Abstract
Ribonuclease Sa and two charge-reversal variants can be converted into amyloid in vitro by the addition of 2,2,2-triflouroethanol (TFE). We report here amyloid fibril formation for these proteins as a function of pH. The pH at maximal fibril formation correlates with the pH dependence of protein solubility, but not with stability, for these variants. Additionally, we show that the pH at maximal fibril formation for a number of well-characterized proteins is near the pI, where the protein is expected to be the least soluble. This suggests that protein solubility is an important determinant of fibril formation.
Keywords: Amyloid, amyloid fibrils, pI, protein solubility, protein stability
The formation of amyloid fibrils is of medical interest because it is associated with diseases such as Alzheimer’s disease, Parkinson’s disease, and cystic fibrosis (Dobson 2001). Fibrils are formed from the extracellular deposition of aggregated protein (Cohen 1967a,b; Glenner 1980a,b; Rochet and Lansbury Jr. 2000). These insoluble aggregates originate from a number of proteins that are structurally diverse; some 20 disease-causing proteins have been identified in humans. Amyloid formation is also of interest to the protein chemist because it has been shown that many proteins can be induced to form amyloid fibrils in vitro, suggesting that amyloid fibril formation is not limited to a select few proteins or by specific physiological factors, but rather it may be a general feature of all polypeptides (Lai et al. 1996; Chiti et al. 1999).
Recently, proteins such as human muscle acylphosphatase, hen egg white lysozyme, the B1 binding domain of protein G, the SH3 domain of PI3, and myglobin have been shown to form fibrils in vitro (Chiti et al. 1999; Krebs et al. 2000; Ramirez-Alvarado et al. 2000; Fändrich et al. 2001). These fibrils show ultrastructure and dye-binding characteristics identical to fibrils formed from the disease-causing proteins (Wetzel 2002). In most cases, fibrils were obtained from solution conditions where the native conformation was destabilized by pH, temperature, and/or the addition of cosolvents like alcohols, salts, or metal ions. These results suggest that the ability of a protein to form fibrils in vitro depends upon the conformational stability of the protein, defined as the difference in free energy (ΔG) between the folded and unfolded conformations (Kelly 1996; Chiti et al. 1999, 2000; Ramirez-Alvarado et al. 2000). It further implies that as proteins are destabilized, either through alterations in amino acid sequence or changes in the solutions conditions, the formation of amyloid fibril is enhanced. Here, we wish to expand these conclusions to include the idea that the solubility of the folded (or partially folded) conformation(s) of a protein is important, and perhaps the major factor in amyloid fibril formation.
A significant population of partially unfolded protein is likely necessary for fibril formation to occur in many of the proteins studied to date (Chiti et al. 1999, 2000; Ramirez-Alvarado et al. 2000; Khurana et al. 2001a,b; Taddei et al. 2001). For example, fibrils of acylphosphatase fibrils are formed in mixtures of water and 2,2,2-trifluoroethanol (TFE), an organic solvent that has been shown to promote secondary structure formation at low concentrations. Because TFE is known to increase the solubility of hydrophobic groups while decreasing the solubility of peptide groups (Luo and Baldwin 1997; Buck 1998), structures that have exposed hydrophobic groups and buried peptide groups are favored. Other studies of fibril formation with transthyretin (TTR; Jiang et al. 2001), human lysozyme (Pepys et al. 1993), the light chain of IgG (Souillac et al. 2002), methionine aminopeptidase (Nielsen et al. 2001), and fibronectin (Litvinovich et al. 1998) emphasize that conditions that favor partial unfolding of the protein can significantly enhance fibril formation. Together, these results also support the hypothesis that any protein can form amyloid under certain conditions, and fibril formation is accelerated when the conditions favor partial unfolding of the native conformation (Dobson 1999).
Although protein stability is certainly important for fibril formation, the association between protein solubility and fibril formation has not been directly correlated. The solubility of globular proteins is determined by the amino acid content, the pK values of the ionizable residues, and environmental factors such as temperature, pH, and the presence of cosolvents (Schein 1990). Proteins and polypeptides are generally least soluble at pH values near their isoelectric point (pI), where the overall net charge is zero (Riès-Kraut and Ducruix 1997).
The purpose of this study is to investigate the relationship between stability, solubility, and fibril formation using ribonuclease Sa (RNase Sa) from Streptomyces aureofaciens as a model system. RNase Sa is a small acidic protein (96 residues, pI = 3.5) with a mixed α + β structure. There are no lysine residues, and the protein contains a single disulfide bond (Shaw et al. 2001). We have recently made variants that replaced solvent-exposed acidic residues with lysine residues (Shaw et al. 2001). The mutant with three such charge reversals is denoted 3K, and that with five charge reversals is 5K. By reversing the charge at these sites, we have changed the pI of RNase Sa from 3.5 to 6.4 (3K) or 10.2 (5K; Shaw et al. 2001). Thus, we are able, here, to study the relationship between stability, solubility, and fibril formation in a single protein over an exceptionally wide pH range.
Results and Discussion
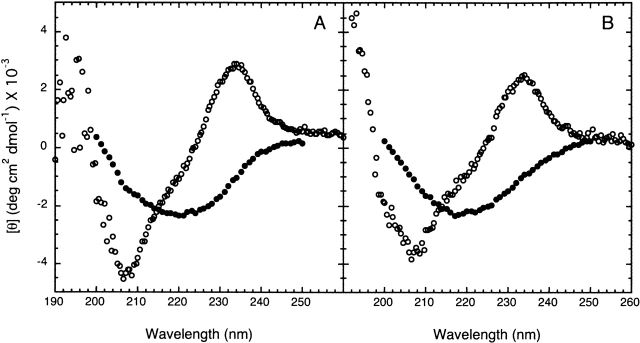
We have investigated the propensity of RNase Sa and two variants (3K and 5K) to form fibrils in the presence of TFE and as a function of pH. Figure 1 ▶ shows the CD spectra for RNase Sa in water and 30% TFE. The presence of TFE induces significant β-structure in the protein at both pH 7 and 3.5. As shown previously, the formation of β-structure appears to be a hallmark of amyloid formation (Kallberg et al. 2001), and it is clear that 30% TFE induces substantial β-structure in RNase Sa.
Figure 1.
Far-UV CD spectra for RNase Sa. Representative spectra are shown for the protein in 0% TFE (open circles) or 30% TFE (filled circles) at pH 7 (A) and pH 3.5 (B). The spectra were obtained after 5-h incubation at 25°C in 10 mM (citrate, phosphate, and borate) buffer. The presence of TFE clearly induces a substantial amount of β-structure in RNase Sa at both pH values.
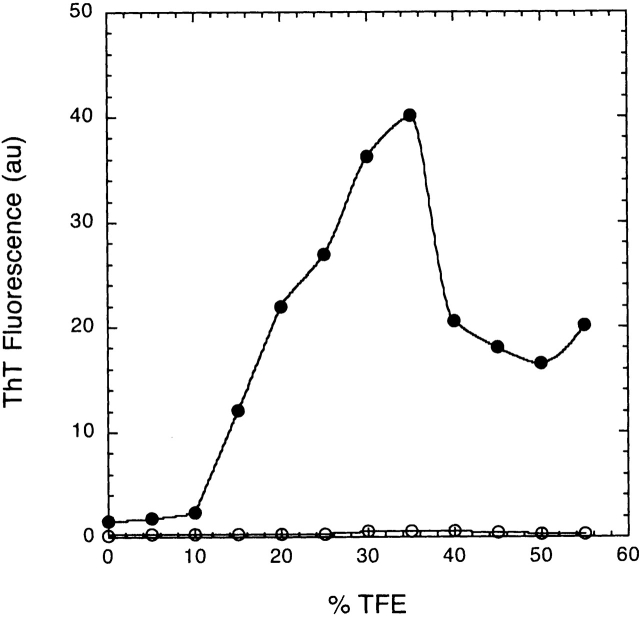
The enhancement in the fluorescence intensity of Thioflavin T (ThT) upon binding to ordered protein aggregates is one method to show the presence of fibril formation (Chiti et al. 2000). Figure 2 ▶ shows ThT fluorescence as a function of TFE concentration for wild-type RNase Sa. At pH 5.5, no fluorescence is observed regardless of TFE concentration. At pH 3.5, the fluorescence increases by ≈15- to 20-fold from 10% TFE to 35% TFE. Above 35% TFE, ThT fluorescence drops markedly, and CD spectra of the samples show a change from predominantly β-structure to substantial α-helical structure (data not shown). This change in secondary structure at high TFE concentrations has also been observed with other proteins (Chiti et al. 2000).
Figure 2.
Fibril formation (monitored by ThT fluorescence) as a function of TFE concentration. The curves represent the changes of fluorescence with TFE concentrations at pH 3.5 (filled circles) and pH 5.5 (open circles) after 18 h of incubation. ThT binding was monitored after removing an aliquot of the aggregation mixture (10 μL) and diluting it with 990 μL of 50 mM sodium phosphate, pH 6.0, containing 3 μM ThT. Fluorescence intensity was determined at 485 nm after excitation at 440 nm using a 96-well plate reader equipped with cutoff filters as previously described (Wood et al. 1996).
When RNase Sa in placed in 30% TFE at pH 3.5, ThT fluorescence was not observed for the first 2–3 h. After 6 h, the fluorescence intensity reached a maximum and did not change over a period of several days, at which time fibril formation was clearly evident. The isolated fibrils have the same morphology, dye-binding properties, nucleation-dependent kinetics, and ultrastructure as those found for a number of other proteins. The figure in the supplemental material shows electron micrographs of the fibrils formed from RNase Sa. In contrast, at pH 5.5 in 30% TFE, no ThT fluorescence was observed and no fibrils formed, even after several weeks of incubation.
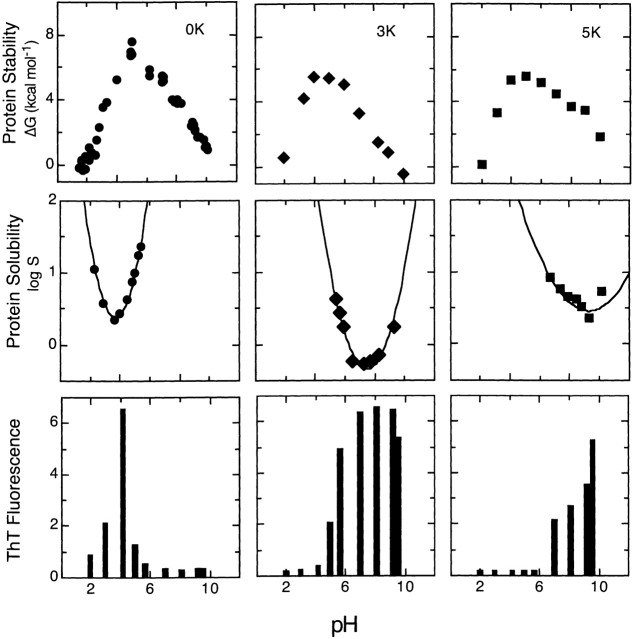
The results shown in Figure 2 ▶ led us to study the effects of pH on fibril formation with our charge-reversal variants. RNase Sa, 3K, and 5K were incubated in solutions of 30% TFE ranging in pH from 2 to 9.6. Figure 3 ▶ shows the correlation between fibril formation, conformational stability, and solubility for these proteins as a function of pH. All three variants are maximally stable near pH 5, but the minimum solubility of the proteins shifts with their pI, from near pH 3.5 for RNase Sa to pH > 9 for the 5K variant (Shaw et al. 2001). Fibril formation is most prominent at the pH where solubilty is minimal, that is, near the pI of the protein. These results show that fibril formation correlates with the pH-dependence of the protein solubility and not with conformational stability for all these RNase Sa variants. These results very strongly show that protein solubility is a major factor in fibril formation.
Figure 3.
The conformational stability, solubility, and fibril formation are shown as a function of pH for wild-type RNase Sa, and the 3K and 5K variants. The proteins were purified as described previously (Hebert et al. 1997), and the pH dependence of stability, the pI determination, and solubility have been described in more detail in an earlier report (Shaw et al. 2001). The solubility data are expressed as logS, where S is the solubility in mg mL−1. The lines through the data are meant only to guide the eye. The amyloid formation data were obtained from the ThT fluorescence assay as described in Figure 2 ▶. The samples were incubated at room temperature in 30% TFE at the indicated pH before measuring ThT fluorescence.
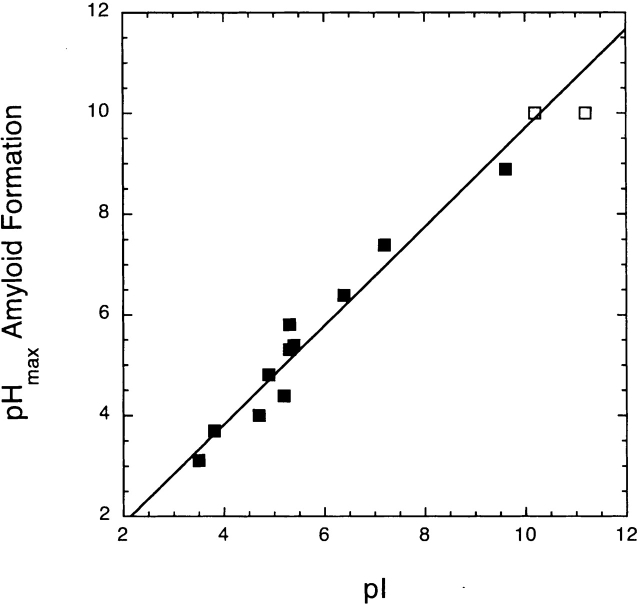
We have measured the pH dependence of fibril formation for a number of other proteins and summarized data from other studies. Table 1 shows the pH values and the maximum conformational stability, pI, and the pH of maximum fibril formation for these proteins. Figure 4 ▶ shows the correlation between the pH of maximal amyloid formation and the pI of the protein. The agreement between pI and pH of maximal fibril formation (pHmax) is excellent, whereas there is little correlation between the pH dependence of fibril formation and conformational stability. The data for α-synuclein, transthyretin, and the Aβ peptide are especially significant, since these proteins are known to form amyloid fibrils in vivo (Wood et al. 1996; Jiang et al. 2001; Hoyer et al. 2002).
Table 1.
The relationship between the pH-dependence of stability, pI, and the pH of maximum fibril formation for several proteins
| Protein | ΔGmax (pH)i (kcal mole−1) | ΔGpij (kcal mole−1) | pIk | pHmaxl |
| RNase Saa,g | 7.0 (5.0) | 4.0 | 3.5 | 3.1 |
| RNase T1b | 8.8 (4.5) | 8.0 | 3.8 | 3.7 |
| α-synucleinc | NA | NA | 4.7 | 4.0 |
| bsHPrd | 4.5 (7.0) | 3.5 | 4.9 | 4.8 |
| TTRe | NA | NA | 5.2 | 4.4 |
| RNase Sa2a | 4.7 (4.5) | 4.2 | 5.3 | 5.8 |
| Aβ peptidef | — | — | 5.3 | 5.3 |
| ecHPrd | 4.5 (7.0) | 3.5 | 5.4 | 5.4 |
| RNase Sa 3Kg | 5.5 (5.0) | 4.2 | 6.4 | 6.4 |
| RNase Sa3a | 8.9 (5.5) | 4.0 | 7.2 | 7.4 |
| RNase Ab | 9.2 (7.0) | 9.0 | 9.6 | 8.9 |
| RNase Sa 5Kg | 5.5 (5.0) | 2.0 | 10.2 | >10 |
| HEWLh | NA | NA | 11.2 | >10 |
a Stability and pI values are from Pace et al. (1998) and the pH of maximum fibril formation was determined as described in the text.
b Stability and pI values are from Pace et al. (1990) and the pH of maximum fibril formation was determined as described in the text.
c The pI was calculated as described in Shaw et al. (2001), and the pH of maximum fibril formation is from Hoyer et al. (2002). There are no entries for stability, because this is not a globular protein.
d Unpublished results from this laboratory (J.P.S. and J.M.S.).
e The pI was calculated as described in Shaw et al. (2001), and pH of maximum fibril formation is from Jiang et al. (2001).
f The pI and pH of maximum amyloid formation are from Wood et al. (1996).
g Stability and pI values are from Shaw et al. (2001), and the pH of maximum fibril formation are from Figure 3 ▶.
h Stability and pI are from Pfeil and Privalov (1976), and the pH of maximum stability was determined as described in the text.
i ΔGmax (pH) is the maximum conformational stability, which occurs at the indicated pH, expressed in kcal mole−1.
j ΔGpI is the conformational stability at pH = pI for the indicated protein, expressed in kcal mole−1.
k The pI value for the indicated protein, either measured or calculated as indicated.
l pHmax is the pH where fibril formation is maximal, based on the ThT assay described in the text.
Figure 4.
The correlation between the pH of maximal amyloid formation and the pI of the proteins listed in Table 1. The open squares represent the 5K variant of RNase Sa and hen egg white lysozyme (HEWL). These two proteins have pHmax values greater than 10, and could not be determined. The correlation coefficient for the 11 proteins shown as solid squares is 0.98.
In conclusion, our results suggest that the solubility of a polypeptide chain is a major factor that determines the in vitro conversion of globular proteins into amyloid fibrils under nonpathological conditions and may contribute to fibril formation in vivo. The results also suggest that increasing the solubility of a protein should prevent the formation of fibrils. We are in the process of testing this hypothesis with RNase Sa and other proteins.
Acknowledgments
This work was supported by grants from the NIH (GM 52483) and from the Robert A. Welch Foundation (BE-1281). J.M.S. is an Established Investigator of the American Heart Association. We thank Nick Pace and his group for providing some of the proteins used in this study and for helpful discussions; Christos Savva of the Texas A&M University Microscopy and Imaging Center for help obtaining the EM images; and Gerald Grimsley for critical comments on the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material: See www.proteinscience.org
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03152903.
References
- Buck, M. 1998. Trifluoroethanol and colleagues: Cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31 297–355. [DOI] [PubMed] [Google Scholar]
- Chiti, F., Webster, P., Taddei, N., Clark, A., Stefani, M., Ramponi, G., and Dobson, C.M. 1999. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. 96 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti, F., Taddei, N., Bucciantini, M., White, P., Ramponi, G., and Dobson, C.M. 2000. Mutational analysis of the propensity for amyloid formation by a globular protein. EMBO J. 19 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, A.S. 1967a. Amyloidosis. N. Engl. J. Med. 277 522–530. [DOI] [PubMed] [Google Scholar]
- ———. 1967b. Amyloidosis (concluded). N. Engl. J. Med. 277 628–638. [DOI] [PubMed] [Google Scholar]
- Dobson, C.M. 1999. Protein misfolding, evolution and disease. Trends Biochem. Sci. 24 329–332. [DOI] [PubMed] [Google Scholar]
- ———. 2001. The structural basis of protein folding and its links with human disease. Philos. Trans. R. Soc. Lond. B 356 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fändrich, M., Fletcher, M.A., and Dobson, C.M. 2001. Amyloid fibrils from muscle myoglobin. Nature 410 165–166. [DOI] [PubMed] [Google Scholar]
- Glenner, G.G. 1980a. Amyloid deposits and amyloidosis. The β-fibrilloses (first of two parts). N. Engl. J. Med. 302 1283–1292. [DOI] [PubMed] [Google Scholar]
- ———. 1980b. Amyloid deposits and amyloidosis: The β-fibrilloses (second of two parts). N. Engl. J. Med. 302 1333–1343. [DOI] [PubMed] [Google Scholar]
- Hebert, E.J., Grimsley, G.R., Hartley, R.W., Horn, G., Schell, D., Garcia, S., Both, V., Sevcik, J., and Pace, C.N. 1997. Purification of ribonucleases Sa, Sa2, and Sa3 after expression in Escherichia coli. Protein Expr. Purif. 11 162–168. [DOI] [PubMed] [Google Scholar]
- Hoyer, W., Antony, T., Cherny, D., Heim, G., Jovin, T.M., and Subramaniam, V. 2002. Dependence of α-synuclein aggregate morphology on solution conditions. J. Mol. Biol. 322 383–393. [DOI] [PubMed] [Google Scholar]
- Jiang, X., Smith, C.S., Petrassi, H.M., Hammarstrom, P., White, J.T., Sacchettini, J.C., and Kelly, J.W. 2001. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry 40 11442–11452. [DOI] [PubMed] [Google Scholar]
- Kallberg, Y., Gustafsson, M., Persson, B., Thyberg, J., and Johansson, J. 2001. Prediction of amyloid fibril-forming proteins. J. Biol. Chem. 276 12945–12950. [DOI] [PubMed] [Google Scholar]
- Kelly, J.W. 1996. Alternative conformations of amyloidogenic proteins govern their behavior. Curr. Opin. Struct. Biol. 6 11–17. [DOI] [PubMed] [Google Scholar]
- Khurana, R., Gillespie, J.R., Talapatra, A., Minert, L.J., Ionescu-Zanetti, C., Millett, I., and Fink, A.L. 2001a. Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates. Biochemistry 40 3525–3535. [DOI] [PubMed] [Google Scholar]
- Khurana, R., Uversky, V.N., Nielsen, L., and Fink, A.L. 2001b. Is Congo red an amyloid-specific dye? J. Biol. Chem. 276 22715–22721. [DOI] [PubMed] [Google Scholar]
- Krebs, M.R., Wilkins, D.K., Chung, E.W., Pitkeathly, M.C., Chamberlain, A.K., Zurdo, J., Robinson, C.V., and Dobson, C.M. 2000. Formation and seeding of amyloid fibrils from wild-type hen lysozyme and a peptide fragment from the β-domain. J. Mol. Biol. 300 541–549. [DOI] [PubMed] [Google Scholar]
- Lai, Z., Colon, W., and Kelly, J.W. 1996. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry 35 6470–6482. [DOI] [PubMed] [Google Scholar]
- Litvinovich, S.V., Brew, S.A., Aota, S., Akiyama, S.K., Haudenschild, C., and Ingham, K.C. 1998. Formation of amyloid-like fibrils by self-association of a partially unfolded fibronectin type III module. J. Mol. Biol. 280 245–258. [DOI] [PubMed] [Google Scholar]
- Luo, P. and Baldwin, R.L. 1997. Mechanism of helix induction by trifluoroethanol: A framework for extrapolating the helix-forming properties of peptides from trifluoroethanol:water mixtures back to water. Biochemistry 36 8413–8421. [DOI] [PubMed] [Google Scholar]
- Nielsen, L., Khurana, R., Coats, A., Frokjaer, S., Brange, J., Vyas, S., Uversky, V.N., and Fink, A.L. 2001. Effect of environmental factors on the kinetics of insulin fibril formation: Elucidation of the molecular mechanism. Biochemistry 40 6036–6046. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Laurents, D.V., and Thomson, J.A. 1990. pH dependence of the urea and guanidine hydrochloride denaturation of ribonuclease A and ribonuclease T1. Biochemistry 29 2564–2572. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Hebert, E.J., Shaw, K.L., Schell, D., Both, V., Krajcikova, D., Sevcik, J., Wilson, K.S., Dauter, Z., Hartley, R.W., et al. 1998. Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. J. Mol. Biol. 279 271–286. [DOI] [PubMed] [Google Scholar]
- Pepys, M.B., Hawkins, P.N., Booth, D.R., Vigushin, D.M., Tennent, G.A., Soutar, A.K., Totty, N., Nguyen, O., Blake, C.C., Terry, C.J., et al. 1993. Human lysozyme gene mutations cause hereditary systemic amyloidosis. Nature 362 553–557. [DOI] [PubMed] [Google Scholar]
- Pfeil, W. and Privalov, P.L. 1976. Thermodynamic investigations of proteins. I. Standard functions for proteins with lysozyme as an example. Biophys. Chem. 4 23–32. [DOI] [PubMed] [Google Scholar]
- Ramirez-Alvarado, M., Merkel, J.S., and Regan, L. 2000. A systematic exploration of the influence of the protein stability on amyloid fibril formation in vitro. Proc. Natl. Acad. Sci. 97 8979–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riès-Kraut, M. and Ducruix, A. 1997. Inferences drawn from physicochemical studies of crystallogenesis and precrystalline state. Methods Enzymol. 276 23–59. [DOI] [PubMed] [Google Scholar]
- Rochet, J.C. and Lansbury Jr., P.T. 2000. Amyloid fibrillogenesis: Themes and variations. Curr. Opin. Struct. Biol. 10 60–68. [DOI] [PubMed] [Google Scholar]
- Schein, C.H. 1990. Solubility as a function of protein structure and solvent components. Biotechnology 8 308–317. [DOI] [PubMed] [Google Scholar]
- Shaw, K.L., Grimsley, G.R., Yakovlev, G.I., Makarov, A.A., and Pace, C.N. 2001. The effect of net charge on the solubility, activity, and stability of ribonuclease Sa. Protein Sci. 10 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souillac, P.O., Uversky, V.N., Millett, I.S., Khurana, R., Doniach, S., and Fink, A.L. 2002. Effect of association state and conformational stability on the kinetics of immunoglobulin light chain amyloid fibril formation at physiological pH. J. Biol. Chem. 277 12657–12665. [DOI] [PubMed] [Google Scholar]
- Taddei, N., Capanni, C., Chiti, F., Stefani, M., Dobson, C.M., and Ramponi, G. 2001. Folding and aggregation are selectively influenced by the conformational preferences of the α-helices of muscle acylphosphatase. J. Biol. Chem. 276 37149–37154. [DOI] [PubMed] [Google Scholar]
- Wetzel, R. 2002. Ideas of order for amyloid fibril structure. Structure 10 1031–1036. [DOI] [PubMed] [Google Scholar]
- Wood, S.J., Maleeff, B., Hart, T., and Wetzel, R. 1996. Physical, morphological and functional differences between ph 5.8 and 7.4 aggregates of the Alzheimer’s amyloid peptide Ab. J. Mol. Biol. 256 870–877. [DOI] [PubMed] [Google Scholar]