Abstract
BaP1 is a 22.7-kD P-I-type zinc-dependent metalloproteinase isolated from the venom of the snake Bothrops asper, a medically relevant species in Central America. This enzyme exerts multiple tissue-damaging activities, including hemorrhage, myonecrosis, dermonecrosis, blistering, and edema. BaP1 is a single chain of 202 amino acids that shows highest sequence identity with metalloproteinases isolated from the venoms of snakes of the subfamily Crotalinae. It has six Cys residues involved in three disulfide bridges (Cys 117–Cys 197, Cys 159–Cys 181, Cys 157–Cys 164). It has the consensus sequence H142E143XXH146XXGXXH152, as well as the sequence C164I165M166, which characterize the “metzincin” superfamily of metalloproteinases. The active-site cleft separates a major subdomain (residues 1–152), comprising four α-helices and a five-stranded β-sheet, from the minor subdomain, which is formed by a single α-helix and several loops. The catalytic zinc ion is coordinated by the Nɛ2 nitrogen atoms of His 142, His 146, and His 152, in addition to a solvent water molecule, which in turn is bound to Glu 143. Several conserved residues contribute to the formation of the hydrophobic pocket, and Met 166 serves as a hydrophobic base for the active-site groups. Sequence and structural comparisons of hemorrhagic and nonhemorrhagic P-I metalloproteinases from snake venoms revealed differences in several regions. In particular, the loop comprising residues 153 to 176 has marked structural differences between metalloproteinases with very different hemorrhagic activities. Because this region lies in close proximity to the active-site microenvironment, it may influence the interaction of these enzymes with physiologically relevant substrates in the extracellular matrix.
Keywords: Snake venom, zinc-dependent metalloproteinases, metzincins, hemorrhagic toxins, amino acid sequence, crystal structure
Snake venoms are a rich source of zinc-dependent metalloproteinases (Bjarnason and Fox 1994; Hati et al. 1999). Apart from playing an important role in the digestion of prey tissues, these enzymes also participate in the pathophysiology of envenoming by inducing local and systemic bleeding (Ownby 1990; Kamiguti et al. 1996a), as well as other tissue-damaging activities and hemostatic alterations (Kamiguti et al. 1996a; Gutiérrez and Rucavado 2000). Snake venom metalloproteinases (SVMPs) and the ADAMs form the “reprolysin” group of zinc-dependent metalloproteinases, which, together with astacins, matrix metalloproteinases, and serralysins, comprise the “metzinczin” superfamily (Bode et al. 1993; Stöcker et al. 1995). All these enzymes contain the sequence HEXXHXXGXXH in their zinc-binding region, as well as a Met-turn that serves as a base for the three active histidine residues (Bode et al. 1993; Stöcker et al. 1995). Depending on the domain composition, SVMPs can be classified as belonging to one of four classes: (1) P-I, which includes enzymes possessing only the metalloproteinase domain; (2) P-II, enzymes that possess both the metalloproteinase and a disintegrin-like domain; (3) P-III, enzymes that possess these two domains in addition to a high-cysteine domain; and (4) P-IV, enzymes that comprise all the aforementioned domains and are linked by a disulfide bridge to a type-C lectin subunit (Hite et al. 1994; Bjarnason and Fox 1994).
The ability of SVMPs to induce hemorrhage has been associated with their capacity to hydrolyze extracellular matrix proteins present in the basement membrane, a structural scaffold that surrounds endothelial cells in the microvasculature (Ohsaka 1979; Bjarnason and Fox 1994; Hati et al. 1999). P-III SVMPs induce a greater hemorrhagic effect than those belonging to the P-I class (Bjarnason and Fox 1994). It has been proposed that the disintegrin-like and high-Cys domains contribute to this enhanced activity by influencing the substrate specificity (Jeon and Kim 1999) by targeting the enzymes to physiologically relevant sites in capillaries (Jia et al. 1997; Gutiérrez and Rucavado 2000), and additionally by inhibiting platelet aggregation (Kamiguti et al. 1996b; Jia et al. 1997; Moura-da-Silva et al. 1999).
Large variations in hemorrhagic potency are observed even within P-I SVMPs; some are quite active, whereas others have no hemorrhagic activity (Bjarnason and Fox 1994; Tsai et al. 2000). Comparative sequence studies between hemorrhagic and nonhemorrhagic P-I venom metalloproteinases have led to various hypotheses concerning the structural determinants involved in the expression of hemorrhagic activity (Takeya et al. 1989, 1990; Gasmi et al. 2000; Tsai et al. 2000), although some of these hypotheses need to be revised in the light of new sequence and structural information (Gomis-Ruth et al. 1994). Recent proposals indicate that variations in a stretch of residues located at the C terminus (Tsai et al. 2000) or in three regions lying on the periphery of the active site (Bolger et al. 2001) are associated with differences in hemorrhagic activity. Only the crystal structures of six P-I SVMPs have been described: adamalysin II from Crotalus adamanteus venom (Gomis-Ruth et al. 1993, 1994), atrolysin C from Crotalus atrox venom (Zhang et al. 1994), H2-proteinase from Trimeresurus flavoviridis venom (Kumasaka et al. 1996), acutolysins A and C from Agkistrodon acutus venom (Gong et al. 1998; Zhu et al. 1999), and TM-3 from Trimeresurus mucrosquamatus venom (Huang et al. 2002). Because these enzymes vary greatly in their ability to induce hemorrhage, the comparative structural analysis of these and new enzymes will undoubtedly contribute to improve our understanding of the structural determinants of hemorrhagic activity.
BaP1 is a P-I basic SVMP isolated from the venom of the crotaline snake Bothrops asper (Gutiérrez et al. 1995; Gutiérrez and Ovadia 1998). This species is responsible for the vast majority of snakebites in Central America and southern Mexico (Gutiérrez 1995). Envenoming by B. asper is characterized, among other clinical features, by severe local tissue damage, often associated with permanent disability and sequelae (Gutiérrez 1995). Local pathology induced by this venom involves muscle necrosis, hemorrhage, edema, and blistering, a complex series of events mediated by venom phospholipases A2 and metalloproteinases (Gutiérrez and Lomonte 1995; Gutiérrez and Rucavado 2000). Among various SVMPs isolated from B. asper venom, BaP1 exerts a wide variety of local pathological alterations, including hemorrhage (Gutiérrez et al. 1995; Rucavado et al. 1995), myonecrosis (Rucavado et al. 1995), dermonecrosis and blistering (Rucavado et al. 1998), and edema (Gutiérrez et al. 1995). In addition, it induces a prominent inflammatory response characterized by complement activation (Farsky et al. 2000) and synthesis of cytokines and matrix metalloproteinases (Rucavado et al. 2002). Because of this wide toxicological profile, BaP1 is a relevant tissue-damaging component in B. asper venom that warrants better characterization. The present study therefore describes its amino acid sequence and crystal structure and compares them with those of related venom enzymes. This is the first crystal structure described of a SVMP isolated from the genus Bothrops; these snakes cause the highest impact in public health in Latin America (Fan and Cardoso 1995; Gutiérrez 1995). Our observations highlight a molecular region in P-I SVMPs that could be correlated with the variation in hemorrhagic activity within this group of enzymes.
Results and Discussion
Amino acid sequence
BaP1 is a single-chain protein consisting of 202 amino acids and a molecular weight of 22,735, as determined by MALDI-TOF-MS. As described for several SVMPs, the N-terminal residue is blocked, and the determination of the N-terminal sequence was possible only after treatment with pyroglutamyl aminopeptidase, with the consequent release of the pyroGlu residue. Such a residue is conventionally assigned as residue −1 (Gomis-Ruth et al. 1994). Following this treatment, the first 26 amino acids were sequenced by direct Edman degradation. Overlapping peptides generated by digestion with CNBr, lysyl peptidase, clostripain, endoproteinase Asp-N, and asparaginyl peptidase were also sequenced. Figure 1 ▶ presents the complete sequence of BaP1. The identity of Asn at position 151 was tentatively assigned based on Edman degradation of a peptide generated by asparaginyl peptidase digestion, and confirmed by examination of the electron density maps. The protein sequence data were deposited in the SWISS-PROT and TrEMBL knowledge base under the accession number P83512. Mature BaP1 comprises only the metalloproteinase domain, confirming its classification within the P-I class of SVMPs (Hite et al. 1994). Owing to its basic pI (≈8.5; Gutiérrez and Ovadia 1998) and its weak hemorrhagic activity (Gutiérrez et al. 1995), BaP1 can be placed within the subclass P-IB (weakly hemorrhagic enzymes) proposed by Bjarnason and Fox (1995). It possesses the characteristic zinc-chelating sequence H142E143XXH146XXGXXH152, and the sequence C164I165M166, associated with the Met-turn. These structural determinants are characteristic of the “metzincin” superfamily of zinc-dependent metalloproteinases (Bode et al. 1993), and are highly conserved in a large number of SVMPs (Fig. 2 ▶). BaP1 has six Cys residues at positions 117, 157, 159, 164, 181, and 197, thus belonging to the group of SVMPs with three disulfide bonds (see structural details below).
Figure 1.
Amino acid sequence of metalloproteinase BaP1. The first 26 residues were determined by direct Edman degradation after the protein was deblocked with pyroglutamyl aminopeptidase. Overlapping peptides were generated by digestion of the protein with CNBr, lysyl peptidase (K), clostripain (CL), endoproteinase Asp-N (D), and asparaginyl peptidase (N). Only the sequences of the peptides required to show overlap coverage for the whole sequence are presented. The cleavage sites are depicted by the symbol /. The sequence of each peptide includes only the residues that were confidently identified.
Figure 2.
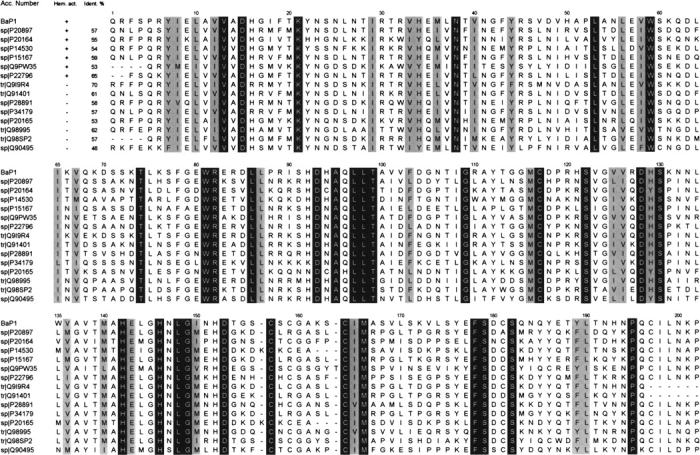
Sequence alignment of metalloproteinase BaP1 with other SVMPs from species of the family Viperidae whose hemorrhagic activity has been characterized. The seven top sequences correspond to SVMPs that are hemorrhagic, and the eight bottom sequences correspond to enzymes devoid of hemorrhagic activity. The numbers at the top correspond to those of the BaP1 sequence. Accession numbers in SWISS-PROT (sp) or trEMBL (tr) databases are indicated on the left. Invariant and conserved residues are presented against black and gray backgrounds, respectively. (P20897) Hemorrhagic metalloproteinase HT-2 (ruberlysin) from Crotalus ruber ruber; (P20164) metalloproteinase HR1B (Trimerelysin I) from Trimeresurus flavoviridis; (P14530) hemorrhagic metalloproteinase HR2A (Trimerelysin II) from Trimeresurus flavoviridis; (P15167) hemorrhagic toxin d (atrolysin-d) from Crotalus atrox; (Q9PW35) acutolysin A from Agkistrodon (Deinakistrodon) acutus; (P22796) hemorrhagic factor II (LHF-II) from Lachesis muta; (Q919R4) nonhemorrhagic fibrinolytic metalloprotease from Bothrops neuwiedi; (Q91401) atroxase from Crotalus atrox; (P28891) fibrolase from Agkistrodon contortrix contortrix; (P34179) adamalysin II from Crotalus adamanteus; (P20165) H2 metalloproteinase from Trimeresurus flavoviridis; (Q98995) lebetase from Vipera lebetina; (Q98SP2) bothrostatin from Bothrops jararaca; (Q90495) ecarin from Echis carinatus.
The sequence identity of BaP1 with various SVMPs of snakes of the family Viperidae, including both hemorrhagic and non-hemorrhagic enzymes, ranged from 70% to 48% (Fig. 2 ▶). BaP1 displays the highest sequence identity with enzymes placed within the clusters of “basic hemorrhagins” and “non-hemorrhagic fibrinogenases” in the classification of P-I SVMPs proposed by Tsai et al. (2000; Fig. 2 ▶). The identity percentage with SVMPs from Naja mossambica and Atractaspis engaddensis, classified in the families Elapidae and Atractaspididae, was 50% and 48%, respectively (data not shown).
Overall crystallographic structure
The three-dimensional structure of BaP1 is very similar to those described for other P-I snake venom metalloproteinases (Gomis-Ruth et al. 1993, 1994; Zhang et al. 1994; Kumasaka et al. 1996; Gong et al. 1998; Zhu et al. 1999; Huang et al. 2002). Atomic coordinates and structure factors were deposited in the Protein Data Bank, Brookhaven National Laboratory (reference: 1ND1). BaP1 is an ellipsoidal molecule with a shallow active-site cleft that separates two subdomains: a major subdomain (residues 1–152) with secondary structure characteristic of α/β proteins, comprising four α-helices (A, B, C, and D) and a five-stranded β-sheet. Strands I, II, III, and V are parallel, whereas strand IV is antiparallel (Fig. 3 ▶). The minor subdomain (residues 153–202) is formed by an α-helix and various loops. The molecule has three disulfide bridges (Cys 117–Cys 197, Cys 159–Cys 181, Cys 157–Cys 164) that stabilize the structure. The former bond links the two subdomains, whereas the latter two occur within the minor subdomain. Thus, BaP1 belongs to the three-disulfide-bridge subgroup of P-I metalloproteinases; in this regard, it is therefore similar to acutolysins A and C (Gong et al. 1998; Zhu et al. 1999) and TM-3 (Huang et al. 2002). In contrast, adamalysin II and atrolysin C present only two disulfide bridges (Gomis-Ruth et al. 1994; Zhang et al. 1994). Both N and C termini of BaP1 are located on the surface of the molecule, as in the other enzymes studied, and the C-terminal helix (helix E) is stabilized by the disulfide bridge Cys 157–Cys 164. The N-terminal residue was identified as pyroglutamate (residue −1), as observed in the electron density maps, in agreement with the results described above for sequence analysis.
Figure 3.
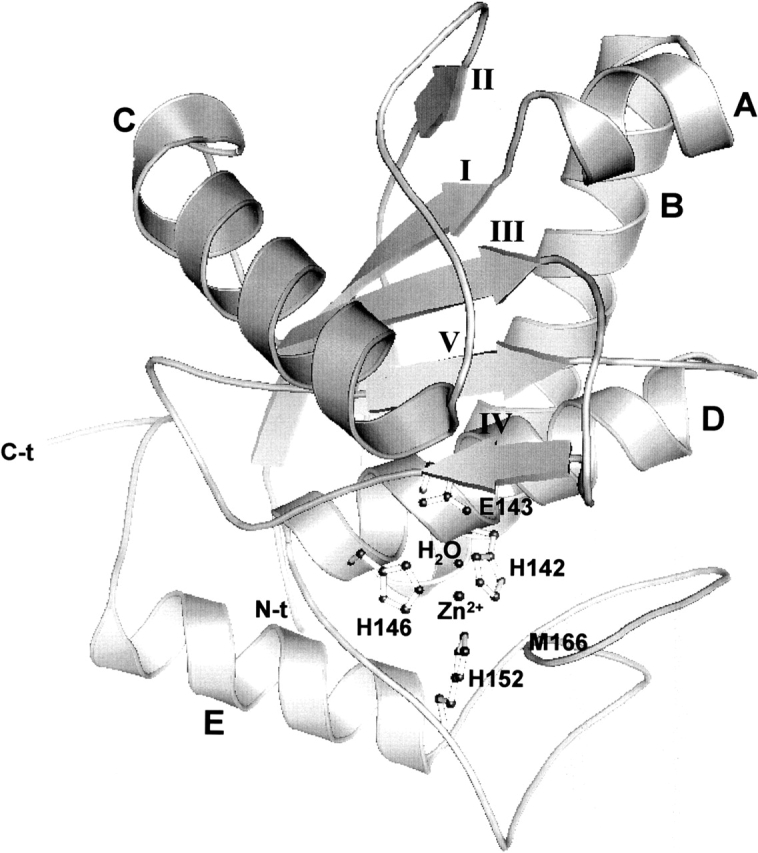
Ribbon diagram of the overall structure of metalloproteinase BaP1, depicted in standard orientation. The catalytic zinc atom is highlighted, together with the three histidines, a glutamic acid, and a water molecule forming the active-site environment. Secondary structures as well as N and C termini are also labeled.
The active site
As described for the other six P-I SVMPs whose structures have been studied, the zinc ion is tetrahedrally coordinated in BaP1 by the Nɛ2 nitrogen atoms of His 142, His 146, and His 152, in addition to a water solvent molecule, which, in turn, is bound to Glu 143 (Fig. 4 ▶). His 142, Glu 143, and His 146 are located on helix D. The Met-turn permits His 152 to be in close contact with the catalytic zinc, and the side chain of Met 166 comprises a hydrophobic base for the active-site groups. The bonds in the active site of BaP1 are detailed in Table 1. In analogy with the catalytic mechanism proposed for the bacterial metalloproteinase thermolysin (Matthews 1988), it is assumed that Glu 143 polarizes the catalytic water molecule, an event that is followed by the nucleophilic attack of the polarized water on the carbonyl carbon of the scissile peptide bond in the substrate. A hydrophobic pocket is present in the S1′ site of SVMPs. Residues Phe 178, Val 138, Ala 141, Tyr 176, and Ile 165 form part of the hydrophobic pocket (Gomis-Ruth et al. 1994) and are conserved in many SVMPs (Fig. 2 ▶). However, there are variations in the depth of this pocket among various SVMPs, depending on the presence of the nonconserved disulfide bridge Cys 159–Cys 181 and on the particular amino acid composition lining the pocket (Gong et al. 1998; Huang et al. 2002). The structural characteristics of this pocket influence the cleavage specificity and the binding of inhibitors (Zhang et al. 1994; Botos et al. 1996).
Figure 4.
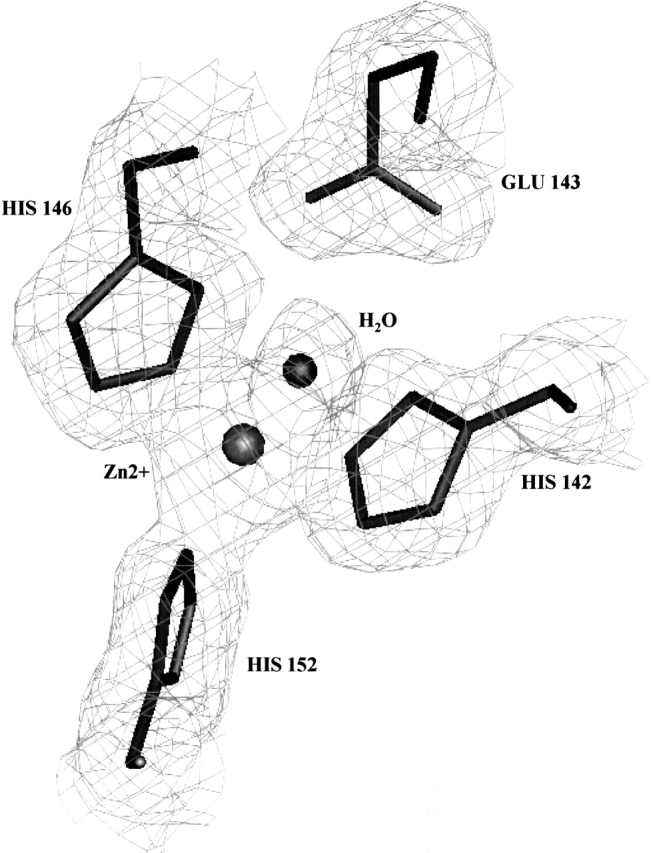
Details of the zinc-binding site of BaP1, superimposed on the electron density map (2Fo − Fc map contoured at 1 σ).
Table 1.
Bond distances at the active site in BaPl
| Bonds | Distance (Å) |
| His 142-Zn2+ | 2.05 |
| His146-Zn2+ | 2.12 |
| His152-Zn2+ | 2.00 |
| H2Ol-Zn2+ | 2.25 |
| H2O2-Glu140Oɛ1 | 3.32 |
| H2O1-Glu143Oɛ2 | 3.01 |
Structural calcium ion
The presence of a calcium ion located on the surface has been reported for several SVMPs and is considered to be important for structural stabilization (Gomis-Ruth et al. 1994). The ion is coordinated by the carbonyl oxygen atom of Glu 9, both carboxylate oxygens of Asp 93, the carbonyl oxygen of Cys 197, and the carboxiamide oxygen of Asn 200, in addition to a structural water molecule (Gomis-Ruth et al. 1994; Gong et al. 1998). Although these four residues are conserved, no calcium ion was found in the crystal structure of BaP1. This can be attributed to the fact that, because of the crystal packing, the Nɛ group of Lys 131 from a symmetry equivalent molecule interacts with Asp 93, Cys 197, and the water, thereby precluding the binding of calcium in that region in the crystal structure. Additionally, Asn 200 Nδ2 and Lys 131 Nɛ are bridged via a water molecule. It is not known if such a Ca2+ ion is present when BaP1 is in solution. Absence of the calcium ion has been also described for H2-proteinase (Kumasaka et al. 1996) and acutolysin-C (Zhu et al. 1999). A role has been proposed for this calcium in stabilizing the structure of these enzymes (Gomis-Ruth et al. 1994), not only of the metalloproteinase domain, but also of the additional domains present in P-II, P-III, and P-IV SVMPs (Gomis-Ruth et al. 1994; Gong et al. 1998), particularly the disintegrin-like domain that follows the catalytic domain in these enzymes. Calcium ions are known to stabilize the tertiary structure of collagenase, a matrix metalloproteinase (Lovejoy et al. 1994).
Structural determinants involved in hemorrhagic activity
SVMPs greatly differ in their ability to induce hemorrhage, which is one of the main manifestations of envenoming by vipers (Bjarnason and Fox 1994; Gutiérrez and Rucavado 2000). P-III metalloproteinases, containing disintegrin-like and high-Cys domains in addition to the metalloproteinase domain, usually induce more intense hemorrhage than the P-I metalloproteinases, which consist of only the metalloproteinase domain (Bjarnason and Fox 1994). It has been proposed that these additional domains may be responsible for this enhanced hemorrhagic activity because of their ability to direct the enzymes to physiologically relevant targets in the microvessels (Zhou et al. 1995; Jia et al. 1997; Gutiérrez and Rucavado 2000), probably resulting in proteolytic degradation of key structural proteins at the basement membrane (Baramova et al. 1990, 1991). Moreover, disintegrin-like and high-Cys domains in P-III SVMPs may influence substrate specificity (Jeon and Kim 1999); they have been shown to inhibit collagen-induced platelet aggregation (Kamiguti et al. 1996b; Jia et al. 1997 Jia et al. 2000; Moura-da-Silva et al. 1999), an effect that might potentiate hemorrhagic activity. In addition to this well-demonstrated difference in hemorrhagic potency between large and small SVMPs, a wide range of hemorrhagic activities is also found in the P-I group of enzymes, indicating that structural features within the metalloproteinase domain play a role in this effect. Catalytic activity is an absolute requirement for the development of hemorrhage, because zinc chelation abrogates this effect (Bjarnason and Fox 1994). Owing to the fact that the catalytic center is identical in these molecules, the structural basis for their different biological activity must depend on structural determinants located in other regions, probably in the region of the catalytic center.
Comparative sequence analyses have led to various proposals concerning the residues and molecular regions putatively involved in hemorrhagic activity (Takeya et al. 1989, 1990; Gasmi et al. 2000; Tsai et al. 2000). Some of these proposals have been refuted as new sequences and crystal structures have become available (Gomis-Ruth et al. 1994). Comparison of the sequences of hemorrhagic and nonhemorrhagic P-I SVMPs demonstrated three regions with high variation between these enzymes: (1) amino acid residues 63–90, (2) residues between 179 and 193, and (3) residues between 150 and 176 (Tsai et al. 2000; Bolger et al. 2001). These regions surround the active-site cleft and are located on the surface of the molecule; it seems likely that they are involved in modulating the interaction with substrates. A phylogenetic analysis of 30 P-I SVMPs permitted their grouping into three different subtypes, these being the highly hemorrhagic acidic enzymes, moderately hemorrhagic basic proteinases, and nonhemorrhagic enzymes (Tsai et al. 2000). Analysis of the C-terminal regions of these SVMPs revealed that both acidic and basic hemorrhagic metalloproteinases are rich in residues with carboxyl- and hydroxyl-containing side chains, as well as hydrophobic residues, whereas nonhemorrhagic proteinases possess in that region predominantly residues with small or cationic side chains (Tsai et al. 2000). On the basis of its sequence in that particular region, BaP1 can be placed within the basic (BH) subgroup of metalloproteinases, in agreement with its basic pI (Gutiérrez and Ovadia 1998) and its relatively low potency in inducing hemorrhage (Gutiérrez et al. 1995). However, as shown in Figure 2 ▶, the comparative sequence analysis of BaP1 with other SVMPs in which hemorrhagic activity, or its absence, have been well established, did not evidence any clear picture of possible residues or positions specifically conserved among hemorrhagic enzymes. Therefore, comparative analysis of three-dimensional structures is required to identify the structural motifs determining hemorrhagic activity in SVMPs.
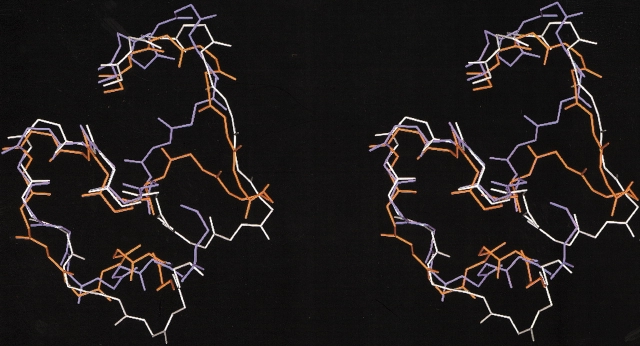
We superimposed the structures of three SVMPs exhibiting an identical pattern of disulfide bonds but displaying large variations in hemorrhagic potency: acutolysin A (highly hemorrhagic), BaP1 (weakly hemorrhagic), and H2-proteinase (nonhemorrhagic). Structural differences among them do not depend on a different disulfide bond arrangement, as occurs when proteinases from the groups having two and three disulfide bonds are compared. Superimposition of these structures indicated a high degree of structural identity in most regions, with the exception of a stretch in the region of residues 153 to 176, which corresponds to a loop located near the catalytic site in the crystal structure (Fig. 5 ▶). This observation supports a previous structural comparison between acutolysin A, H2-proteinase, and adamalysin II, which showed variations in the segments 153–164 and 173–176 (Gong et al. 1998). Within this region, there is a highly conserved sequence Cys–Ile–Met involved in the Met-turn. However, major structural variations in the stretches located before and after this conserved sequence are observed. In addition, there are some amino acid substitutions, insertions, and deletions that also occur around the disulfide bond Cys 159–Cys 164; these contribute to structural divergences in this local microenvironment, as described for H2-proteinase and acutolysin A (Gong et al. 1998). Here we show that BaP1 also differs from these two enzymes in this region, showing that, despite an identical disulfide bonding pattern in these three enzymes, there are conspicuous structural differences in this loop. Interestingly, BaP1 and acutolysin, both of which are hemorrhagic, have more similarities in this region than H2-proteinase, which is not hemorrhagic (Fig. 5 ▶). Because this region lies close to the active site, it is implied that this loop may be involved in the interaction with extracellular matrix substrates. This might influence their ability to bind and hydrolyze basement membrane components, a key step in the weakening and disruption of capillary vessel structure leading to hemorrhage.
Figure 5.
Stereo figure of the superimposition of the main chains of the segment 153–176 in the structures of three P-I SVMPs that have an identical pattern of disulfide bonds but differ in their hemorrhagic potencies. BaP1, a weakly hemorrhagic SVMP (white); H2-proteinase, a SVMP devoid of hemorrhagic activity from the venom of Trimeresurus flavoviridis (blue); and acutolysin A, a SVMP with high hemorrhagic activity from the venom of Agkistrodon (Deinakgistrodon) acutus (orange).
It has been shown that structural motifs determining functions of many proteins are often located in flexible loops. Amino acid substitutions, deletions, and insertions in such motifs may significantly modify the function of a protein without affecting the structural stability of the molecule (Overall 2002). In addition to hemorrhage, BaP1 induces a wide variety of local pathological effects (Gutiérrez et al. 1995; Rucavado et al. 1995, 1998, 2002). It is not known if the various toxicological activities depend on a single structural motif or if different regions are involved. Such a multiple functional profile makes BaP1 a valuable model for investigating structure–function relationships associated with various toxic effects in a single SVMP.
Materials and methods
Isolation of BaP1
Metalloproteinase BaP1 was isolated from a venom pool obtained from >40 adult specimens of B. asper collected in the Pacific region of Costa Rica. The isolation protocol previously described by Gutiérrez et al. (1995) and Rucavado et al. (1998) included ion-exchange chromatography on CM-Sephadex C-25, gel filtration on Sephacryl S-200, and affinity chromatography on Affi-Gel Blue. Molecular mass was determined by MALDI-TOF mass spectrometry on a PE Voyager DE Pro equipment.
Reduction and alkylation
For reduction and alkylation, 20 μg of protein was dissolved in 50 μL of a 0.64 M Tris-HCl (pH 8.0), 8 M urea, 0.16 M methylamine-HCl solution. Following the addition of 5 μL of a 2-mercaptoethanol solution (7 μL/mL), the mixture was incubated at 60°C for 1.5 h. Then, 7 μL of an N-isopropyliodoacetamide solution (2.3 mg in 20 μL of methanol and 80 μL of water) was added to the protein solution, and the mixture was incubated at room temperature for 30 min before the addition of 10 μL of 2-mercaptoethanol solution.
Digestion with cyanogen bromide and various enzymes
For deblocking BaP1, 10 μg of the metalloproteinase in a volume of 5 μL was added to 50 μL of 0.1 M phosphate, 5% glycerol, 10 mM EDTA (pH 8.0) buffer, followed by the addition of 3 μL of 0.1 M dithiothreitol. Then 2 μg of pyroglutamate aminopeptidase was added, and the mixture was incubated at 4°C overnight, followed by incubation at 25°C for 4 h. The protein was then desalted on a Phenomenex C18 column. For CNBr digestion, 5 μg of BaP1 was incubated with 50 μL of a 50 mg/mL CNBr solution. After incubation at room temperature overnight, the mixture was diluted with 400 μL of 0.1% trifluoroacetic acid (TFA) for chromatographic separation of the peptides. Lysyl peptidase digestion was performed by incubating 5 μg of reduced and alkylated BaP1 with 0.5 μg of the enzyme (Wako Biochemicals). The mixture was incubated at 37°C for 4.5 h with shaking. For clostripain digestion, 10 μg of reduced and alkylated BaP1 was desalted on a Brownlee C8 column. After neutralization of TFA with triethylamine and concentration by drying, 0.2 μg of clostripain (Sigma), in 50 mM Tris-HCl, 1 mM CaCl2, and 2.5 mM dithiothreitol (pH 7.5) was added and incubated at room temperature overnight. Endoproteinase Asp-N digestion was carried out by incubating 10 μg of reduced and alkylated BaP1, previously dialyzed against Tris-HCl buffer (pH 8.0), with 0.092 μg of endoproteinase Asp-N. Incubation was performed at 37°C for 18 h. The reaction was stopped with 2 μL of TFA. Asparaginyl peptidase digestion was performed by diluting 10 μg of reduced and alkylated BaP1 with 50 mM acetate buffer (pH 5.0). Then, 0.08 mU of asparaginyl peptidase (Pan Vera) was added, and the mixture was incubated at room temperature overnight.
Amino acid sequence determination and similarity searches
The N-terminal amino acid sequence of BaP1 was determined by direct Edman degradation on a Applied Biosystems 494 amino acid sequencer, operated according to the manufacturer’s instructions, once the enzyme was deblocked by pyroglutamate aminopeptidase. Whole protein was adsorbed on PVDF in a ProSorb cartridge (Applied Byosystems) and analyzed using the manufacturer’s instructions for PVDF samples. Peptides resulting from the various digestions were separated by reverse phase HPLC chromatography. All separations used a gradient of 0.1% TFA in water (solvent A) and 0.09% TFA in acetonitrile (solvent B), on an Applied Biosystems 120A or 140 chromatography system, with detection at 215 nm. The flow rate was 200 μL/min. The columns used were Phenomenex Jupiter C18, Vydac C18, Zorbax CN, and Zorbax SB-C18. The sequences of the peptides resulting from the hydrolysis of BaP1 by the CNBr and enzymatic treatments were determined in an Applied Biosystems 494 sequencer after chromatographic separation of the peptides. The peptides were applied to a Polybrene-coated glass fiber filter and analyzed with the manufacturer’s pulsed liquid cycle. Similarity searches were performed using FASTA (Pearson and Lipman 1988), and amino acid sequences were aligned using the program CLUSTAL W (Higgins et al. 1996).
Protein crystallization
Crystallization was performed by the hanging-drop vapor diffusion method, as described earlier (Watanabe et al. 2002). Briefly, single crystals were obtained when a 3-μL protein droplet, at a protein concentration of 10 mg/mL, was equilibrated over a reservoir containing Bicine buffer (pH 9.0), 10% PEG 20.000, and 2% (v/v) dioxane.
X-ray diffraction data collection
Three-dimensional X-ray diffraction data were collected from a single crystal (maximum dimensions of 0.4 mm) at room temperature at the crystallographic beamline (Polikarpov et al. 1998a,b) of the Laboratório Nacional de Luz Síncrotron (LNLS, Campinas, Brazil). The synchrotron-radiation source was set to a wavelength of 1.54 Å. The diffraction intensities were measured using an MAR 345 imaging plate detector (MAR Research) and an oscillation range of 1°. The crystal diffracted to a nominal resolution of 1.9 Å. The X-ray diffraction data were processed, scaled, and integrated using DENZO/SCALEPACK (Otwinowski and Minor 1997).
Molecular replacement was carried out using the program AmoRe (Navaza 1994) with a homology-built model for BaP1 that was based on the crystal structure of acutolysin-C from the venom of Agkistrodon (Deinakgistrodon) acutus (PDB code 1QUA; Zhu et al. 1999). The rotation function provided a clear solution with a correlation coefficient of 17.9 (the next highest correlation was 9.5) in the resolution range of 20.0–2.5 Å. In the translation search, the correlation increased to 48.1 (R-factor = 45.5%) for data in the same resolution range. This solution was subjected to rigid-body refinement, which resulted in a correlation coefficient of 67.6 and an R-factor of 46.1%. The refinement was carried out using the program CNS (Brunger et al 1998), and the electron density maps were examined using the program package O (Jones et al. 1991). The refinement converged to a crystallographic residual of 19.08% (Rfree = 21.59%) for all data between 500.0 and 1.9 Å with no outliers in the Ramachandran Plot (Laskowski et al. 1993) and with excellent stereochemistry (Table 2). Data collection and refinement statistics are presented in Table 2. The figures of 3D structures were prepared using the PyMOL molecular graphics system (Delano 2002; http://www.pymol.org).
Table 2.
Diffraction data collection and refinement statistics for BaP1
| Crystal type formed | Orthorhombic |
| Space group | P212121 |
| Cell dimension (Å) | a = 38.42, b = 60.36, c = 86.26 |
| Maximum resolution (Å) | 1.90 |
| Percentage solvent (%) | 40.51 |
| No. of observations | 49,843 |
| No. of unique reflections | 14,941 |
| Rsym (%)a | 7.2 |
| Completeness (%) | 92.1 |
| VM (Å3/D) | 2.08 |
| Molecules per asymmetric unit | 1 |
| Refinement resolution (Å) | 500–1.9 |
| Rfactorb (%) | 19.08 |
| Rfreec (%) | 21.59 |
| No. of solvent molecules | 110 |
| Average B valuesd (Å2) RMSD from ideal valuese | |
| Bond length (Å) | 0.00995 |
| Bond angle (°) | 2.2566 |
aRsym = 100 × ∑|I (h) − 〈I (h)〉|/∑I (h), where I (h) is the observed intensity and 〈I (h)〉 is the mean intensity of reflection h over all measurements of I (h).
bRfactor = 100 × ∑|Fo − Fc|/∑(Fo), the sums being taken over all reflections with F/σ(F) > 0 the cutoff.
cRfree = Rfactor for 10% of the data, which were not included during crystallographic refinement.
dB values are average B values for all nonhydrogen atoms.
e RMSD, root-mean-square deviation.
Acknowledgments
This study was supported by grants from the Wellcome Trust (grant no. 062043) to J.M.G. and R.D.G.T., the International Foundation for Science (grant no. F-2707-2), FAPESP (grant 99/09162-4), CNPq (grant 520081/95-NV), SMOLBNet (grant 01/07537-2), FUNDUNESP (Brazil), and Vicerrectoría de Investigación, Universidad de Costa Rica (projects 741-A1-504 and 741-A2-036). L.W. was the recipient of an FAPESP fellowship. Atomic coordinates and structural factors have been deposited in the Protein Data Bank, Brookhaven National Laboratory (accession code 1ND1).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03102403.
References
- Baramova, E.N., Shannon, J.D., Bjarnason, J.B., and Fox, J.W. 1990. Identification of the cleavage sites by a hemorrhagic metalloproteinase in type IV collagen. Matrix 10 91–97. [DOI] [PubMed] [Google Scholar]
- Baramova, E.N., Shannon, J.D., Fox, J.W., and Bjarnason, J.B. 1991. Proteolytic digestion of non-collagenous basement membrane proteins by the hemorrhagic metalloproteinase HT-e from Crotalus atrox venom. Biomed. Biochim. Acta 50 763–768. [PubMed] [Google Scholar]
- Bjarnason, J.B. and Fox, J.W. 1994. Hemorrhagic metalloproteinases from snake venoms. Pharmac. Ther. 62 325–372. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Snake venom metalloendopeptidases: Reprolysins. Meth. Enzymol. 248 345–368. [DOI] [PubMed] [Google Scholar]
- Bode, W., Gomis-Ruth, F.X., and Stöckler, W. 1993. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins.’ FEBS Lett. 331 134–140. [DOI] [PubMed] [Google Scholar]
- Bolger, M.B., Swenson, S., and Markland, F.S. 2001. Three-dimensional structure of fibrolase, the fibrinolytic enzyme from southern copperhead venom, modeled from the X-ray structure of adamalysin II and atrolysin C. AAPS Pharmsci. 3 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botos, I., Scapozza, L., Zhang, D., Liotta, L.A., and Meyer, E.F. 1996. Batimastat, a potent matrix metalloproteinase inhibitor, exhibits an unexpected mode of binding. Proc. Natl. Acad. Sci. 93 2749–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., Delano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N.S., et al. 1998. Crystallography and NMR System (CNS): A new software system for macromolecular structure determination. Acta Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- Delano, W.S. 2002. The PyMOL molecular graphics system. Delano Scientific, San Carlos, CA. http://www.pymol.org.
- Fan, H.W. and Cardoso, J.L. 1995. Clinical toxicology of snake bites in South America. In Clinical toxicology of animal venoms and poisons (eds. J. Meier and J. White), pp. 667–688. CRC Press, Boca Raton, Florida.
- Farsky, S.H.P., Goncalves, L.R.C., Gutiérrez, J.M., Correa, A.P., Rucavado, A., Gasque, P., and Tambourgi, D.V. 2000. Bothrops asper snake venom and its metalloproteinase BaP-1 activate the complement system. Role in leucocyte recruitment. Med. Inflamm. 9 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasmi, A., Srairi, N., Karoui, H., and El Ayeb, M. 2000. Amino acid sequence of VIF: Identification in the C-terminal domain of residues common to non-hemorrhagic metalloproteinases from snake venoms. Biochim. Biophys. Acta 1481 209–212. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth, F.X., Crees, L.F., and Bode, W. 1993. First structure of a snake venom metalloproteinase: A prototype for matrix metalloproteinases/collagenases. EMBO J. 12 4151–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth, F.X., Kress, L.F., Kellermann, J., Mayr, I., Lee, X., Huber, R., and Bode, W. 1994. Refined 2 Å X-ray crystal structure of the snake venom zinc-endopeptidase adamalysin II. Primary and tertiary structure determination, refinement, molecular structure and comparison with astacin, collagenase and thermolysin. J. Mol. Biol. 239 513–544. [DOI] [PubMed] [Google Scholar]
- Gong, W., Zhu, X., Liu, S., Teng, M., and Niu, L. 1998. Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus. J. Mol. Biol. 283 657–668. [DOI] [PubMed] [Google Scholar]
- Gutiérrez, J.M. 1995. Clinical toxicology of snakebite in Central America. In Clinical toxicology of animal venoms and poisons (eds. J. Meier and J. White), pp. 645–665. CRC Press, Boca Raton, Florida.
- Gutiérrez, J.M. and Lomonte, B. 1995. Phospholipase A2 myotoxins from Bothrops snake venoms. Toxicon 33 1405–1424. [DOI] [PubMed] [Google Scholar]
- Gutiérrez, J.M. and Ovadia, M. 1998. Bothrops asper hemorrhagic proteinases BaH1 and BaP1. In Handbook of proteolytic enzymes (eds. A.J. Barret et al.), pp. 1295–1297. Academic Press, London.
- Gutiérrez, J.M. and Rucavado, A. 2000. Snake venom metalloproteinases: Their role in the pathogenesis of local tissue damage. Biochimie 82 841–850. [DOI] [PubMed] [Google Scholar]
- Gutiérrez, J.M., Romero, M., Díaz, C., Borkow, G., and Ovadia, M. 1995. Isolation and characterization of a metalloproteinase with weak hemorrhagic activity from the venom of the snake Bothrops asper (terciopelo). Toxicon 33 19–29. [DOI] [PubMed] [Google Scholar]
- Hati, R., Mitra, P., Sarker, S., and Bhattacharyya, K.K. 1999. Snake venom hemorrhagins. Crit. Rev. Toxicol. 29 1–19. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Thompson, J.D., and Gidson, T.J. 1996. Using CLUSTAL for multiple sequence alignments. Meth. Enzymol. 266 383–402. [DOI] [PubMed] [Google Scholar]
- Hite, L.A., Jia, L.G., Bjarnason, J.B., and Fox, J.W. 1994. cDNA sequences for four snake venom metalloproteinases: Structure, classification and their relationship to mammalian reproductive proteins. Arch. Biochem. Biophys. 38 182–191. [DOI] [PubMed] [Google Scholar]
- Huang, K.F., Chiou, S.H., Ko, T.P., Yuann, J.M., and Wang, A.H. 2002. The 1.35 Å structure of cadmium-substituted TM-3, a snake-venom metalloproteinase from Taiwan habu: Elucidation of a TNFα-converting enzyme-like active-site structure with a distorted octahedral geometry of cadmium. Acta Crystallogr. D 58 1118–1128. [DOI] [PubMed] [Google Scholar]
- Jeon, O.K. and Kim, D.S. 1999. Molecular cloning and functional characterization of a snake venom metalloprotease. Eur. J. Biochem. 263 526–533. [DOI] [PubMed] [Google Scholar]
- Jia, L.G., Wang, X.M., Shannon, J.D., Bjarnason, J., and Fox, J.W. 1997. Function of disintegrin-like/cysteine-rich domains of atrolysin A. Inhibition of platelet aggregation by recombinant protein and peptide antagonists. J. Biol. Chem. 272 13094–13102. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Inhibition of platelet aggregation by the recombinant cysteine-rich domain of the hemorrhagic snake venom metalloproteinase, atrolysin A. Arch. Biochem. Biophys. 373 281–286. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.-Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kamiguti, A.S., Hay, C.R., Theakston, R.D.G., and Zuzel, M. 1996a. Insights into the mechanism of haemorrhage caused by snake venom metalloproteinases. Toxicon 34 627–642. [DOI] [PubMed] [Google Scholar]
- Kamiguti, A.S., Hay, C.R.M., and Zuzel, M. 1996b. Inhibition of collagen-induced platelet aggregation as the result of cleavage of α2β1-integrin by the snake venom metalloproteinase jararhagin. Biochem. J. 320 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumasaka, T., Yamamoto, M., Moriyama, H., Tanaka, N., Sato, M., Katsube, Y., Yamakawa, Y., Omori-Satoh, T., Iwanaga, S., and Ueki, T. 1996. Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis. J. Biochem. 119 49–57. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26 283–291. [Google Scholar]
- Lovejoy, B., Hassell, A.M., Luther, M.A., Weigl, D., and Jordan, S.R. 1994. Crystal structures of recombinant 19-kDa human fibroblast collagenase complexed to itself. Biochemistry 33 8207–8217. [DOI] [PubMed] [Google Scholar]
- Matthews, B.W. 1988. Structural basis of the action of thermolysin and related zinc peptidases. Acct. Chem. Res. 21 333–340. [Google Scholar]
- Moura-da-Silva, A.M., Línica, A., Della-Casa, M.S., Kamiguti, A.S., Ho, P.L., Crampton, J.M., and Theakston, R.D.G. 1999. Jararhagin ECD-containing disintegrin domain: Expression in Escherichia coli and inhibition of the platelet-collagen interaction. Arch. Biochem. Biophys. 369 295–301. [DOI] [PubMed] [Google Scholar]
- Ohsaka, A. 1979. Hemorrhagic, necrotizing and edema-forming effects of snake venoms. In Snake venoms (ed. C.Y. Lee), pp. 480–546. Springer-Verlag, Berlin.
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Overall, C.M. 2002. Molecular determinants of metalloproteinase substrate specificity. Matrix metalloproteinase substrate binding domains, modules and exosites. Mol. Biotech. 22 51–86. [DOI] [PubMed] [Google Scholar]
- Ownby, C.L. 1990. Locally-acting agents: Myotoxins, hemorrhagic toxins and dermonecrotic factors. In Handbook of toxinology (eds. W.T. Shier and D. Mebs), pp. 601–653. Marcel Dekker, New York.
- Pearson, W.R. and Lipman, D.J. 1988. Improved tools for biological sequence comparisons. Proc. Natl. Acad. Sci. 85 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikarpov, I., Oliva, G., Castellano, E.E., Garratt, R.C., Arruda, P., Leite, A., and Craievich, A. 1998a. The protein crystallography beamline at LNLS, the Brazilian National Synchrotron Light source. Nucl. Inst. Methods Phys. Res. A 405 159–164. [Google Scholar]
- Polikarpov, I., Perles, L.A., de Oliveira, R.T., Oliva, G., Castellano, E.E., Garratt, R.C., and Craievich, A. 1998b. Set-up and experimental parameter of the protein crystallography beamline at the Brazilian National Synchrotron Laboratory. J. Synch. Rad. 5 72–76. [DOI] [PubMed] [Google Scholar]
- Rucavado, A., Lomonte, B., Ovadia, M., and Gutiérrez, J.M. 1995. Local tissue damage induced by BaP1, a metalloproteinase isolated from Bothrops asper (terciopelo) snake venom. Exp. Mol. Pathol. 63 186–199. [DOI] [PubMed] [Google Scholar]
- Rucavado, A., Núñez, J., and Gutiérrez, J.M. 1998. Blister formation and skin damage induced by BaP1, a haemorhagic metalloproteinase from the venom of the snake Bothrops asper. Int. J. Exp. Pathol. 79 245–254. [PMC free article] [PubMed] [Google Scholar]
- Rucavado, A., Escalante, T., Teixeira, C.F.P., Fernandes, C.M., Díaz, C., and Gutiérrez, J.M. 2002. Increments in cytokines and matrix metalloproteinases in skeletal muscle after injection of tissue-damaging toxins from the venom of the snake Bothrops asper. Med. Inflamm. 11 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöcker, W., Grams, F., Baumann, U., Reinemer, P., Gomis-Ruth, F.X., McKay, D.B., and Bode, W. 1995. The metzincins—Topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 4 823–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya, H., Arakawa, M., Miyata, T., Iwanaga, S., and Omori-Satoh, T. 1989. Primary structure of H2-proteinase, a non hemorrhagic metalloproteinase isolated from the venom of the habu snake Trimeresurus flavoviridis. J. Biochem. 106 151–157. [DOI] [PubMed] [Google Scholar]
- Takeya, H., Onikura, A., Nikai, T., Sugihara, H., and Iwanaga, S. 1990. Primary structure of a hemorrhagic metalloproteinase, HT-2, isolated from the venom of Crotalus ruber ruber. J. Biochem. 108 711–719. [DOI] [PubMed] [Google Scholar]
- Tsai, I.H., Wang, Y.M., Chiang, T.Y., Chen, Y.L., and Huang, R.J. 2000. Purification, cloning and sequence analyses for pro-metalloprotease-disintegrin variants from Deinagkistrodon acutus venom and subclassification of the small venom metalloproteinases. Eur. J. Biochem. 267 1359–1367. [DOI] [PubMed] [Google Scholar]
- Watanabe, L., Rucavado, A., Kamiguti, A., Theakston, R.D.G., Gutiérrez, J.M., and Arni, R.K. 2002. Crystallization and preliminary diffraction data of BaP1, a haemorrhagic metalloproteinase from Bothrops asper snake venom. Acta Crystallogr. D 58 1034–1035. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Botos, I., Gomis-Ruth, F.X., Doll, R., Blood, C., Njoroge, F.G., Fox, J.W., Bode, W., and Meyer, E.F. 1994. Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d). Proc. Natl. Acad. Sci. 91 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., Smith, J.B., and Grossman, M.H. 1995. Molecular cloning and expression of catrocollastatin, a snake-venom protein from Crotalus atrox (Western diamondback rattlesnake) which inhibits platelet adhesion to collagen. Biochem. J. 307 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Teng, M., and Niu, L. 1999. Structure of acutolysin-C, a haemorrhagic toxin from the venom of Agkistrodon acutus, providing further evidence for the mechanism of the pH-dependent proteolytic reaction of zinc metalloproteinases. Acta Cryst. D 55 1834–1841. [DOI] [PubMed] [Google Scholar]