Abstract
A comparison of the contributions and position dependence of cross-strand electrostatic and aromatic side-chain interactions to β-sheet stability has been performed by using nuclear magnetic resonance in a well-folded β-hairpin peptide of the general sequence XRTVXVdPGOXITQX. Phe–Phe and Glu–Lys pairs were varied at the internal and terminal non–hydrogen-bonded position, and the resulting stability was measured by the effects on α-hydrogen and aromatic hydrogen chemical shifts. It was determined that the introduction of a Phe–Phe pair resulted in a more folded peptide, regardless of position, and a more tightly folded core. Substitution of the Glu–Lys pair at the internal position results in a less folded peptide and increased fraying at the terminal residues. Upfield shifting of the aromatic hydrogens provided evidence for an edge-face aromatic interaction, regardless of position of the Phe–Phe pair. In peptides with two Phe–Phe pairs, substitution with Glu–Lys at either position resulted in a weakening of the aromatic interaction and a subsequent decrease in peptide stability. Thermal denaturation of the peptides containing Phe–Phe indicates that the aromatic interaction is enthalpically favored, whereas the folding of hairpins with cross-strand Glu–Lys pairs was less enthalpically favorable but entropically more favorable.
Keywords: β-Hairpin peptides, aromatic interactions, salt bridges, cross-strand interactions
Since the report of the first monomeric β-hairpin peptide in 1993 (Blanco et al. 1993), determining the factors that stabilize β-hairpin structure has been an active area of research (Searle 2001). β-Hairpins represent excellent model systems for studying the relationship between sequence and secondary structure, allowing for a hierarchical approach to understanding protein folding. In addition, β-hairpins provide a soluble structured model system for studying factors that influence misfolding of proteins, which is believed to occur through a β-sheet type of structure. Thus, a detailed understanding of the factors that influence folding and stability is warranted.
Factors that have been found to influence the stability of a β-hairpin include the turn sequence (de Alba et al. 1997a,b; Haque and Gellman 1997; Ramirez-Alvarado et al. 1997; Syud et al. 1999), the β-sheet propensity of the residues in the strand (Griffiths-Jones et al. 1999; Russell and Cochran 2000; Santiveri et al. 2000), and cross-strand interactions between side chains in opposite strands (Ramirez-Alvarado et al. 1996, 2001; Maynard et al. 1998; Searle et al. 1999; Kobayashi et al. 2000), including both lateral (Santiveri et al. 2000; Tatko and Waters 2002) and diagonal (Syud et al. 2001) interactions. A hydrophobic cluster is typically necessary for stabilizing a β-hairpin (Maynard et al. 1998; Espinosa and Gellman 2000), and previous work has shown that a hydrophobic cluster near the turn provides greater stability to the hairpin than when it is at the terminus (Espinosa et al. 2001). In addition, although terminal residues often appear to be frayed based on Hα chemical shifts, there is some evidence that even they contribute to the overall stability of the hairpin (Stanger et al. 2001). A two-state model is generally assumed for the folding of these peptides, and in some cases, a two-state model has been demonstrated (Searle 2001); however, this is not the case for all β-hairpin peptides (Santiveri et al. 2002).
Most of the information regarding the contribution of cross-strand pairs to β-sheet stability comes from studies of proteins, including statistical analyses and mutation studies (Smith and Regan 1995, 1997; Wouters and Curmi 1995; Blasie and Berg 1997; Cootes et al. 1998; Zaremba and Gregoret 1999; Mandel-Gutfreund et al. 2001). In particular, the mutation study by Smith and Regan (1995) provided the first information on the relative contributions of cross-strand interactions and β-sheet propensities. They found that although salt bridges formed one of the most stabilizing cross-strand interactions, the overall contribution to protein stability was low because of the low sheet propensities of Glu and Lys. In contrast, a cross-strand Phe–Phe interaction provided significant stability to the protein because both the sheet propensity and the cross-strand interaction were favorable. More recently, we have shown that a cross-strand Phe–Phe interaction is stabilizing in a designed β-hairpin due to a favorable edge-face interaction (Tatko and Waters 2002). In the current study, we have compared the contribution of a salt bridge and an aromatic pair at different positions in the strand and determined their contributions to hairpin stability and cooperativity.
Results
Peptide design
We investigated a set of four 14-residue peptides based on Gellman’s sequence, in which two cross-strand pairs at positions 1/14 and 5/10 were varied (Fig. 1 ▶; Stanger and Gellman 1998; Syud et al. 1999). These positions were chosen to study the influence of cross-strand pairs near the turn residues relative to those at the termini. Two additional 12-residue peptides were investigated to determine the contribution of the terminal cross-strand pair. The peptides can be grouped into two different series, the XFFZ series, including FFFF, EFFK, and *FF* (where the asterisk indicates a deleted residue), and the XEKZ series, including FEKF, EEKK, and *EK*. We chose a DPro-Gly turn sequence because of its excellent nucleating properties (Stanger and Gellman 1998). The two cross-strand pairs are in non–hydrogen-bonded (NHB) sites, placing them on the same face of the hairpin, and there is a pair of threonines in the NHB site between them, such that diagonal interactions need not be considered (Syud et al. 2001). The peptides contain a net charge of at least +2 to maintain water solubility and to prevent aggregation. The peptides were investigated at pH 4.2, acetate buffer. In addition, all peptides containing an Glu–Lys pair were investigated at pH 7.1, phosphate buffer, to confirm that Glu was fully deprotonated. No significant difference was observed in peptide stability or structure at these two pHs.
Figure 1.
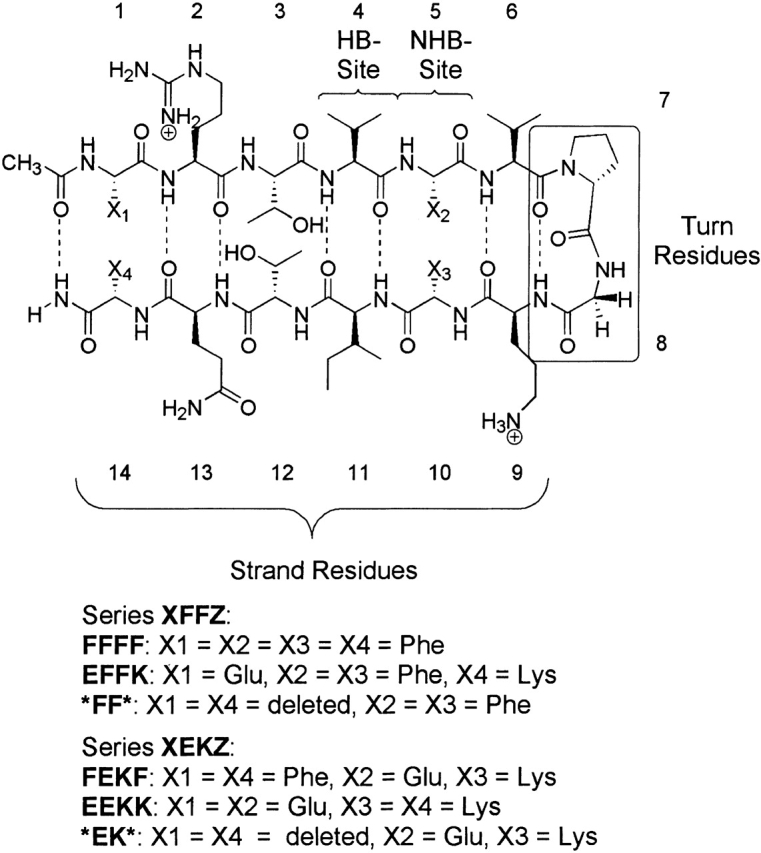
Sequences of the 14- and 12-residue peptides in series XFFZ and XEKZ. HB-site indicates hydrogen-bonded site; NHB-site, non–hydrogen-bonded site. Dashed lines indicate hydrogen bonds.
Structure determination
The peptides were characterized by matrix-assisted laser desorption ionization (MALDI) mass spectrometry and NMR, including correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), and rotating frame nuclear Overhauser spectroscopy (ROCSY). β-Hairpin structure was confirmed by long-range nuclear Overhauser effect peaks observed between cross-strand residues. NOEs shown in Figure 2 ▶ clearly indicate a β-hairpin structure, with long-range NOEs between residues near the N and C termini. Only those NOEs that were unambiguous are shown. The extent of folding was determined qualitatively by comparing Hα chemical shifts relative to random coil values, as determined by the control peptides in which DPro has been replaced by LPro. LPro has been shown to disrupt hairpin formation in short peptides (Stanger and Gellman 1998). The Hα resonances in a β-hairpin are shifted downfield relative to the random coil chemical shifts due to proximity to the carbonyls in an extended sheet conformation (Sharman et al. 2001). Shifting of ≥0.1 ppm for three or more consecutive residues is typically taken as evidence for significant β-sheet population (Wishart et al. 1991, 1992). The spacing between the diastereotopic Gly Hα resonances has also been shown to correlate with extent folding of β-hairpin peptides (Searle et al. 1999).
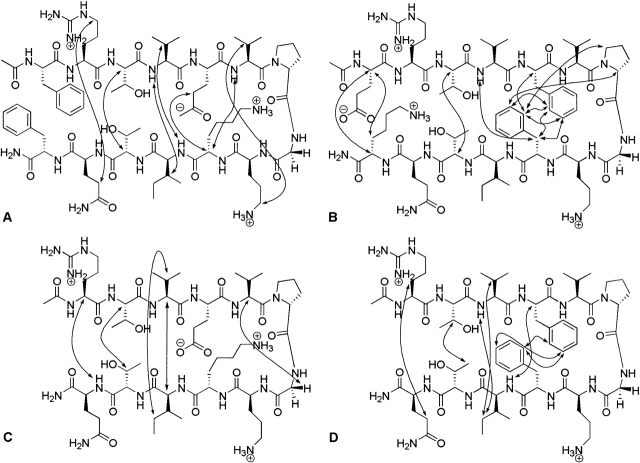
Figure 2.
Long-range NOEs of representative peptides: (A) FEKF, (B) EFFK, (C) *EK*, and (D) *FF*. The NOEs shown are those that were unambiguous but not necessarily the only NOEs observed.
β-Hairpin stability has been quantified by NMR in which control peptides were used to determine the chemical shifts in the unfolded and fully folded states (equation 1). The LPro peptides were used for the unfolded state, and the cyclic peptides in Figure 3 ▶ were used for the fully folded state (Syud et al. 1999). To generate the cyclic peptides, four additional residues, Val-DPro-Gly-Orn, were incorporated to covalently link the N and C termini of the original 14-residue sequences. The validity of the assumption that the cyclic peptides represent the fully folded β-hairpin was investigated by ROESY NMR. NOEs were observed between the same cross-strand residues as the uncyclized peptides along the entire strand, indicating a β-hairpin structure.
Figure 3.
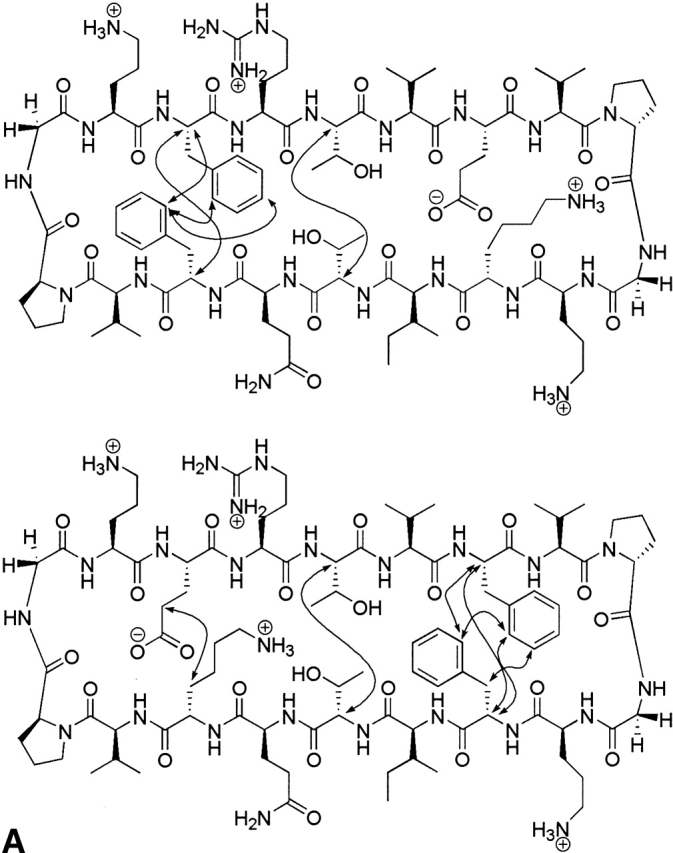
(A) Sequence and NOEs of the cyclic control peptides cyc-FEKF and cyc-EFFK. (B) Downfield shifting of the cyclic control peptides cyc-FEKF and cyc-EFFK.
 |
(1) |
Qualitative analysis of hairpin stability
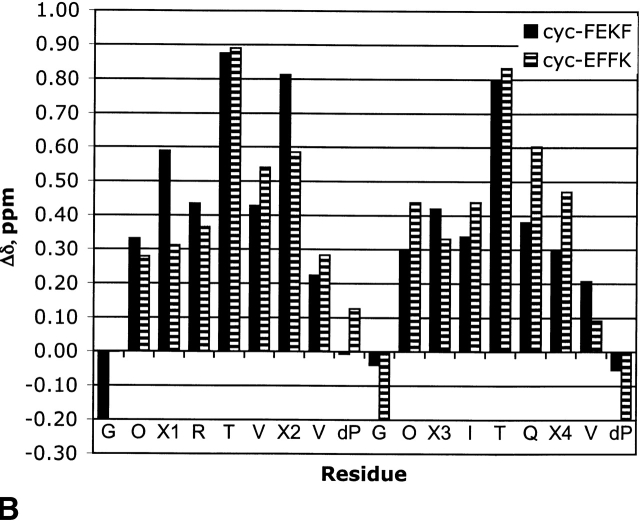
All the peptides in series XFFZ and XEKZ appear to be well folded based on their Hα chemical shifts relative to random coil values, with all but the terminal residues having >0.1 ppm shift (Fig. 4 ▶). This type of fraying is typical for terminal residues in β-hairpins. Based on this data, FFFF appears to be the most stable hairpin, and *EK* appears to be the least folded.
Figure 4.
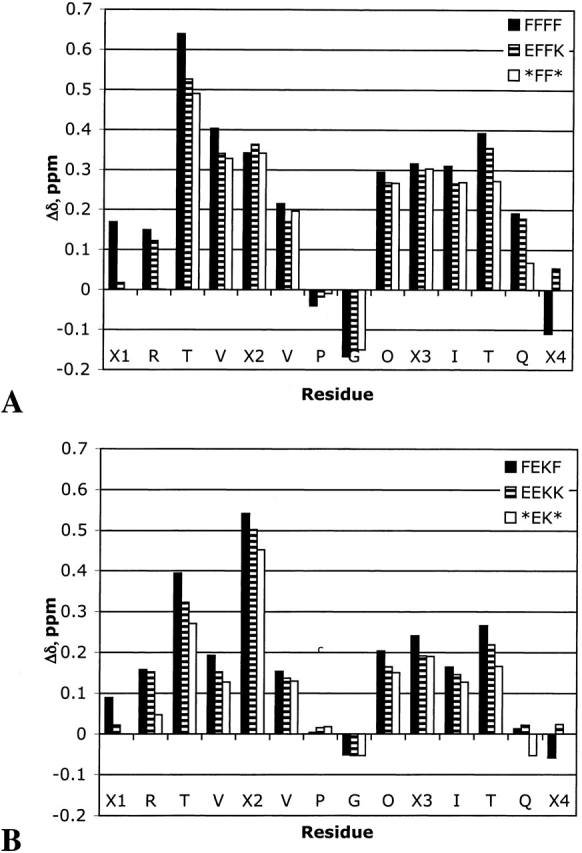
Comparison of Δδ Hα for series XFFZ (A) and series XEKZ (B).
Differences in location of aromatic residues in the XFFZ and XEKZ series can have a significant influence on NMR chemical shifts of neighboring residues. Thus, qualitative analysis cannot be used to make direct comparisons between these two series of peptides. For this reason, quantitative analysis using equation 1 was performed by using a cyclic peptide for the fully folded state (see Materials and Methods).
Quantitative analysis of hairpin stability
Comparison of the XFFZ and XEKZ series
Through use of the LPro and cyclic control peptides, the extent of folding for each of the peptides in series XFFZ and XEKZ was determined by NMR. In this way, a more detailed analysis of the relative contributions of cross-strand pairs can be made. Comparison of FFFF and EEKK indicates that the Phe residues stabilize the hairpin more than the salt bridges (Fig. 5A ▶). This is due to differences in sheet propensities in conjunction with differences in the cross-strand interactions (vide infra). The truncated peptides *FF* and *EK* are both less stable than are their 14-residue parent peptides, but have a stability profile similar to FFFF and EEKK: *FF* is more stable than *EK* along the entire hairpin (Fig. 5B ▶). Again, this is attributed to differences in sheet propensities and side-chain–side-chain interactions. Peptides FEKF and EFFK appear to be similarly folded, with no clear difference in stability over the length of the strand (Fig. 5C ▶). This is surprising given the results of Gellman (Espinosa et al. 2001), which indicated that a hydrophobic cluster was more stabilizing near the turn than at the termini. The fact that FEKF appears to be more folded near the turn than is EFFK may be a result of a competition between the favored geometry of the turn residues versus the preferred geometry between two Phe residues (vide infra).
Figure 5.
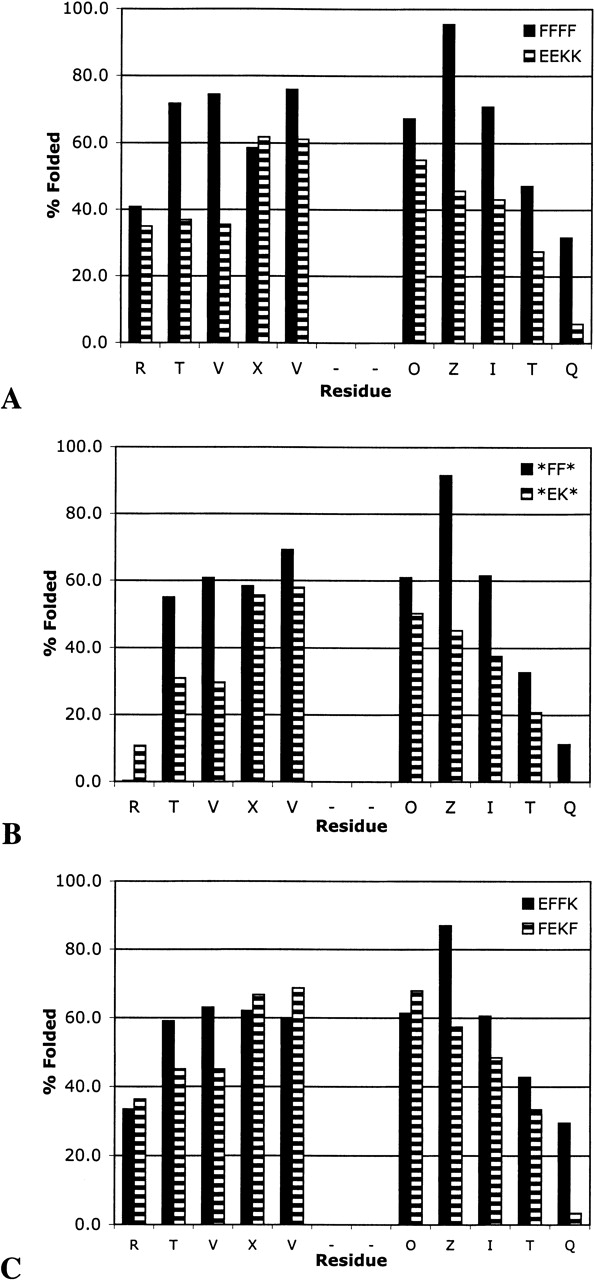
Extent of folding as determined from downfield shifting of the Hα resonances by NMR: (A) FFFF and EEKK, (B) *FF* and *EK*, and (C) EFFK and FEKF.
Comparison of terminal residues
Comparison of FFFF with EFFK and *FF* demonstrates that the terminal residues contribute to the overall stability of the hairpin despite extensive fraying, but the terminal aromatic pair contributes more than does the terminal salt bridge (Fig. 6 ▶). Interestingly, the terminal salt bridge in EFFK seems to have little impact on the extent of folding of the internal positions relative to the truncated peptide, *FF*, but the salt bridge does help to reduce fraying at the termini (Fig. 6A ▶). In contrast, deletion of the terminal Phe–Phe pair influences the stability of the entire hairpin.
Figure 6.
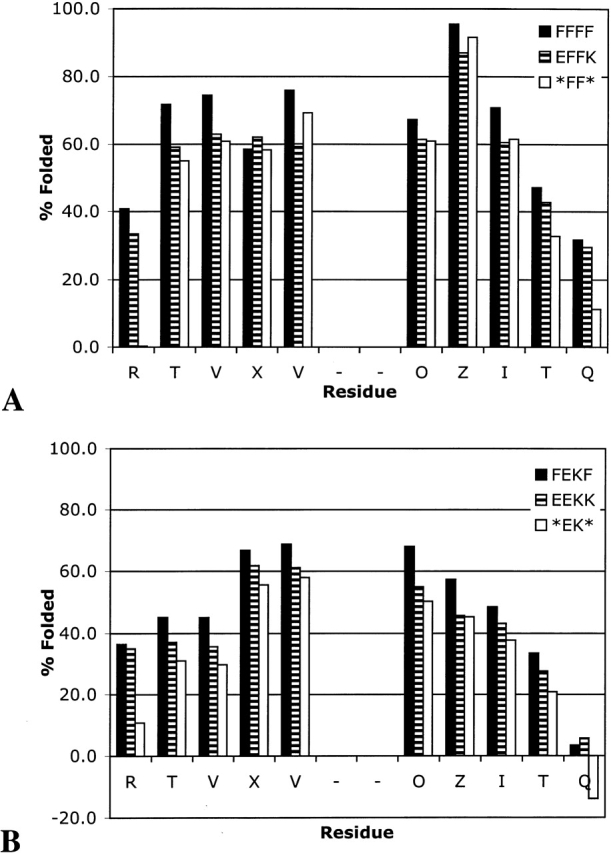
Effect of terminal residues on extent of folding: XFFZ series (A) and XEKZ series (B).
FEKF, EEKK, and *EK* provide an interesting comparison to FFFF, EFFK, and *FF*. The relative contributions of the terminal Phe–Phe pair and the terminal Glu–Lys pair are similar in both series. Substitution of the aromatic pair in FEKF with the salt bridge causes significant loss in stability throughout the length of the hairpin, whereas deletion of the terminal salt bridge increases fraying but does not greatly impact the core of the hairpin (residues EVDPGOK).
Comparison of internal positions
The relative contribution of a salt bridge or an aromatic pair at an internal position was investigated by comparing FFFF to FEKF and EFFK to EEKK (Fig. 7 ▶). Replacement of the internal aromatic pair in FFFF with a salt bridge destabilizes the β-sheet, as was seen when the same substitution was made at the terminus (cf. Figs. 7A ▶ and 6A ▶). Replacing the internal Phe–Phe pair with an Glu–Lys pair in EFFK causes similar destabilization of the hairpin as in FFFF (Fig. 7B ▶). In both cases, the residues immediately adjacent to the turn remain well folded, but destabilization occurs along the strand.
Figure 7.
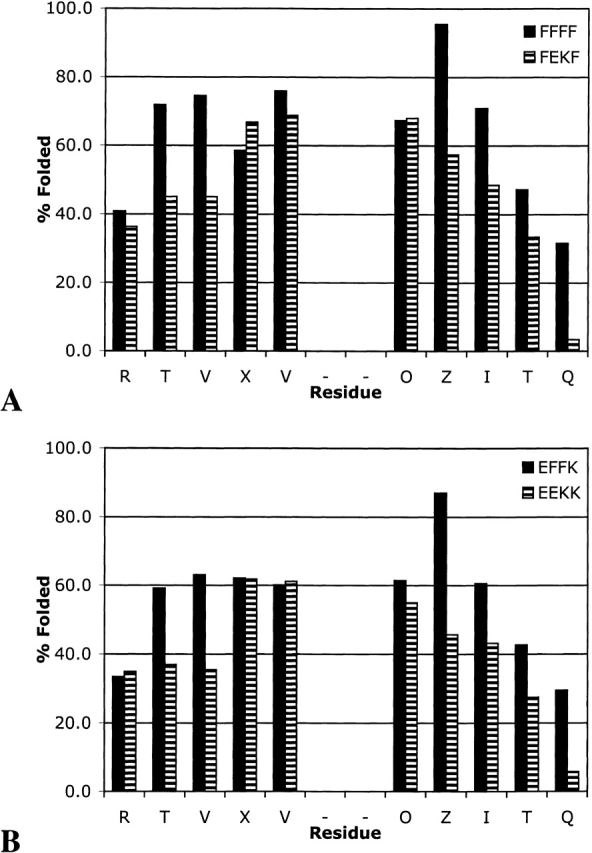
Comparison of the aromatic pair (A) and salt bridge (B) at internal positions.
Contributions of the backbone and side chain in terminal pairs
By replacing each of the terminal residues with alanine (Ala), we probed the importance of the backbone relative to side-chain–side-chain interactions to the stability of the hairpin. Deletion of the Ala residue subsequently removes the backbone and hydrogen-bond functionality. As shown in Figure 8A ▶, replacement of the N-terminal Phe with Ala in FEKF to give AEKF destabilizes the entire hairpin, with a greater impact on the terminus than on the turn. Deletion of the Ala at the N terminus has much less effect on the extent of folding (AEKF → *EKF). Replacement of the C-terminal Phe with Ala also destabilizes the hairpin (Fig. 8B ▶). Interestingly, deletion of Ala at the C terminus (FEKA → FEK*) has a greater impact than does the corresponding Ala at the N terminus. It may be that the methyl group on Ala interacts with the cross-strand Phe in a hydrophobic manner. Support for this comes from the observation that NOEs between the β-hydrogens on the C-terminal Phe and the ring of the N-terminal Phe are commonly observed (see Figs. 2B ▶, 3B ▶).
Figure 8.
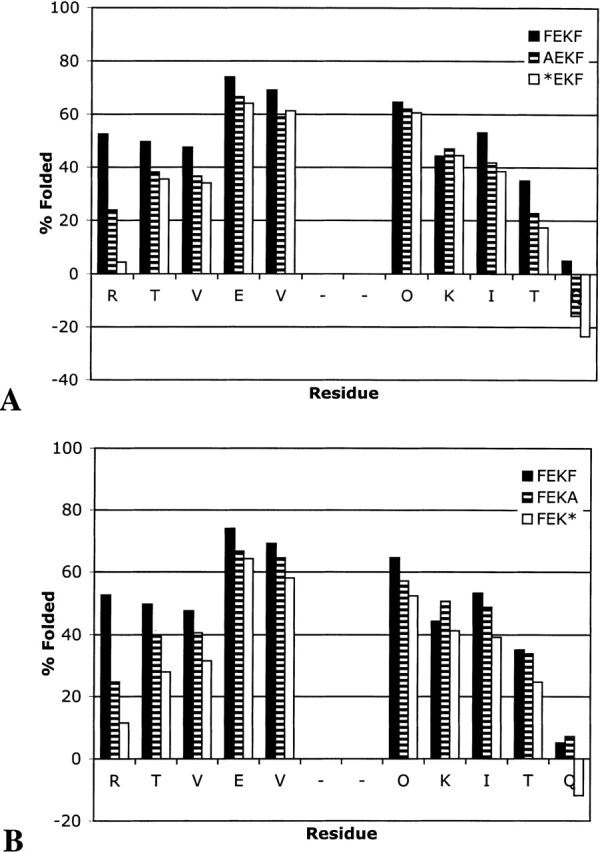
Extent of folding for FEKF, AEKF, and *EKF (A) and for FEKF, FEKA, and FEK* (B).
Replacement of the N-terminal glutamic acid in EFFK with Ala causes an increase in fraying but has little impact at the turn (Fig. 9A ▶). Deletion of the Ala residue to give *FFK has only minimal effect on the hairpin stability at either the turn or the termini (Fig. 9A ▶). Similarly, replacement or deletion of the C-terminal Lys has almost no effect on the hairpin stability, with the exception of an increase in terminal residue fraying (Fig. 9B ▶).
Figure 9.
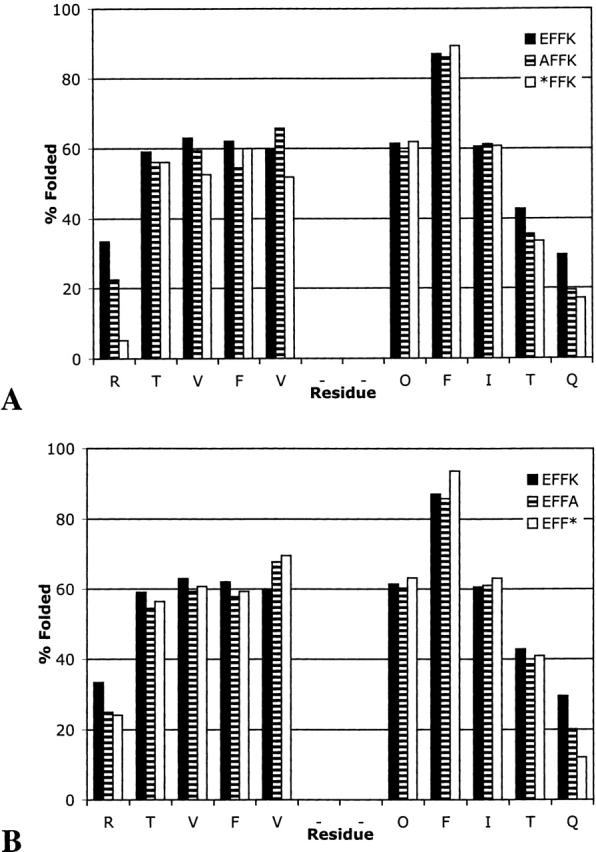
Extent of folding for EFFK, AFFK, and *FFK (A) and for EFFK, EFFA, and EFF* (B).
Comparison of folding populations determined from Hα and from Gly splitting
In many cases, the splitting of the Gly Hα protons in the turn of a β-hairpin has been used to determine β-hairpin populations (Searle 2001; Tatko and Waters 2002). This is convenient because the Gly Hα protons typically fall in a “clean” region of the NMR spectrum, and so, an estimate of hairpin stability can be easily obtained from a simple 1H NMR spectrum of the peptide. However, as can be seen in Table 1, in the peptides studied here, Gly splitting consistently overestimates hairpin population as determined by Hα. This appears to be a trait of the DPro-Gly turn, because the Gly splitting seems to be quite reliable for determining hairpin populations in peptides with Asn-Gly turns (Searle 2001; Tatko and Waters 2002). The fact that the Gly splitting overestimates the hairpin population in these peptides is consistent with the trend observed with Hα that the residues near the turn repeatedly indicate higher hairpin populations than those nearer the terminus (Table 1).
Table 1.
β-Hairpin population as determined by the Hα residues in the hydrogen-bonded sites compared with the population determined by the Gly splitting
| % Foldeda | |||||||
| Entry | Peptide | Val4 | Val6 | Orn9 | Ile11 | Averageb | Gly |
| 1 | FFFF | 75 | 76 | 67 | 71 | 72 (4) | 85 |
| 2 | EFFK | 63 | 60 | 61 | 61 | 61 (1) | 78 |
| 3 | AFFK | 60 | 66 | 59 | 61 | 62 (3) | 75 |
| 4 | EFFA | 59 | 68 | 60 | 61 | 62 (4) | 75 |
| 5 | *FF* | 61 | 69 | 61 | 62 | 63 (4) | 76 |
| 6 | FEKF | 45 | 69 | 68 | 49 | 58 (13) | 90 |
| 7 | EEKK | 36 | 61 | 55 | 43 | 49 (11) | 74 |
| 8 | AEKF | 37 | 59 | 62 | 42 | 50 (12) | 75 |
| 9 | FEKA | 41 | 65 | 57 | 49 | 53 (10) | 75 |
| 10 | *EK* | 30 | 58 | 50 | 38 | 44 (12) | 71 |
Determined in 10 mM acetate buffer (pH 5.0) or 10 mM phosphate buffer (pH 7.1) at 298 K.
a Errors in the fraction folded as determined by Hα chemical shifts is 1%.
b Numbers in parentheses are standard deviations from the average.
Analysis of aromatic interactions
In previous studies, cross-strand Phe–Phe pairs have been shown to interact in an edge-face geometry, with the ortho-hydrogen (H2) of the C-terminal Phe directed at the face of the N-terminal Phe (Das et al. 2001; Tatko and Waters 2002). This results in significant upfield shifting of H2. Thus, the chemical shift of the H2 can be used as another means of determining the β-hairpin population. Comparison of the chemical shifts of H2 on Phe 10 in FFFF and EFFK indicates that changes at the terminal positions influence the extent of folding at the internal Phe pair (Table 2, entries 2 and 3), in qualitative agreement with Hα data. Comparison of the terminal aromatic pairs in FFFF and FEKF indicates that changes at the internal positions also impact the terminal positions (Table 2, entries 6,7). Moreover, the upfield shifting of H2 of Phe 14 in FFFF and FEKF indicate that the side chains at the termini do indeed interact, despite the fraying indicated by the Hα chemical shifts (Fig. 5 ▶). In contrast, comparison of the upfield shifting of H2 in EFFK and *FF* indicates that the terminal salt bridge has very little effect on hairpin stability near the turn, in agreement with Hα data (Table 2, entries 3,4).
Table 2.
Upfield chemical shifting of the C-terminal aromatic ortho-hydrogen
| Entry | Peptide | δ, ppm | Δδ, ppm | % Folded |
| 1 | cyc-EFFK | 6.35 | −0.86 | 100 |
| 2 | FFFF (internal) | 6.49 | −0.71 | 83 |
| 3 | EFFK | 6.56 | −0.64 | 75 |
| 4 | *FF* | 6.57 | −0.63 | 73 |
| 5 | cyc-FEKF | 6.46 | −0.80 | — |
| 6 | FFFF (terminal) | 7.04 | −0.22 | — |
| 7 | FEKF | 7.12 | −0.14 | — |
Determined in 10 mM phosphate buffer (pH 7.1) at 298 K.
The upfield shifting of H2 of Phe 14 provides information on the geometry of interaction at the terminus. The fact that H2 is upfield shifted in both FFFF and FEKF indicates that the aromatic side chains interact in an edge-face geometry even at the terminus, despite the fraying indicated by the Hα chemical shifts. This indicates that the edge-face geometry provides an attractive interaction, rather than simply resulting from conformational restriction in the β-strand.
The upfield shifting of the aromatic residue is another method for estimating the extent of folding. Comparison of the fraction folded as determined by the Phe residue at position 10 (Table 1) agrees well with the values determined from the Gly splitting used for determination of the extent of folding. Shifting of the H2 resonance in the terminal FF pair was not used to determine the percentage folded due to increased fraying at the peptide termini.
Salt studies
We performed salt studies on FEKF, EFFK, *FF*, and *EK* to investigate the contribution of the salt bridge in more detail (Table 3). The chemical shift dispersion decreases in going from 0 to 500 mM NaCl for all four peptides. This indicates that the hairpin stability decreases but makes it difficult to measure due to spectral overlap of the Hα resonances at high salt concentrations. The Gly residues remain well-resolved, however, and so we determined the extent of folding based on these values in buffer and 500 mM NaCl. As can be seen in Table 3, even hairpins without a salt bridge are less folded in 500 mM NaCl, but peptides FEKF and *EK* are destabilized to a greater extent. Energetically, the change in fraction folded at 500 mM NaCl relative to buffer amounts to ~0.29 kcal/mole for *EK*, versus 0.15 kcal/mole for *FF*, as determined by the Gly splitting. Thus, the salt bridge provides ~−0.1 to −0.2 kcal/mole to the hairpin stability and no stabilization at the terminal position (cf. EFFK to *FF* in Table 3). However, because we have seen that the influence of the strand residues is not fully transmitted to the turn residues, this is best considered a lower limit for the energetic contribution of the salt bridge. In other peptide systems, salt bridges have been found to contribute between −0.1 and −0.4 kcal/mole to β-hairpin stability (de Alba et al. 1995; Searle et al. 1999; Ramirez-Alvarado et al. 2001; Ciani et al. 2003), and up to −0.8 to α-helix stability (Scholtz et al. 1993; Smith and Scholtz 1998).
Table 3.
Effect of salt concentration on β-hairpin stability
| D2O | 500 mM NaCl | ||||
| Gly splitting | % folded | Gly splitting | % folded | % Change | |
| FEKF | 0.220 | 90 | 0.191 | 78 | 12 |
| EEKK | 0.181 | 74 | 0.151 | 62 | 12 |
| *EK* | 0.172 | 70 | 0.145 | 59 | 11 |
| EFFK | 0.503 | 78 | 0.469 | 73 | 5 |
| *FF* | 0.490 | 76 | 0.456 | 71 | 5 |
Determined in 10 mM phosphate buffer (pH 7.1) AT 298 K.
Thermal denaturation studies
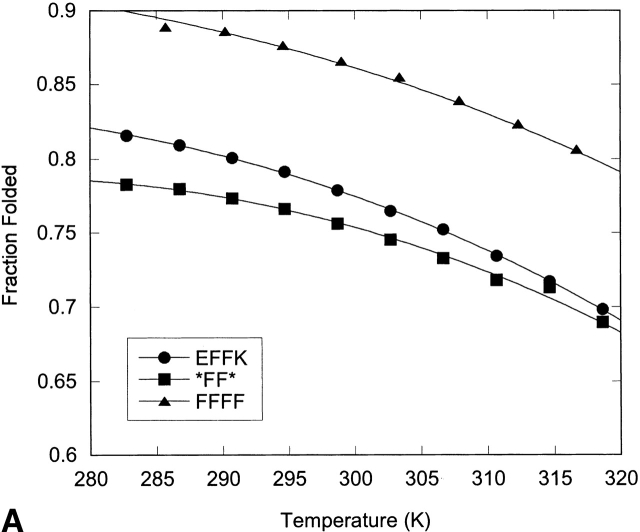
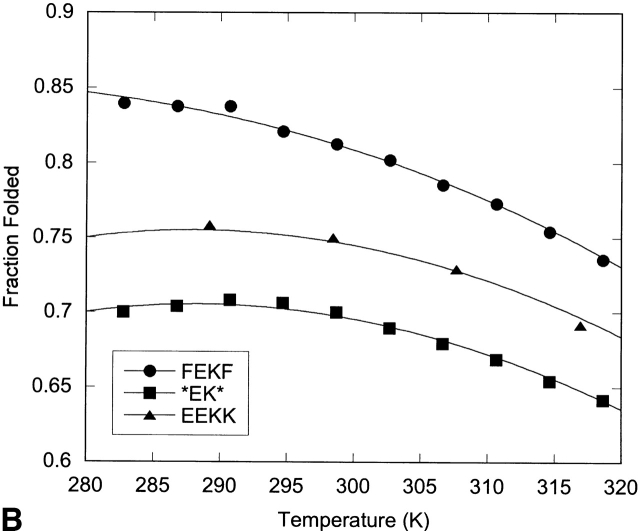
Thermal denaturation of the hairpins was followed by NMR through measurement of the change in glycine splitting with temperature (Fig. 10 ▶; Table 4). Although the fraction folded as determined by Gly splitting is likely an overestimate of the hairpin population, spectral overlap of the Hα resonances precluded their use in thermal denaturation. Thus, although the trends observed for these peptides are meaningful, the absolute values of the enthalpy, entropy, and change in heat capacity may be biased due to the use of the Gly chemical shifts to determine them.
Figure 10.
Fraction folded versus temperature as determined by NMR from the Gly splitting for the XFFZ series (A) and the XEKZ series (B). Curves represent the fit to the data as described in Materials and Methods.
Table 4.
Thermodynamic parameters for folding of seriesXFFZandXEKZat 298 K
| Peptide | ΔH°, kcal/mole | ΔS°, cal mole−1 K−1 | ΔCp°, cal mole−1 K−1 |
| FFFF | −4.0 | −9.7 | −59 |
| EFFK | −3.1 | −7.9 | −85 |
| *FF* | −2.3 | −5.3 | −93 |
| FEKF | −3.0 | −7.3 | −99 |
| EEKK | −1.3 | −2.2 | −131 |
| *EK* | −1.2 | −2.2 | −119 |
Determined from the fitting of the data in Figure 10 ▶. The error is 6% based on 95% confidence limits of the NMR chemical shifts and temperature.
All the peptides investigated display enthalpy-driven folding (Table 4). Peptides FFFF, EFFK, and *FF* display a steady decrease in ΔH and ΔS as the terminal Phe residues are replaced and subsequently deleted. Comparison of EFFK and FEKF indicate that their thermodynamic parameters are similar. Folding of peptides EEKK and *EK* is the least enthalpically favorable but the most entropically favorable, resulting in cold denaturation (Fig. 10 ▶).
Cold denaturation is usually associated with a hydrophobic driving force, yet in the systems studied here, it is only observed when the aromatic pair is deleted. However, cold denaturation has been observed for other peptides with hydrophilic residues in the NHB positions (Searle 2001; Ciani et al. 2003). This may arise from dominance of hydrophobic interactions on the hydrogen-bonded face of the hairpin when the aromatic pair is removed. Enthalpically driven folding has been observed for a number of well-folded β-hairpins with a hydrophobic cluster on the NHB face of the β-hairpin, and has been proposed to be the result of a “tight” interaction, as described by Smithrud and Diederich (1990) and Smithrud et al. (1991). Thus, the decrease in enthalpy and increase in entropy upon replacing or removing an aromatic pair may be a result of decreasing the tightness of the fold (Espinosa and Gellman 2000). This may arise from a decrease in the burial of the backbone or a decrease in the packing of the side chains.
Discussion
We have compared the contributions of an aromatic pair and a salt bridge at an internal and terminal position of a β-hairpin peptide. In both positions, the Phe–Phe pair contributes more to the hairpin stability than does the salt bridge, in agreement with mutation studies in proteins (Smith and Regan 1997). At the internal position, this can be explained in terms of a combination of sheet propensities and side-chain–side-chain interactions. Phe has a higher sheet propensity than does Lys or Glu, and an Phe–Phe cross-strand pair has been shown to form a favorable cross-strand interaction, although the magnitude of the interaction has not been quantified.
At the terminus, where the residues have significantly more conformational freedom, the Phe–Phe pair still stabilizes the hairpin more than does the salt bridge. It is clear that despite the conformational freedom of a terminal position, the terminal Phe–Phe pair interacts in an edge-face geometry, consistent with what has been observed at more restricted sites at internal positions (Tatko and Waters 2002). This provides support for the fact that the edge-face interaction is an attractive interaction and not simply due to geometric restrictions in a β-sheet geometry. The substitution studies in which Phe is replaced with Ala also indicate the presence of a side-chain–side-chain interaction between the Phe residues at the N- and C-terminal positions, in that removal of an aromatic ring causes significant destabilization of the hairpin. In contrast, removal of either the Glu or Lys from a terminal position has very little impact on overall hairpin stability. Salt studies also indicate that the terminal Glu–Lys pair provides little to no stability to the hairpin through salt bridge formation, in that the effect on folding of 500 mM NaCl is the same for EFFK and *FF*. Analysis of the Hα shifts indicates that the Glu–Lys pair reduces fraying, but not to a greater extent than a terminal AK or EA pair (Fig. 9 ▶). Thus, for Glu–Lys, the terminal residues appear to stabilize the hairpin simply by restricting the geometry of the inner residues, not through significant side-chain–side-chain interactions.
We can estimate the strength of a terminal Phe–Phe interaction from the fraction folded data in Table 2. Comparison of FFFF to EFFK or *FF* yields a contribution of the terminal Phe–Phe pair of −0.2 to −0.3 kcal/mole. Comparison of FEKF to EEKK or *EK* also yields values of ~−0.3 kcal/mole. Because replacement of a Phe with Ala results in significant loss of stability, but removal of Ala has little effect on the hairpin stability (Fig. 8 ▶), it is reasonable to estimate that the −0.2 to −0.3 kcal/mole stabilization gained from a terminal Phe–Phe pair is largely due to side-chain–side-chain interactions. As with the Glu–Lys interaction, this is best considered a lower limit because the folding of the peptides in this study is not highly cooperative.
Conclusions
We have investigated the contribution of an Glu–Lys and Phe–Phe pair to β-hairpin stability at two different positions. In this system, we have found the Phe–Phe pair stabilizes the hairpin to a greater extent than does the Glu–Lys pair, independent of position in the strand. Terminal residues contribute to the stability of a β-hairpin, even though they themselves are not well folded. The contribution of the terminal pairs appears to arise from two different sources. The terminal Glu–Lys pair does not appear to form a salt bridge but decreases fraying of the penultimate cross-strand pair. In contrast, there is good evidence that the terminal Phe–Phe pair does indeed form a specific side-chain–side-chain interaction, despite terminal fraying.
Materials and methods
Peptide synthesis and purification
All peptides were synthesized on Fmoc-PAL-PEG-PS amide resin by using standard Fmoc solid-phase protocols on a continuous flow Pioneer Peptide Synthesizer (Applied Biosystems) with HBTU/HOBT [2-(1H-benzotriazol-l-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate/N-Hydroxybenzotriazole] as the coupling reagent. Peptide resin cleavage and deprotection was performed simultaneously by using either 95% trifluoroacetic acid (TFA)/2.5% triisopropylsilane (TIPS)/2.5% H2O or 88% TFA /5% H2O /5% TIPS /2% phenol for 2 to 3 h under N2. The TFA was removed by distillation under reduced pressure. Crude peptides were precipitated with cold ether, extracted into water, and lyophilized. Cyclic peptides were synthesized following the procedure of Syud et al. (1999) using an orthogonally protected glutamic acid derivative and standard solid-phase synthesis.
Crude peptides were purified by reverse-phase HPLC using a Vydac C18 semipreparative column. Peptides were eluted with a linear gradient of 95% H2O /5% acetonitrile (solvent A) with 0.5% TFA and 95% acetonitrile/5% water with 0.5% TFA (solvent B) from 0% to 20% B. Peptides were detected by monitoring at 220 and 280 nm. Molecular weights were determined by MALDI mass spectrometry.
NMR data acquisition
One- and two-dimensional 1H NMR was performed by using INOVA 500- and 600-MHz instruments at 298 K, unless otherwise noted. Spectra were referenced to DSS.
Determination of fraction folded
The extent of folding of these peptides was determined by the Hα chemical shift (Wishart et al. 1992) and glycine chemical shift difference (Searle et al. 1999; Searle 2001; Tatko and Waters 2002) relative to control peptides for the random coil and fully folded states from the equation below:
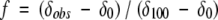 |
(2) |
where f is fraction folded and δ is chemical shift.
Assuming β-hairpin formation is a two-state process, the extent of folding can be related to the equilibrium constant, K, by the following equation:
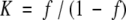 |
(3) |
The free energy change can then be determined from
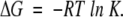 |
For the type II′ DPro-Gly turns, random coil values were determined from the corresponding LPro-Gly peptide because they have been shown not to fold (Stanger and Gellman 1998). α-Hydrogen regions were assigned by using COSY, TOCSY, and ROESY 1H NMR.
Aggregation studies
Peptides were analyzed by NMR using an INOVA 600-MHz spectrometer. The samples were analyzed at a concentration range of 1.3 to 9 mg/mL in D2O buffered with 100 mM sodium acetate-d3 containing 0.5 mM sodium 4,4-dimethyl-4-silapentane-1-sulfonate (DSS) as an internal reference at a pH 3.44. No change in glycine chemical shift difference was observed, indicating that the peptides are monomeric under the conditions studied.
Ionic strength studies
The effect of ionic strength on hairpin folding was studied by NMR using an INOVA 600-MHz spectrometer. The change in glycine chemical shift difference was compared in 100% D2O and 500 mM NaCl in D2O.
Determination of thermodynamic parameters
Variable temperature NMR was used in order to determine the thermodynamic parameters of the peptide folding. A temperature range of 282 to 326 K was explored in five-degree increments using an INOVA 600-MHz spectrometer. Temperature calibration was performed with ethylene glycol and methanol standards by using standard macros in Varian software. The change in glycine chemical shift difference was followed with temperature. The fraction folded of the peptide was plotted against temperature, and the curve was fitted by using the following equation (Maynard et al. 1998):
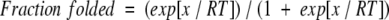 |
(4) |
where
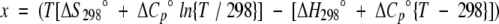 |
Acknowledgments
This work was supported by the National Science Foundation (CHE-0094068).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03215403.
References
- Blanco, F.J., Jimenez, M.A., Herranz, J., Rico, M., Santoro, J., and Nieto, J.L. 1993. NMR evidence of a short linear peptide that folds into a β-hairpin in aqueous-solution. J. Am. Chem. Soc. 115 5887–5888. [Google Scholar]
- Blasie, C.A. and Berg, J.M. 1997. Electrostatic interactions across a β-sheet. Biochemistry 36 6218–6222. [DOI] [PubMed] [Google Scholar]
- Ciani, B., Jourdan, M., and Searle, M.S. 2003. Stabilization of β-hairpin peptides by salt bridges: Role of preorganization in the energetic contribution of weak interaction. J. Am. Chem. Soc. 125 9038–9047. [DOI] [PubMed] [Google Scholar]
- Cootes, A.P., Curmi, P.M.G., Cunningham, R., Donnelly, C., and Torda, A.E. 1998. The dependence of amino acid pair correlations on structural environment. Proteins 32 175–189. [PubMed] [Google Scholar]
- Das, C., Shankaramma, S.C., and Balaram, P. 2001. Molecular carpentry: Piecing together helices and hairpins in designed peptides. Chem. Eur. J. 7 840–847. [DOI] [PubMed] [Google Scholar]
- de Alba, E., Blanco, F.J., Jimenez, M.A., Rico, M., and Nieto, J.L. 1995. Interactions responsible for the β-hairpin conformational population formed by a designed linear peptide. Eur. J. Biochem. 233 283–292. [DOI] [PubMed] [Google Scholar]
- de Alba, E., Jimenez, M.A., and Rico, M. 1997a. Turn residue sequence determines β-hairpin conformation in designed peptides. J. Am. Chem. Soc. 119 175–183. [Google Scholar]
- de Alba, E., Rico, M., and Jimenez, M.A. 1997b. Cross-strand side-chain interactions versus turn conformation in β-hairpins. Protein Sci. 6 2548–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa, J.F. and Gellman, S.H. 2000. A designed β-hairpin containing a natural hydrophobic cluster. Angew Chem. Int. Edit. 39 2330–2333. [DOI] [PubMed] [Google Scholar]
- Espinosa, J.F., Munoz, V., and Gellman, S.H. 2001. Interplay between hydrophobic cluster and loop propensity in β-hairpin formation. J. Mol. Biol. 306 397–402. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S.R., Maynard, A.J., and Searle, M.S. 1999. Dissecting the stability of a β-hairpin peptide that folds in water: NMR and molecular dynamics analysis of the β-turn and β-strand contributions to folding. J. Mol. Biol. 292 1051–1069. [DOI] [PubMed] [Google Scholar]
- Haque, T.S. and Gellman, S.H. 1997. Insights on β-hairpin stability in aqueous solution from peptides with enforced type I′ and type II′ β-turns. J. Am. Chem. Soc. 119 2303–2304. [Google Scholar]
- Kobayashi, N., Honda, S., Yoshii, H., and Munekata, E. 2000. Role of side-chains in the cooperative β-hairpin folding of the short C-terminal fragment derived from streptococcal protein G. Biochemistry 39 6564–6571. [DOI] [PubMed] [Google Scholar]
- Mandel-Gutfreund, Y., Zaremba, S.M., and Gregoret, L.M. 2001. Contributions of residue pairing to β-sheet formation: Conservation and covariation of amino acid residue pairs on antiparallel β-strands. J. Mol. Biol. 305 1145–1159. [DOI] [PubMed] [Google Scholar]
- Maynard, A.J., Sharman, G.J., and Searle, M.S. 1998. Origin of β-hairpin stability in solution: Structural and thermodynamic analysis of the folding of model peptide supports hydrophobic stabilization in water. J. Am. Chem. Soc. 120 1996–2007. [Google Scholar]
- Ramirez-Alvarado, M., Blanco, F.J., and Serrano, L. 1996. De novo design and structural analysis of a model β-hairpin peptide system. Nat. Struct. Biol. 3 604–612. [DOI] [PubMed] [Google Scholar]
- Ramirez-Alvarado, M., Blanco, F.J., Niemann, H., and Serrano, L. 1997. Role of β-turn residues in β-hairpin formation and stability in designed peptides. J. Mol. Biol. 273 898–912. [DOI] [PubMed] [Google Scholar]
- Ramirez-Alvarado, M., Blanco, F.J., and Serrano, L. 2001. Elongation of the BH8 β-hairpin peptide: Electrostatic interactions in β-hairpin formation and stability. Protein Sci. 10 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, S.J. and Cochran, A.G. 2000. Designing stable β-hairpins: Energetic contributions from cross-strand residues. J. Am. Chem. Soc. 122 12600–12601. [Google Scholar]
- Santiveri, C.M., Rico, M., and Jimenez, M.A. 2000. Position effect of cross-strand side-chain interactions on β-hairpin formation. Protein Sci. 9 2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiveri, C.M., Santoro, J., Rico, M., and Jimenez, M.A. 2002. Thermodynamic analysis of β-hairpin–forming peptides from the thermal dependence of H-1 NMR chemical shifts. J. Am. Chem. Soc. 124 14903–14909. [DOI] [PubMed] [Google Scholar]
- Scholtz, J.M., Qian, H., Robbins, V.H., and Baldwin, R.L. 1993. The energetics of ion-pair and hydrogen-bonding interactions in a helical peptide. Biochemistry 32 9668–9676. [DOI] [PubMed] [Google Scholar]
- Searle, M.S. 2001. Peptide models of protein β-sheets: Design, folding and insights into stabilising weak interactions. J. Chem. Soc. Perkin Trans. 2 1011–1020. [Google Scholar]
- Searle, M.S., Griffiths-Jones, S.R., and Skinner-Smith, H. 1999. Energetics of weak interactions in a β-hairpin peptide: Electrostatic and hydrophobic contributions to stability from lysine salt bridges. J. Am. Chem. Soc. 121 11615–11620. [Google Scholar]
- Smith, C.K. and Regan, L. 1995. Guidelines for protein design: The energetics of β-sheet side-chain interactions. Science 270 980–982. [DOI] [PubMed] [Google Scholar]
- ———. 1997. Construction and design of β-sheets. Acc. Chem. Res. 30 153–161. [Google Scholar]
- Smith, J.S. and Scholtz, J.M. 1998. Energetics of polar side-chain interactions in helical peptides: Salt effects on ion pairs and hydrogen bonds. Biochemistry 37 33–40. [DOI] [PubMed] [Google Scholar]
- Smithrud, D.B. and Diederich, F. 1990. Strength of molecular complexation of apolar solutes in water and in organic solvents is predictable by linear free energy relationships: A general model for solvation effects on apolar binding. J. Am. Chem. Soc. 112 339–343. [Google Scholar]
- Smithrud, D.B., Wyman, T.B., and Diederich, F. 1991. Enthalpically driven cyclophane-arene inclusion complexation: Solvent-dependent calorimetric studies. J. Am. Chem. Soc. 113 5420–5426. [Google Scholar]
- Stanger, H.E. and Gellman, S.H. 1998. Rules for antiparallel β-sheet design: D-Pro-Gly is superior to L-Asn-Gly for β-hairpin nucleation. J. Am. Chem. Soc. 120 4236–4237. [Google Scholar]
- Stanger, H.E., Syud, F.A., Espinosa, J.F., Giriatt, I., Muir, T., and Gellman, S.H. 2001. Length-dependent stability and strand length limits in antiparallel β-sheet secondary structure. Proc. Natl. Acad. Sci. 98 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syud, F.A., Espinosa, J.F., and Gellman, S.H. 1999. NMR-based quantification of β-sheet populations in aqueous solution through use of reference peptides for the folded and unfolded states. J. Am. Chem. Soc. 121 11577–11578. [Google Scholar]
- Syud, F.A., Stanger, H.E., and Gellman, S.H. 2001. Interstrand side chain–side chain interactions in a designed β-hairpin: Significance of both lateral and diagonal pairings. J. Am. Chem. Soc. 123 8667–8677. [DOI] [PubMed] [Google Scholar]
- Tatko, C.D. and Waters, M.L. 2002. Selective aromatic interactions in β-hairpin peptides. J. Am. Chem. Soc. 124 9372–9373. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Sykes, B.D., and Richards, F.M. 1991. Relationship between nuclear-magnetic-resonance chemical-shift and protein secondary structure. J. Mol. Biol. 222 311–333. [DOI] [PubMed] [Google Scholar]
- ———. 1992. The chemical-shift index: A fast and simple method for the assignment of protein secondary structure through NMR-spectroscopy. Biochemistry 31 1647–1651. [DOI] [PubMed] [Google Scholar]
- Wouters, M.A. and Curmi, P.M.G. 1995. An analysis of side-chain interactions and pair correlations within antiparallel β-sheets: The differences between backbone hydrogen-bonded and non-hydrogen–bonded residue pairs. Proteins 22 119–131. [DOI] [PubMed] [Google Scholar]
- Zaremba, S.M. and Gregoret, L.M. 1999. Context-dependence of amino acid residue pairing in antiparallel β-sheets. J. Mol. Biol. 291 463–479. [DOI] [PubMed] [Google Scholar]