Abstract
The rapid binding of cytotoxic colicin E3 by its cognate immunity protein Im3 is essential in safeguarding the producing cell. The X-ray structure of the E3/Im3 complex shows that the Im3 molecule interfaces with both the C-terminal ribonuclease (RNase) domain and the N-terminal translocation domain of E3. The association and dissociation rates of the RNase domain and Im3 show drastically different sensitivities to ionic strength, as previously rationalized for electrostatically enhanced diffusion-limited protein–protein associations. Relative to binding to the RNase domain, binding to full-length E3 shows a comparable association rate but a significantly lower dissociation rate. This outcome is just what was anticipated by a theory for the binding of two linked domains to a protein. The E3/Im3 system thus provides a powerful paradigm for the interplay of theory and experiment.
Keywords: Protein, protein association, ribonuclease colicin, immunity protein, electrostatic rate enhancement, flexible linker
Colicin E3 is a muiltidomain protein antibiotic released by Escherichia coli, which kills susceptible competing bacteria that do not produce the same toxin. The killing is afforded by the ribonuclease activity of colicin E3 targeting the 16S ribosomal RNA. A cognate immunity protein Im3 provides protection for the producing cell against the ribonuclease activity by binding to colicin E3 (Masaki et al. 1991). The E3/Im3 association rate constant (ka) and dissociation rate constant (kd) may be delicately controlled for accomplishing the dual task of self protection and cytotoxic action. The effects of electrostatic interactions and bivalent binding on ka and kd have been predicted in previous theoretical models (aZhou 2001a, 2003). These predictions are now confirmed by recent experiments (Soelaiman et al. 2001; Walker et al. 2003).
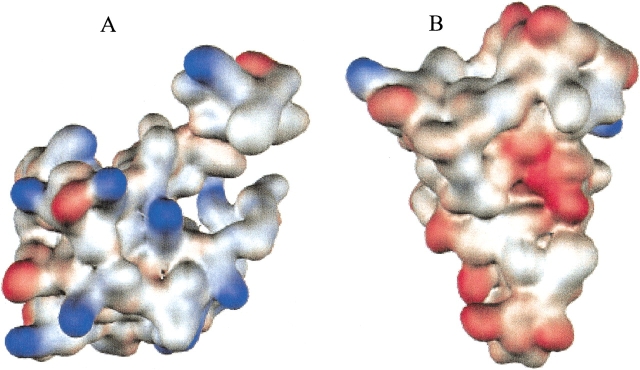
Colicin E3 consists of three domains (see Fig. 1 ▶): a translocation (T) domain f(residues 1–315), a receptor binding (R) domain (residues 316–450), and a ribonuclease (RNase) domain (residues 451–551; Soelaiman et al. 2001). Perhaps unexpectedly, Im3 was found to bind both the RNase and the T domains. The Im3/RNase domain interface, as observed in a previous structure for the complex with the truncated RNase domain (Carr et al. 2000), displays charge complementarity (see Fig. 2 ▶).
Figure 1.
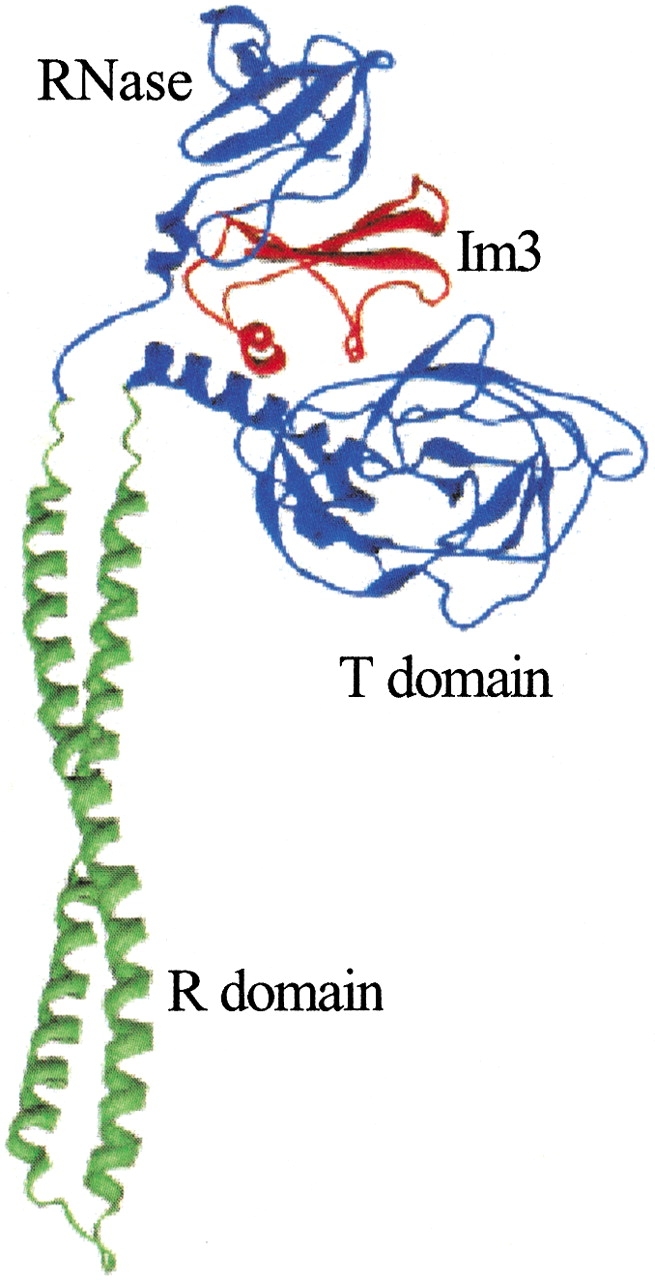
Structure of the colicin E3-Im3 complex (PDB entry 1jch).
Figure 2.
Complementarity of electrostatic potentials at the interface of the colicin E3 RNase domain–Im3 complex (PDB entry 1e44). Positive potential is shown as blue and negative potential is shown as red. (A) Colicin E3 RNase domain, which has mostly positive electrostatic potential facing Im3. (B) Im3, which has mostly negative potential facing the RNase domain.
Disparate ionic-strength dependences of ka and kd for RNase domain and Im3
For a protein complex that has charge complementarity in the interface, the association rate constant ka is expected to be enhanced by electrostatic interactions (Zhou 1993; Gabdoulline and Wade 1997, 2001; Elcock et al. 1999). Electrostatic rate enhancement has been extensively studied by Brownian dynamics simulations (Gabdoulline and Wade 1997, 2001; Elcock et al. 1999). Because of the stereospecificity of the complex and the long-range nature of electrostatic interactions, the electrostatic rate enhancement has been predicted to be given by Zhou (1993, 1997),
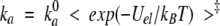 |
(1) |
where ka0 is the rate constant if the electrostatic interactions are turned off, Uel is the electrostatic interaction energy between the associating proteins, kBT is the product of the Boltzmann constant, and the absolute temperature, and the average 〈•••〉‡ is taken over the transition-state ensemble. The transition state for the association of proteins not involving significant conformational changes is expected to be close to the final stereospecific complex (Vijayakumar et al. 1998; Zhou 2001a).
Salt ions screen the electrostatic interactions between the associating proteins. Hence, Uel within the transition state (relative to the unbound state where the two proteins are isolated from each other) will be significantly weakened by an increase in ionic strength. The association rate is thus expected to show a significant decrease with ionic strength. On the other hand, as far as the effect of ionic strength on the dissociation rate constant is concerned, what is relevant is the difference in electrostatic interaction energy between the transition state and the stereospecific bound state. These two states are expected to be geometrically similar for relatively rigid proteins, and thus the effect of ionic strength on kd should be insignificant.
The disparate dependences of ka and kd on ionic strength have been recognized as a common feature of protein–protein associations that are electrostatically rate enhanced and diffusion limited (aZhou 2001a). Table 1 lists the values of ka and kd at low and high ionic strengths for the complexation of nine pairs of proteins, collected from the literature (with references cited within the table). The variations of ka with ionic strength are all much greater than those of kd. The kinetics of the colicin E3 RNase domain binding to Im3, observed recently by Walker et al. (2003), fits nicely into this pattern. When the ionic strength increased from 25 to 525 mM, the association rate constant decreased by three orders of magnitude, from 1.5 × 1010 M−1sec−1 down to 1.6 × 107 M−1sec−1. On the other hand, the dissociation rate constant showed a weak linear dependence on ionic strength, increasing from 1.5 × 10−4 sec−1 to 1.8 × 10−4 and 2.4 × 10−4 sec−1 as the ionic strength increased from 225 to 325 and 525 mM.
Table 1.
Effects of ionic strength on association and dissociation rate constants
| Protein complex | Ionic strength (mM) | ka (107M−1s−1) | kd (s−1) | Reference |
| Barnase/barstar | 25 | 60 | 8 × 10−6 | Schreiber and Fersht 1993 |
| 525 | 1.6 | 40 × 10−6 | ||
| ShB/peptide toxin Lq2 | 25 | 57 | 0.19 | Escobar et al. 1993 |
| 200 | 1.4 | 1.1 | ||
| ShB/N-terminal peptide | 50 | 2 | 15 | Murrell-Lagnado and Aldrich 1993 |
| 600 | 0.1 | 15 | ||
| Colicin E9 DNase/Im9 | 25 | 570 | 0.41 × 10−6 | Wallis et al. 1995 |
| 275 | 5 | 2.5 × 10−6 | ||
| Interleukin 4/IL-4BP | 50 | 2.7 | 1.5 × 10−3 | Shen et al. 1996 |
| 1000 | 0.3 | 2.3 × 10−3 | ||
| Heterodimeric leucine zipper | 74 | 7.2 | 0.38 × 10−3 | Wendt et al. 1997 |
| 525 | 0.37 | 10−3 | ||
| AChE/fasciculin 2 | 10 | 230 | 10 × 10−3 | Radic et al. 1997 |
| 670 | 1.2 | 5 × 10−3 | ||
| Thrombin/thromomodulin | 100 | 1.5 | 0.026 | Baerga-Ortiz et al. 2000 |
| 250 | 0.18 | 0.062 | ||
| Erythropoietin/Epo receptor | 155 | 40 | 0.3 × 10−3 | Darling et al. 2002 |
| 1005 | 0.7 | 0.12 × 10−3 | ||
| Colicin E3 RNase/Im3 | 25 | 1500 | 0.09 × 10−2 a | Walker et al. 2003 |
| 525 | 1.6 | 0.24 × 10−3 |
a Linearly extrapolated from data at ionic strengths of 225, 325, and 525 mM. The dissociation rate constants typically show a weak linear dependence on ionic strengths (e.g., as seen for barnase/barstar, ShB/peptide toxin Lq2, and colicin E9 DNase domain/Im9).
Structural information provides further evidence that the association of the colicin E3 RNase domain and Im3 is electrostatically rate enhanced and diffusion limited. As shown in Figure 2 ▶, the electrostatic potentials on the two sides of the interface of the E3 RNase/Im3 complex show high complementarity, suggesting that the two proteins will have strong electrostatic attraction in the transition state for association. The immunity protein was found not to undergo any significant structural change upon binding with colicin E3 (Li et al. 1999; Soelaiman et al. 2001), likely allowing interprotein relative diffusion to be rate limiting for association. Electrostatic attraction and diffusion control provide optimization of the association rate constant. This optimization may be important for neutralizing the RNase activity of endogenous and exogenous colicin E3.
Difference of Im3-binding kinetics between RNase domain and full-length E3
Both the RNase and the T domains of colicin E3 interface with the bound immunity protein. Bivalency is a well-known mechanism for increasing binding affinity. Recently, a theoretical model has been developed to quantitatively account for the affinity enhancement by bivalency when the two binding domains (RNase and T in the case of colicin E3) are connected by a flexible peptide linker (Zhou 2001b). If the two domains separately have association constants KA and KB, the linked variant has an association constant,
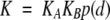 |
(2) |
where p(d) is the probability density for the end-to-end vector of the peptide liner and d is the magnitude of this vector in the bound state. The “effective concentration” p(d) typically is in the mM range; hence, KBp(d) is expected to be much greater than 1 and K much greater than KA.
The linker theory has now been extended to binding kinetics (Zhou 2003). If the two binding domains separately have association rate constants kA+ and kB+ and dissociation rate constants kA− and kB−, then the linked variant has association and dissociation rate constants:
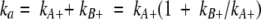 |
(3a) |
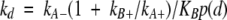 |
(3b) |
These expressions indicate that the affinity enhancement upon linking a second binding domain is primarily manifested as a slow down of the dissociation process.
When Walker et al. (2003) compared the Im3-binding kinetics of the RNase domain and intact colicin E3, they observed just what was predicted by the linker theory. ka of intact colicin E3 at an ionic strength of 225 mM was 5.5 × 107 M−1sec−1, comparable to the counterpart for the RNase domain, 1.1 × 108 M−1sec−1. On the other hand, kd for intact colicin E3 is over two orders of magnitude lower than for the RNase domain, decreasing from 1.5 × 10−4 to 7.6 × 10−7 sec−1.
The RNase and T domains of colicin E3 are connected by the R domain, which forms a coiled-coil, not a flexible peptide linker. However, there is evidence that the linker region (residues 447–454) between the R and RNase domains has a tendency of becoming disordered. In the structure of the truncated RNase domain (starting at residue 447) complexed with Im3, these eight residues are disordered. In addition, because Im3 is wedged between the RNase and T domains in the complex with intact colicin E3, that Im3 can dissociate at all, means that the two domains must open up transiently to let out Im3. Unfortunately, the flexibility of the connection between the RNase and T domains has not been characterized, so a quantitative application of equation 3 is not possible at the present time.
All colicins are known to have a three-domain architecture like colicin E3. It is interesting to compare the immunity protein-binding kinetics of colicin E3 and colicin E9, which has DNase activity. In contrast to the significant slow down in dissociation upon switching from the truncated RNase domain to intact colicin E3, the dissociation (as well as association) rates of the truncated DNase domain and intact colicin E9 are almost the same (Wallis et al. 1995). This dissimilarity suggests that Im9 interfaces solely with the DNase domain of colicin E9. Bivalent binding offers clear advantages to colicin E3: tight binding and slow dissociation of Im3 provide maximal protection against the nuclease activity inside the cell, yet upon binding to the receptor on a susceptible cell and subsequent translocation of the T domain, dissociation of Im3 from the RNase domain becomes relatively fast (Walker et al. 2003). Whether other colicins exploit this strategy and why colicins like E9 do not exploit this strategy remain to be investigated.
In summary, theoretical predictions on the effects of ionic strength and bivalent binding have both been confirmed by experiments on the binding of colicin E3 with its cognate immunity protein. The colicin E3/Im3 system has great potential for further quantitative interplay between theory and experiment on the roles of specific electrostatic interactions and flexibility of interdomain linkers.
Acknowledgments
This work was supported in part by NIH grant GM58187. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03216203.
References
- Baerga-Ortiz, A., Rezaie, A.R., and Komives, E.A. 2000. Electrostatic dependence of the thrombin–thrombomodulin interaction. J. Mol. Biol. 296 651–658. [DOI] [PubMed] [Google Scholar]
- Carr, S., Walker, D., James, R., Kleanthous, C., and Hemmings, A.M. 2000. Inhibition of a ribosome-inactivating ribonuclease: The crystal structure of the cytotoxic domain of colicin E3 in complex with its immunity protein. Struct. Fold. Des. 8 949–960. [DOI] [PubMed] [Google Scholar]
- Darling, R.J., Kuchibhotla, U., Glaesner, W., Micanovic, R., Witcher, D.R., and Beals, J.M. 2002. Glycosylation of erythropoietin affects receptor binding kinetics: Role of electrostatic interactions. Biochemistry 41 14524–14531. [DOI] [PubMed] [Google Scholar]
- Elcock, A.H., Gabdoulline, R.R., Wade, R.C., and McCammon, J.A. 1999. Computer simulation of protein–protein association kinetics: Acetylcholinesterase-fasciculin. J. Mol. Biol. 291 149–162. [DOI] [PubMed] [Google Scholar]
- Escobar, L., Root, M.J., and MacKinnon, R. 1993. Influence of protein surface charge on the bimolecular kinetics of a potassium channel peptide inhibitor. Biochemistry 32 6982–6987. [DOI] [PubMed] [Google Scholar]
- Gabdoulline, R.R. and Wade, R.C. 1997. Simulation of the diffusional association of barnase and barstar. Biophys. J. 72 1917–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Protein–protein association: Investigation of factors influencing association rates by Brownian dynamics simulations. J. Mol. Biol. 306 1139–1155. [DOI] [PubMed] [Google Scholar]
- Li, C., Zhao, D., Djebli, A., and Shoham, M. 1999. Crystal structure of colicin E3 immunity protein: An inhibitor of a ribosome inactivating RNase. Struct. Fold. Des. 7 1365–1378. [DOI] [PubMed] [Google Scholar]
- Masaki, H., Akutsu, A., Uozumi, T., and Ohta, T. 1991. Identification of a unique specificity determinant of the colicin E3 immunity protein. Gene 107 133–138. [DOI] [PubMed] [Google Scholar]
- Murrell-Lagnado, R.D. and Aldrich, R.W. 1993. Energetics of Shaker K channel’s block by inactivation peptides. J. Gen. Physiol. 102 977–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic, Z., Kirchhoff, P.D., Quinn, D.M., McCammon, J.A., and Taylor, P. 1997. Electrostatic influence on the kinetics of ligand binding to acetylcholinesterase. Distinctions between active center ligands and fasciculin. J. Biol. Chem. 272 23266–23277. [DOI] [PubMed] [Google Scholar]
- Schreiber, G. and Fersht, A.R. 1993. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry 32 5145–5150. [DOI] [PubMed] [Google Scholar]
- Shen, B.-J., Hage, T., and Sebald, W. 1996. Global and local determinants for the kinetics of interleukin-4/interleukin-4 receptor α chain interaction. A biosensor study employing recombinant interleukin-4-binding protein. Eur. J. Biochem. 240 252–261. [DOI] [PubMed] [Google Scholar]
- Soelaiman, S., Jakes, K., Wu, N., Li, C., and Shoham, M. 2001. Crystal structure of colicin E3: Implications for cell entry and ribosome inactivation. Mol. Cell 8 1053–1062. [DOI] [PubMed] [Google Scholar]
- Vijayakumar, M., Wong, K.-Y., Schreiber, G., Fersht, A.R., Szabo, A., and Zhou, H.-X. 1998. Electrostatic enhancement of diffusion-controlled protein–protein association: Comparison of theory and experiment on barnase and barstar. J. Mol. Biol. 278 1015–1024. [DOI] [PubMed] [Google Scholar]
- Walker, D., Moore, G.R., James, R., and Kleanthous, C. 2003. Thermodynamic consequences of bipartite immunity protein binding to the ribosomal ribonuclease colicin E3. Biochemistry 42 4161–4171. [DOI] [PubMed] [Google Scholar]
- Wallis, R., Moore, G.R., James, R., and Kleanthous, C. 1995. Protein–protein interactions in colicin E9 DNase-immunity protein complexes. 1. Diffusion-controlled association and femtomolar binding for the cognate complex. Biochemistry 34 13743–13750. [DOI] [PubMed] [Google Scholar]
- Wendt, H., Leder, L., Harma, H., Jelesarov, I., Baici, A., and Bosshard, H.R. 1997. Very rapid, ionic strength-dependent association and folding of a heterodimeric leucine zipper. Biochemistry 36 204–213. [DOI] [PubMed] [Google Scholar]
- Zhou, H.-X. 1993. Brownian dynamics study of the influences of electrostatic interaction and diffusion on protein–protein association kinetics. Biophys. J. 64 1711–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1997. Enhancement of protein–protein association rate by interaction potential: Accuracy of prediction based on local Boltzmann factor. Biophys. J. 73 2441–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— 2001a. Disparate ionic-strength dependence of on and off rates in protein–protein association. Biopolymers 59 427–433. [DOI] [PubMed] [Google Scholar]
- ———. 2001b. The affinity-enhancing roles of flexible linkers in two-domain DNA-binding proteins. Biochemistry 40 15069–15073. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Quantitative account of the enhanced affinity of two linked scFvs specific for different epitopes on the same antigen. J. Mol. Biol. 329 1–8. [DOI] [PubMed] [Google Scholar]