sporulation (CHS5, PEP12, SNF8, TAO3, and THR4), and spore germination (MRPL32, NFU1, and UBP6) as Abf1 targets (Deutschbauer et al., 2002; Deutschbauer and Davis, 2005).
Abf1 Binds to Meiotic Promoters during Mitosis and Meiosis
We next determined mitotic versus meiotic Abf1 promoter occupancy in the presence of a predicted motif for genes specifically expressed during meiosis to ask whether the protein bound only when a locus was actively transcribed. Abf1 was shown to interact with the upstream regions of early meiotic genes involved in control of the onset of meiosis (IME4), SC formation (NDJ1, HFM1, RED1, and ZIP1) and middle meiotic genes required for spore wall synthesis and maturation (SPR3 and SMK1) under all conditions tested (Figure 8). This suggests that Abf1 is present at its site both during mitotic repression and meiotic induction of these promoters. In addition to this clear-cut situation, we also found meiosis-specific genes whose promoters were bound by Abf1 in at least one growth condition and during sporulation (AMA1, DTR1, MAM1, MER1, PFS1, SMA1, SPO19, SPO73, SPS1, SPS100, and SWM1) or only during meiotic development (IME2, MUM1, MEK1, and SPO20), although they contain no UAS (Figure 8).
Figure 8.
Abf1 control of essential meiotic genes. Color-coded heat maps displaying expression levels of meiotic genes in rich (YPD and YPA) and sporulation (SPII) media are shown. Homozygous wild-type (ABF1/ABF1) and mutant strains (abf1-1/abf1-1) and strains containing one wild-type (ABF1/abf1) or mutant allele (abf1-1/abf1) and a GAL-ABF1 transgene are indicated. The temperatures are given at the bottom of the heat map. Dots mark genes whose promoters contain predicted binding sites (UAS) and/or are bound by Abf1 under different mitotic (YPD and YPA) and meiotic (SPII 4h, SPII 8h) conditions. Genes essential for meiosis and sporulation are shown in green. Log2-transformed expression signal intensities are color coded as indicated in the scale bar.
A substantial number of potential target promoters lack a UAS element, whereas sequences that are bound by Abf1 in vivo do not seem to contain any novel conserved Abf1 motif. To test whether Abf1 could interact with base insertion or deletion variants of UAS that might elude our prediction algorithm, we determined their ability to compete a wild-type UAS motif in an EMSA. The results suggest that mutations changing the distance between the highly conserved 5′-TCA and 3′-ACG triplets abolished Abf1 interaction (Schlecht and Primig, unpublished observation). We conclude that Abf1 is unlikely to bind to mutant derivatives of its UAS target sites within the promoters that do not contain the known motif.
DISCUSSION
To fully understand the cellular functions of Abf1, it is necessary to identify its DNA binding patterns and its target genes during distinct stages of growth and development. In this study, we report a comprehensive genome-wide analysis of Abf1. It is based on probabilistic binding site prediction combined with an in vivo binding assay using mitotic and meiotic cells as well as expression profiling of growing or sporulating wild-type and temperature-sensitive mutant strains at both permissive and restrictive temperatures. Among 434 regulatory regions identified, 157 core promoters failed to be normally activated in the absence of fully functional protein (Abf1-1 at 37°C), were bound by Abf1 in vivo, and contained a predicted binding site. This group includes numerous genes not previously identified as Abf1 dependent. Although we found the majority of the promoters to be bound during growth (regardless of the carbon source) and spore development, a subset of regulatory elements were detected in only one or two of the conditions tested.
Correlating Abf1 Target Site Prediction, In Vivo Binding, and Target Gene Deregulation
Comparing computational and molecular biological methods reveals that motif prediction and the in vivo binding assay yield the most concordant results, whereas binding and expression profiling data show the smallest overlap in Abf1 (Supplemental Figure 7, A–C). We conclude that computational predictions of UAS motifs significantly conserved in related budding yeast species are a strong indicator of a functional binding site. Moreover, the experimental approach based on comparing wild-type to mutant alleles at the permissive and restrictive temperatures was yielding, because we identified a cluster of genes that showed the anticipated down-regulation in the mutant at 37°C (Figures 4D, C1 and 7B, C1). It is important to note that this cluster contains the highest percentage of genes (∼50% in YPD, ∼30% in YPA, and ∼40% in SPII) where deregulation in the mutant is confirmed by in vivo binding to the upstream region and the presence of a predicted binding site in the promoter. However, the expected expression pattern is also observed when promoters are bound, although they lack a predicted binding site; when they are not bound, although they do contain a putative Abf1 binding site; and even in the absence of any detectable binding and a target site (Figure 4D). We do not believe that the protein interacts with other as yet unknown elements, because no novel conserved pattern was detected among the intergenic regions identified in the ChIP-Chip assay. It is possible that Abf1 has a nonspecific affinity for DNA during processes such as nucleotide excision repair (Reed et al., 1999) or mRNA transport (Hieronymus and Silver, 2003). This is in keeping with a recent in vitro analysis of the binding site of Abf1 that identified a pool of heterogeneous sequences not containing the UAS motif (Beinoraviciute-Kellner et al., 2005).
We have observed an unanticipated effect of the Abf1-1 allele at the permissive temperature that causes genes to show decreased (C2) or increased (C3) expression compared with the wild type. It is possible that these promoters contain low-affinity binding sites that Abf1-1 cannot interact with even at 25°C and that the overall expression of these genes mostly depends upon Abf1, so they are very sensitive to any perturbation. This is consistent with the finding that Abf1-1 fails to bind to its target site in vitro at room temperature (Rhode et al., 1992; Schlecht and Primig, unpublished observation). However, in most cases this effect seems to be indirect, because ∼80% of the genes in these clusters have promoters that are not bound and lack a binding site.
The abf1-1 Allele as a Tool for Promoter Analysis
The abf1-1 allele was used to study gene expression because the mutant cells stop growing and the Abf1-1 protein fails to bind to its target sites in vivo at 37°C (Miyake et al., 2004; Yarragudi et al., 2007). However, because it was observed that target genes continued to be expressed in the absence of detectable Abf1 binding at restrictive conditions it was postulated that the protein does not need to be continuously present on its target site once gene expression is activated (Schroeder and Weil, 1998). Another possibility is that Abf1-1 could still be able to at least partially interact with DNA at 37°C in the case of high-affinity binding sites or in a favorable protein network context. An assay more sensitive or different in its chemical nature than in vivo footprinting might reveal such residual binding. In this context, we note that FLAG-tagged Abf1-1 showed much reduced but not abolished binding to the RPS28A and SPT15 promoters at the restrictive temperature when examined with a chromatin immunoprecipitation assay (ChIP; Yarragudi et al., 2007). We suspect that this binding signal might even be stronger in the case of the native protein because we found that HA-tagged Abf1 has a sporulation phenotype, indicating that the DNA binding capacity of tagged Abf1 might be reduced. Moreover, our ChIP assays using a polyclonal anti-Abf1 antibody seem to detect native Abf1-1 protein binding to the constitutive UME6 promoter in YPD not only at 25°C but also at 37°C (Schlecht and Primig, unpublished observation).
Abf1 and Cytokinesis
Yeast CDC3, CDC10, CDC11, and CDC12 septins were initially identified as mutants defective in bud-neck filament formation essential for cytokinesis (Hartwell, 1971). All septins except CDC10 are cell cycle regulated and expressed or even highly induced during sporulation (Cho et al., 1998; Primig et al., 2000; www.germonline.org). The meiosis-specific septin SPR3, essential for normal spore development (Fares et al., 1996), is transcriptionally dependent upon Abf1 and Ndt80, the latter being a key regulator of middle meiotic genes (Ozsarac et al., 1997; Pak and Segall, 2002). Our finding that Abf1 is also likely to be directly required for the mitotic and meiotic expression of CDC3 (and CDC10) is consistent with its role as a global regulator of septin-dependent functions such as cytokinesis and spore development. Moreover, it provides a partial explanation for the cytological and meiotic defects observed in the Abf1-1 mutant (Figures 1 and 6; Rhode et al., 1992). It is interesting to note that for mammalian septins such as SEPT3 (CDC10 orthologue), SEPT10 (CDC3), and SEPT4 (CDC12), we have observed testicular expression reminiscent of the one found during mitotic growth and meiotic development in the case of their yeast orthologues (Schlecht et al., 2004; Chalmel et al., 2007). Such an expression pattern suggests an essential role for Septin genes during gametogenesis in mammals. Indeed, it was demonstrated that Sept4 −/− male mice are infertile due to defective spermiogenesis, the postmeiotic process that leads to the production of spermatozoa (Ihara et al., 2005; Kissel et al., 2005).
Role of Abf1 during Meiosis
Abf1 was proposed to be a general activator of many early and middle meiotic genes, including some involved in synaptonemal complex formation (HOP1, RED1, and ZIP1) and spore wall formation (SPR3) based on mutation of its target site in the promoters of these genes (Vershon and Pierce, 2000). Unexpectedly, we detect Abf1 binding to the upstream regions of many meiotic genes (AMA1, HFM1, IME4, MAM1, MND1, NDJ1, PFS1, SMK1, SPO74, SPR3, SPS1, SWM1, and ZIP1) during both growth and development, suggesting that the presence of Abf1 on a meiotic promoter does not prevent its repression during mitosis.
Although ample evidence supports the notion that Abf1 binds and activates the HOP1 promoter during meiosis (Prinz et al., 1995; Gailus-Durner et al., 1996), we and others (Harbison et al., 2004) failed to find supporting evidence by in vivo binding assays carried out using mitotic and meiotic cells. Perhaps Abf1 does not bind to the UASHOP1 target site in living cells, hence, another protein with overlapping sequence specificity might fulfill an activating role at that UAS. An alternative explanation is that the Abf1 binding affinity to its target site in the HOP1 5′-upstream region is rather weak under all physiological conditions tested and therefore remains below our assay's threshold level of detection. This might be the case for a number of promoters that seem not to be bound, although they contain predicted target motifs.
Finally, we found no evidence for a role of Abf1 in the regulation of loci required for meiotic recombination (SPO11, DMC1, REC104, and REC114), chromosome cohesion (REC8) or control of the meiotic divisions (SPO13), although they are repressed during mitosis by Ume6 via its URS1 motif, which is often colocalized with the UAS of Abf1 (Williams et al., 2002). Recent results show that Ume6 is not converted to a coactivator by interaction with the inducer of meiosis Ime1, as thought previously, but rather it is destroyed in a Cdc20-APC/C–dependent manner soon after the onset of meiosis (Mallory et al., 2007). It seems therefore that meiotic activation mechanisms might be more diverse than previously thought, thus raising the question of which factor(s) in addition to Abf1 might be critically involved in the induction of many early meiotic genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Walther for critical reading of the manuscript, H. Leroy and A. Assi for GermOnline system administration, and T. Aust for production of the Abf1 antibody. We acknowledge initial help with array data production and analysis by R. Züst, M. Bellis, C. Niederhauser-Wiederkehr, and F. Chalmel. This project was supported by Swiss National Foundation grant no. 3100A0-105861 (to M.P.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1242) on February 27, 2008.
Author contributions: U.S. designed and performed research and analyzed data. I.E. and E.v.N. provided binding site predictions and analyzed data. P.D. produced the microarray data. N.R. and V.B. supported ChIP-Chip data production and provided analysis procedures. A.N. designed research. M.P. designed research, interpreted data, and wrote the paper.
REFERENCES
- Angermayr M., Oechsner U., Bandlow W. Reb1p-dependent DNA bending effects nucleosome positioning and constitutive transcription at the yeast profilin promoter. J. Biol. Chem. 2003;278:17918–17926. doi: 10.1074/jbc.M301806200. [DOI] [PubMed] [Google Scholar]
- Beinoraviciute-Kellner R., Lipps G., Krauss G. In vitro selection of DNA binding sites for ABF1 protein from Saccharomyces cerevisiae. FEBS Lett. 2005;579:4535–4540. doi: 10.1016/j.febslet.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Borde V., Lin W., Novikov E., Petrini J. H., Lichten M., Nicolas A. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- Chalmel F., Primig M. The Annotation, Mapping, Expression and Network (AMEN) suite of tools for molecular systems biology. BMC Bioinformatics. 2008;9:86. doi: 10.1186/1471-2105-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F., et al. The conserved transcriptome in human and rodent male gametogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:8346–8351. doi: 10.1073/pnas.0701883104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G., Kim J., Rho H. M., Jung G. Structure-function analysis of the DNA binding domain of Saccharomyces cerevisiae ABF1. Nucleic Acids Res. 1995;23:2980–2987. doi: 10.1093/nar/23.15.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R. J., et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B. A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Deutschbauer A. M., Davis R. W. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- Deutschbauer A. M., Williams R. M., Chu A. M., Davis R. W. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S., Dowell S., Diffley J., Rowley A. The Practical Approach Series. New York: Oxford University Press; 1998. [Google Scholar]
- Erb I., van Nimwegen E. Statistical features of yeast's transcriptional regulatory code. IEE Proceedings of the First International Conference on Computational Systems Biology; 20–23 July 2006; Shanghai, China. 2006. pp. 112–118. [Google Scholar]
- Fares H., Goetsch L., Pringle J. R. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B. Transcriptional regulatory networks and the yeast cell cycle. Curr. Opin. Cell Biol. 2002;14:676–683. doi: 10.1016/s0955-0674(02)00391-5. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V., Xie J., Chintamaneni C., Vershon A. K. Participation of the yeast activator Abf1 in meiosis-specific expression of the HOP1 gene. Mol. Cell. Biol. 1996;16:2777–2786. doi: 10.1128/mcb.16.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattiker A., Niederhauser-Wiederkehr C., Moore J., Hermida L., Primig M. The GermOnline cross-species systems browser provides comprehensive information on genes and gene products relevant for sexual reproduction. Nucleic Acids Res. 2007;35:D457–D462. doi: 10.1093/nar/gkl957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Goncalves P. M., Maurer K., Mager W. H., Planta R. J. Kluyveromyces contains a functional ABF1-homologue. Nucleic Acids Res. 1992;20:2211–2215. doi: 10.1093/nar/20.9.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J., Mnaimneh S., Pootoolal J., Robinson M. D., Hughes T. R. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter H., Muller U., Winnacker E. L., Gallwitz D. Isolation and DNA-binding characteristics of a protein involved in transcription activation of two divergently transcribed, essential yeast genes. EMBO J. 1989;8:3029–3037. doi: 10.1002/j.1460-2075.1989.tb08453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C. T., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Hieronymus H., Silver P. A. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- Hochwagen A., Wrobel G., Cartron M., Demougin P., Niederhauser-Wiederkehr C., Boselli M. G., Primig M., Amon A. Novel response to microtubule perturbation in meiosis. Mol. Cell. Biol. 2005;25:4767–4781. doi: 10.1128/MCB.25.11.4767-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M., et al. Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev. Cell. 2005;8:343–352. doi: 10.1016/j.devcel.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kim H. B., Haarer B. K., Pringle J. R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney J. B., Tkacik G., Callan C. G., Jr Precise physical models of protein-DNA interaction from high-throughput data. Proc. Natl. Acad. Sci. USA. 2007;104:501–506. doi: 10.1073/pnas.0609908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel H., Georgescu M. M., Larisch S., Manova K., Hunnicutt G. R., Steller H. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev. Cell. 2005;8:353–364. doi: 10.1016/j.devcel.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Kovari L. Z., Cooper T. G. Participation of ABF-1 protein in expression of the Saccharomyces cerevisiae CAR1 gene. J. Bacteriol. 1991;173:6332–6338. doi: 10.1128/jb.173.20.6332-6338.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzler M., Springer C., Braus G. H. Activation and repression of the yeast ARO3 gene by global transcription factors. Mol. Microbiol. 1995;15:167–178. doi: 10.1111/j.1365-2958.1995.tb02231.x. [DOI] [PubMed] [Google Scholar]
- Lascaris R. F., Groot E., Hoen P. B., Mager W. H., Planta R. J. Different roles for abf1p and a T-rich promoter element in nucleosome organization of the yeast RPS28A gene. Nucleic Acids Res. 2000;28:1390–1396. doi: 10.1093/nar/28.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., DeMarini D. J., Valencik M. L., Al-Awar O. S., Fares H., De Virgilio C., Pringle J. R. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Luscombe N. M., Babu M. M., Yu H., Snyder M., Teichmann S. A., Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Mager W. H., Planta R. J. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol. Cell Biochem. 1991;104:181–187. doi: 10.1007/BF00229818. [DOI] [PubMed] [Google Scholar]
- Mallory M. J., Cooper K. F., Strich R. Meiosis-specific destruction of the Ume6p repressor by the Cdc20-Directed APC/C. Mol. Cell. 2007;27:951–961. doi: 10.1016/j.molcel.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. A., Brandl C. J. GCN4p activation of the yeast TRP3 gene is enhanced by ABF1p and uses a suboptimal TATA element. J. Biol. Chem. 1994;269:15661–15667. [PubMed] [Google Scholar]
- Miyake T., Loch C. M., Li R. Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol. Cell. Biol. 2002;22:505–516. doi: 10.1128/MCB.22.2.505-516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T., Reese J., Loch C. M., Auble D. T., Li R. Genome-wide analysis of ARS (autonomously replicating sequence) binding factor 1 (Abf1p)-mediated transcriptional regulation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:34865–34872. doi: 10.1074/jbc.M405156200. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Berger M. F., Jona G., Wang X. S., Muzzey D., Snyder M., Young R. A., Bulyk M. L. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ozsarac N., Straffon M. J., Dalton H. E., Dawes I. W. Regulation of gene expression during meiosis in Saccharomyces cerevisiae: SPR3 is controlled by both ABFI and a new sporulation control element. Mol. Cell. Biol. 1997;17:1152–1159. doi: 10.1128/mcb.17.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Segall J. Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:6417–6429. doi: 10.1128/MCB.22.18.6417-6429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H., et al. ArrayExpress–a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33:D553–555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce M., Wagner M., Xie J., Gailus-Durner V., Six J., Vershon A. K., Winter E. Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:5970–5980. doi: 10.1128/mcb.18.10.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta R. J., Goncalves P. M., Mager W. H. Global regulators of ribosome biosynthesis in yeast. Biochem. Cell Biol. 1995;73:825–834. doi: 10.1139/o95-090. [DOI] [PubMed] [Google Scholar]
- Primig M., Sockanathan S., Auer H., Nasmyth K. Anatomy of a transcription factor important for the start of the cell cycle in Saccharomyces cerevisiae. Nature. 1992;358:593–597. doi: 10.1038/358593a0. [DOI] [PubMed] [Google Scholar]
- Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., Hwang S. Y., Davis R. W., Esposito R. E. The core meiotic transcriptome in budding yeasts. Nat. Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Primig M., Winkler H., Ammerer G. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 1991;10:4209–4218. doi: 10.1002/j.1460-2075.1991.tb04999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S., Klein F., Auer H., Schweizer D., Primig M. A DNA binding factor (UBF) interacts with a positive regulatory element in the promoters of genes expressed during meiosis and vegetative growth in yeast. Nucleic Acids Res. 1995;23:3449–3456. doi: 10.1093/nar/23.17.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R. M., Hartley P. D., Meneghini M. D., Bao M. Z., Liu C. L., Schreiber S. L., Rando O. J., Madhani H. D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. H., Akiyama M., Stillman B., Friedberg E. C. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev. 1999;13:3052–3058. doi: 10.1101/gad.13.23.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode P. R., Elsasser S., Campbell J. L. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:1064–1077. doi: 10.1128/mcb.12.3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N., Uematsu N., Amiot F., Gidrol X., Barillot E., Nicolas A., Borde V. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:1868–1880. doi: 10.1128/MCB.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schawalder S. B., Kabani M., Howald I., Choudhury U., Werner M., Shore D. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- Schlecht U., Demougin P., Koch R., Hermida L., Wiederkehr C., Descombes P., Pineau C., Jegou B., Primig M. Expression profiling of mammalian male meiosis and gametogenesis identifies novel candidate genes for roles in the regulation of fertility. Mol. Biol. Cell. 2004;15:1031–1043. doi: 10.1091/mbc.E03-10-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht U., Primig M. Mining meiosis and gametogenesis with DNA microarrays. Reproduction. 2003;125:447–456. doi: 10.1530/rep.0.1250447. [DOI] [PubMed] [Google Scholar]
- Schroeder S. C., Weil P. A. Genetic tests of the role of Abf1p in driving transcription of the yeast TATA box binding protein-encoding gene, SPT15. J. Biol. Chem. 1998;273:19884–19891. doi: 10.1074/jbc.273.31.19884. [DOI] [PubMed] [Google Scholar]
- Simchen G. Are mitotic functions required in meiosis? Genetics. 1974;76:745–753. doi: 10.1093/genetics/76.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B. J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K., Shlomi T., Feizi H., Ideker T., Sharan R. Transcriptional regulation of protein complexes within and across species. Proc. Natl. Acad. Sci. USA. 2007;104:1283–1288. doi: 10.1073/pnas.0606914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen E., Zavolan M., Rajewsky N., Siggia E. D. Probabilistic clustering of sequences: inferring new bacterial regulons by comparative genomics. Proc. Natl. Acad. Sci. USA. 2002;99:7323–7328. doi: 10.1073/pnas.112690399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon A. K., Pierce M. Transcriptional regulation of meiosis in yeast. Curr. Opin. Cell Biol. 2000;12:334–339. doi: 10.1016/s0955-0674(00)00104-6. [DOI] [PubMed] [Google Scholar]
- Williams R. M., Primig M., Washburn B. K., Winzeler E. A., Bellis M., Sarrauste de Menthiere C., Davis R. W., Esposito R. E. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl. Acad. Sci. USA. 2002;99:13431–13436. doi: 10.1073/pnas.202495299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarragudi A., Miyake T., Li R., Morse R. H. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:9152–9164. doi: 10.1128/MCB.24.20.9152-9164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarragudi A., Parfrey L. W., Morse R. H. Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:193–202. doi: 10.1093/nar/gkl1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. V., King O. D., Wong S. L., Goldberg D. S., Tong A. H., Lesage G., Andrews B., Bussey H., Boone C., Roth F. P. Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J. Biol. 2005;4:6. doi: 10.1186/jbiol23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Dietrich F. S. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhang M. Q. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.