SUMMARY
We used computational algorithms to find conserved sequences in the 3′ untranslated region (UTR) of transcripts that exhibited rapid decay in primary human T cells and found that the consensus sequence UGUUUGUUUGU, which we have termed a GU-rich element (GRE), was enriched in short-lived transcripts. Using a tet-off reporter system, we showed that insertion of GRE-containing sequences from c-jun, jun B, or TNF receptor 1B, but not mutated GRE sequences, into the 3′UTR of a β-globin transcript conferred instability on the otherwise stable β-globin transcript. CUG-binding protein 1 (CUGBP1) was identified as the major GRE-binding activity in cytoplasmic extracts from primary human T cells based on supershift and immunoprecipitation assays. siRNA-mediated knockdown of CUGBP1 in HeLa cells caused stabilization of GRE-containing transcripts, suggesting that CUGBP1 is a mediator of GRE-dependent mRNA decay. Overall, our results suggest that the GRE mediates coordinated mRNA decay by binding to CUGBP1.
INTRODUCTION
The integration of signals from multiple regulatory proteins that bind to mRNA coordinately regulates selective mRNA decay during development and in response to environmental stimuli (Keene, 2007; Raghavan and Bohjanen, 2004). The best characterized example of coordinate gene regulation through selective mRNA decay is the regulation of mRNA decay by AU-rich elements (AREs). AREs are conserved sequences found in the 3′ untranslated region (UTR) from a variety of short-lived transcripts, including cytokine and proto-oncogene transcripts, which function to mediate rapid mRNA decay (Ogilvie et al., 2005; Raghavan et al., 2004). AREs mediate mRNA decay by interacting with ARE-binding proteins, such as K homology splicing regulatory protein (KSRP), tristetraprolin (TTP), and butyrate response factor 1 (BRF-1), that recruit components of the mRNA decay machinery to specific transcripts (Chen et al., 2001; Gherzi et al., 2004; Hau et al., 2007; Lykke-Andersen and Wagner, 2005), thereby coordinately regulating mRNA decay.
We have previously used microarrays to measure mRNA decay on a genome-wide scale in primary human T cells and identified hundreds of transcripts that exhibited rapid decay (Raghavan et al., 2002; Vlasova et al., 2005). Most of these short-lived transcripts did not contain AREs or other known RNA regulatory elements (Raghavan et al., 2004), leading us to hypothesize that they may contain other regulatory sequences that would allow them to be selectively recognized. In this work, we used computational methods to determine that the conserved 11-mer sequence UGUUUGUUUGU, which we have termed the GU-rich element (GRE), was enriched in the 3′UTRs of short-lived transcripts. Introduction of GRE sequences into the 3′UTR induced destabilization of an otherwise stable β-globin reporter transcript, demonstrating that the GRE is a functional mediator of mRNA decay. The RNA-binding protein CUG-binding protein 1 (CUGBP1), the mammalian homolog of the deadenylation regulator EDEN-BP in Xenopus, bound specifically to the GRE and mediated GRE-dependent mRNA decay. The GRE and CUGBP1 are components of a selective mRNA decay pathway that regulates the expression of an important subset of human transcripts.
RESULTS AND DISCUSSION
Because most transcripts that are regulated at the level of mRNA decay do not contain AREs or other known sequence elements that regulate mRNA decay (Raghavan et al., 2002, 2004), we hypothesized the existence of other regulatory elements that had not yet been discovered. We used computational methods to search for conserved oligomer sequences that were enriched within short-lived transcripts which were induced following T cell activation (see the Experimental Procedures) and found that the 11-mer consensus sequence UGUUUGUUUGU, which we have termed a GRE, was enriched in the 3′UTR of short-lived transcripts. We retrieved the 3′UTR sequences from 4812 transcripts for which we had measured mRNA half-lives in T cells (Raghavan et al., 2002), and searched within these 3′UTR sequences for the 11-mer consensus GRE, allowing one mismatch. We then compared the abundance of this GRE 11-mer sequence in the set of 384 transcripts that exhibited very rapid decay (upper limit of half-life 95% confidence interval of less than 60 min) to the set of 1795 transcripts that were very stable (lower limit of the half-life 95% confidence interval of greater than 360 min), and found significant enrichment (p < 0.0001) of the GRE in short-lived transcripts, with 11.98% of short-lived and 5.79% of long-lived transcripts containing this sequence. As a positive control, we evaluated the abundance of AREs in the 3′UTR of these sets of transcripts and found significant enrichment in short-lived transcripts with 9.90% of short-lived and 5.91% of long-lived transcripts containing a class I ARE (p < 0.01) and 5.21% of short-lived and 2.51% of long-lived transcripts containing a class II ARE (p < 0.01). Thus, the GRE exhibited a similar degree of abundance and enrichment in short-lived transcripts as these AREs. Overall, we found that the 11-mer GRE was present in ∼5% of the 4812 transcripts expressed in T cells for which we had measured mRNA decay rates. A list of these GRE-containing transcripts and their mRNA half-lives is included in Table S1 available online. The 11-mer GRE sequence was present in the 3′UTRs of ∼100 short-lived transcripts with half-lives of less than 60 min. A subset of these transcripts is listed in Table 1. Many of these GRE-containing transcripts encode important regulatory proteins in the cell, including transcription factors, proto-oncogenes, apoptosis regulators, signal transduction regulators, and regulators of metabolism.
Table 1.
Examples of Short-Lived Transcripts that Contain the Consensus 11-mer GRE
| Anti-CD3 + Anti-CD28-Stimulated T Cells |
||||
|---|---|---|---|---|
| Affymetrix Probe Set ID |
Title | Symbol | Median Half-Lifea |
95% Confidence Interval |
| Transcriptional Regulators | ||||
| 32583_at | v-jun sarcoma virus 17 oncogene | JUN | 17 | [1,22] |
| 39421_at | runt-related transcription factor 1 | RUNX1 | 18 | [1,4851] |
| 40511_at | GATA binding protein 3 | GATA3 | 30 | [22,36] |
| 39257_at | Kruppel-like factor 12 | KLF12 | 33 | [3,4979] |
| 32087_at | heat shock transcription factor 2 | HSF2 | 56 | [43,6585] |
| 1519_at | v-ets erythroblastosis virus E26 oncogene homolog 2 | ETS2 | 44 | [18,53] |
| 2049_s_at | jun B proto-oncogene | JUNB | 11 | [1,28] |
| Apoptosis Regulators | ||||
| 38871_at | B cell CLL/lymphoma 10 | BCL10 | 25 | [3,35] |
| 1327_s_at | mitogen-activated protein kinase kinase kinase 5 | MAP3K5 | 52 | [43,85] |
| 36463_at | BCL2-associated athanogene 5 | BAG5 | 46 | [43,49] |
| 33813_at | tumor necrosis factor receptor superfamily, member 1B | TNFRSF1B | 54 | [47,69] |
| 973_at | serum/glucocorticoid-regulated kinase | SGK | 37 | [9,54] |
| 948_s_at | peptidylprolyl isomerase D (cyclophilin D) | PPID | 59 | [43,118] |
| Ubiquitin Cycle and Proteolysis Regulators | ||||
| 41205_at | ubiquitin protein ligase E3A | UBE3A | 52 | [37,69] |
| 37662_at | ubiquitin-conjugating enzyme E2G | UBE2G1 | 59 | [48,70] |
| Metabolism Regulators | ||||
| 38834_at | topoisomerase (DNA) II binding protein | TOPBP1 | 41 | [32,61] |
| 39882_at | translocase of inner mitochondrial membrane 8 | TIMM8A | 53 | [43,404] |
| Signal Transduction Regulators | ||||
| 37645_at | CD69 antigen (p60, early T cell activation antigen) | CD69 | 47 | [39,56] |
| 38311_at | papillary renal cell carcinoma (translocation associated) | TGIF2 | 40 | [31,53] |
Transcript half-lives were measured as described in Raghavan et al. (2002).
The finding that the GRE is more abundant in short-lived transcripts compared to stable transcripts led us to hypothesize that this conserved sequence may function as a mediator of mRNA decay. To test this hypothesis, HeLa Tet-off cells were transiently transfected with tet-repressible β-globin reporter constructs in which the GRE-containing sequences from c-jun, jun B, or TNF receptor superfamily member 1B (TNFRSF1B) shown in Figure 1A were inserted into the 3′UTR. The c-jun GRE sequence was chosen for these experiments because it was contained within a previously characterized 150 nucleotide functional decay element that was classified as a class III ARE based on its overall AU richness (Peng et al., 1996). A green fluorescence protein (GFP) expression construct that was not tetracycline regulated was cotransfected along with the tetracycline-repressible β-globin reporter constructs to control for transfection efficiency. After 48 hr, doxycycline was added to the medium to stop transcription from the tet-responsive promoter and then β-globin and GFP mRNA levels were measured over time by northern blot (Figures 1B and 1C). In the absence of an insert, the β-globin transcript was very stable with a half-life of 3188 ± 200 min (mean ± standard error of the mean). Insertion of the GRE-containing sequences from c-jun, jun B, and TNFRSF1B led to accelerated decay of the β-globin reporter with half-lives of 189 ± 53 min, 200 ± 50 min, and 196 ± 34 min, respectively. In contrast, insertion of the mutated sequences into 3′UTR had relatively little effect on mRNA decay rates with a half-life of 1031 ± 298 min, 1019 ± 230 min, and 1689 ± 542 min for the mutated c-jun, jun B, and TNFRSF1B GRE, respectively. Thus, the introduction of short GRE-containing sequences into the 3′UTR of β-globin mRNA is sufficient for enhancing transcript decay, suggesting that the GRE is a functional mediator of mRNA decay.
Figure 1. GREs Are Functional Mediators of mRNA Decay.
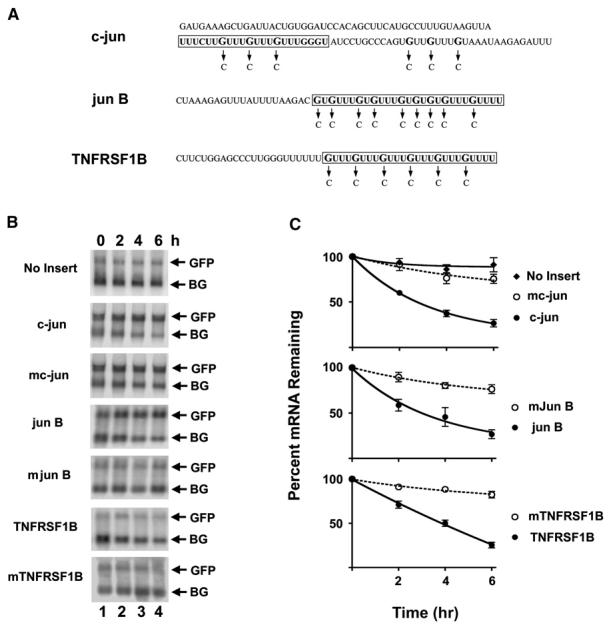
(A) The shown GRE-containing sequences from the 3′UTR of c-jun, jun B, and TNFRSF1B were cloned into the 3′UTR of the pTetBBB β-globin reporter construct. The boxed sequences indicate the sequences of the ribo-oligonucleotides used for the binding reactions shown in Figure 2. The arrows indicate the single-nucleotide mutations (G to C) that were introduced to create the mutated GRE sequences.
(B) HeLa Tet-off cells were transfected with the pTracer GFP expression construct and the pTetBBB β-globin reporter construct (No Insert) or with reporter constructs in which GRE-containing sequences (c-jun, jun B, and TNFRSF1B) or mutated sequences (mc-jun, mjun B, and mTNFRSF1B) shown in (A) were inserted into the 3′UTR. Doxycycline was added to the medium to stop transcription from the tet-responsive promoter, and total cellular RNA was collected at 0, 2, 4, and 6 hr time points. Northern blot analyses were performed to monitor GFP and β-globin (BG) mRNA levels.
(C) The experiment shown in (B) was performed three times, and the northern blots were quantified using a phosphorimager. For each point, the intensity of the β-globin reporter band was normalized to the intensity of the GFP band, and the band intensity at the 0 hr time point was set at 100%. The percent of mRNA remaining was plotted over time. The error bars indicate the standard error of the mean (SEM) from the three experiments.
Because AREs and other RNA elements that regulate mRNA decay function through their interactions with specific RNA-binding proteins, we hypothesized that the GRE may also function by interacting with specific RNA-binding proteins. We performed gel shift assays using cytoplasmic extracts from primary human T cells to search for GRE-specific RNA-binding activities (Figure 2A). A 22 nucleotide radiolabeled ribo-oligonucleotide probe that contained the boxed c-jun GRE sequence shown in Figure 1A was mixed with T cell cytoplasmic extracts, and the mixtures were separated by electrophoresis on a nondenaturing polyacrylamide gel. A major c-jun GRE-binding activity was observed (lane 1), and binding by this activity was competed by the addition of excess of unlabeled ribo-oligonucleotides that contained the boxed c-jun (lane 2), jun B (lane 4), and TNFRSF1B (lane 6) GRE sequences shown in Figure 1A, but not by the indicated mutated sequences (lanes 3, 5, and 7). Binding by this activity was also not competed by unlabeled ribo-oligonucleotides containing a 22 nucleotide GM-CSF ARE sequence or by a 22 nucleotide poly U sequence (data not shown). These data suggested that the GRE-binding activity was sequence specific.
Figure 2. CUGBP1 Binds Specifically to GRE Sequences with High Affinity.
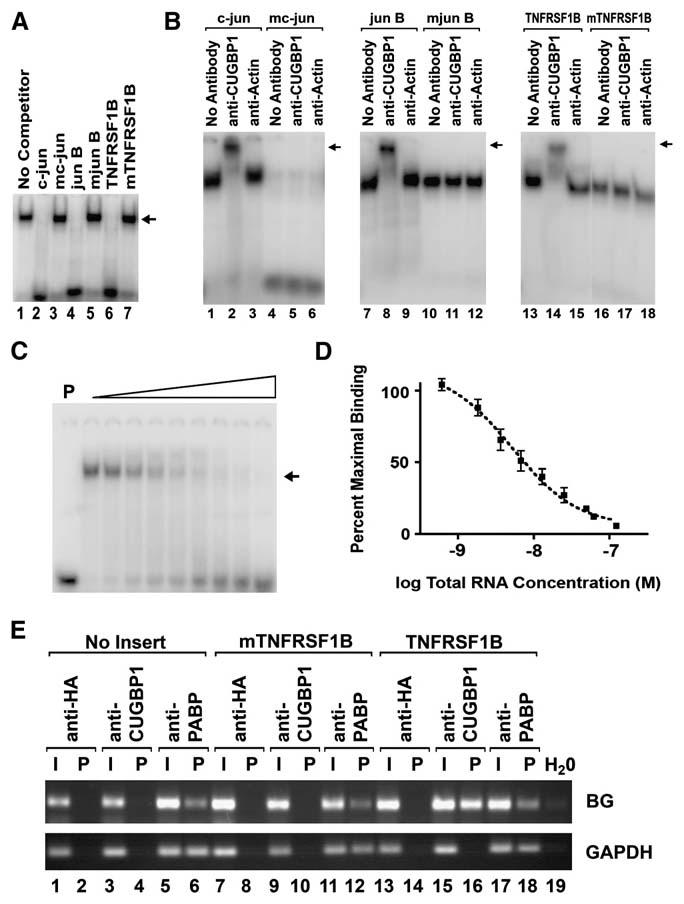
(A) Cytoplasmic extracts from primary human T cells were mixed with a 32P-end-labeled ribo-oligonucleotide probe that contained a GRE sequence from the 3′UTR of the c-jun transcript in the absence (No Competitor) or presence of a 100-fold molar excess of the indicated unlabeled competitor ribo-oligonucleotides. The sequences of the GRE-containing ribo-oligonucleotides and mutated oligonucleotides are indicated as the boxed sequences in Figure 1A. Bands were visualized using a phosphorimager, and the position of migration of the predominant RNA-protein binding complex is indicated with an arrow.
(B) RNA-protein gel shift assays were performed by mixing cytoplasmic extracts from primary human T cells with radiolabeled ribo-oligonucleotide probes that contained GRE sequences from the 3′UTR of c-jun, jun B, and TNFRSF1B or mutated GRE sequences (mc-jun, mjun B, and mTNFRSF1B) in the absence of antibody (No Antibody) or the presence of anti-CUGBP1 or anti-actin antibodies. The binding reactions were then separated by electrophoresis on 10% polyacrylamide gels under nondenaturing conditions. For each panel, the position of the supershifted band is indicated with an arrow.
(C) Cytoplasmic extracts from primary human T cells (10 μg of protein) were incubated with 12 fmol of a 32P end-labeled c-jun GRE probe in the absence of unlabeled RNA or the presence of increasing amounts of unlabeled c-jun GRE RNA (12–2400 fmol in 2-fold increments). The binding reactions were then separated by electrophoresis on a 10% polyacrylamide gel under non-denaturing conditions. The lane marked “P” was loaded with probe alone. The position of migration of the probe bound to CUGBP1 is indicated with an arrow.
(D) The experiment shown in (C) was performed three times, and the amount of bound c-jun GRE probe was quantified with a phosphorimager. The percent of maximal bound radiolabeled RNA was plotted against the concentration of total RNA in each reaction. Each point represents the mean and SEM from the three experiments.
(E) HeLa Tet-off cells were transfected with the pTetBBB β-globin reporter construct (No Insert) or with reporter constructs in which the TNFRSF1B GRE sequence (TNFRSF1B) or the mutated sequence (mTNFRSF1B) shown in Figure 1A was inserted into the 3′UTR. Lysates from these transfected cells were incubated with protein G Sepharose beads that were precoated with anti-HA, anti-CUGBP1, or anti-PABP antibodies. RNA isolated from the input (I) and from the immunoprecipitation pellet (P) was assayed for the presence of β-globin (BG) or GAPDH transcripts using RT-PCR. As a control to detect possible contamination, lane 19 (H2O) contained all components of the RT-PCR reaction except for an RNA sample.
In order to identify this RNA-binding activity, we performed supershift assays using antibodies against RNA-binding proteins that have specificity for U-rich or GU-rich sequences, including CUGBP1, CUGBP2, HuR, TTP, and KSRP. Supershift assays (Figure 2B) were performed using T cell cytoplasmic extracts and radiolabeled GRE or mutated GRE sequences from c-jun, jun B, and TNFRSF1B, and we found that RNA-binding complexes that bound to the GRE sequences were specifically supershifted with an anti-CUGBP1 antibody (lanes 2, 8, and 14), suggesting that they contained CUGBP1. In contrast, complexes that bound to the mutated sequences were not super-shifted with anti-CUGBP1 antibodies (lanes 5, 11, and 17), suggesting that they did not contain CUGBP1 but were composed of other cellular proteins. Antibodies against actin (lanes 3, 9, and 15) or against other RNA-binding proteins including HuR, TTP, CUGBP2, and KSRP did not supershift the GRE-containing complexes (data not shown). Using an RNA-protein UV crosslinking assay we identified a major RNA-binding activity that recognized the c-jun GRE sequence and had a molecular weight of ∼55 kDa, and binding by this activity was competed by the addition of excess unlabeled c-jun, jun B, or TNFRSF1B GRE ribo-oligonucleotides but was not competed by mutated oligonucleotides (Figure S1A), suggesting that this 55 kDa RNA-binding activity specifically recognizes GRE sequences. An anti-CUGBP1 antibody specifically immunoprecipitated the GRE-specific 55 kDa RNA-protein complex (Figure S1B), suggesting that this complex contains CUGBP1. Overall, these results indicate that the major GRE-binding activity that we identified in T cell cytoplasmic extracts is CUGBP1.
CUGBP1, a member of the CELF family of RNA-binding proteins, was first described as a protein that bound to the abnormally extended CUG mRNA repeats that occur in patients with type I myotonic dystrophy (Timchenko et al., 1996) and has since been implicated as a regulator of translation (Timchenko et al., 1999) and alternative splicing (Philips et al., 1998). A search for preferential RNA-binding sites using systemic evolution of ligands exponential enrichment (SELEX) revealed that CUGBP1 bound to UGUU-rich sequences with high affinity (Marquis et al., 2006). We performed competitive RNA-binding experiments and determined the apparent affinity of the CUGBP1-containing complex for the c-jun GRE to be 5.2 ± 1.3 nM (Figures 2C and 2D). We also produced recombinant CUGBP1 in E. coli and tested its specificity for binding to GRE sequences (Figure S1C). Binding by recombinant CUGBP1 to radiolabeled c-jun GRE RNA was competed by the addition of increasing amounts of unlabeled c-jun RNA, but not mutated c-jun RNA, indicating that CUGBP1 binding to the GRE is specific. Based on competitive binding results, we calculated the affinity of recombinant CUGBP1 for the c-jun GRE sequence to be 10.6 ± 2.7 nM. This affinity is similar to the previously reported affinity of CUGBP1 for artificially selected GU-rich sequences (Marquis et al., 2006), and is similar to the affinity of functional ARE-binding proteins for their target sequences (Hau et al., 2007). Overall, these data demonstrate that CUGBP1 binds specifically to GRE sequences with high affinity.
Because CUGBP1 bound specifically to short GRE sequences in vitro, we performed RNA immunoprecipitation assays to determine whether CUGBP1 also bound specifically to GRE-containing full-length reporter transcripts expressed in cells. HeLa Tet-off cells were transiently transfected with a β-globin reporter construct or β-globin reporter constructs in which the TNFRSF1B GRE or a mutated sequence was inserted into its 3′UTR. Lysates from these cells were immunoprecipitated using an anti-CUGBP1 antibody, an antibody against poly(A)-binding protein (PABP), or an anti-HA antibody. RNA was isolated from the immunoprecipitated material and reverse transcriptase-polymerase chain reaction (RT-PCR) was performed to detect β-globin or GAPDH transcripts. As seen in Figure 2E, the anti-CUGBP1 antibody immunoprecipitated the β-globin transcript only when the GRE insert was present (lane 16), but not when no insert (lane 4) or a mutated GRE insert (lane 10) was present. In contrast, an anti-PABP antibody immunoprecipitated the β-globin transcripts regardless of whether or not a GRE insert was present (lanes 6, 12, and 18). The anti-PABP antibody also immunoprecipitated endogenous GAPDH transcripts (lanes 6, 12, and 18), whereas the anti-CUGBP1 antibody did not (lanes 4, 10, and 16). The anti-HA antibody, which served as a negative control, did not effectively immunoprecipitate either transcript (lanes 2, 8, and 14). These results suggest that CUGBP1 is capable of selectively recognizing GRE-containing transcripts in cells.
Based on our findings that the GRE functions to mediate mRNA decay and that CUGBP1 binds to the GRE with high affinity, we hypothesized that CUGBP1 functions as a mediator of mRNA decay. CUGBP1 is evolutionarily conserved, and its homolog in Xenopus, embryo deadenylation element-binding protein (EDEN-BP), serves dual functions as a translational repressor in oocytes and a deadenylation factor following fertilization (Paillard et al., 1998). EDEN-BP regulates deadenylation by binding to the embryonic deadenylation element (EDEN), a sequence that is rich in uridine and purine dinucleotides (Paillard et al., 1998). Interestingly, CUGBP1 was able to functionally substitute for EDEN-BP to mediate transcript deadenylation in Xenopus extracts (Paillard et al., 2003), suggesting that it may play a role in transcript deadenylation. Tethering of CUGBP1 to the 3′UTR of mRNA through an interaction with the MS2 coat protein led to decreased steady-state levels of reporter transcripts that contained an MS2 RNA-binding site while reporter protein levels increased (Barreau et al., 2006), suggesting that CUGBP1 could play a dual role in translation and/or mRNA decay. Recently, CUGBP1 was shown to associate with poly(A) ribonuclease and to stimulate poly(A) tail shortening in HeLa cell S100 extracts (Moraes et al., 2006), suggesting that CUGBP1 may play a role in transcript deadenylation.
To determine whether CUGBP1 is a regulator of GRE-mediated mRNA decay, we evaluated the effect of siRNA-mediated knockdown of CUGBP1 on the decay of GRE-containing reporter transcripts. HeLa tet-off cells were treated with two independent siRNAs (siCUGBP1-A and siCUGBP1-B) designed to target CUGBP1 or a control siRNA that targets red fluorescence protein. Both of the siRNAs directed against CUGBP1 or a pool of siRNAs against CUGBP1 led to a decrease in CUGBP1 protein level by 80%–99% as assessed by western blot (see Figure S1). These same cells were used to assess the decay of β-globin reporter transcripts that contained a GRE insertion (Figures 3A and 3B). In cells that were transfected with the red fluorescence protein control siRNA, the decay rate of the β-globin reporter transcript that contained the TNFRSF1B GRE insertion was 183 ± 159 min. In contrast, this transcript was stabilized (p < 0.05) in cells treated with siRNAs directed against CUGBP1 with half-lives of 1689 ± 635 min or 1102 ± 202 min following treatment with siCUGBP1-A or siCUGBP1-B, respectively. We found similar results when we used a pool of siRNAs directed against CUGBP1 (Figures 3C and 3D). In cells that were transfected with a pool of nontargeting siRNAs, the decay rates of β-globin reporters that contained TNFRSF1B GRE and jun B GRE insertions were 151 ± 37 min and 193 ± 22 min, respectively. These transcripts were stabilized (p < 0.05) in cells treated with siRNAs directed against CUGBP1 with a half-life of the TNFRSF1B GRE-containing transcript of 451 ± 70 min and a half-life of the jun B GRE-containing transcript of 708 ± 56 min. These findings suggest that CUGBP1 is a mediator of GRE-directed mRNA decay and that insertion of a GRE into the 3′UTR of the β-globin transcript is sufficient to allow CUGBP1 to function.
Figure 3. siRNA-Mediated Knockdown of CUGBP1 Induced Stabilization of GRE-Containing Transcripts.
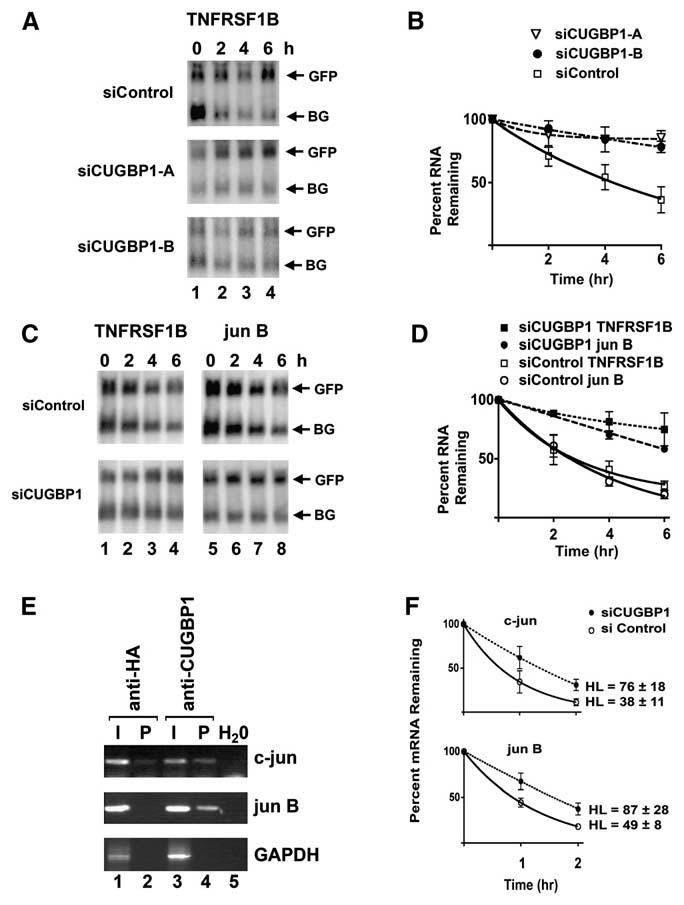
In (A) and (B), HeLa Tet-off cells were treated in two rounds with two independent siRNAs directed against CUGBP1 (siCUGBP1-A and siCGBP1-B) or a control siRNA directed against red fluorescence protein (siControl). In (C) and (D), HeLa Tet-off cells were treated in three rounds with smart pool siRNAs directed against CUGBP1 (siCUGBP1) or nontargeting siRNA (siControl). These cells were then transfected with the pTracer GFP expression construct and a pTetBBB β-globin reporter construct in which GRE-containing sequences (TNFRSF1B or jun B) were inserted into the 3′UTR. Doxycycline was added to the medium to stop transcription from the tet-responsive promoter, and total cellular RNA was collected at 0, 2, 4, and 6 hr time points. Northern blots were performed to monitor GFP, and β-globin (BG) mRNA levels. The experiments in (A) and (C) were performed three times, the northern blots were quantified using a phosphorimager, and the graphed results are shown in (B) and (D), respectively. For each point, the intensity of the β-globin reporter band was normalized to the intensity of the GFP band, and the band intensity at the 0 hr time point was set at 100%. The percent of mRNA remaining following the addition of doxycycline was plotted over time. The error bars indicate the standard error of the mean from the three experiments. (E) Lysates from HeLa cells were incubated with protein G Sepharose beads that were precoated with anti-HA or anti-CUGBP1 antibodies. RNA isolated from the input (I) and from the immunoprecipitation pellet (P) was assayed for the presence of c-jun, jun B, or GAPDH transcripts using RT-PCR. As a control to detect possible contamination, lane 5 (H2O) contained all components of the RT-PCR reaction except for an RNA sample. (F) HeLa cells were treated in two rounds with pooled siRNAs directed against CUGBP1 (siCUGBP1) or non-targeting siRNA (siControl). Actinomycin D was added to stop transcription and total cellular RNA was harvested at 0, 1, and 2 hr time points. The mRNA levels of c-jun and jun B were measured by real-time RT-PCR using transcript-specific primers, and transcript levels were normalized to the level of the HPRT transcript. The normalized level of each transcript was set at 100% at time zero, and the other time points were graphed relative to that value. Each point represents the mean and SEM from three independent experiments.
The RNA immunoprecipitation assay was also performed to determine whether CUGBP1 was able to bind to endogenous GRE-containing transcripts in cells. HeLa cell lysates were immunoprecipitated with an anti-CUGBP1 antibody or an anti-HA antibody, and RT-PCR was used to assess the presence of endogenous c-jun, jun B, or GAPDH transcripts in RNA isolated from the immunoprecipitated material (Figure 3E). The anti-CUGBP1 antibody immunoprecipitated c-jun and jun B transcripts, but not the GAPDH transcripts, whereas the anti-HA antibody did not effectively immunoprecipitate any of these transcripts. These results suggest that CUGBP1 was able to bind to endogenous GRE-containing transcripts in cells. To determine the functional consequences of CUGBP1 binding to endogenous RNA in cells, we also evaluated the effect of siRNA-mediated knockdown of CUGBP1 on the decay of endogenous c-jun and jun B transcripts. HeLa cells were treated with siRNA directed against CUGBP1 or control siRNA, then actinomycin D was added to stop transcription by RNA polymerase II, and the decay of c-jun and jun B transcripts was measured using real-time RT-PCR (Figure 3F). siRNA-mediated knockdown of CUGBP1 led to the stabilization of these transcripts, suggesting that in addition to mediating the decay of GRE-containing reporter transcripts, CUGBP1 also mediates the decay of endogenous GRE-containing transcripts.
Overall, we have identified the GRE as an mRNA decay element and have defined CUGBP1 as a functional mediator of GRE-dependent mRNA decay. Our results demonstrate that CUGBP1 is a regulator of mammalian gene expression at the level of mRNA decay and that CUGBP1 and the GRE define a conserved pathway that selectively regulates mRNA decay.
EXPERIMENTAL PROCEDURES
Identification and Analysis of Conserved GU-Rich Sequences
We previously used microarrays to measure the mRNA decay rates of ∼6000 transcripts expressed in resting and activated primary human T cells (Raghavan et al., 2002; Vlasova et al., 2005). We analyzed the set of transcripts that were induced greater than 2-fold (p ≤ 0.05) following stimulation of primary human T cells with anti-CD3 and anti-CD28 antibodies and divided these transcripts into two groups: one group contained transcripts with half-lives less than or equal to 60 min and the other group contained transcripts with half-lives greater than 360 min. We examined the frequencies of all possible 12-mer sequences (412) within the 3′UTR of these transcripts and compiled the top 1000 12-mers that had a higher frequency in the short-lived transcripts compared to the long-lived transcripts. Using a 1000×1000 similarity matrix, we performed agglomerative clustering using the program CLUTO (Rasmussen et al., 2003). The similarity matrix was computed simply by calculating the alignment score between two 12-mers. The match was scored as 1 and mismatch 0, and no gaps were allowed. CLUTO output provided for each cluster, the number of objects, average similarity between the objects of each cluster (internal similarity), and the average similarity of the objects of each cluster and the rest of the objects (external similarity). A promising cluster contained the sequence UGUUUGUUUGUU.
Further evaluation of all short-lived transcripts expressed in primary human T cells revealed that the related 11-mer sequence UGUUUGUUUGU occurred frequently in the 3′UTR of these transcripts, and therefore this conserved sequence was analyzed in more detail. Sequences and annotations, including the positions of the 3′UTRs, for 4812 transcripts for which we had measured mRNA decay rates were retrieved from the RefSeq database, and the 3′UTR sequences were extracted using scripts written in the Practical Extraction and Report Language (PERL). FindPatterns from GCG (Version 11.0, Accelrys, San Diego) was used to search for the 11-mer sequence UGUUUGUUUGU, permitting one mismatch. Transcripts containing an ARE were extracted from the ARE 3.0 database (Bakheet et al., 2006). ARE-containing transcripts were further separated into transcripts containing class I or class II AREs using PERL scripts. We compared the abundance of the GU-rich 11-mer sequence or the ARE sequences in short-lived transcripts defined as transcripts whose half-life 95% confidence interval upper limit was less than 60 min to their abundance in long-lived transcripts defined as transcripts whose half-life 95% confidence interval lower limit was greater than 360 min. Enrichments were tested for significance using Fisher's exact test in R (http://www.r-project.org).
Tet-Off mRNA Decay Assay
The decay of β-globin reporter constructs was performed as described previously (Ogilvie et al., 2005), with minor modifications. HeLa tet-off cells (1.6 × 106 cells) were transfected with 3.0 μg of the tet-responsive reporter construct that encoded the rabbit β-globin transcript or rabbit β-globin transcripts that contained the additional sequences shown in Figure 1A inserted at the unique BglII restriction site in its 3′UTR. Tet-responsive rabbit β-globin constructs, pTetBBB and pTetBBB/c-jun (Peng et al., 1996), were gifts from Dr. Ann-Bin Shyu (University of Texas-Houston). The c-jun, mutated c-jun (mc-jun), jun B, mutated jun B (mjun B), TNFRSF1B, and mutated TNFRSF1B (mTNFRSF1B) sequences were inserted into the pTetBBB plasmid using the GeneTailor site-directed mutagenesis system (Invitrogen, Carlsbad, CA). The HeLa tet-off cells were cotransfected with 1 μg of the pTracerC-EF/V5-His/lacZ construct (Invitrogen Life Technologies), which produces GFP, to control for transfection efficiency. Transfections were performed with 2.5 U of TransIT-LT1 reagent (Mirus, Madison, WI) per microgram of plasmid DNA. After 48 hr, 300 ng/ml of doxycycline was added to stop transcription, and total RNA was isolated after 0, 2, 4, and 6 hr. RNA isolation and northern blotting to assess expression of β-globin and GFP transcripts were performed as described previously (Ogilvie et al., 2005). For each point, the hybridization intensity of the β-globin transcript was normalized to the hybridization intensity of the GFP transcript, and the normalized values were used to calculate half-lives using GraphPad Prism 4.03 software based on a one-phase exponential model of decay.
Gel Shift, Supershift, and UV-Crosslinking Assays
Ribo-oligonucleotides were purchased commercially (Dharmacon Research, Boulder, CO). The sequences for each ribo-oligonucleotide are shown as the boxed sequences in Figure 1A. Radiolabeled ribo-oligonucleotide probes were end-labeled with [γ-32P]ATP (6000 Ci/mmol) using T4 polynucleotide kinase (Invitrogen) to produce a radiolabeled probe with a specific activity of 4 × 106 cpm/μg. Gel shift and UV crosslinking assays were conducted as described previously (Raghavan et al., 2001). Each reaction contained 8–10 μg of cytoplasmic protein and 10–15 fmol of radiolabeled RNA probe in a total volume of 20–24 μl. For supershift assays, an anti-CUGBP1 mouse monoclonal antibody (3B1, Santa Cruz Biotechnologies) or an anti-actin mouse monoclonal antibody (C-2, Santa Cruz Biotechnologies) was added to the reaction mixtures. The gels were dried and analyzed on a phosphorimager (Molecular Dynamics). For competitive binding assays, GraphPad Prism version 4.03 (GraphPad Software, San Diego, CA) was used to graph binding data and calculate apparent affinities and standard errors using a homologous competition with depletion one-site binding model.
RNA Immunoprecipitation Assays
RNA immunoprecipitation assays were performed as described (Tenenbaum et al., 2002). RNA was extracted from the immunoprecipitated samples and the input samples using the RNeasy kit (QIAGEN) following the manufacturer's protocol, including a DNase digest. Superscript II reverse transcriptase (Invitrogen) was used to convert 4 μg of total RNA or a corresponding amount of immunoprecipitated RNA using an oligo dT15 primer. Products were amplified for 38 cycles from cDNA using the following primers: β-globin forward 5′-GT CTACCCATGGACCCAGAGG-3′, β-globin reverse 5′-GTGAGCGGCATTGGC CACACC-3′;c-jun forward 5′-CCCCAAGATCCTGAAACAGA-3′,c-jun reverse 5′-CCGTTGCTGGACTGGATTAT-3′; jun B forward 5′-TGGAACAGCCCTTC TACCAC-3′, jun B reverse 5′-CTCAGGAGCATGGGGATAAA-3′; GAPDH forward 5′-TGATGGTACATGACAAGGTGC-3′, GAPDH reverse 5′-ACAGTC CATGCCATCACTGC-3′.
siRNA Transfection
The following siRNA duplexes were purchased commercially from Dharmacon Research (Boulder, CO): siCUGBP1-A (5′-GAGCCAACCUGUUCAUCUA-3′; catalog number J-020166-05), siCUGBP1-B (5′-GCUGUUUAUUGGUAUGA UU-3′; catalog number D-020166-04), siRNA targeting red fluorescence protein (5′-AAAGGACGGAGGACATTAT-3′), smart pool siRNAs representing a mixture of four distinct RNA duplexes directed against CUGBP1 (catalog number M-020166), and a control pool of nontargeting siRNAs. For the Tet-off mRNA decay reporter assay, HeLa tet-off cells (4 × 105 cells) were transfected sequentially two or three times at 24 hr intervals with 50–100 nmol of siRNAs directed against CUGBP1 or control siRNAs according to the manufacturer's instructions. For Tet-off mRNA decay assays, 24 hr after the last siRNA transfection, the cells were then transfected with β-globin and GFP constructs and mRNA decay was measured as described above. For actinomycin D mRNA decay assays, actinomycin D (10 ug/ml) was added to the culture media, and total RNA harvested after 0, 1, and 2 hr was analyzed by real-time RT-PCR using transcript-specific primers to evaluate mRNA decay as described previously (Vlasova et al., 2005). In each experiment, aliquots of siRNA-transfected cells were used to prepare total cellular extracts (Atasoy et al., 2003) for western blotting with an anti-CUGBP1 mouse monoclonal antibody (Santa Cruz Biotechnologies) or an anti-actin mouse monoclonal antibody (Calbiochem) to assess the effectiveness of CUGBP1 knockdown.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants AI49494 and AI52170 from the NIH to P.R.B. and by a research grant from the Minnesota Medical Foundation (to P.R.B. and I.A.V.). I.A.V. was funded through awards from the Minnesota Supercomputing Institute and the Lymphoma Research Foundation. N.M.T. was funded by the Pediatric Infectious Diseases Training Program (NIH grant T32 HD007381). O.L. was supported by a postdoctoral fellowship from the Swedish Research Council. D.F. and P.B.B. were supported by HL076779 and HL073719 from the NIH. We thank Dr. Ann-Bin Shyu and Dr. Thomas Cooper for providing plasmids.
Footnotes
Supplemental Data include two figures and one table and can be found with this article online at http://www.molecule.org/cgi/content/full/29/2/263/DC1/.
REFERENCES
- Atasoy U, Curry SL, Lopez de Silanes I, Shyu AB, Casolaro V, Gorospe M, Stellato C. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J. Immunol. 2003;171:4369–4378. doi: 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Watrin T, Beverley Osborne H, Paillard L. Protein expression is increased by a class III AU-rich element and tethered CUGBP1. Biochem. Biophys. Res. Commun. 2006;347:723–730. doi: 10.1016/j.bbrc.2006.06.177. [DOI] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J. Cell. Biochem. 2007;100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard L, Legagneux V, Beverley Osborne H. A functional deadenylation assay identifies human CUG-BP as a deadenylation factor. Biol. Cell. 2003;95:107–113. doi: 10.1016/s0248-4900(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Shyu AB. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol. Cell. Biol. 1996;16:1490–1499. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Raghavan A, Bohjanen PR. Microarray-based analyses of mRNA decay in the regulation of mammalian gene expression. Brief. Funct. Genomic. Proteomic. 2004;3:112–124. doi: 10.1093/bfgp/3.2.112. [DOI] [PubMed] [Google Scholar]
- Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J. Biol. Chem. 2001;276:47958–47965. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]
- Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan A, Dhalla M, Bakheet T, Ogilvie RL, Vlasova IA, Khabar KS, Williams BR, Bohjanen PR. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics. 2004;84:1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Rasmussen MD, Deshpande MS, Karypis G, Johnson J, Crow JA, Retzel EF. wCLUTO: a Web-enabled clustering toolkit. Plant Physiol. 2003;133:510–516. doi: 10.1104/pp.103.024885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, McNabb J, Raghavan A, Reilly C, Williams DA, Bohjanen KA, Bohjanen PR. Coordinate stabilization of growth-regulatory transcripts in T cell malignancies. Genomics. 2005;86:159–171. doi: 10.1016/j.ygeno.2005.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


