Abstract
In sulfatases a Cα-formylglycine residue is found at a position where their cDNA sequences predict a cysteine residue. In multiple sulfatase deficiency, an inherited lysosomal storage disorder, catalytically inactive sulfatases are synthesized which retain the cysteine residue, indicating that the Cα-formylglycine residue is required for sulfatase activity. Using in vitro translation in the absence or presence of transport competent microsomes we found that newly synthesized sulfatase polypeptides carry a cysteine residue and that the oxidation of its thiol group to an aldehyde is catalyzed in the endoplasmic reticulum. A linear sequence of 16 residues surrounding the Cys-69 in arylsulfatase A is sufficient to direct the oxidation. This novel protein modification occurs after or at a late stage of cotranslational protein translocation into the endoplasmic reticulum when the polypeptide is not yet folded to its native structure.
A novel amino acid derivative, Cα-formylglycine (FGly; 2-amino-3-oxopropionic acid), has been found in eukaryotic sulfatases at a position, where their cDNAs predict a cysteine. By mass spectrometry this was shown for two human sulfatases and for one from the green alga Volvox carteri (1, 2). The FGly residue is part of the highly conserved hexapeptide L/V-FGly-X-P-S-R, which is found in the N-terminal region of all eukaryotic sulfatases and which mostly carries a hydroxyl or thiol group on residue X (2). The FGly is critical for the catalytic activity of sulfatases. In multiple sulfatase deficiency (MSD), an autosomal recessively transmitted lysosomal storage disorder, the activity of all sulfatases known so far is severely decreased (3). Analysis of the sulfatase polypeptides synthesized in MSD fibroblasts revealed that they lack the FGly residue and contain a cysteine instead, as predicted by the cDNA (1). Moreover, expression of sulfatase cDNAs in MSD fibroblasts led to the synthesis of inactive sulfatases, while expression of the same cDNAs in non-MSD fibroblasts yielded active sulfatases (4). Recent crystallographic analysis of two human sulfatases, arylsulfatase A (ASA) and arylsulfatase B, has shown that the FGly residue is part of the catalytic site. The aldehyde is likely to serve as an acceptor for sulfate during catalysis (ref. 5; G. Lukatela, N. Krauss, T. Selmer, V. Gieselmann, K.v.F., and W. Saenger, unpublished work). These observations have shown that sulfatases share a unique FGly residue that is likely to be generated posttranslationally from a cysteine residue and that is required for cleavage of sulfate ester bonds. MSD, on the other hand, is likely to be caused by mutations affecting the conversion of cysteine to FGly.
In the present study we have examined in an in vitro system, whether cysteine is incorporated into newly synthesized ASA polypeptides and whether the cysteine is converted posttranslationally into FGly. Furthermore, we asked where and when the FGly residue is generated and which determinants in ASA specify this novel protein modification.
MATERIALS AND METHODS
Site-Directed Mutagenesis.
Mutagenesis of a cDNA coding for human ASA (6) was carried out by PCR methods using appropriate primers.
Protein Expression and Purification.
The cDNAs of wild-type ASA and ASA-F59M, respectively, were cloned into the pMPSVEH vector (7) downstream of the myeloproliferative sarcoma virus promoter. The resulting plasmid and PGK-hygro as selection marker were used for stable transfection of mouse embryonic fibroblasts deficient in both mannose-6-phosphate receptors, as described (8). The expressed ASA protein was purified from the secretions of the cells by affinity chromatography (9). It mainly contained an FGly residue in position 69. Due to overexpression a minor fraction was unmodified carrying Cys-69 (1).
For in vitro expression, an optimized translation initiation site (AACATG) was created in the cDNAs of wild-type ASA and ASA-F59M, respectively, and a stop-codon was introduced 3′ of codon 200. This product was cloned into pBluescript II (Stratagene) downstream of the T7-promoter or, for high-level expression (see Figs. 2, 3, 4, 5), into the pSP64-derived vector pTD1 (see below) downstream of the SP6-promoter. To exchange the ASA signal peptide for that of preprolactin an oligonucleotide (ACTCCGGAC) comprising a BspEI-site was added 5′ of codon 19 and the BspEI/EcoRI-fragment encoding ASA-F59M residues 19–200 was cloned into pTD1 in frame with a sequence encoding the signal peptide of preprolactin (residues 1–30). The connecting BspEI-oligonucleotide codes for the tripeptide TPD, which is similar to the N terminus of mature prolactin (TPV, residues 31–33); the resulting plasmid was designated pTD3. pTD1 contained the complete cDNA of preprolactin-V33D. pTD1 was obtained by replacing in pCA37 (10) the NheI/BspEI-fragment encoding the signal peptide of preprocecropin A by the corresponding fragment of pCA38 (10) encoding the signal peptide of preprolactin. ASA-F59M fragments furthermore were fused to the signal peptide of preprocecropin A by cloning the corresponding part of the cDNA into pCA37.
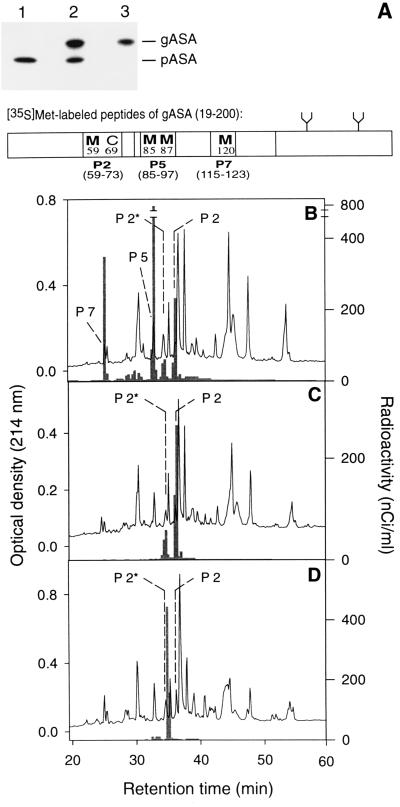
Figure 2.
In vitro modification of peptide 2 of ASA in the endoplasmic reticulum. (A) A cDNA coding for a fusion of the signal peptide of preprolactin and the N-terminal residues 19–200 of mature ASA-F59M was expressed in vitro in the presence of [35S]methionine and dog pancreas microsomes. The translation products were analyzed by SDS/PAGE and fluorography. The translation product imported into and glycosylated by the microsomes (gASA) was separated from the nonimported precursor (pASA) by sedimentation of microsomes. The supernatant is shown in lane 1 and the pellet in lane 2. Precursor remaining unspecifically bound to the surface of microsomes was digested by proteinase K and the proteolytic fragments were removed by two further centrifugation steps leading to purified gASA (lane 3). The distribution of labeled methionines within gASA and its tryptic peptides 2, 5, and 7 is shown in the scheme. (B) Radiolabeled gASA (≈60 nCi per methionine) was mixed with 40 μg of unlabeled ASA-F59M protein and subjected to analysis of its tryptic peptides, as described in Fig. 1. In the HPLC chromatogram the positions of the unmodified and the modified peptide 2 (P2 and P2*, respectively) are indicated as well as those of peptide 5 and 7, as identified by mass spectrometry and N-terminal sequencing. The labeled peptides were localized by liquid scintillation counting (see histogram) and identified by radiosequencing on the basis of the position of their methionines (not shown). Labeled and unlabeled peptides 2, 5, and 7, respectively, coeluted in the same fractions. (C) HPLC analysis of tryptic peptides derived from a mixture of unlabeled ASA-F59M protein and a labeled gASA, in which the three methionines in peptides 5 and 7 were replaced by threonine (position 85) or leucine (positions 87 and 120). As observed in B, the labeled peptide 2 appeared in two forms. (D) HPLC analysis of tryptic peptides derived from a mixture of unlabeled ASA-F59M protein and a labeled gASA, in which methionines 85, 87, and 120 were replaced by threonine or leucine (see C) and Cys-69 by serine.
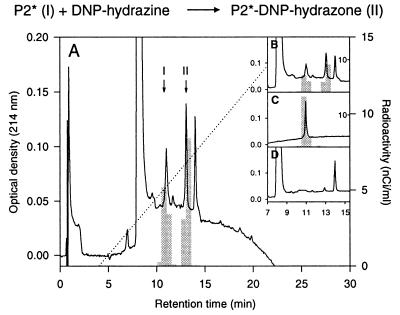
Figure 3.
Generation of an aldehyde group in peptide 2 upon in vitro translocation of ASA into microsomes. The [35S]methionine-labeled peptide 2 coeluting with unlabeled P2* (see Fig. 2B) was assayed for the presence of an aldehyde group by reaction with DNP-hydrazine. (A and B) After incubation of the peptide with the reagent, P2* (I) and its DNP-hydrazone derivative (II) were separated by RP-HPLC and identified by mass spectrometry. Based on absorbance at 214 nm, 61% of the unlabeled P2* were recovered as hydrazone. The 35S-labeled P2* was converted into the DNP-hydrazone with an efficiency of 57%. (C) RP-HPLC of the peptide incubated only with the solvent of DNP-hydrazine. (D) RP-HPLC of an incubation mixture lacking the peptide, thus showing the UV-signals associated with the reagent.
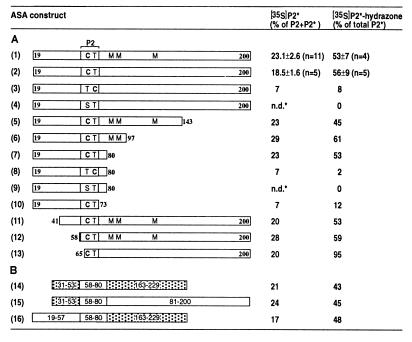
Figure 4.
Modification of Cys-69 in ASA depends on adjacent sequence information. In vitro modification efficiencies for ASA constructs 1–16 are expressed as percentage of [35S]P2* of the sum of labeled P2 plus P2*, as determined after HPLC of tryptic peptides (see Fig. 2). In addition, the fraction of [35S]P2* that was converted into the corresponding DNP-hydrazone (see Fig. 3) is given. The SD calculated from independent experiments is given only for constructs 1 and 2 and was similar for all other constructs. (A) The constructs were synthesized in vitro in the presence of microsomes (Fig. 2A) as fusions of preprolactin signal peptide and ASA fragments with different C and N termini, which are given by the numbers. In peptide 2 amino acid residues 69 and 70 are shown. Where present, the methionines in positions 85, 87, and 120 are indicated. In other constructs they were mutated (see Fig. 2C). The methionine introduced in position 59 (constructs 1–12) or 64 (construct 13) is not shown. C-terminal truncations were generated by introducing stop-codons into the cDNA. The N terminus was shortened by fusing the signal peptide encoding sequence to various codons of ASA cDNA. In constructs 3 and 8 the sequence of Cys-69 and Thr-70 was inverted and in constructs 4 and 9 Cys-69 was replaced by serine. (B) Chimeras were analyzed in which residues 19–57 and/or residues 81–200 of ASA construct 2 (A) were replaced by residues 31–53 and/or 163–229 of preprolactin, as indicated. In the scheme the sequences derived from preprolactin are stippled. All constructs retain the tryptic peptide 2 of ASA (residues 59–73), which contains the only methionine (Met-59) present in the chimeric polypeptides. n.d.*: After HPLC, the nonmodified Ser-69 containing form of [35S]P2 eluted partially overlapping with the modified form of unlabeled peptide 2, P2* (see Fig. 2D). A calculation of [35S]P2* was therefore not possible. The labeled material, assayed fraction by fraction, did not react with DNP-hydrazine.
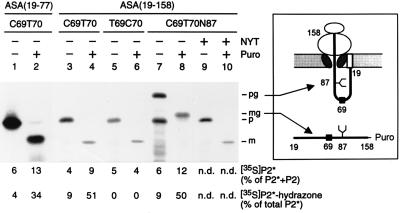
Figure 5.
In vitro modification of nascent ASA polypeptides occurs at a posttranslational or late cotranslational stage. In vitro translation reactions in the presence of [35S]methionine and rough microsomes (see Fig. 2) were programmed with run-off transcripts coding for a fusion of the preprolactin signal peptide and the N-terminal residues 19–77 (lanes 1–2) or 19–158 (lanes 3–10) of the following ASA constructs shown in Fig. 4: construct 2 (lanes 1–4), construct 3 (lanes 5–6) or construct 2 carrying the N-glycosylation signal N87Y88T89 (lanes 7–10). After translation the microsomes were isolated and one half was kept on ice, while the other half was incubated with puromycin (Puro), as indicated. One tenth of each sample was analyzed by SDS/PAGE and fluorography; note that lanes 1 and 2 were run in a different gel system (see Methods). The remaining part was stopped with guanidine hydrochloride and analyzed for modification of peptide 2, as explained in Fig. 4 (n.d., not determined). The polypeptides visible in the fluorograms represent the precursor (p), mature (m), glycosylated precursor (pg), or glycosylated mature (mg) form of the translation products. The identification of the pg (lane 7) and the mg form (lane 8) is confirmed by the observation that in the presence of 0.1 mM glycosylation inhibitor peptide NYT (16) the nonglycosylated p (lane 9) and m (lane 10) forms were synthesized. Note that the mature form contains one methionine and the precursor form contains two methionines. The scheme depicts the topology of the glycosylated precursor arrested during translocation and exposing Cys-69 inside the ER (Upper). The glycosylated mature form is released into the ER-lumen as a peptidyl-puromycin product (Lower). The numbers refer to the ASA residues. The symbols at positions 69 and 87 indicate the sites of modification and N-glycosylation, respectively. The signal peptide is shown as an open box.
To fuse residues 1–53 of preprolactin-V33D to residues 58–200 of ASA, an oligonucleotide (AGACACAGGCACGTAGAAGTCTGTCATCCG) comprising a BsaAI-site and coding for ASA-F59M residues 58–63 was added 3′ of codon 53 of pTD1. The PCR product was cloned as a HindIII/BsaAI fragment into a pTD3-derivative (see Fig. 4A, construct 2) yielding pTD48. To fuse residues 163–229 of preprolactin to residues 58–80 of ASA, an oligonucleotide (CTCCTGACCGGCCGG) comprising an EagI-site and coding for ASA residues 76–80 was added 5′ of codon 163 of pTD1. The PCR product was cloned as an EagI/EcoRI fragment into pTD48.
In vitro synthesis of ASA-derived proteins was carried out in a coupled transcription/translation system (TNT, Promega) according to the instructions of the manufacturer; for translation intermediates see below. The system is based on SP6- or T7-polymerase, reticulocyte lysate, and on either [35S]methionine or [35S]cysteine (Amersham) and was programmed with circular DNA. Rough microsomes from dog pancreas (11) were added at 7.5 equivalents (12) per 50 μl translation mixture. After incubation for 45 min at 30°C the microsomes were sedimented for 5 min at 75,000 × g and treated with 170 μg/ml proteinase K for 45 min at 0°C. Then 0.2 mg/ml phenylmethylsulfonyl fluoride was added and after 5 min on ice the microsomes were sedimented again two times. In vitro translation products were chilled on ice and aliquots were boiled immediately with electrophoresis sample buffer. SDS/PAGE and fluorography were as described (11). High-Tris gels containing 17.5% acrylamide/0.23% bis-acrylamide or (Fig. 5, lanes 1–2), high-Tris/urea gels containing 19.5% acrylamide/0.26% bis-acrylamide were used. The major part of the samples was stopped with carboxymethylation buffer (1) at a final concentration of 4 M guanidine hydrochloride and heating to 80°C. This material was used for peptide analysis.
Generation of Translation/Translocation Intermediates.
Nascent polypeptide chains attached to ribosomes were synthesized upon programming the translation system with run-off transcripts lacking a stop-codon. Such transcripts were obtained by in vitro transcription, using the Ribomax system (Promega), of the ASA cDNA containing expression vectors (see above) cleaved in the coding region with BsrFI (3′ of codon 77), AvaI (3′ of codon 95) or with AlwNI (3′ of codon 158). After translation of these transcripts for 15 min at 30°C in the presence (or absence) of microsomes, the membrane-associated (or ribosome-associated) polypeptides were sedimented at 75,000 × g for 5 min (200,000 × g for 15 min) and, where indicated, released from the ribosomes by incubation with 1.5 mM puromycin for 15 min at 37°C.
Peptide Analysis.
Purified in vitro translation/translocation products were mixed with 30–40 μg of unlabeled ASA or ASA-F59M carrier protein and subjected to peptide analysis. Reductive carboxymethylation and generation of tryptic peptides was carried out as described (1). However, the concentrations of DTT and iodoacetic acid were reduced 5-fold and trypsin was increased to 2–4% (wt/wt) of total protein. Separation of tryptic peptides by reversed phase (RP)-HPLC, mass spectrometry, and sequencing of peptides also were described earlier (1). To assay for the presence of an aldehyde group, purified P2* was lyophilized and dissolved in 50 μl 0.5% trifluoroacetic acid. One half of the peptide sample was incubated with 25 μl of a freshly prepared saturated solution of dinitrophenylhydrazine in 0.5% trifluoroacetic acid/50% acetonitrile for 30 min at 37°C, the other half with the solvents alone. Unreacted P2* and its dinitrophenylhydrazone derivative were separated by RP-HPLC on a C2/C18-μPeak column (Pharmacia) and identified by mass spectrometry using 2,3-dimethoxy-4-hydroxycinnamic acid as a matrix.
RESULTS
The Primary Translation Product of ASA Contains Cysteine at the Position of FGly Present in the Mature Enzyme.
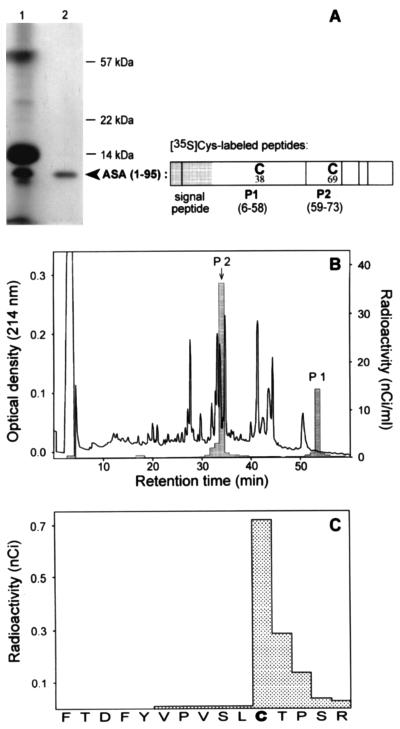
The possibility was taken into consideration that FGly formation occurs pretranslationally at the level of the amino acyl-tRNA, as is known for the incorporation of selenocysteine into proteins (13). To find out whether the primary translation product of human ASA contains a cysteine or a FGly in position 69, the N-terminal 95 amino acid residues were synthesized in vitro in the presence of [35S]cysteine. The translation system was programmed with an ASA transcript terminating at codon 95, leading to peptidyl-tRNAs that remain associated with the ribosomes due to the lack of a stop-codon. The nascent polypeptides were purified by sedimentation of ribosomes, from which they were released by addition of puromycin (Fig. 1A). The peptidyl-puromycin product was mixed with carrier ASA protein comprising both the modified and the unmodified form of ASA. This mixture was subjected to reductive carboxymethylation of cysteines and tryptic digestion. Residue 69 is part of the tryptic peptide 2 of ASA comprising residues 59–73. After separation of tryptic peptides by RP-HPLC two [35S]cysteine-labeled peptides were recovered, namely peptide 1 containing Cys-38 and peptide 2 containing Cys-69 (Fig. 1B). The labeled peptide 2 was found to coelute with the unmodified form of peptide 2 (P2) of the added ASA carrier protein. This was verified by mass spectrometry of P2 and radiosequencing of [35S]P2 (Fig. 1C).
Figure 1.
Incorporation of cysteine into the primary ASA translation product at position 69. The N-terminal 95 residues of ASA were expressed in vitro in the presence of [35S]cysteine using a truncated run-off transcript as a message. The translation products were analyzed by SDS/PAGE and fluorography (A). From the crude translation products of the reticulocyte lysate (lane 1) the ASA (1–95) nascent polypeptide was purified by sedimentation of ribosomes (lane 2). The distribution of cysteines within ASA (1–95) and its tryptic peptides 1 and 2 is shown in the scheme. Following release from the ribosomes by addition of puromycin, radiolabeled ASA (1–95) (about 10 nCi per cysteine; 1 Ci = 37 GBq) was mixed with 30 μg of unlabeled ASA protein, serving as carrier, and subjected to reductive carboxymethylation, digestion with trypsin and separation of tryptic peptides by RP-HPLC (B). The cysteine containing form of peptide 2 (position indicated by an arrow) and the FGly containing form of peptide 2 were identified by mass spectrometry. The 35S-labeled peptides were localized by liquid scintillation counting of HPLC fractions (see histogram). [35S]P2 was identified by radiosequencing demonstrating that radioactivity was incorporated in position 69 (sequencing cycle 11) (C). The sequence of P2 is given on the abscissa. [35S]P2 of the translation product and the cysteine containing P2 of the carrier protein coeluted in the same fraction. 35S-labeled peptide 1 eluted at an 8% higher acetonitrile concentration as compared with peptide 1 of the carrier protein. This can be explained by the N-terminal extension of [35S]peptide 1 by the hydrophobic residues 6–18, which are part of the signal peptide that is absent in the carrier ASA.
Because the 35S-label of Cys-69 is expected to be eliminated during FGly formation, a modified form of peptide 2 might have been overlooked in the experiment shown in Fig. 1. Therefore, in vitro translation also was carried out in the presence of [35S]methionine. The translation system was programmed with a transcript encoding the N-terminal 77 residues of ASA-F59M. Phe-59 was replaced by methionine to incorporate a label into peptide 2 during translation. ASA protein with the same F59M mutation and showing a catalytic activity similar to the wild-type was added as carrier to the purified translation product prior to carboxymethylation and tryptic digestion. The FGly-containing form of peptide 2 (P2*) is not carboxymethylated and elutes from the RP-column at a 1.5% lower acetonitrile concentration than the unmodified P2 containing carboxymethyl-cysteine (see Fig. 2C). A single [35S]methionine-labeled peptide was recovered from the ASA (1–77) translation product, which by radiosequencing was identified as peptide 2 and which coeluted exclusively with the unmodified P2 of ASA (not shown). These data demonstrate that a cysteine is incorporated into the nascent ASA polypeptide chain in position 69, as predicted by the cDNA. The conversion of this cysteine to FGly therefore represents a novel co- or posttranslational protein modification.
Modification of Cys-69 Occurs in the Endoplasmic Reticulum (ER).
Since all sulfatase cDNAs encode a signal peptide directing translocation of sulfatase proteins into the ER and since several of them are known to be secreted as active enzymes (1, 14, 15), the ER in particular and the secretory pathway in general are candidate sites for FGly formation. The ability of microsomes to catalyze the modification of Cys-69 of ASA was examined under in vitro conditions. A cDNA encoding an N-terminal fragment (residues 19–200) of ASA-F59M followed by a stop-codon was subjected to coupled in vitro transcription and translation in the presence of [35S]methionine and dog pancreas microsomes. To ensure efficient translocation into the microsomes the ASA signal peptide (residues 1–18) was replaced by the signal peptide of preprolactin. The translation product imported into and glycosylated by the microsomes was purified (see Fig. 2A), mixed with carrier ASA-F59M protein and subjected to carboxymethylation and tryptic digestion. Four major [35S]methionine-labeled peptides were recovered after RP-HPLC (Fig. 2B), which were identified as the tryptic peptides P2 and P2* (containing Met-59), P5 (Met-85 and Met-87), and P7 (Met-120) by radiosequencing and by their coelution with the respective unlabeled peptides. To examine for the presence of an aldehyde function each of the labeled peptides was subjected to reaction with dinitrophenylhydrazine (DNPH). The parent peptides and their hydrazone derivatives were separated by RP-HPLC. Only P2* reacted with DNPH. Fifty to sixty percent of [35S]P2*, as well as of the unlabeled P2* carrier, were converted into the hydrazone (Fig. 3 and Fig. 4A, construct 1). The P2*-hydrazone showed an absorbance at 450 nm due to the dinitrophenyl group (not shown) and had the predicted mass of 1879 Da, i.e., 180 Da more than P2*. Hydrazone formation was almost quantitative after reaction of a shorter, more hydrophilic P2* derivative with DNPH (see below; Fig. 4A, construct 13).
When the methionine residues 85, 87, and 120 of the ASA fragment were replaced by threonine or leucine, P2 and P2* were the only labeled peptides recovered after RP-HPLC (Fig. 2C). The modification efficiency (P2* as a percentage of P2 plus P2*) was similar as for the ASA fragment containing methionines 85, 87, and 120 (Fig. 4A, construct 2). When Cys-69 was replaced by serine in the ASA construct lacking the three methionines, a single labeled tryptic peptide was recovered, which eluted from the RP-column between P2* and P2 (Fig. 2D). This peptide did not react with DNPH (Fig. 4A, construct 4). When the positions of Cys-69 and Thr-70 were inverted, the modification of peptide 2 was severely decreased, but not abolished (Fig. 4A, construct 3).
When microsomes were added after inhibition of translation by cycloheximide and puromycin, [35S]methionine- or [35S]cysteine-labeled peptide 2 was recovered only in the carboxymethylated form, as was observed in the absence of microsomes (see Fig. 1). In the presence of microsomes the [35S]cysteine label was eliminated from P2* but not from P2, as was determined by radiosequencing of the peptide labeled with both [35S]cysteine and [35S]methionine. Incorporation of [35S]cysteine in position 69 was normalized for that of [35S]methionine in position 59. The 35S-label in position 69 of P2* was at the level of detection, i.e., <10% as compared with P2 (data not shown). Taken together these results demonstrate that during translation a cysteine is incorporated in position 69, a fraction of which is converted into FGly upon translocation into the ER. Furthermore, serine cannot substitute for cysteine in the formation of FGly.
On average 23.1% of the labeled peptide 2 were recovered as P2* (Fig. 4A, construct 1). The P2* fraction could not be increased by prolonging translation and translocation. It increased, however, when less ASA fragment was imported into the microsomes, as was observed under conditions of limiting template for transcription or when expression was under control of a T7- instead of a SP6-promoter. In the latter case the amount of translocated protein was reduced 20-fold and the P2* fraction increased to 38–41% (not shown). This indicates that the extent of Cys-69 modification is limited by a saturable component in the ER.
Modification Depends on Linear Sequence Information in the Vicinity of Cys-69.
The modification of Cys-69 in the ASA fragment 19–200 revealed that the recognition motif for the modifying machinery is confined to the N-terminal 200 amino acids. To determine the minimal length of an ASA fragment supporting cysteine modification, the ASA was progressively truncated at the C terminus by introducing stop-codons at positions 144, 98, 81, and 74. The minimal fragment supporting the modification comprised residues 19–80 (Fig. 4A, construct 7). Replacing in this construct Cys-69 by serine abolished the modification. A severe reduction of modification was also observed after inverting the sequence Cys-69/Thr-70 or after further truncation of the C-terminus following residue 73 (Fig. 4A, constructs 8–10). Deleting the N-terminal residues 19–40 or 19–57 did not affect the modification (Fig. 4A, constructs 11 and 12). Furthermore, a fragment retaining residues 65–200 of ASA construct 2 and containing a methionine in position 64 was analyzed. After tryptic digestion a labeled peptide 2 derivative was obtained, in which the sequence TPDM was followed by residues 65–73 of ASA. Of this peptide 20% were found to be modified and to react efficiently with DNPH (Fig. 4A, construct 13). We conclude from these results that the information required for converting Cys-69 to FGly is located within residues 65–80 of ASA.
We furthermore examined whether the linear sequence directing the modification in ASA is sufficient to do so when inserted into an unrelated polypeptide. A chimera was constructed in which ASA-F59M residues 58–80 were placed between residues 31–53 and 163–229 of preprolactin (see Fig. 4B). FGly formation could be analyzed as for the ASA constructs, since the chimera retained the entire sequence of peptide 2 of ASA flanked by its tryptic cleavage sites. Twenty-one percent of the labeled peptide 2 were recovered as P2* (Fig. 4B, construct 14). FGly formation in the chimeric polypeptide was as efficient as in constructs in which one or both of the flanking sequences were derived from ASA (Fig. 4, constructs 1 and 2, and 15 and 16).
Cysteine Modification Occurs After or at a Late Stage of Protein Translocation.
Next we tested whether Cys-69 becomes already modified, when translation and translocation are still going on. For this purpose the translation/translocation system was programmed with ASA transcripts terminating at codons 77 or 158, leading to peptidyl-tRNAs that remain associated with the ribosomes (see Fig. 1). Peptidyl-tRNAs inserted into the microsomal translocation apparatus are released as peptidyl-puromycin products into the lumen of the microsomes upon addition of puromycin. In the peptidyl-tRNA terminating at residue 77 the Cys-69 is located within the ribosome and should become accessible to the modifying machinery only after addition of puromycin. Fig. 5 (lane 1) shows that after translation the microsome-associated products comprised exclusively the precursor polypeptide. RP-HPLC of its tryptic peptides revealed that 6% of the radioactivity associated with P2 and P2* coeluted with unlabeled P2*. Of this material only 4% reacted with DNPH. This residual modification is attributed to leakage of translocation arrest (see below). Upon chase with puromycin all precursor was processed to the mature form (Fig. 5, lane 2). In this case peptide analysis revealed that 13% of the radioactivity coeluted with P2* and 34% of this material were converted into the hydrazone. Thus, Cys-69 in the peptidyl-puromycin product was modified following release from the translocation arrest. The relatively low modification efficiency, which also was observed for longer peptidyl-puromycin products (see below) indicates that the transient arrest of translocation or the C-terminal puromycin may affect modification.
When the translation/translocation system was programmed with a transcript terminating at codon 158, it was anticipated that residues 65–80 (which are sufficient to ensure modification of Cys-69; Fig. 4) would be exposed at the lumenal side of microsomes, even if the translation product remains attached to the ribosome. To control for this assumption, a derivative of this construct was made with an N-glycosylation site at position 87. Glycosylation occurs in the ER-lumen when the glycosylation site has reached a distance of at least 12 residues from the plane of the ER-membrane (17). When translation products carrying no glycosylation site were analyzed, only precursor was recovered (Fig. 5, lane 3). The same held true when the glycosylation site was present. In this case 60% of the precursor were glycosylated (Fig. 5, lane 7). The incompleteness of glycosylation is ascribed to a restricted accessibility of the glycosylation site located only 71 residues from the C terminus (17). Both glycosylated and nonglycosylated translation products could be precipitated by the cationic detergent cetyltrimethylammonium bromide (18) indicating the presence of a C-terminal tRNA (not shown). Thus, the polypeptides were fixed at the ribosome, while Cys-69 was accessible inside the microsomes. The efficiency of FGly-69 formation (4–6%) was as low as for the construct terminating at residue 77 (Fig. 5, lanes 3, 7, and 1). Upon chase with puromycin the polypeptides were quantitatively translocated and processed by the signal peptidase. A significant fraction received the aldehyde group reacting efficiently with DNPH (Fig. 5, lanes 4 and 8). In controls, in which the sequence Cys-69/Thr-70 was inverted (see Fig. 4A), no aldehyde formation was detected (Fig. 5, lanes 5 and 6). This strongly suggests that the background level of modification observed in samples not chased with puromycin was due to leakage of the translocation arrest. Taken together these results indicate that modification of Cys-69 occurs after N-glycosylation and when protein synthesis has advanced beyond residue 158.
DISCUSSION
The present study demonstrates that the conversion of a thiol into an aldehyde group, which occurs at a unique cysteine residue conserved among all eukaryotic sulfatases, is catalyzed in the ER. During protein synthesis a cysteine is incorporated into the ASA polypeptide at position 69 (Fig. 1), which is converted into FGly upon translocation into the ER. This is based on the characterization of ASA fragments obtained after in vitro translation and translocation into microsomes. The tryptic peptide containing residue 69 during RP-HPLC coeluted with the authentic FGly-69 containing peptide (Fig. 2), it was converted into a hydrazone upon reaction with DNPH (Fig. 3), and it had lost its 35S-label when generated from [35S]cysteine-labeled ASA.
Formation of FGly occurs either after or at a late stage of cotranslational protein translocation. Exposure of Cys-69 in the ER lumen while the nascent polypeptide was still attached to the ribosome did not lead to FGly formation in the translocation intermediates tested (Fig. 5). The observation of a nascent polypeptide that was glycosylated in position 87 but remained unmodified in position 69 indicates that FGly formation occurs after passing the active site of the oligosaccharyltransferase located at the lumenal surface of the ER membrane. However, FGly formation prior to N-glycosylation cannot rigorously be excluded. Since chase with puromycin not only led to FGly formation but also to more efficient glycosylation (Fig. 5, compare lanes 7 and 8), in theory it is possible that the modified species originated exclusively from the nonglycosylated but not from the glycosylated precursor. This, however, would not explain why the glycosylated precursor formed in the absence of puromycin was not modified (Fig. 5, lane 7) and why nonglycosylated precursor and a mixture of glycosylated and nonglycosylated precursor were modified with similar efficiency after addition of puromycin (Fig. 5, compare lanes 4 and 8).
The temporal relation of FGly formation to another cotranslational event, i.e., signal peptide cleavage, could not be resolved. Unexpectedly, cleavage of the preprolactin signal peptide, which occurs when nascent preprolactin exceeds a length of 140–150 residues (refs. 19 and 20; data not shown), was not observed when fused to ASA residues 19–158, unless puromycin was added after translation (Fig. 5). The mature part of ASA seems to be responsible for the delayed signal peptide cleavage, since it was also observed when ASA (19–158) was fused to the signal peptides of ASA and preprocecropin A (not shown).
The generation of FGly is determined by linear sequence information located in the vicinity of the cysteine to be modified. Residues 58–80 of ASA are sufficient to direct the modification of Cys-69, as was demonstrated by transferring this sequence to a heterologous protein (Fig. 4B). Since a deletion N-terminal of residue 65 did not affect modification (Fig. 4A), we conclude that residues 65–80 of ASA (PVSLCTPSRAALLTGR) comprise an autonomous signal for generation of FGly. This sequence includes the L/V-C-X-P-S-R hexapeptide (see Introduction) and an L/M-T-G-R/K tetrapeptide, which both are fully conserved among human sulfatases (2, 21). While the position of that sequence within the translation product can be varied, the position of the cysteine within the sequence is critical. Furthermore, folding of the polypeptide is unlikely to be a prerequisite for the modification because the various recombinant constructs and chimeras tested (see Fig. 4) are not expected to fold into any structure resembling native ASA. The importance of this novel protein modification for the catalytic activity of sulfatases is emphasized by the recent finding that the FGly residue is localized in the active site cavity of ASA and serves as an acceptor for sulfate during sulfate ester cleavage (G. Lukatela, N. Krauss, T. Selmer, V. Gieselmann, K.v.F., and W. Saenger, unpublished data).
Acknowledgments
We thank P. Schlotterhose for skillful technical assistance, D. Isbrandt for providing the starting cDNA, and K. Neifer for sequencing. T. Selmer gave valuable advice in peptide analysis. The pCA-plasmids were a gift of R. Zimmermann (Homburg, Germany). This work was supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ASA, arylsulfatase A (cerebroside-3-sulfate 3-sulfohydrolase, EC 3.1.6.8); MSD, multiple sulfatase deficiency; DNPH, dinitrophenylhydrazine; ER, endoplasmic reticulum; FGly, Cα-formylglycine; RP-HPLC, reversed-phase HPLC; P2, Cys-containing form of peptide 2; P2*, FGly-containing form of peptide 2.
References
- 1.Schmidt B, Selmer T, Ingendoh A, von Figura K. Cell. 1995;82:271–278. doi: 10.1016/0092-8674(95)90314-3. [DOI] [PubMed] [Google Scholar]
- 2.Selmer T, Hallmann A, Schmidt B, Sumper M, von Figura K. Eur J Biochem. 1996;238:341–345. doi: 10.1111/j.1432-1033.1996.0341z.x. [DOI] [PubMed] [Google Scholar]
- 3.Kolodny E H, Fluharty A L. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 2693–2741. [Google Scholar]
- 4.Rommerskirch W, von Figura K. Proc Natl Acad Sci USA. 1992;89:2561–2565. doi: 10.1073/pnas.89.7.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond C S, Clements P R, Ashby S J, Collyer C A, Harrop S J, Hopwood J J, Guss J M. Structure (London) 1997;5:277–289. doi: 10.1016/s0969-2126(97)00185-8. [DOI] [PubMed] [Google Scholar]
- 6.Stein C, Gieselmann V, Kreysing J, Schmidt B, Pohlmann R, Waheed A, Meyer H E, O’Brien J S, von Figura K. J Biol Chem. 1989;264:1252–1259. [PubMed] [Google Scholar]
- 7.Artelt P, Morelle C, Ausmeier M, Fitzek M, Hauser H. Gene. 1988;68:213–219. doi: 10.1016/0378-1119(88)90023-6. [DOI] [PubMed] [Google Scholar]
- 8.Kasper D, Dittmer F, von Figura K, Pohlmann R. J Cell Biol. 1996;134:615–623. doi: 10.1083/jcb.134.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommerlade H J, Selmer T, Ingendoh A, Gieselmann V, von Figura K, Neifer K, Schmidt B. J Biol Chem. 1994;269:20977–20981. [PubMed] [Google Scholar]
- 10.Schlenstedt G, Gudmundsson G H, Boman H G, Zimmermann R. J Biol Chem. 1992;267:24328–24332. [PubMed] [Google Scholar]
- 11.Dierks T, Volkmer J, Schlenstedt G, Jung C, Sandholzer U, Zachmann K, Schlotterhose P, Neifer K, Schmidt B, Zimmermann R. EMBO J. 1996;15:6931–6942. [PMC free article] [PubMed] [Google Scholar]
- 12.Walter P, Blobel G. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- 13.Stadtman T C. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 14.Anson D S, Muller V, Bielicki J, Harper G S, Hopwood J J. Biochem J. 1993;294:657–662. doi: 10.1042/bj2940657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallmann A, Sumper M. Eur J Biochem. 1994;221:143–150. doi: 10.1111/j.1432-1033.1994.tb18723.x. [DOI] [PubMed] [Google Scholar]
- 16.Brunke M, Dierks T, Schlotterhose P, Escher A, Schmidt B, Szalay A A, Lechte M, Sandholzer U, Zimmermann R. J Biol Chem. 1996;271:23487–23494. doi: 10.1074/jbc.271.38.23487. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson I, von Heijne G. J Biol Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- 18.Gilmore R, Collins P, Johnson J, Kellaris K, Rapiejko P. Methods Cell Biol. 1991;34:223–239. doi: 10.1016/s0091-679x(08)61683-0. [DOI] [PubMed] [Google Scholar]
- 19.Rapoport T A, Jungnickel B, Kutay U. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 20.Martoglio B, Dobberstein B. Trends Cell Biol. 1996;6:142–147. doi: 10.1016/0962-8924(96)10001-5. [DOI] [PubMed] [Google Scholar]
- 21.Franco B, Meroni G, Parenti G, Levilliers J, Bernard L, Gebbia M, Cox L, Maroteaux P, Sheffield L, Rappold G A, Andria G, Petit C, Ballabio A. Cell. 1995;81:15–25. doi: 10.1016/0092-8674(95)90367-4. [DOI] [PubMed] [Google Scholar]