Abstract
The Hsp70 family of molecular chaperones participates in a number of cellular processes, including binding to nascent polypeptide chains and assistance in protein (re)folding and degradation. We present the solution structure of the substrate binding domain (residues 393–507) of the Escherichia coli Hsp70, DnaK, that is bound to the peptide NRLLLTG and compare it to the crystal structure of DnaK(389–607) bound to the same peptide. The construct discussed here does not contain the α-helical domain that characterizes earlier published peptide-bound structures of the Hsp70s. It is established that removing the α-helical domain in its entirety does not affect the primary interactions or structure of the DnaK(393–507) in complex with the peptide NRLLLTG. In particular, the arch that protects the substrate-binding cleft is also formed in the absence of the helical lid. 15N-relaxation measurements show that the peptide-bound form of DnaK(393–507) is relatively rigid. As compared to the peptide-free state, the peptide-bound state of the domain shows distinct, widespread, and contiguous differences in structure extending toward areas previously defined as important to the allosteric regulation of the Hsp70 chaperones.
Keywords: Hsp70, DnaK, allosteric, solution structure, dynamics, NMR
The Hsp70 family of molecular chaperones participates in a wide range of biological processes including regulation of the heat shock response, prevention and disruption of protein aggregates, translocation across membranes, and binding to nascent peptide chains to promote correct folding (Bukau and Horwich 1998). Included in this family are DnaK from Escherichia coli, the yeast proteins SSA and SSB (Ziegelhoffer et al. 1995), the mammalian cytosolic proteins Hsp70 and Hsc70 (Bukau and Horwich 1998), and BiP from the endoplasmic reticulum (Craig et al. 1993). The eukaryotic chaperones have functions similar to those of DnaK, but also assist in protein trafficking. The Hsp70 chaperones were initially identified as proteins that are overexpressed under stress conditions such as heat shock (e.g., Hsp70 and DnaK; Lindquist 1986), but they can also be constitutively produced (Hsc70; Bukau and Horwich 1998).
The Hsp70 proteins consist of three domains. Residues 1–392 constitute the 44-kD regulatory domain, which binds ADP and binds and hydrolyzes ATP. The 13-kD substrate-binding domain (residues 393–507) consists of a β-sandwich comprised of two antiparallel β-sheets of four strands each. The peptidic substrate-binding site, which lies between the two β-sheets, is hydrophobic in nature and contains a deep pocket with specificity for hydrophobic residues, particularly leucine (Richarme and Kohiyama 1993; Gragerov et al. 1994; Zhu et al. 1996; Rüdiger et al. 1997a,b; Wang et al. 1998; Morshauser et al. 1999). The substrate binding domain is followed by a less conserved 10-kD α-helical domain, which lies like a lid over the substrate-binding cleft. Intrinsic to the function of the Hsp70s is the allosteric control of substrate binding by the 44-kD regulatory domain (McCarty et al. 1995). Peptidic substrate is bound with high affinity in the ADP-bound state, whereas ATP binding decreases affinity for substrate by approximately two orders of magnitude. Conversely, substrate binding stimulates the hydrolysis of ATP. In vivo, nucleotide binding and release by DnaK is controlled in part by two cofactors, GrpE, which enhances the exchange of nucleotides, and the cochaperone DnaJ, which stimulates substrate binding and ATP hydrolysis (Misselwitz et al. 1998; Pierpaoli et al. 1998).
Despite extensive structural and biochemical studies, the fundamental question of how nucleotide binding modulates substrate binding affinity remains largely unanswered. Previous studies have shown the C-terminal α-helical domain to be important in the kinetics of peptide binding. In particular, studies on constructs in which most or all of the α-helical domain has been removed reveal an increase in the peptide dissociation rate, but the association rate remains largely unchanged (Buczynski et al. 2001). Structural data have suggested a model in which the α-helical domain controls access to the peptide-binding site by moving, perhaps in its entirety (Morshauser et al. 1999). These observations suggested that communication from the ATPase domain may occur through the region containing residues 502–538, thus mediating the allosteric response to nucleotide state. However, this cannot be the extent of allosteric regulation of substrate binding as constructs of both DnaK and an Hsp 70 homolog BiP, in which the α-helical domains have been partially or entirely removed, continue to be at least modestly functional in vivo and in vitro. In particular, an allosteric response to nucleotide state is still observed for DnaK(1–517) and DnaK(1–507) (Pellecchia et al. 2000; Buczynski et al. 2001). Therefore, it appears that the α-helical domain is not solely responsible for the allosteric control of substrate binding.
In this report, we provide a detailed comparison between the solution structure of the substrate binding domain (β-domain, DnaK[393–507]) in complex with the peptide NRLLLTG with the crystal structure of the substrate-binding unit DnaK(389–607)in complex with the same peptide (Zhu et al. 1996) and with the solution structure of the substrate-binding domain (β-domain, DnaK[393–507]) in absence of the peptide (Pellecchia et al. 2000). Through this comparison, we have firmly established that the α-helical domain is not required for peptide recognition and binding. Moreover, we show, unexpectedly, that the arch that protects the substrate-binding cleft is also formed in the absence of the helical lid when peptide is present. 15N-relaxation measurements show that the peptide-bound form of DnaK(393–507) is relatively rigid throughout. The study affords further insight in the global changes in structure and dynamics of the β-domain as caused by peptide binding, in relation to allosteric regulation in the Hsp70 chaperone family.
Results and Discussion
Structure
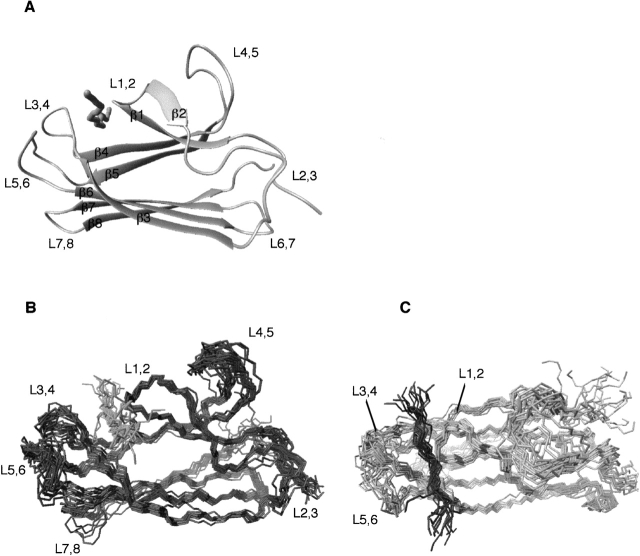
The three-dimensional (3D) solution structure of the β-domain (DnaK[393–507]) in complex with the peptide NRLLLTG was obtained by multinuclear, multidimensional NMR spectroscopy (Fig. 1A ▶). The fold consists of a two-layer β-sandwich, each sheet containing four antiparallel strands consisting of strands β3, β6, β7, and β8 and an irregular sheet containing strands β5, β4, β1, and β2. The peptide-bound form of the β-domain is monomeric in solution as indicated by 15N NMR relaxation experiments, from which a correlation time of 7.7 ns was calculated. The final ensemble of 20 structures was calculated with the program ARIA (Linge et al. 2001) using 2147 unambiguous NOE restraints, and 120 dihedral and 92 hydrogen-bonding restraints. Superposition of the secondary structural elements of the 15 lowest energy structures showed a root mean squared deviation (r.m.s.d.) of ∼0.57 Å for the backbone atoms of the secondary structure elements. Analysis of the ensemble of the complex structures by PROCHECK (Laskowski et al. 1993) revealed that, on average, 93.3% of the backbone angles lay in regions of Ramachandran space classified as favorable/allowed with 5.3% classified as generously allowed (see Table 1 for statistics).
Figure 1.
(A) Ribbon diagram of the average structure of the DnaK β-domain (DnaK[383–507]) bound to the substrate peptide NRLLLTG. (B,C) Twenty energy-minimized conformers representing the solution structure of β-domain (DnaK393–507) bound to the peptide NRLLLTG are depicted. The conformers were superimposed using the secondary structure elements. The two views are rotated by 90°.
Table 1.
NMR statistics of DnaK(393–507) + NRLLLTG
| Restraints | |
| NOE distance restraints | 2147 |
| Intraresidue | 1208 |
| Short range (≤i, i + 4) | 390 |
| Long range | 507 |
| Intermolecular | 42 |
| Hydrogen bond restraintsa | 92 |
| Dihedral restraintsb | |
| φ | 60 |
| ξ | 60 |
| Structure statistics | |
| Average total energy (kcal) | 460 ± 125 |
| NOE violationsc (Å) | 0.0076 ± 0.0022 |
| NOE violations >0.5 Å | 4.5 ± 2.5 |
| Dihedral violations (>5°, 2° − 5°) | 0, 4.3 ± 1.8 |
| PROCHECK Ramachandrand (% favorable, generously allowed, disallowed) | 93.3, 5.3, 1.4 |
| R.m.s.d. to averaged coordinates (Å) | |
| Backbone (excluding peptide)e | 1.07 |
| Secondary structure elements,f backbone | 0.57 |
| Secondary structure elements,f all atoms | 1.33 |
a Two restraints were included per hydrogen bond for linearity.
b Derived from Chemical Shift Index.
c Average NOE violation divided by total number of NOE restraints.
d The average statistics from the 15 lowest energy structures determined with the software PROCHECK NMR (Laskowski et al. 1993).
e Sequence including residues 395–502.
f Sequence including residues 398–403, 407–412, 420–423, 435–442, 452–460, 472–478, 484–489, and 496–501.
Comparison with the crystal structure
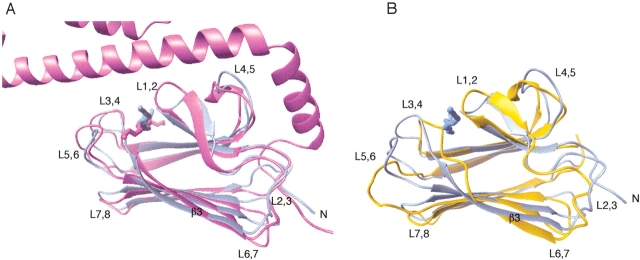
The solution structure of DnaK(393–507) in complex with the peptide NRLLLTG was found to be essentially the same as the corresponding subdomain in the crystal structure of the longer construct DnaK(389–607) in complex with the same peptide (Zhu et al. 1996). The r.m.s.d value comparing the two structures is 1.66 Å for residues 394–502 and 1.07 Å when aligning the secondary structure elements (Fig. 2A ▶). However, there are a few subtle differences between the structures that merit discussion.
Figure 2.
(A) Comparison of the average solution structure of DnaK393–507 (blue) and the crystal structure of DnaK389–607 (PDB code 1DKZ; violet) in complex with the peptide NRLLLTG. The molecules were aligned based on the backbone atoms of the β-strands. The r.m.s.d value for these elements is 1.07 Å. (B) Comparison of the average solution structure of DnaK393–507 in complex with the peptide NRLLLTG (blue) and the average solution structure of DnaK393–507 without peptide (PDB code 1DG4; orange). The molecules were aligned based on the backbone atoms of the β-strands. The r.m.s.d value for these elements is 1.70 Å.
The crystal structure of DnaK(389–607) indicated the presence of salt bridges between the α-helical domain and Arg 445 and Lys 446 of loop L4,5 and between the α-helical domain and Val 407 and Met 404 of loop L1,2. These salt bridges were expected to stabilize the structure of these loops (Zhu et al. 1996; Wang et al. 1998). In the present solution structure, the conformation of loop L4,5 is defined with limited precision. However, its average conformation is, even without the helix, virtually identical to that in the crystal, arguing that the stabilization of loop L4,5 by these interactions is not critical.
Loops L1,2 and L3,4, flanking the bound peptide, are in solution equally well defined as the core of the protein (Fig. 1B,C). This is most probably due to the numerous NOEs that report on interaction with the substrate. Loop L1,2 is located in a slightly shifted position, even though the interactions with the peptide are similar to those observed in the crystal structure (Fig. 2A ▶). The position of loop L3,4 at the opposite side of the peptide-binding cleft coincides with the position in the crystal structure. Loops L5,6 and L7,8 are among the least defined areas of the protein in solution. Nevertheless, they still retain an average structure very close to that observed in the crystal structure. A salt bridge between Arg 467 of L5,6 and residue Asp 540 of the α-helical domain was observed in the crystal structure of DnaK(389–607) (Zhu et al. 1996). Our results thus suggest that the absence of this interaction does not influence the average conformation of the loop. Figure 2A ▶ shows that the average conformation adopted by the loop L2,3 is very similar in the solution and crystal structures. Therefore the structure appears to be inherent to the β-domain itself, and is not induced by interactions with nor is influenced by movement of the α-helical domain. In addition, the N terminus of the β-domain appears to be relatively structured in solution (see Fig. 1 ▶) and corresponds to the position in the crystal structure (Fig. 2A ▶). These areas are thought to be very important to the allosteric mechanism of the Hsp70 proteins.
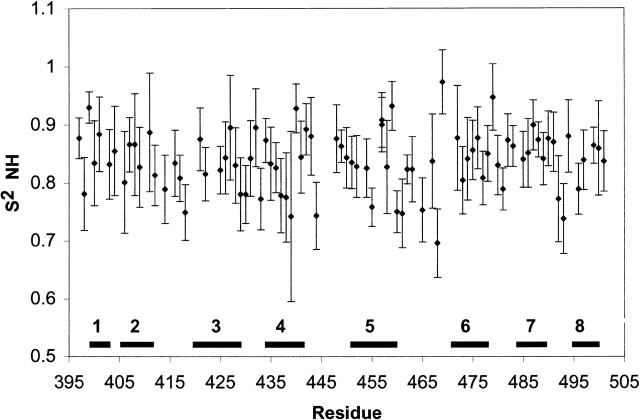
15N relaxation measurements were obtained for DnaK(393–507). The derived order parameters for the peptide-bound form fluctuate somewhat over the sequence, but not in a manner that clearly delineates the secondary structure elements and loops (Fig. 3 ▶). The exceptions are residues 492 and 493 in loop L7,8, residue 468 in loop L5,6, and residue 444 in loop L4,5, which show somewhat reduced order due to motions on the nanosecond–picosecond time scale. The other loop areas are found to be immobile on the nanosecond–picosecond time scale. The relaxation data for most residues could be fitted with models 1 or 2 from the Modelfree program (Mandel et al. 1995), indicating overall absence of conformational exchange under CPMG refocusing conditions. The exception is residue 468, for which a statistically significant conformational exchange broadening of 2 s−1 was derived (see Electronic Supplementary Material). No differences were obtained for R2 rates obtained with or without application of CPMG refocusing (results not shown), from which we conclude that the loops, and indeed the entire protein, do not display pervasive dynamics on the millisecond time scale either (Stevens et al. 2001). The combination of NMR structural and dynamical data thus indicates that the peptide-bound β-domain is a relatively rigid structure that does not require the α helix for its integrity.
Figure 3.
Amide NH vector order parameters derived from Modelfree analysis (Mandel et al. 1995) of R1 and R2CPMG 15N-spin relaxation data and 1H → 15N NOE. The secondary structure elements are indicated. Details are listed in the Electronic Supplemental Material.
The peptide binding
Binding of the peptide NRLLLTG to the DnaK(393–507) was followed by 15N, 1H, and 13C, 1H correlation spectroscopy. Binding kinetics is slow on the NMR time scale as indicated by the disappearance of the NMR resonances corresponding to the ligand free state and the appearance of new resonances representative of the ligand bound state. The Kd from this titration was estimated to be 600 ± 200 μM (Pellecchia et al. 2000), which is approximately 30-fold higher than that observed for wild-type DnaK (24 μM; Pierpaoli et al. 1998).
In the current work, 42 unambiguous intermolecular NOEs were observed between the peptide and the β-domain, all but three involving the three central leucines in the peptide. As with the DnaK(389–607) crystal structure, the highest concentration of intermolecular interactions involved Leu 4 of the peptide. This residue is fixed in a conserved subpocket of the binding cleft as defined by 20 NOEs with residues in strands β1, β3, β5, β6, and β7. The peptide could be oriented in the binding site due to several NOEs between the protein and Arg 2 of the peptide. It was found to be the same as observed in the crystal structure and previous NMR structures of chaperone domains that bound to their own C terminus (Wang et al. 1998; Morshauser et al. 1999).
The current structure is the first view of a solution structure of the protein with an intermolecularly bound peptide. Only the three center residues of peptide are well defined by NOEs (Fig. 1C ▶). This suggests that the structural "footprint" of Hsc70 chaperones is limited to the center of the peptide, corresponding to the central area of 3–4 residues that convey specificity as defined by systematic biochemical studies (Rüdiger et al. 1997a). Indeed, the B-factors for the peptide in the crystal structure of DnaK(389–607) increase from the center of the peptide (16 Å2) to the termini (40–60 Å2). According to the Debye–Waller equation (Creighton 1993), these factors could correspond to 0.8 Å and 1.5 Å r.m.s. fluctuations respectively, indicating increased mobility of the peptide termini in the crystal as well. We can thus conclude that the α-helical domain is not necessary for the peptide-binding function of DnaK.
Comparison with the ligand-free form: Inferences for the allosteric mechanism
15N relaxation rates were measured for both the ligand-free (results not shown) and peptide-bound forms of the β-domain (Fig. 3 ▶, Electronic Supplemental Material). Data analysis yields a rotational correlation time of 7.7 ns for the peptide-bound form and 14 ns for the apo form. Together with translational diffusion NMR experiments and light-scattering experiments (results not shown), this indicates that the apo form of the protein is dimeric and that ligand binding causes the dimer to dissociate into monomers. The average (one-dimensional [1D]) 15N relaxation rates and the two-dimensional (2D) 15N-HSQC spectrum of the apo form do not change in the protein concentration range of 10–500 μM, indicating that the dimerization is very tight with a dissociation constant not larger than 1 μM. It has been reported by several workers that Hsp70 proteins are prone to aggregation in the absence of substrate (Blond-Elguindi et al. 1993; Benaroudj et al. 1995; Burkholder et al. 1996). This is generally assumed to be mediated by the substrate-binding domain. However, the substrate-binding cleft of apo DnaK(393–507) itself is not directly involved in dimerization to the best of our knowledge; that is, the cleft contains water (Cai et al. 2003), but is otherwise empty, as evidenced by a lack of NOEs indicating otherwise (Pellecchia et al. 2000). The dimerization interface is not yet known and is subject of further investigations. We do point out that corresponding DnaK constructs including the nucleotide-binding domain but lacking part or all of the α-helical domain are functional (DnaK[1–517], Buczynski et al. 2001; DnaK[1–507], Pellecchia et al. 2000). Part of the structural differences between the apo form and peptide-bound form described below might thus be due to the substrate-linked dimer–monomer equilibrium. Although a monomer–dimer equilibrium has been implicated in the activity of Hsp70s in general (Blond-Elguindi et al. 1993; Benaroudj et al. 1995), at this time we cannot be certain that the dimer observed with this construct represents behavior reported for full-length chaperones.
In the ligand-free β-domain, there are several significant differences in structure as compared to the structure of the ligand-bound form (Fig. 2B ▶). In absence of ligand, strand β3 can only be defined through His 422 because the HN resonances of residues Gln 424-Ser 427 could not be detected, likely due to exchange broadening. Our current data show that, upon complexation with the peptide substrate, the resonances reappear in conjunction with the diagnostic NOEs that indicate that β3 participates in the lower β-sheet structure through residue Ser 427, corresponding to the crystal structure. These observations are confirmed by analysis with the Chemical Shift Index (data not shown), which also predicts a β-strand for β3 through Ser 427.
A hydrophobic arch over the substrate-binding site is present throughout the Hsp70 family (Zhu et al. 1996; Rüdiger et al. 2000). A closed arch is proposed to be a characteristic of the ADP form of the molecular chaperones, hindering peptide release and affording an apparent increase in affinity for peptide. In the case of DnaK, this arch is formed by residues Met 404 and Ala 429. In the ligand-free β-domain, Met 404 from loop L1,2 and Thr 428 and Ala 429 of loop L3,4 have shifted to partially occlude the hydrophobic pocket of the binding site, rendering it less accessible to substrate (Pellecchia et al. 2000). Therefore, the peptide must compete for the interaction between the loops L1,2 and L3,4 to make the binding site accessible. Our current study shows that the disruption of the arch in the apo form of the protein can be reversed by substrate binding as evidenced by multiple NOEs between the two arch residues. Thus, the conserved arch structure can also be formed in the absence of the helical lid. This important finding confirms that the arch plays a key role, independent of the α-helical domain, in the regulation of substrate binding and release.
The HN resonances of residues Thr 417 and Ile 418 are exchange broadened in the ligand-free β-domain, suggesting that loop L2,3 is conformationally flexible in this state (Pellecchia et al. 2000). Upon binding the peptide substrate in the spatially remote binding cleft, the line broadening is quenched, and loop L2,3 is found to adopt a structure that is essentially identical to that observed for the DnaK(389–607) (Fig. 2A,B). In the peptide-bound form, loop L2,3 is not very dynamic at the nanosecond–picosecond time scale either (Fig. 3 ▶). We conclude that the dynamic behavior of this region in the ligand-free form is due not to the lack of the α-helical domain, but to the absence of ligand.
A network of NOEs is observed between Ile 478 of strand β6 and residues contained in loop L2,3 in both forms of the molecule. However, the NOEs observed are different for the two ligation states. In the ligand-free β-domain, NOEs are observed to the backbone of Thr 416 and Thr 417 from the side chain of Ile 478. These NOEs are absent in the peptide bound form of the molecule. In the latter state, a set of NOEs between the side chain of Ile 418, the residue that is exchange broadened in the 1HN dimension, and the backbone of Ile 478 is observed. This change in observed contacts indicates that peptide binding induces a substantial change in conformation (Fig. 2B ▶) in addition to the previously reported change in dynamics in this area.
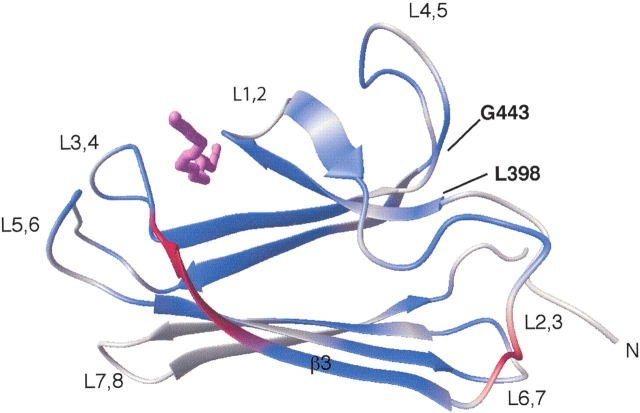
Chemical shift changes upon substrate binding occur throughout the molecule and extend from the substrate-binding cleft toward the area of loop L2,3 (Fig. 4 ▶). The fact that many of these changes are observed in the core of the protein is suggestive of a pathway that can transmit the conformational changes from cleft to loop. Limitations in the precision of the structure determinations in these intervening areas preclude the identification of the (subtle) concomitant structural changes. Some of the chemical shifts observed are likely due to dimer dissociation. Until the interface has been characterized fully, we cannot ascertain the biological relevance of the dimer formation or its role in the allostery of DnaK. Therefore, it is not possible to relate the observed chemical shifts and structural and dynamical changes exclusively with the allosteric mechanism. However, mutagenesis studies have also identified loop L2,3 to be important for the allosteric regulation of substrate binding by the nucleotide-binding domain. Mutating Pro 419 to serine produced a molecule severely deficient in allosteric behavior (Burkholder et al. 1996; Vosine et al. 1999). Changing Lys 414 of DnaK to isoleucine afforded a lower peptide-binding affinity and an increase in ATP hydrolysis (Montgomery et al. 1999) that was consistent with a role of this residue in the allosteric response of the molecule (Buchberger et al. 1995). These observations suggest that our findings of conformational and dynamical changes in loop L2,3 in response to the peptide binding may be of relevance to the allosteric process.
Figure 4.
Chemical shift differences observed upon titration of NRLLLTG form a path from loop L2,3 to the binding site. The residues in β3 that are unobserved due to exchange broadening (Gln 424–Ser 427) in the ligand-free state are colored red. The nuclei that exhibit chemical shift changes greater than or equal to 0.05 ppm for 1HN and/or greater than or equal to 0.30 ppm for 15N are colored blue. The peptide NRLLLTG is violet.
The N-terminal region containing residues 393–398 lies at the interface between β-domain and the beginning of the α-helical domain in the crystal structure of DnaK(389–607). The region is structured in a similar way in both the solution and crystal structures of the peptide-bound β-domain, despite the fact that the entire α-helix domain is missing in the former (Fig. 2A ▶). Large chemical shift changes were observed for residues in this area upon peptide binding (Gly 443, Glu 444, Leu 397, and Ser 398; see Fig. 4 ▶). These observations strongly indicate a ligand-linked conformational change in this area. All of these residues are very conserved throughout the Hsp70s. Our structural observations again correspond with mutagenesis studies that indicate that the interaction between these residues is important to the allosteric mechanism of DnaK (Burkholder et al. 1996). Mutation of Gly 443 to either aspartic acid or serine leads to a dramatic decrease in peptide affinity and allosteric function as evaluated by peptide stimulation of ATPase activity. Gly 443 adopts a backbone conformation that is glycine specific (φ,ξ = 157°, −178°); thus any mutation will alter the local conformation. DnaK function was decreased to a lesser degree by mutation of Glu 444 to lysine (Burkholder et al. 1996). The structural and biochemical results thus reinforce each other in suggesting that this region is important for the allosteric communication of nucleotide state between the ATPase domain and the β-domain.
Conclusions
Through comparison of the different binding states of the β-domain of DnaK, we deepened our understanding of the structural and dynamical properties of the Hsp70 chaperones. We observed that removal of the α-helical domain in its entirety does not affect the primary interactions or structure of the β-domain in complex with the peptide NRLLLTG. Significantly, we find that the arch over the substrate binding cleft can also be formed in the absence of the helix, confirming that it plays a primary role in modulating substrate-binding activity. Our data also indicate that the structure of loop L2,3 and residues 393–398 of the N-terminal area, which is close to the α-helix–β-domain interface area, is independent of the presence or absence of the α domain. Our NMR data shows that peptide release causes changes in structural and dynamical parameters in this remote area, which has been previously identified from mutagenesis experiments as important for the allosteric mechanism of this class of proteins. The NMR relaxation data indicates that the peptide-bound form of the isolated β-domain DnaK is a relatively rigid molecule, whereas the ligand-free form contains several areas that are exchange broadened. Peptide binding also causes a dramatic shift in dimer–monomer equilibrium. Taken together, our data indicate that many changes take place in structure and dynamics of the isolated substrate-binding domain upon peptide binding and release. Several of the changes are suggestive of long-range transduction of an allosteric signal, without (primary) requirement of the helical domain.
Materials and methods
Construction and purification of DnaK(393 –507) (β-domain)
DnaK(393–507) was constructed by PCR amplification of DNA, subcloned into the plasmid pET15b (Novagen), and subsequently purified. The β-domain was expressed in the E. coli strain BL21(DE3) and contains a His-tag MGSSHHHHHHGLVPRGSHM393DnaK507. The protein was purified using a Ni-chelating column (Qiagen). The N-terminal His-tag was not cleaved from the protein.
NMR resonance assignments
The NMR resonance assignments of DnaK(393–507) in complex with NRLLLTG were performed with 0.66 mM 13C/15N labeled protein with 4.66 mM peptide a solution of 90% H2O/10% D2O or 100% D2O containing 10 mM sodium phosphate buffer (pH 7.4). Unless specified differently, the spectra were measured on a Varian Inova spectrometer operating at 800 MHz 1H frequency at 30°C. Resonance assignments were based on scalar connectivities with 3D HNCA, 3D HN(CA)HA, and 3D HNCACB. Aliphatic side chain assignments were obtained from 3D H(C)CH-TOCSY (τm = 19 ms) and (H)CCH-TOCSY (τm = 19 ms), 3D [1H, 13C, 1H] NOESY-HSQC (τm = 70 ms) and 3D [13C, 13C, 1H] HMQC-NOESY-HSQC (τm = 70 ms; for a review of methods used, see, e.g., Wüthrich 1986, Fesik and Zuiderweg 1990, Grzesiek and Bax 1993, and Cavanagh et al. 1996). Additionally, a 13C-resolved NOESY spectrum with 13C spectral width of 24 ppm and carrier at 123 ppm was recorded to assign NOEs arising from aromatic protons. The resonances of bound NRLLLTG were assigned from the amide region of a double filtered 1H -1H NOESY experiment. Intermolecular NOEs were obtained from a 3D NOESY-HSQC 12C-filtered, 13C-edited experiment as well as from the standard 3D 13C-resolved NOESY spectra. The spectra were processed with the software PROSA (Güntert et al. 1992) or NMRPipe (Delaglio et al. 1995). Spectral analysis was performed with the program XEASY (Bartels et al. 1995). The assignments of backbone atoms and Cβ are 95% complete for the nonprolyl residues of the β-domain in complex with the peptide NRLLLTG. The 1Hα, 13Cα, and 13Cβ chemical shift values were used to predict secondary structure by the Chemical Shift Index (Wishart and Sykes 1994), the results of which were used as dihedral restraints in the structure calculation. Complete side-chain assignments, excluding labile side-chain protons were obtained for approximately 90% of the residues.
Restraints identification and structure calculation
Three-dimensional [1H, 15N, 1H] NOESY-HSQC, 3D [1H, 13C, 1H] NOESY-HSQC, and 3D [13C, 13C, 1H] HMQC-NOESY-HSQC spectra recorded with 0.66 mM protein 4.66 mM NRLLLTG, each measured at 800 MHz with a mixing time of 70 ms, were exhaustively analyzed. The input for calculation of the DnaK(393–507) structure with the program ARIA (Linge et al. 2001) consisted of upper limit constraints derived from integrated NOE cross-peaks, hydrogen bond constraints, and dihedral angle constraints derived from CSI calculations. A final set of 2115 upper distance limits were manually assigned (1208 intra, 390 short range, 507 long range; see Table 1), 121 dihedral angle constraints (61 for phi, 60 for psi) were obtained and used as input for structure calculation. The standard simulated annealing protocol was used for the ARIA structure calculations to determine the protein fold in the absence of ligand. Virtually identical structures were obtained whether starting from an extended structure or the crystal structure of DnaK(389–607). The lowest energy conformer from the extended calculation was used as the starting structure for incorporating the peptide into the structure calculation. Forty-two intermolecular NOEs were added with a weight of >1 to ensure that they were not changed during the course of the ARIA calculation. The 20 conformers with the lowest energy were subjected to energy minimization. The 15 lowest-energy structures with the best structural statistics are presented here and were used to calculate an average structure. The statistics of the structure determination are listed in Table 1. Structure analysis was carried out with Procheck (Laskowski et al. 1993) and color figures were made with the program MOLMOL (Koradi et al. 1996).
NMR spectroscopy: Peptide titration experiments
The titration experiments were carried out with the peptide NRLLLTG using a solution of 0.66 mM DnaK(393–507). The peptide was synthesized by the University of Michigan Medical School Core Facility. A concentrated solution of NRLLLTG was added to the protein in peptide/protein ratios of 0, 0.1, 0.2, 0.4, 1.0, 2.1, 3.0, and 5.0. Peptide concentration was determined by comparison of 1D NMR spectroscopy on an arginine standard. The binding was monitored by 15N-1H and 13C-1H HSQC spectroscopy at 11.7 T (500 MHz 1H). The intensities of all shifting resonances changed simultaneously upon peptide titration.
NMR spectroscopy: Relaxation experiments
The relaxation experiments were performed on the same peptide-bound sample. Experiments with relaxation periods of 2, 12, 22, 32, 42, 52, and 62 ms were recorded with and without CPMG (Orekhov et al. 1994; Akke and Palmer 1996; Stevens et al. 2001). The 15N CPMG experiments were carried out using an echo repetition rate of 2 ms. For the 15N T2 experiment, a single composite 15N 180° refocusing pulse was used in the relaxation period to perturb the exchange broadening as little as possible. Differences between the rates measured by these experiments indicate the presence of conformational/chemical exchange broadening at a time scale that can be suppressed by the CPMG cycle (i.e., slower than 500 s−1). Complete relaxation experiments, including 15N R1 rates and 1H → 15N NOE experiments were obtained for the peptide-bound form only. R2, R2 CPMG, and R1 relaxation experiments were also carried out for the ligand-free state, using a 0.3-mM sample of the β-domain under the same buffer conditions. The relaxation rates were extracted from the data using NMRView (Johnson and Blevins 1994). Order parameters were calculated for the peptide-bound state with the program Modelfree (Mandel et al. 1995), using an NH bond distance of 1.02 Å and a 15N CSA of 172 ppm. A value of 7.9 ns was calculated for the averaged rotational correlation time, and a value of 1.17 for the ratio Dpar/Dperp of the rotational diffusion tensor. These values were obtained when the program did not take Rex terms in consideration (i.e., only models 1 and 2 were used). The rotational correlation time corresponds closely to the value of 7.7 ns calculated from the ratio of the trimmed averages of R2 and R1 and is reasonable for the 12-kD protein. The program returned an optimized rotational correlation time of 4.6 ns and very large global exchange broadening (Rex) when it was allowed to also consider Rex terms (i.e., Model 3). Because a 4.6-ns correlation time is quite unreasonable for this protein and because we could not detect any (overall) differences in R2 rates measured with or without CPMG, we decided that the program is unstable in this mode with our data. We thus elected to fix τc to 7.7 ns (isotropic) to calculate the order parameters and Rex values presented in Figure 3 ▶ and the Electronic Supplemental Material.
Coordinates
The atomic coordinates of DnaK(393–507) in complex with NRLLLTG have been deposited at the Protein Data Bank (PDB code 1QSL).
Electronic supplemental material
The electronic supplemental material consists of one table listing order parameters, local correlation times, and exchange values for DnaK(393–507) in complex with NRLLLTG, calculated using the Modelfree program from 15N R2, R1, and {1H}15N NOE data acquired at 500 MHz and 30°C.
Acknowledgments
This work was supported by NIH grant GM52421. The W.M. Keck Foundation, the NIH, and the NSF are gratefully acknowledged for financial support toward the 500- and 800-MHz NMR instruments. We thank Dr. L. Gierasch (U. Massachusetts) for the construct and procedures to obtain DnaKβ. Drs. T. Wang and K.J. Smith are acknowledged for assistance with relaxation and isotope-edited experiments, respectively.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03269103.
References
- Akke, M. and Palmer III, A.G. 1996. Monitoring macromolecular motions on microsecond to millisecond time scales by R1-R1 constant relaxation time NMR spectroscopy. J. Am. Chem. Soc. 118 911–912. [Google Scholar]
- Bartels, C., Xia, T., Billeter, M., Güntert, P., and Wüthrich, K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6 1–10. [DOI] [PubMed] [Google Scholar]
- Benaroudj, N., Matelier, G., Triniolles, F., and Ladjimi, M.M. 1995. Self-association of the molecular chaperone HSC70. Biochemistry 34 15282–15290. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi, S., Fourie, A.M., Sambrook, J.F., and Gething, M.-J.H. 1993. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J. Biol. Chem. 268 12730–12735. [PubMed] [Google Scholar]
- Buchberger, A., Theyssen, H., Schröder, H., McCarty, J.S., Virgallita, G., Milkereit, P., Reinstein, J., and Bukau, B. 1995. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J. Biol. Chem. 270 16903–16910. [DOI] [PubMed] [Google Scholar]
- Buczynski, G., Slepenkov, S.V., Sehorn, M.G., and Witt, S.N. 2001. Characterization of a lidless form of the molecular chaperone DnaK. J. Biol. Chem. 276 27231–27236. [DOI] [PubMed] [Google Scholar]
- Bukau, B. and Horwich, A.L. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92 351–366. [DOI] [PubMed] [Google Scholar]
- Burkholder, W.F., Zhao, X., Zhu, X., Hendrickson, W.A., Gragerov, A., and Gottesman, M.E. 1996. Mutations in the C-terminal fragment of DnaK affecting peptide binding. Proc. Natl. Acad. Sci. 93 10632–10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, S., Stevens, S.Y., Budor, A.P., and Zuiderweg, E.R.P. 2003. Solvent interaction of a Hsp70 chaperone substrate-binding domain investigated with Water-NOE NMR experiments. Biochemistry (in press). [DOI] [PubMed]
- Cavanagh, J., Fairbrother, W.J., Palmer III, A.G., and Skelton, N.J. 1996. Protein NMR spectroscopy. Academic Press, San Diego, CA.
- Craig, E.A., Gambill, B.D., and Nelson, R.J. 1993. Heat-shock proteins—Molecular chaperones of protein biogenesis. Microbiol. Rev. 57 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton, T.E. 1993. Proteins, 2nd ed. W.H. Freeman and Co., New York.
- Delaglio, F., Grzesiek, W., Vuister, G., Zhu, G., Pfeiffer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Fesik, S.W. and Zuiderweg, E.R.P. 1990. Heteronuclear three-dimensional NMR spectroscopy of isotopically labeled biological macromolecules. Q. Rev. Biophys. 23 97–131. [DOI] [PubMed] [Google Scholar]
- Gragerov, A., Zeng, L., Zhao, X., Burkholder, W., and Gottesman, M.E. 1994. Specificity of DnaK-peptide binding. J. Mol. Biol. 235 848–854. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S. and Bax, A. 1993. Methodological advances in protein NMR. Acc. Chem. Res. 26 131–138. [Google Scholar]
- Güntert, P., Dotsch, V., Wider, G., and Wüthrich, K. 1992. Processing of multi-dimensional NMR data with the new software PROSA. J. Biomol. NMR 2 619–629. [Google Scholar]
- Johnson, B.A. and Blevins, R.A. 1994. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14 52–55. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26 283–291. [Google Scholar]
- Lindquist, S. 1986. The heat-shock response. Annu. Rev. Biochem. 55 1151–1191. [DOI] [PubMed] [Google Scholar]
- Linge, J.P., O’Donoghue, S.I., and Nilges, M. 2001. Assigning ambiguous NOEs with ARIA. Methods Enzymol. 339 71–90. [DOI] [PubMed] [Google Scholar]
- Mandel, A.M., Akke, M., and Palmer III, A.G. 1995. Backbone dynamics of Escherichia coli ribonuclease HI: Correlations with structure and function in an active enzyme. J. Mol. Biol. 246 144–163. [DOI] [PubMed] [Google Scholar]
- McCarty, J.S., Buchberger, A., Reinstein, J., and Bukau, B. 1995. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249 126–137. [DOI] [PubMed] [Google Scholar]
- Misselwitz, B., Staeck, O., and Rapoport, T.A. 1998. J-proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell. 2 593–603. [DOI] [PubMed] [Google Scholar]
- Montgomery, D.L., Morimoto, R.I., and Gierasch, L.M. 1999. Mutations in the substrate binding domain of the E. coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J. Mol. Biol. 286 915–932. [DOI] [PubMed] [Google Scholar]
- Morshauser, R.C., Wang, H., Hu, W., Pang, Y., Flynn, G., and Zuiderweg, E.R.P. 1999. High resolution NMR solution structure of the substrate-binding domain of the mammalian chaperone protein Hsc70. J. Mol. Biol. 289 1387–1403. [DOI] [PubMed] [Google Scholar]
- Orekhov, V.Y., Pervushin, K.V., and Arseniev, A.S. 1994. Backbone dynamics of (1–71)bacterioopsin studied by two-dimensional 1H-15N NMR spectroscopy. Eur. J. Biochem. 219 887–896 [DOI] [PubMed] [Google Scholar]
- Pellecchia, M., Montgomery, D.L., Stevens, S.Y., Vander Kooi, C.W., Feng, H.-P., Gierasch, L.M., and Zuiderweg, E.R.P. 2000. Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Struct. Biol. 7 298–303. [DOI] [PubMed] [Google Scholar]
- Pierpaoli, E.V., Sandmeier, E., Schönfeld, H.-J., Gisler, S., and Christen, P. 1998. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J. Biol. Chem. 273 6643–6649. [DOI] [PubMed] [Google Scholar]
- Richarme, G. and Kohiyama, M. 1993. Specificity of the E. coli chaperone DnaK (70 kDa heat shock protein) for hydrophobic amino acids. J. Biol. Chem. 268 24074–24077. [PubMed] [Google Scholar]
- Rüdiger, S., Germeroth, L., Schneider-Mergener, J., and Bukau, B. 1997a. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger, S., Buchberger, A., and Bukau, B. 1997b. Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4 342–349. [DOI] [PubMed] [Google Scholar]
- Rüdiger, S., Mayer, M.P., Schneider-Mergener, J., and Bukau, B. 2000. Modulation of substrate specificity of the DnaK chaperone by alteration of a hydrophobic arch. J. Mol. Biol. 304 245–251. [DOI] [PubMed] [Google Scholar]
- Stevens, S.Y., Sanker, S., Kent, C., and Zuiderweg, E.R.P. 2001. Delineation of the allosteric mechanism for a cytidylyltransferase exhibiting negative cooperativity. Nat. Struct. Biol. 8 947–952. [DOI] [PubMed] [Google Scholar]
- Vosine, C., Craig, E.A., Zufall, N., von Ahsen, O., Pfanner, N., and Voos, W. 1999. The protein import motor of mitochondria: Unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 97 565–574. [DOI] [PubMed] [Google Scholar]
- Wang, H., Pang, Y., Kurochkin, A.V., Hu, W., Flynn, G.C., and Zuiderweg, E.R.P. 1998. The solution structure of the chaperone protein DnaK substrate binding domain: A preview of chaperone–protein interaction. Biochemistry 37 7929–7940. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S. and Sykes, B.D. 1994. A simple method for the identification of protein secondary structure using C-13 chemical shift data. J. Biomol. NMR 4 171–180. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. 1986. NMR of proteins and nucleic acids. Wiley, New York.
- Zhu, X., Zhao, X., Burkholder, W.F., Gragerov, A., Ogata, C.M., Gottesman, M.E., and Hendrickson, W.A. 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelhoffer, T., Lopez-Buesa, P., and Craig, E.A. 1995. The dissociation of ATP from hsp70 of Saccharomyces cerevisiae is stimulated by both Ydj1p and peptide substrates. J. Biol. Chem. 270 10412–10419. [DOI] [PubMed] [Google Scholar]