Abstract
Splicing regulates gene expression and contributes to proteomic diversity in higher eukaryotes. However, in yeast only 283 of the 6000 genes contain introns and their impact on cell function is not clear. To assess the contribution of introns to cell function, we initiated large-scale intron deletions in yeast with the ultimate goal of creating an intron-free model eukaryote. We show that about one-third of yeast introns are not essential for growth. Only three intron deletions caused severe growth defects, but normal growth was restored in all cases by expressing the intronless mRNA from a heterologous promoter. Twenty percent of the intron deletions caused minor phenotypes under different growth conditions. Strikingly, the combined deletion of all introns from the 15 cytoskeleton-related genes did not affect growth or strain fitness. Together, our results show that although the presence of introns may optimize gene expression and provide benefit under stress, a majority of introns could be removed with minor consequences on growth under laboratory conditions, supporting the view that many introns could be phased out of Saccharomyces cerevisiae without blocking cell growth.
INTRODUCTION
Most eukaryotic genomes seem very inefficient when compared with prokaryotes where almost every sequence contributes to gene expression (Herbert and Rich, 1999a,b). Unlike the situation in bacteria, the majority of eukaryotic genes consist of small islands of protein coding regions (exons) in a sea of noncoding sequences (introns). Introns need to be removed through splicing to generate mature mRNA carrying the information needed for protein synthesis. The number, length and abundance of introns vary greatly between organisms. Intron sequences constitute 24% of mammalian genomes (Venter et al., 2001) and more than 95% of the human genes sequence (Lander et al., 2001). In contrast, the genome of baker's yeast Saccharomyces cerevisiae contains only 296 introns present in 283 genes accounting for 5% of the yeast genes (Chervitz et al., 1999; Hodges et al., 1999; Lopez and Seraphin, 1999; Spingola et al., 1999; Davis et al., 2000; Clark et al., 2002; Kellis et al., 2003; Juneau et al., 2007). In addition, both intron distribution and splicing regulation seem to be much simpler in yeast compared with higher eukaryotes (Spingola et al., 1999). Human genes often contain multiple large introns with sizes greater than 10,000 nucleotides (nt) and the majority are alternatively spliced producing several transcripts from a single gene (Grasso et al., 2004; Nagasaki et al., 2005; Chen et al., 2006). In addition, the introns of many human genes contain RNA elements with distinct biological functions like snoRNA, miRNA, and other potentially functional RNA motifs (Tycowski et al., 1996; Hirose and Steitz, 2001; Hirose et al., 2003; Zhou et al., 2004). In yeast, in the vast majority of cases, only one intron is found per gene, and their average size is about 100 and 400 nt and only a minority carries functionally distinct RNA motifs (Ooi et al., 1998; Villa et al., 1998; Spingola et al., 1999; Mattick, 2004). Alternative splicing plays an important role in regulating growth and development of mammalian cells, and defects in the splicing program are associated with many human diseases (Faustino and Cooper, 2003; Baralle and Baralle, 2005; Buratti et al., 2006; Licatalosi and Darnell, 2006). In contrast, there are only a few examples of alternative splicing in yeast, and the role of introns to yeast biology is unclear.
Several hypotheses were put forward to explain the uncharacteristic simplicity and rarity of splicing in yeast (Ares et al., 1999, 2000; Bon et al., 2003). One hypothesis proposes that introns in yeast have appeared recently and that their full function did not have time to evolve (intron-in model), whereas another and more supported hypothesis (Bon et al., 2003) suggests that introns are on their way out of the yeast genomes (intron-out model). Recently, genome-wide analysis of splicing and global surveillance of intron expression suggested that yeast introns may improve and regulate gene expression (Juneau et al., 2006, 2007), thereby fuelling the debate about yeast intron function and the fundamental value of splicing in eukaryotes.
To understand the basic functions of introns and directly assess the possibility of creating splicing-independent eukaryotic cells, we initiated a large-scale intron deletion effort in S. cerevisiae. We classified all intron-containing genes in yeast with respect to their functions, individually removed one-third of all yeast introns and tested for impact on cell functions. In addition, we have removed all introns from genes related to the cytoskeleton, freeing an entire metabolic pathway from splicing, and assessed the impact of this intervention on cell function. The results indicate that although some introns are required for optimal growth, the majority could be removed without major consequences.
MATERIALS AND METHODS
Strains and Plasmids
Two independent colonies of the strain 10I (MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 lys2Δ0/lys2Δ0 ADE2/ade2Δ::hisG HIS3/his3Δ200) were used in parallel for intron deletions, these strains were created by mating the strains LLY34 (MATa ura3Δ0 leu2Δ0 lys2Δ0 ade2Δ::hisG) and LLY35 (MATα ura3Δ0 leu2Δ0 lys2Δ0 his3Δ200). Strains LLY34 and LLY35 were obtained by mating the strains BY4700 (MATa ura3Δ0) and BY4705 (MATα ura3Δ0 leu2Δ0 lys2Δ0 ade2Δ::hisG his3Δ200 trp1Δ63 met15Δ0; Brachmann et al., 1998). Cells lacking the MTR2 gene (JPY500) were created by replacing the coding sequence as well as 5′ and 3′ UTRs with URA3 gene using standard PCR-based gene replacement (Sikorski and Hieter, 1989) in the 10I2 strain. The PCR fragments were generated using the primers D-MTR2_for, 5′-GCACGCTTGGCGTCAATATCCTAAAACGGAAAACTAATCAGTTAGATGTGAGATTGTACTGAGAGTGCAC-3′ and D-MTR2_rev, 5′-GTACCCGTGCAGCCGTTTCCGTGCCTCGGTTCCTCCGAGATATCCTTAGGCTGTGCGGTATTTCACACCG-3′ (the underlined bases are complementary to pRS plasmids; Sikorski and Hieter, 1989). Strains lacking YRA1 and the TAD3 genes were obtained from the yeast knockout strains of Open Biosystems (Open Biosystems, Huntsville, AL, record no. 24217: MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 his3Δ1/his3Δ1 met15Δ0/MET15 yra1Δ::KMX4/YRA1 and record no. 25225: MATa/α ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 his3Δ1/his3Δ1 met15Δ0/MET15 tad3Δ::KMX4/TAD3, respectively). The plasmid pISCE was generated by inserting a 3.85-kb EcoRI fragment of pPEX7 (Fairhead and Dujon, 1993) containing the I-SceI endonuclease under the control of the inducible GAL1-CYC1 promoter into the EcoRI site of pRS424 (Christianson et al., 1992). The expression plasmid pRS425-SCE was constructed by inserting the 3.94-kb SacI-ApaI fragment of pISCE into the 6753-base pair SacI-ApaI fragment of pRS425 (Christianson et al., 1992). The plasmid pURA3-SCE that contains the recognition site of I-SceI in front of the URA3 marker was constructed by inserting two annealed oligonucleotides (Nde.SCE for: TACCGTAGGGATAACAGGGTAATCGG and Nde.SCE rev: TACCGATTACCCTGTTATCCCTACGG, where the underlined bases are the I-SceI site) in the NdeI site of pRS306 (Sikorski and Hieter, 1989). To amplify the yeast ACT1 promoter from wild-type genomic DNA, we used the following primers for PCR: XhoI-ACT1p-F 5′-CTCCTCGAGCTCACCCTAACATATTTTCCAATTA-3′ (the underlined bases are complementary to position −450 to −435 relative to the ACT1 translational start site, and the 5′ extension contains an XhoI site) and HindIII-ACT1p-rev 5′-CCCAAGCTTTGTTAATTCAGTAAATTTTCGATCT-3′ (the underlined bases are complementary to position −25 to −1 relative to the ACT1 translational start site, and the 5′ extension contains an HindIII site). The PCR product was digested with XhoI and HindIII and cloned into pRS316 (Sikorski and Hieter, 1989) to generate pACT1pr. The mtr2Δ1i, mtr2Δ2i, mtr2Δ3i, mtr2Δ4i, mtr2Δ5i, and mtr2Δ6i genes were amplified by PCR from their respective heterozygous diploid yeast strain with primers created against regions −262 to −237 and +1681 to +1706 relative to the MTR2 start codon defines by the Saccharomyces genome database (SGD). The PCR products were digested with the appropriate restriction enzymes and cloned into pACT1pr. The yeast gene YRA1 with or without intron (Δi) was amplified by PCR from wild-type or yra1Δi DNA, respectively, with primers created against regions 0 (without promoter) or 430 base pairs (with its own promoter) upstream of the start codon and 153 base pairs downstream of the stop codon. The PCR products were digested with the appropriate restriction enzymes and cloned into pACT1pr or pRS316 (Sikorski and Hieter, 1989). The pACT1pr-TAD3Δ2i was constructed by inserting the TAD3 cDNA into the pACT1pr plasmid. Yeast cells were transformed by a modification (Gietz and Woods, 2002) of the lithium acetate method (Ito et al., 1983) and were grown in standard yeast media (Zakian and Scott, 1982; Rose et al., 1990). All plasmids were propagated in bacteria using standard Escherichia coli strains and growth conditions (Sambrook et al., 1989).
Direct Introns Displacement Strategy
To remove an intron, we used direct intron displacement strategy described in Figure S1A. A fragment containing the endonuclease I-SceI recognition site in front of the URA3 marker gene was amplified from the plasmid pURA3-SCE. The forward primer used to amplify URA3 included 80 nucleotides complementary to the exon 2 sequence of the targeted genes, whereas the reverse primer included 45 nucleotides complementary to the targeted introns sequence. The generated PCR products were used in a second round of PCR amplification as template. The reverse primer was the same as that used in the first round, whereas the forward primer included sequence representing the perfect junction between exon 1 (45 nt) and exon 2 (55 nt). This resulting PCR product was transformed in the diploid 10I1 and 10I2 yeast strains harboring the pRS425-SCE plasmid. After verifying the fragment insertion by PCR, the production of the I-SceI enzyme was induced for 8 h at 30°C by growing the cells in the presence of galactose (final concentration, 2%) to favor the pop-out of the URA3 gene (Plessis et al., 1992; Lukacsovich et al., 1994; Cohen-Tannoudji et al., 1998). Loss of the URA3 marker was screened by growing cells on media containing 5-fluoroorotic acid (5-FOA; Boeke et al., 1987). Cells containing the correct replacements were sporulated and dissected, and haploid strains containing the mutated allele were mated together to obtained homozygote diploid yeast strains. Each step of the intron direct displacement strategy was verified by PCR amplification using primers upstream and downstream of the exons 1 and 2 junction. The list of the primers used can be supplied upon request.
Growth Assays
The yeast strains were grown to ∼4 × 106 cells/ml in rich growth medium (YPD). For drugs assays, 50 μl of yeast cells suspension was added to 50 μl of each drug diluted in YPD (see Table S1 for final concentration; drug concentrations were adjusted to decrease the growth of wild-type cells by 50%). For carbon sources assays, the yeast cells were washed twice and resuspended in rich medium without glucose (YEP). Assays were carried by mixing equal volume of washed yeast cells and YEP. The carbon source was added separately and adjusted to a final concentration of 2% except for the glycerol that was 3%. Samples were prepared in triplicate and the assay were performed in Costar 96-well polystyrene not treated microplate (Fisher Scientific Company, Ottawa, ON, Canada), and growth was monitored using PowerWave microplate spectrophotometer reader (BioTek Instruments, Winooski, VT; Toussaint et al., 2006). Methyl methanesulfonate (MMS), hydrogen peroxide (H2O2), benomyl, ketoconazole, camptothecin, calcofluor white, cycloheximide, and caffeine were obtained from Sigma-Aldrich Canada (Oakville, ON, Canada). Hygromycin B was obtained from Roche Diagnostics (Laval, QC, Canada). Staurosporine and cyclosporin A were obtained from Alexis (Alexis Biochemicals, San Diego, CA). Latrunculin A was obtained from Calbiochem (VWR CANLAB, Mississauga, ON, Canada).
Growth Curves Analysis
The maximal growth rate (μm) of each growth curve was calculated as described previously (Toussaint et al., 2006). For each 96-well microplate, the mutant cells μm was divided by that calculated for wild-type growing in the same plate.
Fitness Test
Competitive growth assays were performed by adding equal quantity (6.5 × 105 cells/ml) of wild-type (ade2Δ background) and Δi (ADE2 background) strains into 25 ml of YPD containing 100 μg/ml adenine to avoid the accumulation of the red pigmentation of ade2 mutants. The mixed cultures were grown at 30°C and diluted each day (at 6.5 × 105 cells/ml) until 50 generations (±2) were reached. The number of generations was calculated using this equation: [G = ln (Cf/Ci)/ln (2)], where G represents the number of generations and Cf and Ci are the final and the initial concentration of cells of the mixed cultures, respectively. At the beginning of the experiment (generation 0) and after 50 divisions, ∼300 cells of the mixed cultures were plated on SC plates containing a low concentration of adenine (20 μg/ml) to permit the red pigmentation of ade2Δ cells and grown at 30°C. The white (Δi) and red (wild type) cells were counted to calculate the percentage of each strain at the beginning and at the end of the experiment.
Northern Blot
Total RNA from exponentially growing cells was isolated and blotted as described earlier (Abou Elela et al., 1996; Abou Elela and Ares, 1998). The levels of each mRNA were quantified using a random priming probe complementary to the last exon of the corresponding gene. Quantification for all signals was obtained by storage phosphorimaging with Storm and ImageQuant software (GE Healthcare Bio-Sciences, Baie d'Urfé, QC, Canada). The mRNA levels of the intronless genes was normalized to HSC82 or RPL8A that were used as internal control. Loading of the SAC6 mRNA was normalized using the 25S rRNA as reference.
RESULTS
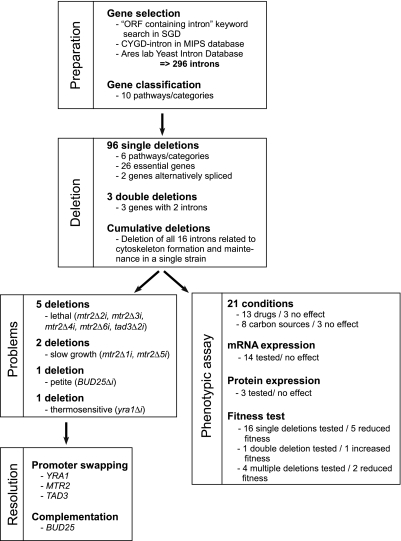
Gene Selection and Classification
Intron-containing genes were identified using S. cerevisiae database (Dwight et al., 2002), Ares Lab Yeast Intron Database (Spingola et al., 1999), and the Munich Information Center for Protein Sequences Comprehensive Yeast Genome Database (Guldener et al., 2005). In addition, we also added nine recently discovered introns (Juneau et al., 2007) that were not annotated in these databases. The intron-containing genes were grouped by function into 10 classes (cytoskeleton, RNA metabolism, chromosome segregation and meiosis, metabolism, mitochondria, protein folding and degradation, transcription and translation, endoplasmic reticulum and Golgi, ribosomal proteins, and uncharacterized genes) according to MIPS CYGD (Comprehensive Yeast Genome Database) classification (Guldener et al., 2005). For this first study, we avoided classes containing many genes with multiple cellular functions (e.g., transcription and translation) or high concentration of predicted splicing sensitive genes (e.g., ribosomal proteins genes). Instead, we selected six metabolic pathways grouping 87 genes with well-defined housekeeping functions (Table 1) as representatives of intron-containing genes in yeast (Figure 1). In this set, one-third of the genes are essential: three contain two introns, and two are alternatively spliced. The sizes of the targeted introns vary from 56 to 1002 nt and most are found within an ORF, except for 7, which were located in the 5′ untranslated region (5′ UTR).
Table 1.
Pathways and categories of intron-containing genes studied in this article
| Class of genes | Genes |
|---|---|
| Cytoskeleton | ACT1,TUB1, SAC6,COF1, GIM5, CIN2,ARP9,PFY1, TUB3,ARP2, YSC84, LSB3,MOB1, MOB2, DYN2(2) |
| RNA metabolism | LSM2, LSM7, MUD1,MTR2 (6AS), STO1,POP8,TAD3 (2), NMD2,NSP1,SMD2,DBP2,YRA1, TAN1, MPT5,YSF3 |
| Chromosome segregation and meiosis | MEI4, HOP2, MND1, DMC1, HFM1, AMA1, SAE3, REC107,RFA2, SPO1, BUD25, ECM33, PCH2, MCM21, SPO22, RAD14, REC102, REC114, SRC1(2AS) |
| Metabolism | YBR220C,PMI40,GLC7,BIG1, EPT1, APE2, CPT1,GCR1, HNT1, HNT2,GPI15, VMA9(2), PHO85,SPT14, VMA10, SCS22, IMD4, VPS75, YOP1 |
| Mitochondria | RIM1,ERV1, MRPL44, YML6, COX4, QCR9, QCR10, COX5B |
| Protein folding and degradation | UBC4, UBC5, UBC8,UBC9, UBC12, UBC13, RUB1,PRE3, MRK1, MMS2, DCN1 |
Underlined, essential genes; bold, intron overlapped with another ORF. If the gene contains more than one intron, the number of introns is written in parentheses and AS indicates alternatively spliced.
Figure 1.
Pipeline for the deletion and functional analysis of yeast introns. All known intron-containing genes were identified from public databases (Dwight et al., 2002; Guldener et al., 2005; Spingola et al., 1999) and classified in 10 pathways using gene ontology (Guldener et al., 2005). Introns were either individually deleted in separate strains or sequentially removed from a single strain to create a yeast strain carrying multiple intron deletions. The effect of intron deletions on growth was measured in rich media and strains with growth defects were identified. All intron deletion strains were subjected to a battery of 21 different phenotypic assays in liquid media and cells carrying multiple deletions were tested for fitness.
Efficient Deletion of Yeast Introns Requires Expression of the Endonuclease I-SceI
The general strategy used to evaluate intron function is described in Figure 1, and the method used to delete the intron is described in Supplementary Figure S1A. Intron deletions were obtained using a standard two-step (pop-in/pop-out) method (Boeke et al., 1987). To increase the frequency of pop-outs without losing targeted deletions, we included within the inserted fragments a cleavage site for the double-strand break enzyme I-SceI (Fairhead and Dujon, 1993) upstream of the URA3 gene (Figure S1A). The double-strand break was introduced by expressing a plasmid-born copy of I-SceI before the selection for pop-outs in media containing 5-FOA (Figure S1A). Using this method, we have deleted 96 introns in 87 genes (Table 1). All intron removals were conducted in diploid cells and verified by PCR amplifications (data not shown). Cells with the correct intron deletions were sporulated and the haploid cells were tested for growth (Figure S1B).
Most Introns Can Be Removed with Little Impact on Growth under Normal Conditions
The effect of intron deletions on cell viability was first assessed by monitoring growth on rich media (for an overview, see Figure 1). None of the deletions affected the growth of the heterozygous diploid strain on rich media except the intron deletion of the YRA1 gene (data not shown), whereas 9 of the 99 intron deletions tested slowed or inhibited growth of haploid cells on rich media (Figure 1). The growth defects were caused by intron deletions in four genes containing a total of nine introns, three of which related to RNA metabolism (MTR2, YRA1, and TAD3) and one that was implicated in chromosome segregation and meiosis (BUD25; Figures 2 and 3 and Figure S2).
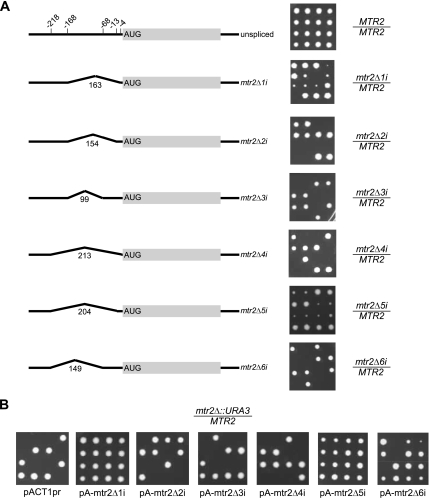
Figure 2.
Deletion of the alternatively spliced MTR2 gene introns is lethal. (A) Tetrad dissection of cells carrying different intron deletions in MTR2 gene. Left, schematic representation of the gene structure and the position of introns in relation to the translation start codon; right, tetrad dissection on rich media. The intron length in nucleotides is indicated above each intron. (B) MTR2 gene carrying deletion in the first or fifth intron support growth when expressed from a heterologous promoter. Empty plasmid (pACTpr) or plasmids expressing mtr2Δ1i, mtr2Δ2i, mtr2Δ3i, mtr2Δ4i, mtr2Δ5i, or mtr2Δ6i alleles from ACT1 promoter were transformed in a diploid strain carrying deletion of the MTR2 gene. After these strains were sporulated, the tetrads were dissected and tested for growth. The pictures shown were taken after 3 d of incubation at 30°C.
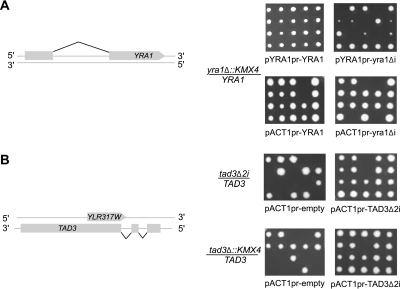
Figure 3.
Splicing of YRA1 and TAD3 is required for growth in a promoter-dependent manner. (A) Diploid yeast strains heterozygous for the deletion of the YRA1 gene were transformed with a plasmid expressing the native YRA1 (pYRA1pr-YRA1) or YRA1-deleted intron (pYRA1pr-yra1Δi) mRNA from the native YRA1 promoter (top panel), or plasmid expressing the YRA1 (pACT1pr-YRA1) or YRA1-deleted intron (pACT1pr-yra1Δi) mRNA from ACT1 promoter (bottom panel). After sporulation the tetrads were dissected on YPD plates and pictures were taken after 4 d of incubation at 23°C. (B) Diploid yeast strains heterozygous for the deletion of TAD3 introns (tad3Δ2i; top panel) or heterozygous for the complete deletion of the TAD3 gene (bottom panel) were transformed with empty plasmid (pACT1pr-empty) or plasmid expressing intronless TAD3 mRNA from ACT1 promoter (pACT1pr-TAD3Δ2i). Tetrad dissections were carried as described above, and pictures were taken after 3 d of incubation at 30°C. In some cases spores did not grow because of the loss of pACT1pr-YRA1, pACT1pr-yra1Δi, or pACT1pr-TAD3Δ2i when grown on rich media. The left panels illustrate the structure and position of YRA1 and TAD3 genes.
Six intron deletions causing growth defects were found in the alternatively spliced MTR2 gene (Kadowaki et al., 1994; Santos-Rosa et al., 1998) implicated in the regulation of mRNA transport. The gene harbors six overlapping introns in the 5′ UTR that could produce six isoforms (Davis et al., 2000). Evidence for the use of five of the six possible splice site combinations had been provided by the sequencing of cloned RT-PCR products (Davis et al., 2000). As shown in Figure 2A, the deletion of introns 1 and 5 slows growth, whereas the deletion of introns 2, 3, 4, and 6 are lethal. Growth defects were also observed upon the deletion of introns from the constitutively spliced genes YRA1 and TAD3, which carry one or two introns, respectively (Figure 3). Consistent with previous studies (Preker et al., 2002; Rodriguez-Navarro et al., 2002), deleting the intron of the mRNA transport gene YRA1 rendered cells temperature sensitive (Figure 3A and data not shown). On the other hand, although deleting any single intron of TAD3, which encodes a tRNA specific adenosine deaminase, did not inhibit cell growth, the deletion of both introns was lethal (Figure 3B). Deleting the introns of TAD3 (tad3Δ2i) removes 15 nucleotides of a dubious ORF (YLR317W) residing on the opposite strand of the introns. However, expressing the entire YLR317W gene from a plasmid did not restore growth, and point mutations disrupting the YLR317W reading frame had no effect on growth (data not shown). Thus, at least one TAD3 intron is essential for growth. In contrast to the intron deletions in RNA metabolism genes, the growth defect associated with the chromosome segregation and meiosis BUD25 gene was found to be intron-independent and was caused by the disruption of an overlapping gene (Figure S2).
MTR2, YRA1, and TAD3 Introns Are Required for Growth in a Promoter-dependent Manner
To understand why deleting introns from MTR2, YRA1, and TAD3 genes inhibits growth, we ectopically expressed intronless versions of these genes from native or heterologous promoters and monitored their ability to support cell growth. Expression of MTR2 versions carrying deletions in any of its six introns from a plasmid under the control of its native promoter failed to restore growth to cells lacking the MTR2 gene (data not shown). In contrast, transforming mtr2Δ cells with plasmid-expressing MTR2 genes lacking the first or the fifth intron from the ACT1 promoter restored normal growth, whereas transformation of other MTR2 variants did not (Figure 2B). We conclude that the alternatively spliced introns of MTR2 contribute differently to cell growth probably by differentially modulating gene expression.
Like MTR2, YRA1 codes for an essential protein required for the nuclear export of mRNA (Sträßer and Hurt, 2000). However, unlike MTR2, YRA1 contains only one constitutively spliced intron, the deletion of which was previously shown to render cells temperature-sensitive (Zenklusen et al., 2001; Preker et al., 2002; Rodriguez-Navarro et al., 2002; Preker and Guthrie, 2006; Dong et al., 2007). We tested the ability of an intronless copy of YRA1 expressed from a panel of different promoters. Only the ACT1 promoter supported normal growth (Figure 3A). This result indicates that splicing is not required for YRA1 expression and suggests that the intron is used to set optimal expression from the native YRA1 promoter. Similarly, expression of an intronless version of TAD3 from the ACT1 promoter restored the growth defects caused by expressing tad3Δ2i from its native promoter (Figure 3B). Thus, genes of most yeast metabolic pathways do not require splicing for normal function with the exception of a few genes implicated in RNA metabolism.
The Deletion of Several Yeast Introns Alters Growth under Special Conditions
Because most intron deletions were tolerated under normal growth conditions, we examined their impact when cells were exposed to stress. All intron-deleted strains growing normally on rich media were tested for growth in different media containing seven different carbon sources and 13 drugs affecting different metabolic activities (Table S1 and Figure S3). As shown in Figure 4A, 48 of 90 mutant strains grew slower or faster than the wild-type strain by 20% or more in at least one growth condition. However, only four deletions (HNT1, SAE3, GLC7, and HOP2) reduced growth in two different conditions and only one (VMA9-1) reduced growth in three conditions. In most cases, the growth defect was observed in conditions related to the known or predicted function of the mutated gene. For example, a growth phenotype was observed in the presence of camptothecin, cyclosporine, and MMS, which affect chromatin organization, transcription, and DNA repair (Chlebowicz and Jachymczyk, 1979; Hemenway and Heitman, 1996; Halas et al., 1997; Capranico et al., 2007) after deleting introns from certain genes involved in DNA synthesis, transcription, and mitochondrial integrity. On the other hand, sensitivity to staurosporine, which affects cell wall integrity, was observed when deleting introns from genes affecting meiosis and chromosome segregation (Chai et al., 2002). However, intron features (e.g., size or position) or gene ontology classification did not correlate with drug sensitivity (Figure S4). Notably, the percentage of intron deletions affecting growth under stress was not equally distributed among the different classes of genes. Almost all the intron deletions in genes included in RNA metabolism, meiosis, and general metabolism affected growth at least in one condition, whereas only a minority of the genes in other pathways had an effect on growth (Figure 4B). Differences between pathways are not related to the percentage of essential and nonessential genes in each pathway or the number of genes per pathway (Figures 4B and S4). We conclude that although introns may be largely dispensable for basic cellular functions, they play an important role in attuning growth to various conditions.
Figure 4.
Phenotypic analysis of different intron deletion strains. (A) Heatmap of the maximum growth rate (μm) of intron-deleted strains in 21 growth conditions. The μm of each mutated strain has been measured and compared with the μm of the wild-type strain growing under the same condition. The heatmap was generated using mutant strains with a growth rate that differs from that of the wild-type strain by 20% or more. Right, the gene names; left, the metabolic pathway of each gene and its requirement for growth. Genes related to cytoskeleton, RNA metabolism, chromosome segregation and meiosis, metabolism, mitochondria, and protein folding and degradation are shown in red, green, blue, light blue, pink, and yellow, respectively. The drugs or carbon sources used in the growth assays are shown on the bottom. The arrow indicates growth in glucose, which is used as normal growth conditions. Dark blue, blue, and light blue indicate cells growing faster, similar, or slower than wild-type cells, respectively. (B) Histogram showing the percentage of essential (light gray) and nonessential (dark gray) genes that reduce cell growth within a given pathway when introns are deleted (left panel). The percentages of essential (light gray) and nonessential (dark gray) genes that induce growth are shown on the right. Note that certain intron-deleted genes can provoke a reduction of cell growth in a condition and an induction in another.
Introns Are Not Required for Cytoskeleton Generation and Maintenance
If yeast introns are generally dispensable for basic functions, it may be possible to relieve an entire pathway from splicing with little effect on growth. To test this hypothesis, we cumulatively deleted introns from all genes of the cytoskeleton pathway and monitored the effect on cell growth and function. The cytoskeleton pathway was selected because it contains genes with well-established housekeeping functions and tools are available to examine the expression of associated proteins and follow the impact on related molecular functions. In yeast, there are 15 cytoskeleton related genes containing 16 introns (Table 1). About half of the genes are essential for function and one contains two introns. To remove multiple introns in a single strain, each intron deletion was introduced individually, and the resulting haploid strains were mated to generate cumulative mutations in sets of four as illustrated in Figure S1B. Once haploid cells containing four intronless genes were obtained (Δ4i), they were backcrossed three times with wild-type cells to remove or dilute deleterious mutations in the mutant cells. Clean cells were mated to obtain homozygous diploid strains carrying all four deletions and the process was repeated until all introns in the cytoskeleton pathway were deleted (Figure S1B).
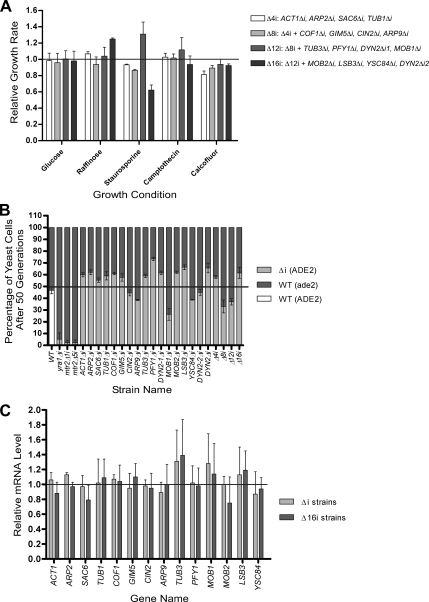
We tested the effect of deleting 4 (Δ4i), 8 (Δ8i), 12 (Δ12i), and 16 (Δ16i) introns of cytoskeleton-related genes on growth under different conditions (Figure 5 and Figure S5). All strains grew on rich media, and none of the multiple-deletion strains displayed a phenotype not observed with strains carrying single deletion (Figure 5A and Figure S5). This indicates that eliminating introns from an entire metabolic pathway can be tolerated. However, intron contribution might only be revealed when cells are in competition for limited resources. Therefore, we monitored the capacity of the different single and multiple mutant strains to compete with wild-type cells for growth in the same culture. The distribution of the mutant and wild-type cells were counted in the beginning of the experiment and after 50 generations. As shown in Figure 5B, three individual intron deletions reduced the cell fitness by 10–20%, whereas 10 single intron deletions increased fitness by 10–20%. In general, the fitness of cells carrying multiple deletions reflected the largest effect conferred by an individual intron deletion, and no multiple deletion-specific effects were observed. Strikingly, deleting all introns from the cytoskeleton-related genes conferred a slight net growth advantage over wild-type cells (Figure 5B). We also considered the possibility that intron deletions may affect gene expression without visible effects on phenotype. For this reason, we assessed the expression level of intron-containing cytoskeleton-related mRNAs and monitored the protein level of selected genes. As shown in Figures 5C and Figure S6, intron deletions did not significantly change the expression levels of the intronless genes even in cells with reduced fitness (e.g., MOB1, ARP9, or YSC84). In addition, no changes in the cell division or microtubule formation and functions were observed by microscopic evaluation (Figure S7). We conclude that the deletion of individual introns of cytoskeleton-related genes does not alter their gene expression and that freeing at least one metabolic pathway from splicing in yeast confers a growth advantage in a competitive environment.
Figure 5.
Phenotypic analysis and expression profiling of yeast strain carrying multiple intron deletions in genes associated with cytoskeleton. (A) Maximum growth rate of different strains carrying multiple intron deletions in cytoskeleton-related genes was obtained and plotted relative to that of a wild-type strain. Growth conditions inducing the greatest variance between the wild-type and mutant strains are shown (X axis). The data are an average of at least three different experiments. Strain carrying intron deletions in ACT1, ARP2, SAC6, and TUB1 genes (Δ4i) is indicated with white bars. Light gray bars, the strain carrying Δ4i deletions plus additional ones in COF1, GIM5, CIN2, and ARP9 genes (Δ8i); gray bars, Δ12i strain that carries Δ8i deletion plus deletions in TUB3, PFY1, DYN2_1 and MOB1 genes; dark gray bars, Δ16i that carries the Δ12i deletions plus additional one in MOB2, LSB3, YSC84 and DYN2_2 genes. (B) Competitive growth assay of wild type and intron-deletion strains in rich media (fitness test). Mixed cultures were started by adding equal quantities of cells from wild-type (ade2Δ) and intron deletion strains (Δi; ADE2) in YPD containing 100 μg/ml adenine to avoid the accumulation of the red pigmentation of ade2 mutants. The mixed cultures were grown at 30°C and diluted each day until 50 generations (± 2) were reached. At generation 0 and 50, the number of mutant (white) and wild-type (red) cells was counted by plating 300 cells (±44) on SC plates containing a low concentration of adenine (20 μg/ml), and the initial and final percentages of each strain was calculated. The initial percentages were set to 50:50 (black line), and the final percentages shown in the graph were relative to this ratio. Dark gray bars, the percentage of wild-type cells; light gray bars, the percentage of cells carrying intron deletion. Competitive growth assays were performed with two wild-type strains that have different background (ADE2, white bar; and ade2Δ) as negative control and also with wild-type and yra1Δi, wild-type and mtr2Δ1i, and wild-type and mtr2Δ5i strains as positive controls. The strains are indicated on the X-axis. n = 3 for the individual experiments, except for the multiple intron deletion strains, where the data represent six different experiments. (C) Expression profiling of cytoskeleton related mRNAs before and after intron deletions. The abundance of each mRNA associated with the cytoskeleton was determined in wild-type cells, cells carrying a single deletion in the gene analyzed (light gray bars), or cells carrying multiple deletions in all cytoskeleton related genes (gray bars). The RNA was quantified using Northern blot, normalized using internal controls, and plotted relative to the expression of the wild-type mRNA. The data represent average values for RNA obtained from two different cultures grown independently in duplicates.
DISCUSSION
In this study, we have shown that a large number of yeast introns can be removed with very little effect on growth in rich media. Only three genes (MTR2, YRA1, and TAD3) required introns for function, and in all cases growth defects caused by intron deletions were rectified by changing the promoter. This suggests that some introns are kept to optimize expression from otherwise suboptimal promoters. Sixteen percent of yeast introns either enhanced or reduced growth under various conditions, suggesting that introns can contribute positively or negatively to cell growth. Surprisingly, deleting 5% of yeast introns in a single yeast strain enhanced cell growth in a competitive environment, suggesting that introns can negatively contribute to cell growth. Together our results indicate that the number of essential yeast introns is small and that the production of an intronless eukaryote through laboratory manipulation might be possible.
It was previously suggested that yeast introns regulate gene expression (Lei and Silver, 2002; Preker et al., 2002; Juneau et al., 2006; Preker and Guthrie, 2006). Deletion of the YRA1 intron increases the expression of YRA1 leading to a dominant negative growth phenotype presumably mediated by depletion of critical mRNA export factors (Lei and Silver, 2002; Preker et al., 2002; Preker and Guthrie, 2006). Our results independently confirm the dominant negative and temperature-sensitive phenotype generated by the deletion of the YRA1 intron (Figure 3 and data not shown). However, we found no strict correlation between the act of splicing and the growth defect generated by intron deletion. Expression of an intronless copy of the YRA1 gene from the native promoter (YRA1pr-yra1Δi) or from the ACT1 promoter (ACT1pr-yra1Δi) generated similar amounts of mRNAs, yet ACT1pr-yra1Δi supports normal growth, whereas YRA1pr-yra1Δi does not (Figure 3A and data not shown). Similarly, the expression of intronless copies of TAD3 and MTR2 from ACT1 promoter dramatically increased RNA expression and restored normal growth. Thus, we did not find a strong correlation between intron removal and RNA expression or with the growth phenotype (Figure S4 and data not shown). These results suggest that intron-dependent growth phenotypes are not directly related to changes in RNA expression. Curiously, the only three genes known so far to require introns for growth encode RNA-binding proteins that influence mRNA metabolism, and two of them (YRA1 and MTR2) are involved in RNA transport (Santos-Rosa et al., 1998; Preker and Guthrie, 2006). It is thus tempting to suggest that the phenotype associated with deleting introns from these three genes results from the perturbation of mRNA transport or from inefficient translation.
Less than 5% of yeast genes contain introns and about half of them are found in RNAs encoding ribosomal proteins, suggesting that splicing may play an important role in ribosome biogenesis (Ares et al., 1999). On the other hand, the contributions of splicing to nonribosomal genes remain unclear. Here we have shown that introns of six metabolic pathways in yeast can be removed with little effects on growth under normal conditions (Figure 4 and Figure S4). This suggests that all yeast introns can possibly be removed to generate a streamlined eukaryotic genome. However, not all introns may be removed without modifying the genome. We have found that deleting the introns in three genes (MTR2, YRA1, and TAD3) can only be tolerated if the native promoters of these genes are substituted by heterologous ones (Figures 2 and 3). In one case, intron removal disrupted the expression of an overlapping gene, and normal growth required modification of the chromosomal locus (Figure S2). Situations like this may prevent intron loss from certain genes during evolution. We did not find a clear example of an intron-specific function associated with splicing that impairs growth under normal conditions. However, it is possible that splicing-specific regulation becomes important only under special conditions. Indeed, about half of the introns affected growth under stress, albeit weakly (Figure 4). It is also possible that many essential introns will be concentrated in a single metabolic pathway that was not examined in this study. For example, ribosome biogenesis is predicted to contain the majority of essential introns because 71% of ribosomal proteins genes contain introns and they represent the bulk of spliced mRNAs in yeast (Ares et al., 1999; Pleiss et al., 2007). Consistent with this view, the ribosomal protein Rpl30p autoregulates the splicing of its own mRNA by binding to a duplex structure containing intron sequences (Vilardell et al., 2000). The results presented here demonstrate that splicing of nonribosomal mRNAs in yeast plays a nonessential role in the regulation of gene expression and function.
Compared with mammals, DNA information in bacteria is transmitted in an uninterrupted manner into protein with little noncoding sequence to maintain (Herbert and Rich, 1999a,b). It was argued that this so called “hard-wired” bacterial genome is most suited to simple single-cell organisms where replication speed and the need for a prompt response to the environment determine success in limiting and fluctuating growth conditions (Herbert and Rich, 1999a,b). On the other hand, the mammalian “soft-wired” genome provides multicellular organisms with the complexity and diversity required to combat diseases and fine-tune metabolic pathways in a relatively stable environment (Herbert and Rich, 1999a,b). If indeed intronless genomes are more suitable for the lifestyle of single-cell organisms, then one might expect simple eukaryotes to be more tolerant to intron deletions. In this study, we provide the first experimental evidence that this may be the case. By removing all introns from the cytoskeleton pathway and reducing the total number of introns by 5%, we increased cell fitness, albeit modestly (Figure 5). It will be interesting to see if removing additional introns of nonribosomal mRNAs further enhances cell fitness. The cumulative removal of introns may increase cell growth by enhancing the expression of otherwise limited mRNA, for example, by improving the splicing of suboptimal introns (Pleiss et al., 2007). This is consistent with previous data showing that different genes react differently to the loss of different splicing factors (Pleiss et al., 2007). It will also be interesting to investigate whether yeast introns contribute to long-term genome stability and whether there is a threshold number of introns below which cells ceases to function. Meanwhile, the results presented here demonstrate that simple eukaryotes may tolerate and in some cases even marginally benefit from multiple intron loss under in vitro growth conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. D. Drubin (University of California, Berkeley) for providing the anti-Sac6p antibodies and L. Laliberté for the construction of the pURA3-SCE plasmid. We also thank members of Abou Elela lab for helpful discussions. This work is supported by a grant from the Canadian Institute of Health Research MOP 74594 to B.C. and S.A. S.A. is a Chercheur Boursier Senior of the Fonds de la Recherche en Santé du Québec. B.C. is a Canada Research Chair in Functional Genomics.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1254) on February 20, 2008.
REFERENCES
- Abou Elela S., Ares M., Jr Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Elela S., Igel H., Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- Aravind L., Watanabe H., Lipman D. J., Koonin E. V. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl. Acad. Sci. USA. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M., Jr, Grate L., Pauling M. H. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle D., Baralle M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bon E., et al. Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Buratti E., Baralle M., Baralle F. E. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capranico G., Ferri F., Fogli M. V., Russo A., Lotito L., Baranello L. The effects of camptothecin on RNA polymerase II transcription: roles of DNA topoisomerase I. Biochimie. 2007;89:482–489. doi: 10.1016/j.biochi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Chai B., Hsu J. M., Du J., Laurent B. C. Yeast RSC function is required for organization of the cellular cytoskeleton via an alternative PKC1 pathway. Genetics. 2002;161:575–584. doi: 10.1093/genetics/161.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. C., Chen C. J., Ho J. Y., Chuang T. J. Identification and evolutionary analysis of novel exons and alternative splicing events using cross-species EST-to-genome comparisons in human, mouse and rat. BMC Bioinformatics. 2006;7:136. doi: 10.1186/1471-2105-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz S. A., et al. Using the Saccharomyces Genome Database (SGD) for analysis of protein similarities and structure. Nucleic Acids Res. 1999;27:74–78. doi: 10.1093/nar/27.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowicz E., Jachymczyk W. J. Repair of MMS-induced DNA double-strand breaks in haploid cells of Saccharomyces cerevisiae, which requires the presence of a duplicate genome. Mol. Gen. Genet. 1979;167:279–286. doi: 10.1007/BF00267420. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Clark T. A., Sugnet C. W., Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Cohen-Tannoudji M., Robine S., Choulika A., Pinto D., El Marjou F., Babinet C., Louvard D., Jaisser F. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol. Cell. Biol. 1998;18:1444–1448. doi: 10.1128/mcb.18.3.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. A., Grate L., Spingola M., Ares M., Jr Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 2000;28:1700–1706. doi: 10.1093/nar/28.8.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Li C., Zenklusen D., Singer R. H., Jacobson A., He F. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell. 2007;25:559–573. doi: 10.1016/j.molcel.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwight S. S., et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO) Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead C., Dujon B. Consequences of unique double-stranded breaks in yeast chromosomes: death or homozygosis. Mol. Gen. Genet. 1993;240:170–178. doi: 10.1007/BF00277054. [DOI] [PubMed] [Google Scholar]
- Faustino N. A., Cooper T. A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Grasso C., Modrek B., Xing Y., Lee C. Genome-wide detection of alternative splicing in expressed sequences using partial order multiple sequence alignment graphs. Pac. Symp. Biocomput. 2004:29–41. doi: 10.1142/9789812704856_0004. [DOI] [PubMed] [Google Scholar]
- Guldener U., et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 2005;33:D364–368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halas A., Baranowska H., Policinska Z., Jachymczyk W. J. Involvement of the RE V3 gene in the methylated base-excision repair system. Co-operation of two DNA polymerases, delta and Rev3p, in the repair of MMS-induced lesions in the DNA of Saccharomyces cerevisiae. Curr. Genet. 1997;31:292–301. doi: 10.1007/s002940050208. [DOI] [PubMed] [Google Scholar]
- Hemenway C. S., Heitman J. Immunosuppressant target protein FKBP12 is required for P-glycoprotein function in yeast. J. Biol. Chem. 1996;271:18527–18534. doi: 10.1074/jbc.271.31.18527. [DOI] [PubMed] [Google Scholar]
- Herbert A., Rich A. RNA processing and the evolution of eukaryotes. Nat. Genet. 1999a;21:265–269. doi: 10.1038/6780. [DOI] [PubMed] [Google Scholar]
- Herbert A., Rich A. RNA processing in evolution. The logic of soft-wired genomes. Ann. NY Acad. Sci. 1999b;870:119–132. doi: 10.1111/j.1749-6632.1999.tb08872.x. [DOI] [PubMed] [Google Scholar]
- Hirose T., Shu M. D., Steitz J. A. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell. 2003;12:113–123. doi: 10.1016/s1097-2765(03)00267-3. [DOI] [PubMed] [Google Scholar]
- Hirose T., Steitz J. A. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA. 2001;98:12914–12919. doi: 10.1073/pnas.231490998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges P. E., McKee A. H., Davis B. P., Payne W. E., Garrels J. I. The Yeast Proteome Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 1999;27:69–73. doi: 10.1093/nar/27.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K., Miranda M., Hillenmeyer M. E., Nislow C., Davis R. W. Introns regulate RNA and protein abundance in yeast. Genetics. 2006;174:511–518. doi: 10.1534/genetics.106.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K., Palm C., Miranda M., Davis R. W. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc. Natl. Acad. Sci. USA. 2007;104:1522–1527. doi: 10.1073/pnas.0610354104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Hitomi M., Chen S., Tartakoff A. M. Nuclear mRNA accumulation causes nucleolar fragmentation in yeast mtr2 mutant. Mol. Biol. Cell. 1994;5:1253–1263. doi: 10.1091/mbc.5.11.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Lander E. S., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lei E. P., Silver P. A. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D. D., Darnell R. B. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Lopez P. J., Seraphin B. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice-site recognition. RNA. 1999;5:1135–1137. doi: 10.1017/s135583829999091x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacsovich T., Yang D., Waldman A. S. Repair of a specific double-strand break generated within a mammalian chromosome by yeast endonuclease I-SceI. Nucleic Acids Res. 1994;22:5649–5657. doi: 10.1093/nar/22.25.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. RNA regulation: a new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Nagasaki H., Arita M., Nishizawa T., Suwa M., Gotoh O. Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene. 2005;364:53–62. doi: 10.1016/j.gene.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Ooi S. L., Samarsky D. A., Fournier M. J., Boeke J. D. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA [In Process Citation] RNA. 1998;4:1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss J. A., Whitworth G. B., Bergkessel M., Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis A., Perrin A., Haber J. E., Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130:451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P. J., Guthrie C. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA. 2006;12:994–1006. doi: 10.1261/rna.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P. J., Kim K. S., Guthrie C. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA. 2002;8:969–980. doi: 10.1017/s1355838202020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., Sträßer K., Hurt E. An intron in the YRA1 gene is required to control Yra1 protein expression and mRNA export in yeast. EMBO Rep. 2002;3:438–442. doi: 10.1093/embo-reports/kvf091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santos-Rosa H., Moreno H., Simos G., Segref A., Fahrenkrog B., Pante N., Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spingola M., Grate L., Haussler D., Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K., Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint M., Levasseur G., Gervais-Bird J., Wellinger R. J., Elela S. A., Conconi A. A high-throughput method to measure the sensitivity of yeast cells to genotoxic agents in liquid cultures. Mutat. Res. 2006;606:92–105. doi: 10.1016/j.mrgentox.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Tycowski K. T., Shu M. D., Steitz J. A. A mammalian gene with introns instead of exons generating stable RNA products [see comments] Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- Venter J. C., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vilardell J., Chartrand P., Singer R. H., Warner J. R. The odyssey of a regulated transcript. RNA. 2000;6:1773–1780. doi: 10.1017/s135583820000145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa T., Ceradini F., Presutti C., Bozzoni I. Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol. 1998;18:3376–3383. doi: 10.1128/mcb.18.6.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A., Scott J. F. Construction, replication, and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol. Cell. Biol. 1982;2:221–232. doi: 10.1128/mcb.2.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., Vinciguerra P., Strahm Y., Stutz F. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol. 2001;21:4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Zhao J., Yu C. H., Luo Q. J., Chen Y. Q., Xiao Y., Qu L. H. Identification of a novel box C/D snoRNA from mouse nucleolar cDNA library. Gene. 2004;327:99–105. doi: 10.1016/j.gene.2003.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.