Abstract
Proteins can adjust their structure and function in response to shifting environments. Functional diversity is created not only by the sequence but by changes in tertiary structure. Here we present evidence that lipid cofactors may enable otherwise unstable protein folding variants to maintain their conformation and to form novel, biologically active complexes. We have identified unsaturated C18 fatty acids in the cis conformation as the cofactors that bind apo α-lactalbumin and form HAMLET (human α-lactalbumin made lethal to tumor cells). The complexes were formed on an ion exchange column, were stable in a molten globule-like conformation, and had attained the novel biological activity. The protein–fatty acid interaction was specific, as saturated C18 fatty acids, or unsaturated C18:1trans conformers were unable to form complexes with apo α-lactalbumin, as were fatty acids with shorter or longer carbon chains. Unsaturated cis fatty acids other than C18:1:9cis were able to form stable complexes, but these were not active in the apoptosis assay. The results demonstrate that stereo-specific lipid–protein interactions can stabilize partially unfolded conformations and form molecular complexes with novel biological activity. The results offer a new mechanism for the functional diversity of proteins, by exploiting lipids as essential, tissue-specific cofactors in this process.
Keywords: α-Lactalbumin, HAMLET, fatty acids, apoptosis, cancer, protein folding
The human genome sequence was recently found to contain much fewer distinctive genes than previously expected (Green and Chakravarti 2001). This finding emphasizes the need to identify mechanisms by which individual proteins may attain functional diversity. One may predict that proteins must be able to sense different environments and adjust their structure and function to meet local demands. Structural diversity may be achieved by various chemical modifications such as methylation, sulphation, or acetylation, but in addition, changes in tertiary structure are becoming recognized as a mechanism potentially allowing proteins to achieve diversity of function (Jeffery 1999). The most striking example is the prion protein, which changes from a mixed α-helical and β-sheet conformation to the β-sheet–rich, disease-causing iso-form Prpsc (Pan et al. 1993; Safar et al. 1993; Pergami et al. 1996). It has been postulated that this shift requires tissue specific cofactors that stabilize Prpsc, but the “factor x” has not yet been identified (Booth et al. 1997; Cohen and Prusiner 1998; Jackson and Collinge 2000).
This study addressed the role of specific lipids as cofactors that may enable protein folding variants to attain new functions. HAMLET (human α-lactalbumin made lethal to tumor cells) was used as a model. HAMLET is a molecular complex that induces in vitro apoptosis in tumor cells but not in healthy differentiated cells (Hakansson et al. 1995; Svanborg et al. 2003). The apoptotic activity of this variant fold was discovered by serendipity, in a fraction of human milk casein obtained by precipitation at low pH (Hakansson et al. 1995). By spectroscopy, the active fraction was shown to contain partially unfolded α-lactalbumin (Svensson et al. 1999), with native-like secondary structure, but lacked specific tertiary packing of the side chains. Unlike apo α-lactalbumin, however, the protein was stable in this unfolded state at physiologic pH and in the presence of Ca2+. The link between apoptosis induction and the folding change was proven by deliberate conversion of native α-lactalbumin to the apoptosis-inducing form (Svensson et al. 2000). The conversion was shown require both the folding change and a lipid cofactor (Svensson et al. 2000).
The aim of the present study was to compare structural variants of oleic acid, and other fatty acids differing in the degree of saturation, carbon chain length and cis/trans conformation. We demonstrate that highly specific interactions guide the formation of HAMLET and that only the cis-unsaturated C18 fatty acids serve as suitable cofactors.
Results
Conversion of α-lactalbumin to HAMLET requires the C18:1 fatty acid cofactor
To form HAMLET from native α-lactalbumin, the strongly bound Ca2+ ion was removed with etylenediamintetra acetic acid (EDTA), and the conformational change was confirmed by spectroscopy. Apo α-lactalbumin was then subjected to ion exchange chromatography on a matrix that had been preconditioned with oleic acid, C18:1:9cis. The protein was retained on the conditioned column, forming a complex that eluted as a sharp peak after 1 M NaCl. The eluted protein maintained a molten globule-like conformation at neutral pH and in the presence of Ca2+, as shown by circular dichroism (CD) and 8-anilinonaphtalene-1-sulfonic acid (ANS) spectroscopy (Svensson et al. 2000). The HAMLET complex induced apoptosis in many types of tumor cells, including the L1210 lymphoma cell line (Svanborg et al. 2003). HAMLET was thus defined as the conversion product of apo α-lactalbumin and oleic acid, with the ability to induce apoptosis in tumor cells.
Comparison with other fatty acids
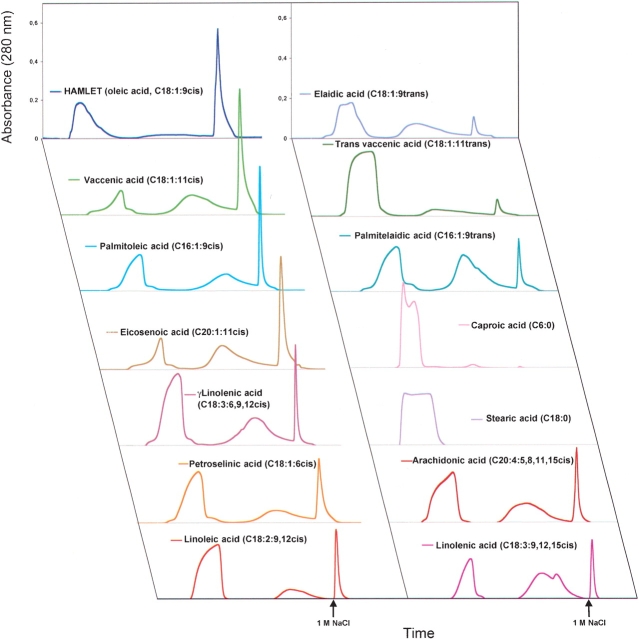
To understand the specificity of the protein–lipid interaction, we used structural variants of oleic acid and other fatty acids. Fatty acids differing in the degree of saturation, carbon chain length, and cis/trans conformation were assayed for their ability to form HAMLET-like complexes with apo α-lactalbumin (Fig. 1 ▶). Clean column matrices were conditioned with the individual fatty acids, apo α-lactalbumin was applied to the columns, and the material eluting after 1 M NaCl was collected for analysis.
Figure 1.
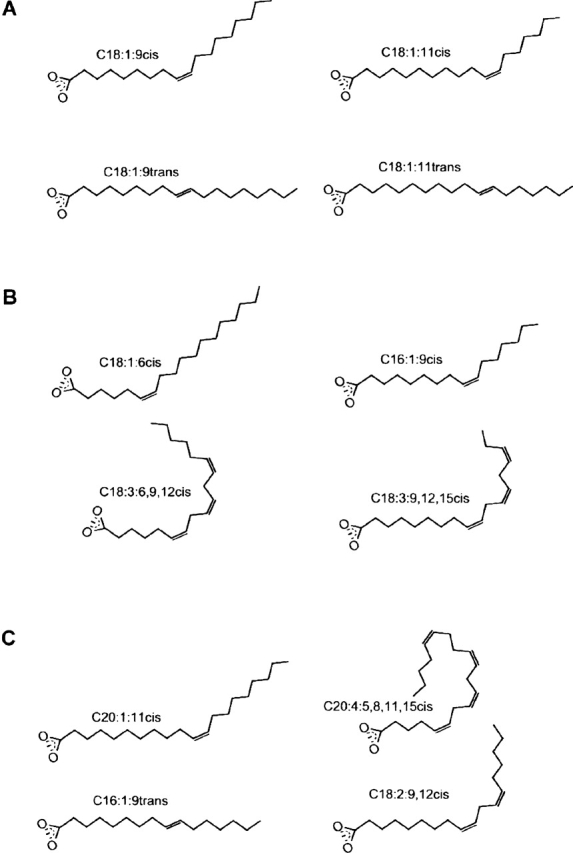
Schematic fatty acid structures. Line drawings of the unsaturated fatty acids, which were investigated for their ability to produce HAMLET-like complexes. (A) C18:1:9cis, oleic acid; C18:1:11cis, vaccenic acid; C18:1:9trans, elaidic acid; and C18:1:11trans, trans vaccenic acid. (B) C18:1:6cis, petroselinic acid; C16:1:9cis, palmitoleic acid; C18:3:6, 9, 12cis, γ linolenic acid; and C18:3:9, 12, 15cis, linolenic acid. (C) C20:1:11cis,eicosenic acid; C20:4:5, 8, 11, 15cis, arachidonic acid; C16:1:9trans, palmitelaidic acid; and C18:2:9, 12cis, linoleic acid.
Results of ion exchange chromatography on fatty acid conditioned matrices are shown in Figure 2 ▶. Unsaturated C18 fatty acids in the cis conformation formed stable complexes with apo α-lactalbumin with a high yield. C18:1:9cis converted >90% of the added apo α-lactalbumin, whereas C18:1:11cis was somewhat less efficient, with a yield of ~70%. Other unsaturated C18 cis fatty acids (C18:1:6cis, C18:2:9, 12cis, C18:3:9, 12, 15cis and γC18:3:6, 9, 12cis) gave lower yields.
Figure 2.
Retention of apo α-lactalbumin on ion-exchange matrices conditioned with individual fatty acids. Apo α-lactalbumin was applied to column matrices that had been preconditioned with each indicated fatty acid, and the eluate after 1M NaCl was collected. Columns conditioned with C18:1:9cis were used as a positive control. All unsaturated fatty acids in the cis conformation retained apo α-lactalbumin on the column, but with varying efficiencies. Unsaturated fatty acids in the trans conformation (C18:1:9trans, C16:1:9trans) or saturated fatty acids (C6:0 and C18:0) failed to retain apo α-lactalbumin on the column.
When exposed to columns conditioned with the trans conformers of C18:1 or with saturated fatty acids, the protein remained largely unconverted. Only small amounts eluted with high salt from columns conditioned with C18:1:9trans, C18:1:11trans, or the saturated C18:0 fatty acid and the eluted complexes were inactive in the apoptosis assay.
Unsaturated cis fatty acids with shorter (C16:1:9cis) or longer (C20:1:11cis and C20:4:5, 8, 11, 15cis) carbon chains formed stable complexes, but with yields lower than C18:1:9cis. The columns conditioned with the saturated fatty acids C6:0, C14:0, or C16:0 retained no apo α-lactalbumin. The results demonstrate that apo α-lactalbumin interacts in a stereo-specific manner with fatty acids in the cis conformation, and the C18 cis fatty acids give the highest yield.
Conformation analysis by CD spectroscopy
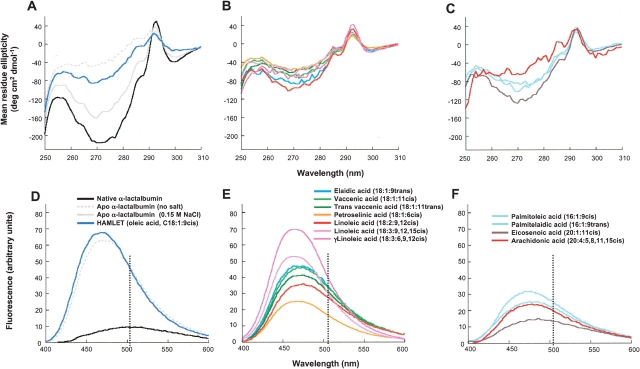
The conformation of each protein fatty acid complex was examined by CD spectroscopy (Fig. 3 ▶). By near-UV CD spectroscopy, HAMLET (C18:1:9cis) was shown to resemble apo α-lactalbumin in the molten globule-like state formed at low salt (Fig. 3A ▶). The other C18cis complexes (C18:1:11cis, γC18:3:6, 9, 12cis, C18:1:6cis, C18:2:9, 12cis, C18:3:9, 12, 15cis) showed a similar spectral pattern (Fig. 3B ▶). Interestingly, the C18:1:9trans complex showed a similar CD spectrum as that of the C18 cis fatty acids complexes, even though very little material was formed. The C20:4:5, 8, 11, 15cis and the C16:1:9cis fatty acid complexes resembled HAMLET, whereas the C20:1:11cis or trans complexes had adopted a more native-like conformation, resembling the apo control at high salt (Fig. 3C ▶). We conclude that the unsaturated fatty acids, which retained apo α-lactalbumin on the column, also stabilized the partially unfolded conformation. By CD spectroscopy, there was no discernable difference between the active HAMLET complex and the inactive complexes (see below).
Figure 3.
Tertiary structure and hydrophobicity of the fatty acid-protein complexes examined by CD and ANS spectroscopy. (A) CD spectrum of native α-lactalbumin showing the characteristics of a well-folded protein at 25°C, with a minimum at 270 nm arising from tyrosine residues and a maximum at 294 nm arising from tryptophan residues. The apo α-lactalbumin control had lost most of the characteristic signals, indicating less restrained tyrosines and tryptophans. (B) The C18 complexes resembled HAMLET with a loss of signal in the tyrosine and tryptophan regions. (C) The C20:4:5, 8, 11, 15cis and the C16:1:9 fatty acid complexes resembled HAMLET, whereas the C20:1:11cis complex showed a more native like conformation. (D) Native α-lactalbumin did not bind ANS as shown by the flat curve, and the low signal at 490 nm at 25°C, but apo-α-lactalbumin showed significant ANS binding with enhanced intensity and a maximum at 470 nm, as expected from the increased hydrophobicity of this fold. All of the α-lactalbumin- fatty acid complexes except C20:1:11cis bound ANS. (E) The C18:1:6cis, C18:3:9, 12, 15cis, and γC18:3:6, 9, 12cis fatty acid complexes, showed a wavelength shift similar to HAMLET, whereas C18:1:11cis, C18:2:9, 12cis, C18:1:9trans, and C18:1:11trans complexes showed a slightly lower wavelength shift. (F) The other complexes (C16:1:9cis, C20:4:5, 8, 11, 15cis, C20:1:11cis) showed a lower shift more similar to the apo α-lactalbumin control.
ANS fluorescence spectroscopy
Exposure of hydrophobic surfaces was probed by ANS spectroscopy (Fig. 3D–F ▶). Native α-lactalbumin failed to bind ANS, as shown by the low intensity with a maximum at 510 nm. HAMLET showed enhanced intensity and a maximum at 470 nm (Fig. 3D ▶). The C18:1:6cis, C18:3:9, 12, 15cis, and γC18:3:6, 9, 12cis fatty acid complexes, bound ANS with similar wavelength shift as HAMLET (Fig. 3E ▶). C18:1:9trans, C18:1:11cis, C18:1:11trans, and C18:2:9, 12cis complexes showed a slightly lower wavelength shift than did HAMLET (Fig. 3E ▶). The other complexes (C16:1:9cis, C20:4:5, 8, 11, 15cis, C20:1:11cis) also showed lower ANS fluorescence intensity than the apo α-lactalbumin control, as well as a lower shift. Despite these differences, all of the fatty acid complexes were blue-shifted compared with native α-lactalbumin. To exclude the direct binding of ANS to the fatty acids in the protein–lipid complexes, mixtures of ANS and fatty acids were subjected to spectroscopy. No ANS–fatty acid interaction was observed (data not shown). We conclude that all eluted fatty acid complexes bind ANS more efficiently than native protein, indicating increased exposure of hydrophobic surfaces.
Biological activity
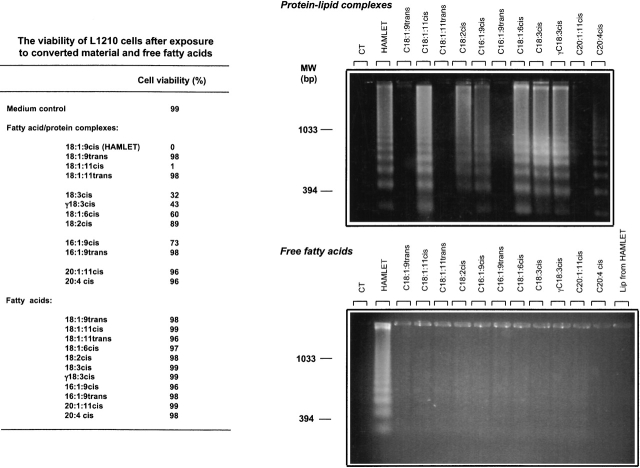
HAMLET has been shown to induce apoptosis in >40 different types of tumor cells (Svanborg et al. 2003). In this study, apoptosis induction was tested by using the L1210 leukemia cell line (Fig. 4 ▶). The C18:1:9cis and C18:1:11cis fatty acid complexes reduced cell viability from 99% to 0% and 1%, respectively, in 6 h at a concentration of 0.3 mg/mL, and DNA fragmentation was observed after 6 h, confirming that the cells were dying by apoptosis. In contrast, the C18:1trans fatty acid complexes were inactive in the cellular assay. Other C18 cis protein–fatty acid complexes showed weaker activity, but cell death was associated with DNA fragmentation (Fig. 4 ▶). The remaining complexes were inactive in the apoptosis assay, as were the free fatty acids. These results demonstrate that C18:1:9cis and C18:1:11cis, are optimal cofactors in the formation of HAMLET.
Figure 4.
Tumor cell apoptosis induced by the lipid–protein complexes. Apoptosis induction in L1210 leukemia cells exposed to the different protein–lipid complexes, including the HAMLET control. HAMLET and the C18:1:11cis complex had killed 99% to 100% of the cells after 6 h and induced DNA fragmentation, but the C18:1trans fatty acid complexes were inactive. The C16 and C20 unsaturated fatty acid complexes caused a small loss of cell viability and intermediate DNA fragmentation. Lipid extracts derived from HAMLET or from each of the other complexes showed no effect on cell viability or DNA fragmentation. The lipid concentrations corresponded to the amount present in 1.0 mg of each complex. At very high lipid concentrations, the cells died of necrosis, but at no time were there evidence of apoptosis in response to lipids.
Stoichiometry of protein and lipid in the HAMLET complexes
The protein content in HAMLET was determined by acid hydrolysis of three different batches, after gel filtration on a PD-10 column to remove unbound fatty acid. Fatty acids were esterified with acidified methanol and quantified by gas chromatography/mass spectrometry (GC/MS). The estimated number of oleic acid residues per α-lactalbumin molecule was 1.3, 0.8, and 0.6 (mean, 0.9; SD, 0.36). These experiments indicated that the HAMLET complexes contain one oleic acid per α-lactalbumin molecule.
NMR spectroscopy of the protein fatty acid complexes
1H-NMR was used in an attempt to better understand the structural basis for the difference in activity between the C18:1cis and the inactive protein–fatty acid complexes (Fig. 5 ▶). The spectrum of the C18:1:9cis and C18:1:11cis complexes showed broad lines and little shift dispersion. The lines in the aromatic region were clustered, and there were no outshifted methyl signals <0.7 ppm. The fatty acid signal remained unchanged after PD-10 gel filtration to remove excess free fatty acid. The trans isomer complexes (C18:1:9trans and C18:1:11trans) differed markedly from the C18:1:9cis or C18:1:11cis complexes. Signals from fatty acids were detected, but they were smaller than for the cis complexes. The protein lines were narrow and more outshifted both in the methyl and in the aromatic regions. A low level of native signals were observed, indicating that the trans fatty acid complexes were more unstable than were the HAMLET complexes.
Figure 5.
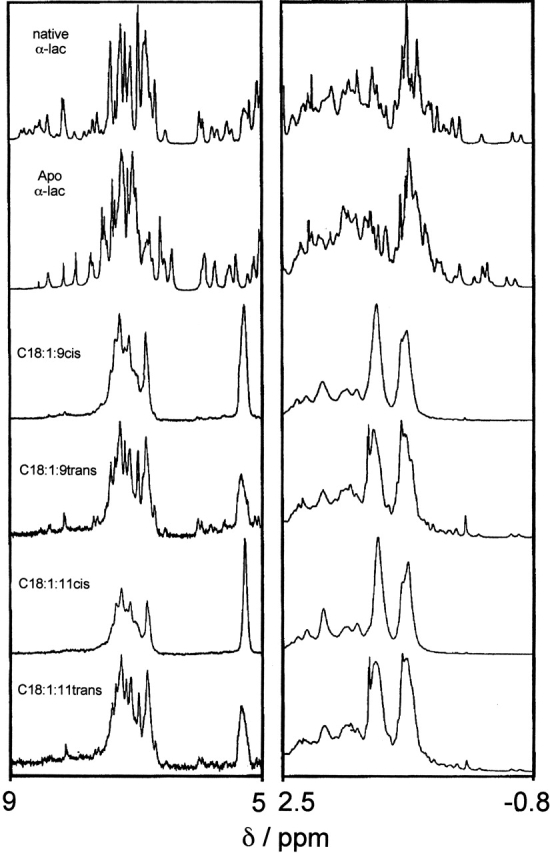
HAMLET complexes examined by NMR. The aromatic and methylated regions are shown in the left and right panels, respectively. Native α-lactalbumin showed the characteristics of a folded and well-ordered protein with narrow lines and significant shift dispersion, a large number of sharp signals in the aromatic region (~7 ppm), and several out-shifted methyl signals (between 0.7 and −0.6 ppm). The apo protein displayed narrow lines and significant shift dispersion with significant variations relative to the native state in the chemical shifts of a large number of resonances. HAMLET and the C18:1:11cis protein complex differed from the native protein, and showed broad lines and the lack of out-shifted methyl signals suggestive of a partially unfolded state. The spectra of the trans fatty acid complexes showed more narrow lines and out-shifted signals, indicating that the trans fatty acid complexes are less stable and that the protein may be reverting to the native state. The NMR spectra were obtained at 37°C.
The results indicate that specific molecular interactions stabilize α-lactalbumin in the HAMLET conformation, and the unsaturated C18:1:9cis or C18:1:11cis fatty acids have the stereo-specific properties required to achieve this conformation. The broad fatty acid signals indicated that they formed an integral part of the complex.
Discussion
Changes in tertiary structure allow proteins to achieve diversity of function. This study demonstrated that lipid cofactors enable proteins to adopt stable novel conformations and, thus, act as partners in protein folding. The change of α-lactalbumin to HAMLET was studied as an example. The native protein acts as a coenzyme in lactose synthesis, but when partially unfolded and coupled to a fatty acid, it changes function and forms HAMLET that kills tumor cells. We have identified the optimal cofactors as C18:1 fatty acids with a double bond in the cis conformation at position 9 or 11. Saturated C18 fatty acids, unsaturated fatty acids in the trans conformation, or fatty acids with shorter carbon chains could not form HAMLET. The results show that the unfolded protein interacts specifically with certain fatty acids, resulting in complex formation and new biological activity.
HAMLET is defined as the conversion product of apo α-lactalbumin and oleic acid, with the ability to induce apoptosis in tumor cells. Three molecular events are involved in the conversion of α-lactalbumin to HAMLET: Protein unfolding, fatty acid binding, and formation of the biologically active complex. First, the protein must be partially unfolded and/or destabilized, as the native protein cannot be converted. This is achieved experimentally by adding EDTA (pH 8.5) to remove the tightly bound Ca2+ ion. Second, the apo-protein must bind the lipid cofactor, and this occurs with high efficiency on an ion exchange matrix preconditioned with oleic acid. Fatty acids bind to the apo protein also in solution, but the yield of HAMLET in such mixtures of α-lactalbumin and oleic acid are much lower than on the column. Third, the protein and the lipid form a complex with apoptosis-inducing activity. This third step may occur on the column, during elution with high salt, or during the dialysis step. In the active complex, α-lactalbumin has a molten globule-like conformation and has exposed hydrophobic surfaces, but additional molecular modifications may have occurred.
The structural basis for biologic activity is not quite clear, but several important observations in this study allow us to speculate about the unique molecular features of cis-unsaturated C18 fatty acids as cofactors in HAMLET. Clearly, α-lactalbumin has a higher affinity for cis-unsaturated fatty acids than for the trans-unsaturated or saturated fatty acids. The yields from fatty acid conditioned ion exchange resins probably reflect this affinity. Regardless of affinity and yield, all unsaturated fatty acids formed complexes that maintained the partially unfolded conformation and were more hydrophobic than was the native protein. However, only C18:1:11cis yielded a complex with biologically activity comparable to that of HAMLET (C18:1:9cis). We conclude that the unsaturated C18:1 cis fatty acids have unique structural features, allowing them to form HAMLET from apo α-lactalbumin. The fatty acids show the correct stereo-specific match for the protein, and yield a conversion product with biological activity.
Apo α-lactalbumin differs from other known lipid-binding proteins in that it contains both α-helical and β-sheet domains. The members of the intracellular lipid-binding protein family have an all β-barrel structure, forming a cavity that binds a range of fatty acids varying in chain length and saturation (Thompson et al. 1997). Typically, the carboxylate head group of the fatty acid interacts with two to four positively charged amino acids, usually arginines, and the carbon chain is coordinated by 6 to 10 hydrophobic amino acids. The crystal structure of human serum albumin has revealed six asymmetrically distributed, fatty acid binding sites within the repeating α-helical domain structure of the protein (Curry et al. 1998). Each hydrophobic pocket is capped at one end by basic or polar side chains, coordinating the fatty acid head group. The binding of fatty acids to human serum albumin causes conformational changes with rotations of the three domains of the protein, and adjustments of side chains to make way for the incoming fatty acid (Curry et al. 1998), but the molecule does not unfold or change function. We may therefore conclude that the role of the lipid cofactor in the conversion of α-lactalbumin HAMLET differs both structurally and functionally from these previously known protein–lipid interactions.
Tentative fatty acid binding sites were identified based on the three-dimensional structures of native and apo-α-lactalbumin. The native α-lactalbumin molecule is a hydrophilic, acidic protein, exposing mainly charged and polar amino acids. Two hydrophobic regions are located in the interior of the globular structure. One is formed by residues from the C and D helices and the β-sheet domain in the interface between the two domains. The second is formed by residues in the A, B, and 310-helices of the α domain (Wu and Kim 1998; Saito 1999). The crystal and NMR structures of bovine apo α-lactalbumin have revealed a significant structural change in the cleft between the two domains (Chrysina et al. 2000; Wijesinha-Bettoni et al. 2001) upon Ca2+ release. The expansion of the Ca2+-binding loop tilts the 310 helix toward the C helix, resulting in a disruption of the aromatic cluster in the interface between the two domains (Trp 60 and 104, Phe 53, and Tyr 103; Chrysina et al. 2000). The α-domain, in contrast, remains structured in both the native and the apo-conformations, with near native side-chain packing. We therefore speculate that the C18:1 fatty acid binds in the interface between the α and β domains, and thus stabilizes a molten globule-like conformation.
The shape of the hydrophobic pocket indicated that it should favor interactions with bent molecules. This may indeed explain the inability of the C18:1trans conformers to form HAMLET. Although fatty acids in the cis conformation are U-shaped around the double bond, with both carbon chains projecting in one direction, trans fatty acids are rod shaped around the double bond due to the carbon chains on opposite sides of the double bond. The saturated fatty acids are most flexible, with no structural constraints due to the lack of double bonds. The results thus indicate that only the cis conformation allows fatty acids a close stereo-specific fit, and the additional critical feature of the fatty acid is the carbon chain length. In addition, the pocket is capped by basic residues, which may coordinate the polar head groups of the fatty acids, thus orienting the lipid. This interaction is, however, not sufficient for activation as the trans and saturated fatty acids, which possess the same charged head group failed to form the active complex. It is highly likely that the stereo-specific fit involves both hydrophobic interactions with the lipid tail and electrostatic interactions of the negatively charged head group with basic side chains. Based on the analogy with other fatty acid binding proteins, the fatty acid may bind to HAMLET by electrostatic interactions between its negatively charged head group and basic side chains in the protein, due to the hydrophobic effect, as well as by van der Waal’s contacts with the tail, which are optimized for the preferred stereo-specific match (C18:1:9cis).
The present study raises the possibility that lipids may act as stabilizing cofactors in a variety of protein folding processes. The prions and amyloid fibrils are examples of unsuccessful protein processing, resulting in protein accumulation in peripheral tissues in which damage is done (McLaurin et al. 2000; Dobson 2001; Pepys 2001). It has been postulated that a cofactor, factor x or “protein x” is required for the transmission of human prions to transgenic mice, to form the nascent scrapie isoforms during prion propagation (Telling et al. 1994; Billeter et al. 1997). By mutational analysis, the interaction with protein x was shown to depend on a discontinuous epitope formed by the C-terminal α helix, with residues 167 and 171 in an adjacent loop (Kaneko et al. 1997), but the molecular nature of factor x has remained elusive. There is evidence that amyloid fibers contain a mixture of lipid species, but their role in the formation of fibrils remains to be defined (Kim et al. 1967; McLaurin et al. 2000). The present study indicates that lipids should be explored as cofactors locking the prions and amyloid proteins in their β-sheet–rich conformations.
Finally, the availability of fatty acid cofactors may facilitate and determine if proteins will adapt to different environments. In case of α-lactalbumin, this mechanism may allow the molecule to be modified in the nursing child. The low pH in the stomach is, for example, known to cause partial unfolding of α-lactalbumin, and lipids are hydrolyzed by acid lipases to release oleic acid (Bernback et al. 1990; Sarles et al. 1992). It is interesting to speculate that the beneficial function of HAMLET explains the special abundance of both α-lactalbumin (2 mg/mL [140 μM]; Heine et al. 1991) and oleic acid (>50% of the fatty acid chains of triglycerides; Jensen 1996) in human milk. Breast-fed children show an overall lower incidence of childhood cancer with an especially strong epidemiological association for lymphomas (crude odds ratio, 8.19; Davis et al. 1988) that might result from the selective purging of unwanted cells in the nursing child.
Materials and methods
α-Lactalbumin purification
Native human α-lactalbumin was purified from human milk by ammonium sulphate precipitation and phenyl-Sepharose chromatography as described (Svensson et al. 2000).
Anion-exchange chromatography
Columns (14 × 1.6 cm) packed with diethylaminoethyl (DEAE)-Trisacryl M (BioSepra) were attached to a Bio-Logic chromatography system (Bio-Rad Laboratories), and eluted with a NaCl gradient (buffer A, 10 mM tris (hydroxymethyl) aminomethane [Tris]/HCl at pH 8.5; buffer B, buffer A containing 1 M NaCl). The matrices were each conditioned with one of the different fatty acids used in the study (Larodan Biochemicals). Ten milligrams was dissolved in 500 μL 99.5% ethanol by sonication using a Branson 2200 bath sonicator (Branson). After addition of 10 mL of 10 mM Tris/HCl (pH 8.5), the lipid solution was applied to a newly packed DEAE-Trisacryl M matrix and dispersed throughout the matrix by using a NaCl gradient.
Ten milligrams of human α-lactalbumin was dissolved in 20 mL of 10 mM Tris/HCl (pH 8.5) with 0.08 mM EDTA and added to the column. The protein fraction eluting after high salt was desalted by dialysis (Spectra/Pore, Spectrum Medical Industries; membrane cutoff, 3.5 kD), against distilled water with at least four changes of water in 100-fold volume excess, and then lyophilized.
Spectroscopic analysis
Stock solutions were prepared by dissolving the lyophilized material in 10 mM potassium phosphate buffer (pH 7.5), and concentrations determined based on the absorbance at 280 nm. CD spectra were obtained on a JASCO J-720 spectro-polarimeter with a JASCO PTC-343 Peltier type thermostated cell holder. Quartz cuvettes were used with 1-cm path length, and spectra were recorded between 240 and 320 nm at 25°C. The wavelength step was 1 nm; the response time, 8 sec; and the scan rate, 10 nm/min. Six scans were recorded and averaged for each spectrum. Baseline spectra were recorded with pure buffer in the cuvette and subtracted from the protein spectra.
The mean residue ellipticity θm was calculated from the recorded ellipticity, θ, as
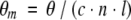 |
where c is the protein concentration in M; n, the number of residues in the protein (123 in this case); l, the path length in m; and θ, the ellipticity in degrees.
ANS fluorescence emission spectra were recorded at 25°C on a Perkin Elmer LS-50B spectrometer by using a quartz cuvette with 1-cm excitation path length, between 400 and 600 nm (step 1 nm) with excitation at 385 nm. Both the excitation and emission bandpass were set to 5 nm. ANS ammonium salt (Fluka) was added stepwise, and the spectra at 1.5 M equivalents are shown.
The 1H NMR spectra were recorded by using an Omega 500 spectrometer at 500 MHz in D2O with 0.15 M NaCl at 37°C, for 1 to 2 mM solutions of protein. Lyophilized HAMLET, native or apo α-lactalbumin (5 mg), or the protein–lipid complexes were dissolved in 500 μL of D2O, and the pH was set to 7.0 by using NaOD. Apo α-lactalbumin was generated by dissolving α-lactalbumin in doubly distilled water containing 10-fold molar excess of ethylene-bis(oxyethyleneitriol)tetraacetic acid (EGTA) at pH 8.0. The sample was applied to a PD-10 gel filtration column after an aliquot of saturated NaCl (calcium-depleted) and eluted by doubly distilled water. The sample was passed through the saturated NaCl to reduce binding of EGTA to the protein, and EGTA-free apo protein was eluted in the water.
Stoichiometry of protein and lipid in the HAMLET complexes
Three batches of HAMLET were subjected to gel filtration on a PD-10 column (Pharmacia AB) for removal of unbound free fatty acids. Ten milligrams of lyophilized HAMLET was dissolved in 0.5 mL phosphate-buffered saline (PBS; 0.14 M NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4 at pH 7.4) and applied on the gel filtration column. Fractions containing protein were eluted with PBS and pooled (~1 mL). The protein concentration was determined by the Amino Acid Analysis Centre at Uppsala University on 100 μL of the sample. The fatty acid content was determined by GC/MS on 200 μL of the sample, using C17:0 and C19:0 as internal standards. The sample solutions were evaporated with nitrogen gas. Internal standards and 1 mL of 2 M acidified methanol were added, and the samples were methanolysed for 1 h at 85°C. Heptane was added, and the samples were centrifuged. The heptane phase, containing the fatty acids methyl esters, was isolated. The nonpolar phases were dried with nitrogen gas, and 1 mL heptane was added to the samples. Two μL was further analyzed on a Varian gas chromatograph (model 3500) equipped with a split/split-less injector and a flame ionization detector, separated on a (0.25-mm inner diameter) fused silica capillary column coated with FFAP (Chrompack). The column temperature was programmed from 140°C/min to 240°C/min at 8°C/min. Chromatograms were evaluated by using a Varian integrator model 4290.
Bioassays of apoptosis
The L1210 (ATCC, CCL 219) cell line was cultured in suspension, as described (Svensson et al. 1999). The cells were harvested by centrifugation (200g for 10 min), resuspended in cell culture medium (RPMI 1640 supplemented without fetal calf serum, nonessential amino acids, sodium pyruvate, and 50 μg gentamicin/mL; Life Technologies, GIBCO BRL), and seeded into 24-well plates (Falcon, Becton Dickinson) at a density of 2 × 106/well. The different substances were dissolved in cell culture medium, without fetal calf serum, and added to the cells (final volume 1 mL/well). Plates were incubated at 37°C in 5% CO2 atmosphere, and 100 μL of fetal calf serum was added to each well only after 30 min of incubation with HAMLET. Cell culture medium served as a control. Cell viability was determined by trypan blue exclusion after 6 h of incubation.
DNA fragmentation
Oligonucleosome length DNA fragments were detected by agarose gel electrophoresis. The cell suspension remaining after trypan blue (970 μL, 2 × 106/mL) was lysed in 5 mM Tris, 20 mM EDTA, 0.5% Triton X-100 (pH 8.0) for 1 h at 4°C and centrifuged at 13,000g for 15 min. DNA was ethanol precipitated over night in −20°C, treated with proteinase K and RNAse, loaded on 1.8% agarose gels, and electrophoresed with constant voltage set at 50 V over night. DNA fragments were visualized with ethidium bromide by using a 305-nm UV-light source and photographed by using Polaroid type 55 positive-negative film.
Acknowledgments
We thank Hanna Nilsson for the purification of α-lactalbumin and Lennart Larsson for the GC/MS analysis of the lipid content in HAMLET. This work was supported by The Swedish Cancer Society (grant number 3807-B97-01XAB [1997–2002] to C.S.), American Cancer Society (grant number SPG-97-157 [1997–2003] to C.S.), The Swedish Medical Research Council (grant number K97-03X-11552-02BK, to S.L.), The Swedish Pediatric Cancer Society, The Segerfalk Foundation, The Österlund Foundation, The Lund Hospital Foundation, and the Swedish Natural Science Research Council (grant number K-AA/KU 10178-300, to S.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
GC/MS, gas chromatography/mass spectrometry
EDTA, ethylenediamine tetra acetic acid
Tris, tris (hydroxymethyl) aminomethane
ANS, 8-anilinonaphtalene-1-sulfonic acid
CD, circular dichroism
DEAE, diethylaminoethyl
EGTA, ethylene-bis(oxyethyleneitriol)tetraacetic acid
PBS, phosphate-buffered saline
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0231103.
References
- Bernback, S., Blackberg, L., and Hernell, O. 1990. The complete digestion of human milk triacylglycerol in vitro requires gastric lipase, pancreatic colipase-dependent lipase, and bile salt- stimulated lipase. J. Clin. Invest. 85 1221–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter, M., Riek, R., Wider, G., Hornemann, S., Glockshuber, R., and Wuthrich, K. 1997. Prion protein NMR structure and species barrier for prion diseases. Proc. Natl. Acad. Sci. 94 7281–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, D.R., Sunde, M., Bellotti, V., Robinson, C.V., Hutchinson, W.L., Fraser, P.E., Hawkins, P.N., Dobson, C.M., Radford, S.E., Blake, C.C., et al. 1997. Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature 385 787–793. [DOI] [PubMed] [Google Scholar]
- Chrysina, E.D., Brew, K., and Acharya, K.R. 2000. Crystal structures of apo- and holo-bovine α-lactalbumin at 2.2 Å resolution reveal an effect of calcium on inter-lobe interactions. J. Biol. Chem. 275 37021–37029. [DOI] [PubMed] [Google Scholar]
- Cohen, F.E. and Prusiner, S.B. 1998. Pathologic conformations of prion proteins. Annu. Rev. Biochem. 67 793–819. [DOI] [PubMed] [Google Scholar]
- Curry, S., Mandelkow, H., Brick, P., and Franks, N. 1998. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 5 827–835. [DOI] [PubMed] [Google Scholar]
- Davis, M.K., Savitz, D.A., and Graubard, B.I. 1988. Infant feeding and childhood cancer. Lancet 2 365–368. [DOI] [PubMed] [Google Scholar]
- Dobson, C.M. 2001. The structural basis of protein folding and its links with human disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, E.D. and Chakravarti, A. 2001. The human genome sequence expedition: Views from the “base camp.” Genome Res. 11 645–651. [DOI] [PubMed] [Google Scholar]
- Hakansson, A., Zhivotovsky, B., Orrenius, S., Sabharwal, H., and Svanborg, C. 1995. Apoptosis induced by a human milk protein. Proc. Natl. Acad. Sci. 92 8064–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine, W.E., Klein, P.D., and Reeds, P.J. 1991. The importance of α-lactalbumin in infant nutrition. J. Nutr. 121 277–283. [DOI] [PubMed] [Google Scholar]
- Jackson, G.S. and Collinge, J. 2000. Prion disease: The propagation of infectious protein topologies. Microbes Infect. 2 1445–1449. [DOI] [PubMed] [Google Scholar]
- Jeffery, C.J. 1999. Moonlighting proteins. Trends Biochem. Sci. 24 8–11. [DOI] [PubMed] [Google Scholar]
- Jensen, R.G. 1996. The lipids in human milk. Prog. Lipid Res. 35 53–92. [DOI] [PubMed] [Google Scholar]
- Kaneko, K., Zulianello, L., Scott, M., Cooper, C.M., Wallace, A.C., James, T.L., Cohen, F.E., and Prusiner, S.B. 1997. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. 94 10069–10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I.C., Shirahama, T., and Cohen, A.S. 1967. The lipid content of amyloid fibrils purified by a variety of methods. Am. J. Pathol. 50 869–886. [PMC free article] [PubMed] [Google Scholar]
- McLaurin, J., Yang, D., Yip, C.M., and Fraser, P.E. 2000. Review: Modulating factors in amyloid-β fibril formation. J. Struct. Biol. 130 259–270. [DOI] [PubMed] [Google Scholar]
- Pan, K.M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., Mehlhorn, I., Huang, Z., Fletterick, R.J., Cohen, F.E., et al. 1993. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. 90 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepys, M.B. 2001. Pathogenesis, diagnosis and treatment of systemic amyloidosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergami, P., Jaffe, H., and Safar, J. 1996. Semipreparative chromatographic method to purify the normal cellular isoform of the prion protein in nondenatured form. Anal. Biochem. 236 63–73. [DOI] [PubMed] [Google Scholar]
- Safar, J., Roller, P., Gajdusek, D., and Gibbs, C.J. 1993. Thermal-stability and conformational transitions of scrapie amyloid (prion) protein correlate with infectivity. Protein Sci. 2 2206–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, M. 1999. Molecular dynamics model structures for the molten globule state of α-lactalbumin: Aromatic residue clusters I and II. Protein Eng. 12 1097–1104. [DOI] [PubMed] [Google Scholar]
- Sarles, J., Moreau, H., and Verger, R. 1992. Human gastric lipase: Ontogeny and variations in children. Acta Paediatr. 81 511–513. [DOI] [PubMed] [Google Scholar]
- Svanborg, C., Agerstam, H., Aronson, A., Bjerkvig, R., Duringer, C., Fischer, W., Gustafsson, L., Hallgren, O., Leijonhuvud, I., Linse, S., et al. 2003. HAMLET kills tumor cells by an apoptosis-like mechanism: Cellular, molecular, and therapeutic aspects. Adv. Cancer Res. 88 1–29. [DOI] [PubMed] [Google Scholar]
- Svensson, M., Sabharwal, H., Hakansson, A., Mossberg, A.K., Lipniunas, P., Leffler, H., Svanborg, C., and Linse, S. 1999. Molecular characterization of α-lactalbumin folding variants that induce apoptosis in tumor cells. J. Biol. Chem. 274 6388–6396. [DOI] [PubMed] [Google Scholar]
- Svensson, M., Hakansson, A., Mossberg, A.K., Linse, S., and Svanborg, C. 2000. Conversion of α-lactalbumin to a protein inducing apoptosis. Proc. Natl. Acad. Sci. 97 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telling, G.C., Scott, M., Hsiao, K.K., Foster, D., Yang, S.L., Torchia, M., Sidle, K.C., Collinge, J., DeArmond, S.J., and Prusiner, S.B. 1994. Transmission of Creutzfeldt-Jakob disease from humans to transgenic mice expressing chimeric human-mouse prion protein. Proc. Natl. Acad. Sci. 91 9936–9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J., Winter, N., Terwey, D., Bratt, J., and Banaszak, L. 1997. The crystal structure of the liver fatty acid–binding protein: A complex with two bound oleates. J. Biol. Chem. 272 7140–7150. [DOI] [PubMed] [Google Scholar]
- Wijesinha-Bettoni, R., Dobson, C.M., and Redfield, C. 2001. Comparison of the structural and dynamical properties of holo and apo bovine α-lactalbumin by NMR spectroscopy. J. Mol. Biol. 307 885–898. [DOI] [PubMed] [Google Scholar]
- Wu, L.C. and Kim, P.S. 1998. A specific hydrophobic core in the α-lactalbumin molten globule. J. Mol. Biol. 280 175–182. [DOI] [PubMed] [Google Scholar]