Abstract
HAMLET (human α-lactalbumin made lethal to tumor cells) is a complex of human α-lactalbumin and oleic acid (C18:1:9 cis) that kills tumor cells by an apoptosis-like mechanism. Previous studies have shown that a conformational change is required to form HAMLET from α-lactalbumin, and that a partially unfolded conformation is maintained in the HAMLET complex. This study examined if unfolding of α-lactalbumin is sufficient to induce cell death. We used the bovine α-lactalbumin Ca2+ site mutant D87A, which is unable to bind Ca2+, and thus remains partially unfolded regardless of solvent conditions. The D87A mutant protein was found to be inactive in the apoptosis assay, but could readily be converted to a HAMLET-like complex in the presence of oleic acid. BAMLET (bovine α-lactalbumin made lethal to tumor cells) and D87A-BAMLET complexes were both able to kill tumor cells. This activity was independent of the Ca2+site, as HAMLET maintained a high affinity for Ca2+ but D87A-BAMLET was active with no Ca2+ bound. We conclude that partial unfolding of α-lactalbumin is necessary but not sufficient to trigger cell death, and that the activity of HAMLET is defined both by the protein and the lipid cofactor. Furthermore, a functional Ca2+-binding site is not required for conversion of α-lactalbumin to the active complex or to cause cell death. This suggests that the lipid cofactor stabilizes the altered fold without interfering with the Ca2+site.
Keywords: α-Lactalbumin, Ca2+-binding site, protein folding, HAMLET, tumor cell death
HAMLET (human α-lactalbumin made lethal to tumor cells) induces apoptosis-like death in tumor cells but spares most healthy differentiated cells (Hakansson et al. 1995; Svensson et al. 2000; Svanborg et al. 2003). After binding to the tumor cell surface, HAMLET is internalized and travels through the cytoplasm to the nucleus, where it disrupts the chromatin structure and causes DNA fragmentation (Hakansson et al. 1999; Svensson et al. 1999). In the process, HAMLET interacts with mitochondria, causing the release of cytochrome c and activation of the caspase cascade (Kohler et al. 1999, 2001). A detailed structural analysis of HAMLET is needed to understand the molecular basis for this multifaceted yet selective activation of death programs in tumor cells.
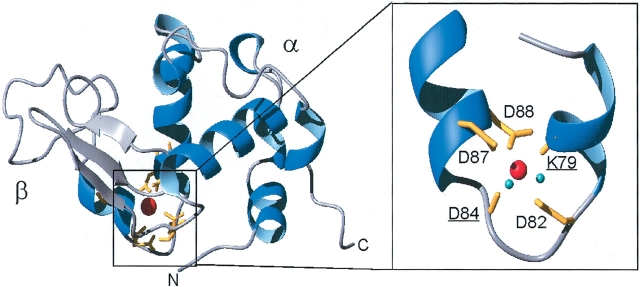
HAMLET is a molecular complex of partially unfolded human α-lactalbumin and oleic acid. α-Lactalbumin is the dominant protein in human milk, where it is present at a concentration of 2 mg/mL (140 μM). The mature protein consists of 123 amino acid residues (14 kD), and its three-dimensional structure has been determined to 1.7 Å resolution (Acharya et al. 1991), demonstrating four α-helices and a triple stranded antiparallel β-sheet (Fig. 1 ▶). Binding of Ca2+ to a single very high affinity Ca2+-binding site is required for the protein to maintain a native conformation. Five of the seven oxygens that ligate the Ca2+ are contributed by side-chain carboxylates of Asp residues at positions 82, 87, and 88 and by carbonyl oxygens of Lys 79 and Asp 84, and two water molecules supply the remaining ligands (Acharya et al. 1991). The bound Ca2+ brings the α-helical region and the β-sheet in close proximity, and two disulfide bonds flanking the Ca2+-binding site, make this part of the molecule fairly inflexible.
Figure 1.
3D structure of the α-lactalbumin Ca2+-binding site. Ribbon representation of the crystal structure of Ca2+-loaded human α-lactalbumin (Harata et al. 1999); PDB accession number 1B90. The Ca2+-binding site is magnified (rotated 180°) with the Ca2+-coordinating groups in yellow (K79 and D84 carbonyls, underlined, and D82, D87, and D88 carboxylate side chains). Ca2+ is red and Ca2+-coordinating water molecules are cyan. Figures were made in MOLMOL (Koradi et al. 1996) and rendered in Pov-Ray.
The Ca2+-bound form of α-lactalbumin cannot be converted to HAMLET, and does not induce cell death (Svensson et al. 2000). Following the removal of Ca2+, however, the destabilized apo state of α-lactalbumin can incorporate oleic acid, form HAMLET, and thus acquire the ability to kill tumor cells. In HAMLET, α-lactalbumin can maintain a partially unfolded conformation also in the presence of Ca2+. This is quite surprising, as the apo state is unstable, and much more sensitive to the solution conditions than the native state. Unfolded states of α-lactalbumin only persist in the absence of metal ions or at low pH, and the protein reverts to the native state when solvent conditions change. Our experiments show that the removal of Ca2+ induces the conformational change needed to form HAMLET, but have not defined the role of the Ca2+-binding site or the unfolded protein per se for the novel biologic activity. The high affinity for Ca2+ and requirement for Ca2+ in the apoptosis assay have precluded experiments testing the activity of the apo protein. It thus remains unclear if a conformational change might be sufficient to attain the new biological activity.
We have used site-directed mutagenesis of Ca2+-ligating amino acids to generate α-lactalbumin that maintains a partially unfolded state regardless of solvent conditions. The mutant proteins were used to examine if the resulting conformational change makes α-lactalbumin able to kill tumor cells. Furthermore, the mutant proteins were used to investigate the Ca2+ affinity of HAMLET and the importance of the Ca2+-binding site for the activity against tumor cells.
Results
Bovine α-lactalbumin can be converted to BAMLET (bovine α-lactalbumin made lethal to tumor cells)
The ability to induce apoptosis in tumor cells is a unique feature of HAMLET, not shared by native human α-lactalbumin. In view of the structural homology between human and bovine α-lactalbumin it should be possible to convert bovine α-lactalbumin to a HAMLET-like molecule, with apoptosis-inducing properties. Hence, we subjected bovine α-lactalbumin to the conversion conditions previously used for the conversion of human α-lactalbumin to HAMLET. A folding change was induced by removal of Ca2+ with EDTA, the Ca2+-free protein was applied to an oleic acid conditioned ion exchange matrix and the eluate after high salt was analyzed.
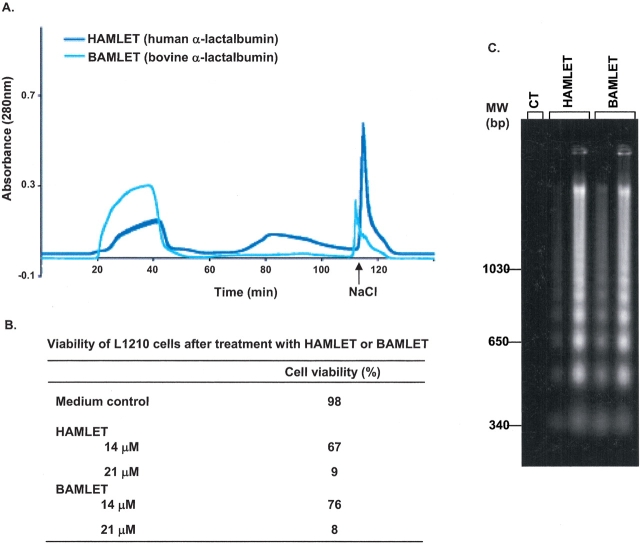
A large proportion of the applied bovine protein eluted in the void (about 60%), but a small sharp peak eluted after 1 M NaCl (Fig. 2A ▶). In the apoptosis assay, the converted bovine protein reduced cell viability from 98% to 8% at 21 μM, after 6 h incubation and induced DNA fragmentation (Fig. 2C ▶). There was no apparent difference in efficiency of apoptosis induction between HAMLET and the bovine equivalent, BAMLET (Fig. 2 ▶). We conclude that bovine apo α-lactalbumin can be converted in the presence of C18:1 to a molecular complex that induces apoptosis, and named this complex BAMLET (bovine α-lactalbumin made lethal to tumor cells). BAMLET cannot be generated from bovine milk, however, due to a lack of C18:1 fatty acids.
Figure 2.
Conversion of bovine α-lactalbumin to BAMLET. (A) Ion exchange chromatography of human (blue) and bovine (cyan) α-lactalbumin on a C18:1 conditioned column eluted with a NaCl gradient. The proteins were treated with EDTA to remove Ca2+. (B) Viability of L1210 cells after 6-h exposure to HAMLET, BAMLET, or medium. (C) Agarose gel electrophoresis of DNA extracts from L1210 cells after 6-h exposure to HAMLET, BAMLET, or medium (CT). The two lanes for each protein correspond to DNA extracts from cells exposed to 14 μM and 21 μM of each protein, respectively.
Spectroscopic characterization of BAMLET and bovine α-lactalbumin in cell culture medium
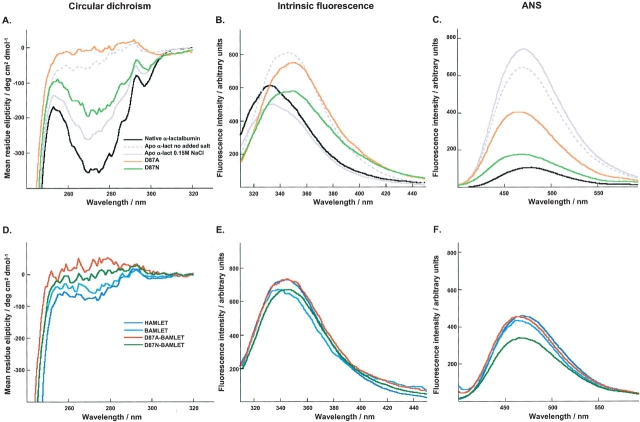
In cell culture medium, native bovine α-lactalbumin shows the characteristic near-UV CD spectrum of a well-folded protein with a negative tyrosine dip and a positive tryptophan peak (Fig. 3A ▶). The intrinsic fluorescence spectrum of the native protein in medium is blue shifted, indicating that the tryptophan residues are buried in the hydrophobic core (Fig. 3B ▶). Native bovine α-lactalbumin does not appear to bind ANS when dissolved in buffer (data not shown), but there is a slight blue shift and fluorescence intensity increase in medium (Fig. 3C ▶).
Figure 3.
Optical spectra. Optical spectra obtained in cellular medium for native bovine α-lactalbumin (black), D87A α-lactalbumin (orange), D87N α-lactalbumin (light green), HAMLET (blue), BAMLET (cyan), D87A-BAMLET (red), D87N-BAMLET (green), and in water at pH 7.3 for apo α-lactalbumin without (dashed gray line) or with 0.15 M NaCl (gray). (A, D) Near UV CD spectra. (B, E) Intrinsic fluorescence spectra. (C, F) ANS fluorescence spectra at 1.5 equivalents of ANS.
The apo control spectra were recorded in water at pH 7 with no added salt or with 0.15 M NaCl. At low salt, the bovine apo protein has very low signal in both the tyrosine and tryptophan regions of the near-UV CD spectrum (Fig. 3A ▶), indicative of a molten globule-like structure with rapidly rotating aromatic side chains. At physiological salt, significant signals are observed, suggesting that under these conditions the apo state has a more native-like organization (Chrysina et al. 2000; Permyakov et al. 2001a). At low salt, the intrinsic fluorescence spectrum of the apo form indicates solvent-exposed tryptophan side chains, whereas addition of physiological salt produces a native-like spectrum with blue-shifted signal indicating that the tryptophan residues are more buried (Fig. 3B ▶). Furthermore, the apo form displays significant ANS binding with a maximum at 470 nm and enhanced intensity both at low and high salt (Fig. 3C ▶). It was not possible to study the apo protein in Ca2+-containing cell culture medium.
The conformation of BAMLET was compared to HAMLET and native bovine α-lactalbumin in medium and to the bovine apo control at low and 0.15 M NaCl (Fig. 3D–F ▶). The bovine complex strongly resembled HAMLET and the apo control at low salt. BAMLET and HAMLET had very similar near-UV CD and tryptophan fluorescence spectra, and both bound ANS (Fig. 3D–F ▶).
Mutations of the Ca2+-binding site of bovine α-lactalbumin
Site-directed mutagenesis of the aspartic acid at position 87 to alanine (D87A) was shown to inactivate the strong Ca2+-binding site (Anderson et al. 1997; Fig. 1 ▶) and the mutant protein remained Ca2+-free under the conditions of the cellular assays. The conformation of D87A in cell culture medium was investigated using optical spectroscopy. By near-UV CD spectroscopy, a nearly complete loss of ellipticity was observed demonstrating loss of tertiary structure (Fig. 3A ▶) and the intensity maximum in the intrinsic tryptophan spectrum was shifted towards higher wavelengths, indicating exposure of tryptophan to solvent (Fig. 3B ▶). The D87A mutant bound ANS, indicating exposed hydrophobic surfaces, as shown by the height of the curve and the shift of the intensity maximum to shorter wavelengths (Fig. 3C ▶). No spectral changes were observed following the addition of excess EDTA or Ca2+ (1 mM), showing that the D87A mutant did not bind Ca2+ under these conditions (Anderson et al. 1997). These results confirm that the D87A mutation locks the protein in a molten globule-like conformation, which is insensitive to the Ca2+ conditions. The spectra of the D87A mutant in cellular medium most closely resemble those of the apo form of the wild-type protein at low ionic strength.
To minimize the structural distortion in the mutant protein, D87 was also replaced by an asparagine (N) (Permyakov et al. 2001b), which lacks the negative charge of a carboxylate group, but has the same side-chain volume and geometry as aspartic acid. The mutant protein (D87N) was induced because it was shown to bind Ca2+ with reduced affinity (Ka = 2 × 105 M−1).
In cell culture medium, the near-UV CD spectrum was intermediate between the native and the D87A spectra, with approximately half the ellipticity of native α-lactalbumin (Fig. 3A ▶), and the intrinsic fluorescence spectrum was red shifted compared to native protein, suggesting exposed tryptophans and a loss of tertiary structure (Fig. 3B ▶). The D87N mutant bound ANS demonstrating exposed hydrophobic surfaces (Fig. 3C ▶). Addition of EDTA or Ca2+ (1 mM) had only a marginal effect on the near-UV CD ellipticity (Permyakov et al. 2001b). We conclude that a larger fraction of the D87N protein is in a conformation with better-defined tertiary structure than D87A, but it is still partially unfolded relative to the native protein. D87N, hence, represents an intermediate case between the wild-type protein and D87A.
The D87A and D87N mutants fail to induce apoptosis
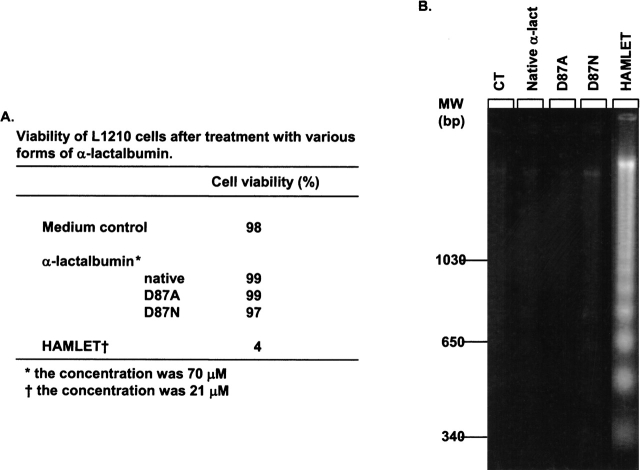
The ability of the mutant proteins to induce apoptosis was tested using the L1210 cell line. The proteins were suspended in cell culture medium at 0.14 mM and the cell viability was determined after 6 h of incubation as was the DNA fragmentation. The HAMLET control induced apoptosis at 21 μM, but the mutant proteins had no effect (Fig. 4 ▶).
Figure 4.
Apo α-lactalbumin does not induce apoptosis. (A) Viability of L1210 cells after 6-h exposure to native α-lactalbumin, D87A-α-lactalbumin, D87N-α-lactalbumin, HAMLET, or medium control. (B) Agarose gel electrophoresis of DNA extracts from the cells described in A.
We concluded that partially unfolded α-lactalbumin without associated oleic acid does not induce apoptosis in tumor cells.
Conversion of the mutant proteins to BAMLET
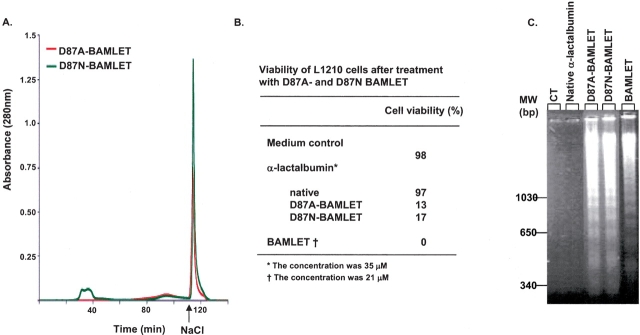
The D87A and D87N α-lactalbumin mutants were subjected to the conversion conditions previously used for bovine α-lactalbumin to BAMLET. The yields were much higher for D87A (>90%) and D87N (>95%; Fig. 5A ▶). D87A–BAMLET and D87N–BAMLET reduced cell viability from 98% to 13% and 17%, respectively, and induced DNA fragmentation (Fig. 5B,C ▶). The LD50 values of the mutant complexes (28 μM) were slightly higher than for BAMLET and HAMLET (14 μM).
Figure 5.
Conversion to D87A- and D87N-BAMLET. (A) Ion exchange chromatography of the D87A mutant (red) and the D87N mutant (green) on a C18:1 conditioned column eluted with a NaCl gradient. D87A was applied without EDTA, while D87N mutant was treated with EDTA to remove residual Ca2+ before it was applied to the column. The material eluted as a sharp peak after high salt (arrow) was named D87A- and D87N-BAMLET. (B) Viability of L1210 cells after 6-h exposure to native α-lactalbumin, D87A- and D87N-BAMLET, or medium. (C) Agarose gel electrophoresis of DNA extracts from the cells described in A.
We concluded that both mutants can be converted to active complexes, albeit with somewhat lower apoptosis-inducing activity than BAMLET. Ca2+ removal prior to oleic acid treatment was not required for the D87A mutant.
Spectroscopic characterization of D87A– and D87N–BAMLET mutants
The conformations of D87A– and D87N–BAMLET in cellular medium were compared to the bovine apo control in water and to BAMLET and native bovine α-lactalbumin in medium (Fig. 3D–F ▶). The CD spectrum of D87A–BAMLET was very similar to the unconverted D87A protein with virtually no ellipticity showing that D87A–BAMLET is in a molten globule-like conformation (Fig. 3D ▶). The spectrum of D87N–BAMLET was virtually identical to that of BAMLET, with reduced ellipticity in both the tyrosine and tryptophan regions (Fig. 3D ▶). By intrinsic fluorescence spectroscopy both D87A– and D87N–BAMLET showed intensity maxima at 345 nm with a shoulder at 355 nm strongly resembling BAMLET and the apo α-lactalbumin control, suggesting that tryptophans are accessible to solvent (Fig. 3E ▶). D87A– and D87N–BAMLET bound ANS with the intensity maxima shifted to 470 nm and an increased quantum yield compared to the native control, indicating exposed hydrophobic surfaces in all proteins. The spectra were similar to BAMLET and to the apo-α-lactalbumin control (Fig. 3F ▶).
We concluded that D87A–BAMLET and D87N– BAMLET are in partially folded states, with structural and functional properties resembling HAMLET and BAMLET. As the mutated BAMLET proteins maintain their biologic activity, we further conclude that a functional Ca2+-binding site was not required for the apoptotic function of this protein.
Ca2+-binding to α-lactalbumin and the HAMLET complex
The Ca2+ affinity of α-lactalbumin has been extensively studied, mainly for the bovine protein (Nitta et al. 1988; Kronman 1989; Aramini et al. 1996; Anderson et al. 1997; Griko and Remeta 1999). The reported values (Table 1) vary between 2.5 × 106 and 5.7 × 108 M−1, probably reflecting the experimental conditions in terms of temperature, buffer, ionic strength, salt, and protein preparation.
Table 1.
Ca2+ association constants (Ka) for α-lactalbumin or HAMLET
| Protein | Ka (M−1) | Buffer | Salt | pH | T (K) | Method | Reference |
| α-lac human | 1.8 • 109 | 2 mM Tris/HCl | — | 298 | Chelator | This study | |
| α-lac human | 8.3 • 106 | 2 mM Tris/HCl | 150 mM | 298 | Chelator | This study | |
| NaCl | |||||||
| HAMLET | 5.9 • 108 | 2 mM Tris/HCl | — | 298 | Chelator | This study | |
| HAMLET | 5.3 • 106 | 2 mM Tris/HCl | 150 mM | 298 | Chelator | This study | |
| NaCl | |||||||
| D87A-BAMLET | n.d.* | 2 mM Tris/HCl | — | 298 | Chelator | This study | |
| α-lac bovine | 4.3 • 108 | 10 mM Tris/HCl | — | 7.5 | 298 | ITC | a |
| α-lac goat | 5.7 • 108 | 10 mM Tris/HCl | — | 7.5 | 298 | ITC | a |
| α-lac bovine | 2.85 • 108 | 10 mM Tris/HCl | — | 8 | — | DSC | b |
| α-lac bovine | 4.8 • 107 | 10 mM Ammoinium Bicarbonate | 7.8 | EGTA, Fluoroscence | c | ||
| α-lac bovine, human | 2.0 • 107 | H2O | 100 mM | 7 | 294 | EDTA, 43Ca-NMR | d |
| KCl | |||||||
| α-lac bovine | 2.7 • 106 | 20 mM Tris | — | 7.5 | 298 | 1) Fluorescence 2) Hummel & Dryer method |
e,f |
| α-lac | 5 mM Tris, 0.1 | — | 7.2 | 298 | CD 270 nm | g | |
| 1) bovine | 1) 2.5 • 108 | mM EDTA | |||||
| 2) human | 2) 3 • 108 | ||||||
| 3) goat | 3) 2.8 • 108 | ||||||
| Bovine | 2.0 • 107 | H2O | 100 mM KCl | 8 | 294–298 | EGTA 13C-NMR | h |
| Bovine | 1) 2.0 • 107 | 50 mM Hepes | — | 8 | 1) 310 | Fluorescence | i |
| 2) 4.0 • 108 | 2) 293 | ||||||
| Bovine | 2.5 • 106 | 20 mM Tris | — | 7.5 | 298 | Microcalorimetry | j |
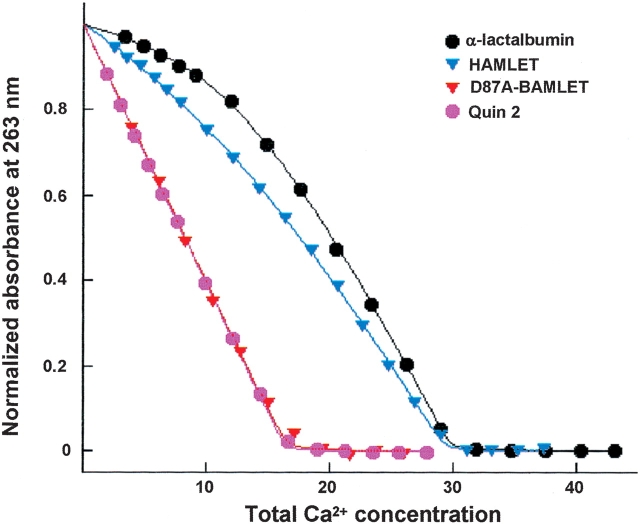
We examined the Ca2+ affinity of the different forms of α-lactalbumin in this study, by titration with Ca2+ in the presence of the chromophoric chelator quin 2 (25°C, pH 7.5). Ca2+-binding to quin 2 is monitored, while α-lactalbumin competes with the chelator for Ca2+, and the absorbance change following Ca2+-binding to the protein is negligible at the wavelength used. The titration curve is very sensitive to the differences in affinity between chelator and protein, so this ratio can be obtained from the data with high precision. Normalized data (absorbance versus total Ca2+ concentration) at low salt are shown in Figure 6 ▶, and the resulting Ca2+-binding constants for the proteins at physiological (0.15 M NaCl) and low (no added) salt are listed in Table 1 (for human α-lactalbumin). We found a Ka of 1.8 • 109 M−1 at low salt (KD = 0.56 • 10−9 M). The Ca2+ ion bound more weakly at physiological salt concentrations (0.15 M NaCl): Ka = 8.3 • 106 M−1 (KD = 1.2 • 10−7 M).
Figure 6.
Ca2+ titrations in the presence of quin 2. The absorbance is shown as a function of total Ca2+ concentration for quin 2 mixed with human α-lactalbumin (black), HAMLET (blue), or D87A-BAMLET (red), and quin 2 alone (purple). The solid lines are the fitted curves. The absorbance is normalized using the fitted values for completely Ca2+-free and Ca2+-loaded system, respectively.
HAMLET was shown to bind Ca2+ (Fig. 6 ▶). The Ca2+-binding constant for HAMLET was Ka = 5.9 • 108 M−1 (KD = 1.7 • 10−9 M) at low salt, and Ka = 5.3 • 106 M−1 (KD = 1.9 • 10−7 M) at physiological salt concentrations. Ca2+-titration data for D87A–BAMLET showed no difference compared to the titration of quin 2 alone (Fig. 6 ▶), confirming that D87A–BAMLET has lost the functional Ca2+-binding site.
We conclude that HAMLET maintains a high Ca2+ affinity in both a low and physiological salt environment. Hence, the Ca2+ affinity is only three times lower for HAMLET than for α-lactalbumin when no salt is added and 1.6 times lower at physiological salt concentration.
Discussion
α-Lactalbumin acts as a tumor-cell killer when partially unfolded and bound to a specific lipid cofactor (Svensson et al. 2000). Release of Ca2+ unfolds the protein during the conversion to HAMLET, but it has remained unclear if unfolding of the protein is sufficient for the change in activity. The high affinity for Ca2+ causes α-lactalbumin to revert to the native state, and this has precluded experiments testing the activity of the apo protein in the cellular assays. This study demonstrated that unfolded α-lactalbumin does not induce cell death. We used the Ca2+ site mutant D87A in bovine α-lactalbumin, which is unable to bind Ca2, and thus maintains a partially unfolded state regardless of solvent conditions. The D87A mutant protein was inactive in the apoptosis assay, but could readily be converted to a HAMLET-like complex in the presence of oleic acid. The mutant protein bound the C18:1 fatty acid cofactor and formed molecular complexes that killed tumor cells with an efficiency similar to HAMLET. We conclude that the unfolded state of the protein and the specific fatty acid are required to form HAMLET. The efficient conversion of the Ca2+-free α-lactalbumin mutant D87A– to D87A–BAMLET further emphasized that Ca2+-binding is not required for HAMLET to be active in the cellular assays.
The Ca2+-binding site is 100% conserved with a few exceptions in α-lactalbumins from different species (Acharya et al. 1991), illustrating the importance of this function for the protein. The Ca2+ ion is coordinated by five different amino acids and two water molecules. The side-chain carboxylate of D87 together with D88 chelate the bound Ca2+ ion and form internal hydrogen bonds that stabilize the structure (Anderson et al. 1997). A loss of either D87 or D88 has been shown to impair Ca2+-binding, and to render the molecule stable in the partially unfolded state (Anderson et al. 1997). This study used two different point mutations in the Ca2+-binding site of bovine α-lactalbumin. Substitution of the aspartic acid at position 87 by an alanine (D87A) totally abolished Ca2+-binding and disrupted the tertiary structure. After substitution of the aspartic acid by asparagine, the protein (D87N) still bound Ca2+ but with lower affinity, and showed a partial loss of tertiary structure, although not as pronounced as for the D87A mutant (Permyakov et al. 2001b). The D87N mutant protein showed a minimal change in packing volume as both amino acids have the similar average volume of 91–96 Å3 (Creighton 1993) and the oxygen of the amide side chain of asparagine allows the protein to coordinate Ca2+, but less efficiently (Permyakov et al. 2001b). Both mutant proteins were stable in partially unfolded conformations at physiologic temperatures, but despite this conformational change they were biologically inactive in the apoptosis assay. The results demonstrate that a conformational change to a partially unfolded state is not sufficient to induce apoptosis.
Native bovine α-lactalbumin and the Ca2+ mutants could be converted to the HAMLET-like complex named BAMLET, showing that the same fatty acid stabilizes bovine α-lactalbumin in the BAMLET conformation. The conversion yield was lower, however, suggesting either that lipid binding to the bovine protein is less efficient, or that the bovine polypeptide chain is less prone to switching conformation. Bovine and human α-lactalbumin show 76% amino acid sequence identity and have similar native conformations (Wijesinha-Bettoni et al. 2001). The divergent sequences are mainly located in the α-helical region (A-helix 57%, B-helix 50%, C-helix 23%, and 310-helix 25% difference) but this region is unlikely to be involved in fatty acid binding (Svensson et al. 2003). We have proposed that the lipid-binding site in human α-lactalbumin is located in the grove between the α-helical and the β-sheet domains, which becomes exposed in the apo-protein. This region of the molecule differs between the bovine and the human proteins, in that one of the three basic amino acids (R70) is changed to S70 in bovine α-lactalbumin, thus eliminating one potential coordinating side chain. The native bovine protein may also be more stable, and less prone to adopting an unfolded conformation than human protein. As a consequence, a smaller fraction of the bovine protein would be converted to BAMLET. The molecular basis for the difference in conversion efficacy between the human and the bovine proteins needs further study.
The obtained Ca2+ affinity of α-lactalbumin at low salt is higher than previously reported, but our value at 0.15 M NaCl falls in the range reported by others at 0.1 M NaCl (Kronman 1989, and references therein; Table 1). The difference may be explained by the lower ionic strength (about 2 mM compared to 10 mM and up) as we observed that the Ca2+ affinity for α-lactalbumin decreased by a factor of 200 between low and physiological ionic strength. This is expected from salt screening of electrostatic interactions, and similar decreases have been obtained with other negatively charged proteins. For example, the Ca2+ affinity of calbindin D9k decreases by a factor of 100 between low and physiological salt (Linse et al. 1991b), and of calmodulin by a factor of 70 (Linse et al. 1991a). The decrease in Ka of α-lactalbumin at physiological salt may also be due to competition between Na+ and Ca2+ for the same site, as Na+ has been reported to bind to and stabilize α-lactalbumin in vitro (Ka = 100 M−1 at 20°C) (Desmet et al. 1987). The lack of near-UV CD signals for D87A in the presence of NaCl is a striking contrast to wild-type α-lactalbumin, suggesting that Na+ binds to the Ca2+ site of the wild-type protein, and that the D87 side chain is also important for coordination of this monovalent ion.
HAMLET retained a high affinity for Ca2+ at both low and physiological salt concentrations, showing that HAMLET can bind Ca2+ without loss of activity. This may appear surprising, as partially unfolded conformations of α-lactalbumin usually are associated with the Ca2+-free state. Two possible explanations may be offered. In the first and most likely scenario, α-lactalbumin converts to HAMLET by unfolding and binding of oleic acid with little disturbance of the α-helical domain. The Ca2+-binding site may then retain a similar affinity as in the absence of oleic acid. A second possibility is that the Ca2+ site is disrupted and that the observed Ca2+-binding is explained by the generation of a new Ca2+ site in HAMLET. The head group of oleic acid might potentially coordinate Ca2+ together with amino acid residues. In this case, the classical Ca2+-binding site in α-lactalbumin would not be needed for Ca2+-binding to HAMLET. We find this a less likely explanation, as D87A–BAMLET has the oleic acid bound to the unfolded protein, and thus should form the new Ca2+ site, but does not bind Ca2+. We conclude that the Ca2+-binding site is not involved in the conversion of α-lactalbumin to an apoptosis-associated conformation, and that the structural changes associated with Ca2+-binding to HAMLET do not hinder the novel function.
In HAMLET, α-lactalbumin retains a partially unfolded conformation as well as a high-affinity Ca2+-binding site. This apparent paradox sheds new light on the molecular characteristics of α-lactalbumin in the complex. The X-ray structure of the native like apo form shows that the α and β regions are largely intact, while the cleft between them is widened (Chrysina et al. 2000). In the accompanying publication (Svensson et al. 2003), we suggest that oleic acid binds in the interface between the α and the β domains, and that the bound oleic acid locks this region of the molecule, while allowing the α-domain to maintain a native-like conformation. This is supported by our finding that HAMLET binds Ca2+ while retaining its activity against tumor cells. HAMLET is therefore in a different molecular state than either the low salt apo α-lactalbumin or the native-like apo form in physiological salt.
Our results demonstrate that the change in biologic function of α-lactalbumin requires not just a conformational change of the protein, but also the lipid cofactor. The need for these two independent events may have evolved to control where and when folding variants arise, and where the new function is needed, thus providing a mechanism for tissue specificity. To promote the new function, local environments must favor the altered protein fold, and provide the lipid cofactor. This role may be to compartmentalize and ensure different functions in different contexts, that is, lactose production in the mammary gland, and HAMLET formation under the conditions in the stomach of the nursing child. The acidic pH of the stomach precipitates casein and Ca2+ is released from α-lactalbumin. The low pH also activates pH-sensitive lipases that release oleic acid from the milk phospholipids. It is interesting to note that α-lactalbumin and oleic acid, respectively, are the most abundant proteins and fatty acid in human milk. We propose that lipids may function as “postsecretion chaperones,” involved in the adaptation of proteins to shifting external environments. The need for both a folding change and a tissue specific lipid makes sense to protect tissues from the protein folding variants that arise in response to metabolic stress or inflammation, and to target the site where the novel function is needed.
Materials and methods
Chemicals
The chromophoric chelator quin 2 and ANS were obtained from Fluka Chemie AG. Other chemicals were of highest obtainable laboratory quality. To produce Ca2+-free buffers, membrane tubing (M.W. cutoff: 3500, Spectrum Medical Industries Inc., boiled four times in doubly distilled water before use) was filled with 10 mL Chelex 100 (BioRad), sealed, and stored in the solutions to absorb Ca2+.
Purification of human and bovine α-lactalbumin and bovine α-lactalbumin mutants
Native human α-lactalbumin was purified from human milk by ammonium sulphate precipitation and phenyl sepharose chromatography as described (Svensson et al. 2000). Bovine α-lactalbumin was both purchased from Sigma, and purified from bovine milk using ammonium sulphate precipitation and phenyl sepharose chromatography (Svensson et al. 2000). The mutated proteins (D87A and D87N) were expressed in Escherichia coli, purified, folded, and lyophilized as described (Anderson et al. 1997; Permyakov et al. 2001b). The purity of the protein was assayed by SDS-PAGE and agarose gel electrophoresis, and by NMR spectroscopy.
Apo human or bovine α-lactalbumin for spectroscopic and Ca2+-binding studies was generated by dissolving α-lactalbumin in doubly distilled water containing a 10-fold molar excess of EGTA at pH 8.0. The sample was applied to a G-25 gel filtration column after an aliquot of saturated NaCl (Ca2+-depleted) and eluted by doubly distilled water. The sample was passed through the saturated NaCl to reduce binding of EGTA to the protein, and protein free from both Ca2+ and EGTA eluted in the water. The residual Ca2+ content was below 0.1 equivalents as estimated from the titration in the presence of quin 2, as described below.
Ca2+- and EDTA-free HAMLET was generated as described before (Svensson et al. 2000) with the following adaptions. All buffers were stored with chelex (preparation described above) on a tipping board for a minimum of 5 days before use. Only plastic vials were used. The FPLC system (BioRad Biologic), including the 20-mL ion exchange column and all tubings, was washed with 2 volumes of 100 mM EDTA, pH 8.5, and then rinsed with at least 10 volumes of Millipore water. This was followed by 2 volumes of buffer before the application of oleic acid. EGTA-free apo α-lactalbumin (20 mg) in 80-mL Ca2+-free buffer was applied to the column in four consecutive runs (20 mL in each run). The collected fractions were pooled, dialyzed, lyophilized, and checked for activity on tumor cells. The Ca2+ content was assayed using atomic absorption spectroscopy or titration in the presence of quin 2 (described below). The product was also characterized by agarose electrophoresis and 1H-NMR.
Anion-exchange chromatography
A column (14 × 1.6 cm) packed with DEAE-Trisacryl M (BioSepra) was attached to a Bio-Logic chromatography system (BioRad Laboratories), and eluted with a NaCl gradient (buffer A, 10 mM Tris/HCl pH 8.5; buffer B, buffer A containing 1 M NaCl). The matrix was conditioned with oleic acid (Sigma). Ten milligrams of oleic acid was dissolved in 100 μL 99.5% ethanol by sonication (3 min using a Branson 2200 bath sonicator; Branson). After addition of 10 mL of 10 mM Tris/HCl, pH 8.5, the lipid solution was applied to a newly packed DEAE-Trisacryl M matrix and dispersed through out the matrix using a NaCl gradient. Ten milligrams of human or bovine α-lactalbumin were dissolved in 20 mL of 10 mM Tris/HCl pH 8.5 containing 0.08 mM EDTA (except for D87A, which does not bind Ca2+) and added to the column. The protein fraction eluting after high salt was desalted by dialysis (Spectra/Pore, Spectrum Medical Industries, membrane cutoff 3.5 kD) against distilled water with at least four changes of water in 100-fold volume excess, and then lyophilized. The EDTA concentration used in the conversion protocol is suboptimal in terms of Ca2+ removal from the protein. At an ionic strength of about 10 mM and at pH 8.5, EDTA binds Ca2+ ca. 4–10-fold more strongly than α-lactalbumin (Skoog and West 1976; Table 1). Assuming that the purified α-lactalbumin contains one equivalent of Ca2+ after extensive dialysis against Ca2+-depleated water, and with 35 μM protein, 80 μM EDTA and a total Ca2+ concentration of about 35 μM, we calculate that 90%–96% of the protein is in the apo state, using simple equilibria. Higher EDTA concentrations have been found to increase the fraction of the protein in the apo state, but decrease the yields in conversion to HAMLET/BAMLET, probably due to EDTA binding to the protein, and/or a slight increase in the ionic strength due to the addition of tetravalent EDTA ions.
Spectroscopic analyses
The proteins or protein fractions were dialyzed against doubly distilled water and lyophilized. Stock solutions were prepared by dissolving the lyophilized material in the cellular medium or in 0.15 M NaCl, and concentrations determined based on the absorbance at 280 nm. CD and fluorescence spectra were obtained at 37°C for proteins dissolved in the same medium as used in the cellular assays (cf. below). The medium is a clear liquid and the major inorganic ions are 0.42 mM Ca2+, 0.41 mM Mg2+, 134 mM Na+, 5.3 mM K+, 112 mM Cl−, 24 mM carbonate, and 5.6 mM phosphate. The baseline obtained with medium alone was of low intensity and subtracted from the spectra. NMR spectra were obtained in 150 mM NaCl at 37°C, because signals from amino acids and nutrients would dominate the NMR spectrum if obtained in medium.
Circular dichroism (CD) spectra were obtained on a JASCO J-720 spectropolarimeter with a JASCO PTC-343 Peltier type thermo stated cell holder. Quartz cuvettes were used with 1-cm path length, and spectra were recorded at 37°C between 240 and 320 nm. The wavelength step was 1 nm, the response time 8 sec and the scan rate was 10 nm per min. Six scans were recorded and averaged for each spectrum. Baseline spectra were recorded with pure buffer in the cuvette and subtracted from the protein spectra.
The mean residue ellipticity θm was calculated from the recorded ellipticity, θ, as
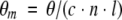 |
where c is the protein concentration in M, n the number of residues in the protein (123 in this case), l the path length in m, and θ is the ellipticity in degrees.
Fluorescence spectra were recorded at 37°C on a Perkin-Elmer LS-50B spectrometer using a quartz cuvette with 1-cm excitation path length. Intrinsic (tryptophan) fluorescence emission spectra were recorded between 305 and 530 nm with excitation at 295 nm. The excitation bandwidth was 3 nm and the emission bandwidth was 5 nm. ANS fluorescence emission spectra were recorded between 400 and 600 nm with excitation at 385 nm. Both the excitation and emission band pass were set to 5 nm. ANS ammonium salt solution (Fluka) was added stepwise and the spectra at 1.5 molar equivalents are shown.
1H-NMR spectroscopy
1H-NMR spectra were recorded using an Omega 500 spectrometer at 500 MHz, or a Varian-Inova Unity Plus spectrometer at 600 MHz, at 37°C, for 1–2 mM (Ca and EDTA-free HAMLET) or 70–140 μM (mutant BAMLETs) solutions of protein. Lyophilized protein was dissolved in D2O with 0.15 M NaCl, and the pH was set to 7.0 using NaOH.
Ca2+-binding studies
To study the Ca2+-binding equilibrium, each protein was titrated with Ca2+ in the presence of a chromophoric chelator, quin 2, for which the absorbance at 263 nm decreases (~ 85%) upon Ca2+-binding. The method (Linse 2002) relies on competition for Ca2+ between the protein and chelator, and can be used to quantitate high-affinity sites (ca. 107−5 • 109 M−1 at low salt, and ca. 5 105–1 • 108 M−1 at 0.15 NaCl) when quin 2 is the chelator. The exact concentration of the chelator solution (in the range of 25–30 μM) was calculated from the absorbance at 239.5 nm in the presence of excess Ca2+ (using ɛ239.5 = 4.2 × 104 L mole−1 cm−1 for quin 2). Lyophilized protein was dissolved in Ca2+-free (<1 μM Ca2+) chelator solution at a concentration of 25–30 μM. The absorbance at 263 nm (A263) was recorded for the protein/chelator solution using a UV/Vis 920 spectrophotometer (GBC Scientific Equipment Pty Ltd). Ca2+ solution (3 mM CaCl2 in 2 mM Tris/HCl, pH 7.5, with or without 0.15 M NaCl) was added in portions of 4 μL. A263 was recorded after each Ca2+ addition. The titration was continued until no absorbance change was seen for the last five additions. A more concentrated Ca2+ stock (10 mM) was used at the end of the titrations of the proteins in 0.15 M NaCl. The data analysis consists of computer fitting directly to the measured quantity: absorbance versus total Ca2+ concentration. The fitting was performed with the CaLigator software (Andre and Linse 2002). Initial Ca2+ concentration and the dilution effect of the Ca2+ additions were taken into account in the fits. The calculated absorbance, Acalc, at all data points for each set of trial parameters was compared to the measured absorbance to calculate the sum of residuals, χ2, which was minimized in an iterative procedure.
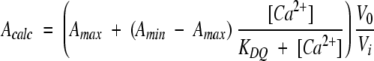 |
(1) |
where the free Ca2+ concentration, [Ca2+], at each titration point i was solved from
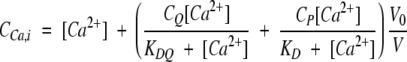 |
(2) |
where CQ is the initial total chelator concentration and KDQ is the equilibrium dissociation constant for the chelator-Ca2+ complex. KD is the equilibrium dissociation constant of the Ca2+-protein complex. CP is the initial total protein concentration. Amax is the absorbance in Ca2+ free solution. Amin is the absorbance in a solution with fully Ca2+ saturated chelator and protein. CCa is the total Ca2+ concentration as calculated from the initial and added Ca2+ and corrected for the dilution by the additions. The volume ratio V0/V is the dilution effect on the concentrations and absorbance. V0 is the initial volume and V, the volume of the titration point.
Bioassays of apoptosis
The L1210 (ATCC, CCL 219) cell line was cultured in suspension, as described (Svensson et al. 2000). The cells were harvested by centrifugation (200 × g for 10 min), resuspended in cell culture medium (RPMI 1640 supplemented without fetal calf serum, nonessential amino acids, sodium pyruvate and 50 μg gentamicin/mL, Life Technologies, Gibco BRL), and seeded into 24-well plates (Falcon, Becton Dickinson) at a density of 2 × 106/well. The different agonists were dissolved in cell culture medium, without fetal calf serum, and added to the cells (final volume 1 mL per well). Plates were incubated at 37°C in 5% CO2 atmosphere and 100 μL of fetal calf serum was added to each well after 30 min. Cell culture medium served as a control.
Cell viability was determined by Trypan blue exclusion after 6 h of incubation. For analysis, 30 μL of the cell suspension was mixed with 30 μL of a 0.2% Trypan blue solution, and the number of stained cells (dead cells) per 100 cells was determined by interference contrast microscopy (Ortolux II, Leitz Wetzlar).
DNA fragmentation
Oligonucleosome length DNA fragments were detected by agarose gel electrophoresis. The cell suspension remaining after Trypan blue (970 μL, 2 × 106/mL) was lysed in 5 mM Tris, 20 mM EDTA, 0.5% Triton X-100 pH 8.0 at 4°C for 1 h and centrifuged at 13,000g for 15 min. DNA was ethanol precipitated over night in −20°C, treated with proteinase K and RNAse, loaded on 1.8% agarose gels, and electrophoresed with constant voltage set at 50 V overnight. DNA fragments were visualized with ethidium bromide using a 305 nm UV-light source and photographed using Polaroid type 55 positive-negative films.
Acknowledgments
This work was supported by The Swedish Cancer Society (grant no. 3807-B97-01XAB [1997–2002] CS), The American Cancer Society (grant no. SPG-97-157 [1997–2003] CS), The Swedish Medical Research Council (grant number K97-03X-11552-02BK, SL), The Swedish Pediatric Cancer Society, The Segerfalk Foundation, The Österlund Foundation, The Lund Hospital Foundation, and the Swedish Natural Science Research Council (grant number K-AA/KU 10178-300, SL). Ingemar André is acknowledged for providing the Caligator software prior to release.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
CD, circular dichroism
ANS, 8-Anilinonaphtalene-1-sulfonic acid
Tris, tris(hydroxymethyl)aminomethane
EDTA, ethylenediamine tetra acetic acid
FPLC, fast protein liquid chromatography
PBS, phosphate-buffered saline
UV, ultraviolet
DEAE, diethylaminoethyl
quin 2, 2–[[2–[bis(carboxymethyl)amino-5-methylphenoxy]methyl]-6-methoxy-8–[bis(carboxymethyl)amino]quinoline
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0231003.
References
- Acharya, K.R., Ren, J.S., Stuart, D.I., Phillips, D.C., and Fenna, R.E. 1991. Crystal structure of human α-lactalbumin at 1.7 Å resolution. J. Mol. Biol. 221 571–581. [DOI] [PubMed] [Google Scholar]
- Anderson, P.J., Brooks, C.L., and Berliner, L.J. 1997. Functional identification of calcium binding residues in bovine α-lactalbumin. Biochemistry 36 11648–11654. [DOI] [PubMed] [Google Scholar]
- Andre, I. and Linse, S. 2002. Measurement of Ca2+-binding constants of proteins and presentation of the CaLigator software. Anal. Biochem. 305 195–205. [DOI] [PubMed] [Google Scholar]
- Aramini, J.M., Drakenberg, T., Hiraoki, T., Ke, Y., Nitta, K., and Vogel, H.J. 1992. Calcium-43 NMR studies of calcium-binding lysozymes and α-lactalbumins. Biochemistry 31 6761–6768. [DOI] [PubMed] [Google Scholar]
- Aramini, J.M., Hiraoki, T., Grace, M.R., Swaddle, T.W., Chiancone, E., and Vogel, H.J. 1996. NMR and stopped-flow studies of metal ion binding to α-lactalbumins. Biochim. Biophys. Acta 1293 72–82. [DOI] [PubMed] [Google Scholar]
- Bratcher, S.C. and Kronman, M.J. 1984. Metal ion binding to the N and A conformers of bovine α-lactalbumin. J. Biol. Chem. 259 10875–10886. [PubMed] [Google Scholar]
- Chrysina, E.D., Brew, K., and Acharya, K.R. 2000. Crystal structures of apo- and holo-bovine α-lactalbumin at 2.2 Å resolution reveal an effect of calcium on inter-lobe interactions. J. Biol. Chem. 275 37021–37029. [DOI] [PubMed] [Google Scholar]
- Creighton, T.E. 1993. Proteins: Structure and molecular properties, 2nd ed.
- Desmet, J., Hanssens, I., and van Cauwelaert, F. 1987. Comparison of the binding of Na+ and Ca2+ to bovine α-lactalbumin. Biochim. Biophys. Acta 912 211–219. [DOI] [PubMed] [Google Scholar]
- Düringer, C., Hamiche, A., Gustafsson, L., Kimura, H., and Svanborg, C. 2004. HAMLET interacts with histones and chromatin in tumor cell nuclei. J. Biol. Chem. (in press). [DOI] [PubMed]
- Gerken, T.A. and Dearborn, D.G. 1984. Carbon-13 NMR studies of native and modified ovine submaxillary mucin. Biochemistry 23 1485–1497. [DOI] [PubMed] [Google Scholar]
- Griko, Y.V. and Remeta, D.P. 1999. Energetics of solvent and ligand-induced conformational changes in α-lactalbumin. Protein Sci. 8 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson, A., Zhivotovsky, B., Orrenius, S., Sabharwal, H., and Svanborg, C. 1995. Apoptosis induced by a human milk protein. Proc. Natl. Acad. Sci. 92 8064–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson, A., Andreasson, J., Zhivotovsky, B., Karpman, D., Orrenius, S., and Svanborg, C. 1999. Multimeric α-lactalbumin from human milk induces apoptosis through a direct effect on cell nuclei. Exp. Cell Res. 246 451–460. [DOI] [PubMed] [Google Scholar]
- Harata, K., Abe, Y., and Muraki, M. 1999. Crystallographic evaluation of internal motion of human α-lactalbumin refined by full-matrix least-squares method. J. Mol. Biol. 287 347–358. [DOI] [PubMed] [Google Scholar]
- Hendrix, T., Griko, Y.V., and Privalov, P.L. 2000. A calorimetric study of the influence of calcium on the stability of bovine α-lactalbumin. Biophys. Chem. 84 27–34. [DOI] [PubMed] [Google Scholar]
- Kohler, C., Hakansson, A., Svanborg, C., Orrenius, S., and Zhivotovsky, B. 1999. Protease activation in apoptosis induced by MAL. Exp. Cell Res. 249 260–268. [DOI] [PubMed] [Google Scholar]
- Kohler, C., Gogvadze, V., Hakansson, A., Svanborg, C., Orrenius, S., and Zhivotovsky, B. 2001. A folding variant of human α-lactalbumin induces mitochondrial permeability transition in isolated mitochondria. Eur. J. Biochem. 268 186–191. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14 51–56. [DOI] [PubMed] [Google Scholar]
- Kronman, M.J. 1989. Metal-ion binding and the molecular conformational properties of α lactalbumin. Crit. Rev. Biochem. Mol. Biol. 24 565–667. [DOI] [PubMed] [Google Scholar]
- Kronman, M.J., Sinha, S.K., and Brew, K. 1981. Characteristics of the binding of Ca2+ and other divalent metal ions to bovine α-lactalbumin. J. Biol. Chem. 256 8582–8587. [PubMed] [Google Scholar]
- Linse, S. 2002. Calcium binding to proteins studied via competition with chromophoric chelators. Methods Mol. Biol. 173 15–24. [DOI] [PubMed] [Google Scholar]
- Linse, S., Helmersson, A., and Forsen, S. 1991a. Calcium binding to calmodulin and its globular domains. J. Biol. Chem. 266 8050–8054. [PubMed] [Google Scholar]
- Linse, S., Johansson, C., Brodin, P., Grundstrom, T., Drakenberg, T., and Forsen, S. 1991b. Electrostatic contributions to the binding of Ca2+ in calbindin D9k. Biochemistry 30 154–162. [DOI] [PubMed] [Google Scholar]
- Nitta, K., Tsuge, H., Shimazaki, K., and Sugai, S. 1988. Calcium-binding lysozymes. Biol. Chem. Hoppe Seyler 369 671–675. [DOI] [PubMed] [Google Scholar]
- Permyakov, E.A., Yarmolenko, V.V., Kalinichenko, L.P., Morozova, L.A., and Burstein, E.A. 1981. Calcium binding to α-lactalbumin: Structural rearrangement and association constant evaluation by means of intrinsic protein fluorescence changes. Biochem. Biophys. Res. Commun. 100 191–197. [DOI] [PubMed] [Google Scholar]
- Permyakov, S.E., Uversky, V.N., Veprintsev, D.B., Cherskaya, A.M., Brooks, C.L., Permyakov, E.A., and Berliner, L.J. 2001a. Mutating aspartate in the calcium-binding site of α-lactalbumin: Effects on the protein stability and cation binding. Protein Eng. 14 785–789. [DOI] [PubMed] [Google Scholar]
- ———. 2001b. Mutating aspartate in the calcium-binding site of α-lactalbumin: Effects on the protein stability and cation binding. Protein Eng. 14 785–789. [DOI] [PubMed] [Google Scholar]
- Schaer, J.J., Milos, M., and Cox, J.A. 1985. Thermodynamics of the binding of calcium and strontium to bovine α-lactalbumin. FEBS Lett. 190 77–80. [DOI] [PubMed] [Google Scholar]
- Segawa, T. and Sugai, S. 1983. Interactions of divalent metal ions with bovine, human, and goat α-lactalbumins. J. Biochem. (Tokyo) 93 1321–1328. [DOI] [PubMed] [Google Scholar]
- Skoog, D.A. and West, D.M. 1976. Fundamentals of analytical chemistry, 3rd ed., p. 804. Holt Rinehart and Winston, New York.
- Svanborg, C., Agerstam, H., Aronson, A., Bjefkvig, R., Düringer, C., Fischer, W., Gustafsson, L., Hallgren, O., Leijonhuvud, I., Linse, S., et al. HAMLET kills tumor cells by an apoptosis-like mechanism—cellular, molecular, and therapeutic aspects. Adv. Cancer Res. 88 1–29. [DOI] [PubMed]
- Svensson, M., Sabharwal, H., Hakansson, A., Mossberg, A.K., Lipniunas, P., Leffler, H., Svanborg, C., and Linse, S. 1999. Molecular characterization of α-lactalbumin folding variants that induce apoptosis in tumor cells. J. Biol. Chem 274 6388–6396. [DOI] [PubMed] [Google Scholar]
- Svensson, M., Hakansson, A., Mossberg, A., Linse, S., and Svanborg, C. 2000. Conversion of α-lactalbumin to a protein inducing apoptosis. Proc. Natl. Acad. Sci. 97 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, M., Mossberg, A., Pettersson, J., Linse, S., and Svanborg, C. 2003. Lipids as cofactors in protein folding: Stereo-specific lipid–protein interactions are required to form HAMLET (human α-lactalbumin made lethal to tumor cells. Protein Sci. (this issue). [DOI] [PMC free article] [PubMed]
- Vanhooren, A., Vanhee, K., Noyelle, K., Majer, Z., Joniau, M., and Hanssens, I. 2002. Structural basis for difference in heat capacity increments for Ca(2+) binding to two α-lactalbumins. Biophys. J. 82 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesinha-Bettoni, R., Dobson, C.M., and Redfield, C. 2001. Comparison of the denaturant-induced unfolding of the bovine and human α-lactalbumin molten globules. J. Mol. Biol. 312 261–273. [DOI] [PubMed] [Google Scholar]