Abstract
Nine genetically inherited neurodegenerative diseases are linked to abnormal expansions of a polyglutamine (polyQ) encoding region. Over the years, several structural models for polyQ regions have been proposed and confuted. The cross-β-spine steric zipper motif, identified recently for the GNNQQNY peptide, represents an attractive model for amyloid fibers formed by polyQ fragments. Here we report a detailed molecular dynamics investigation of polyQ models assembled by cross-β-spine steric zipper motifs. Our simulations indicate clearly that these assemblies are very stable. Glutamine side chains contribute strongly to the overall stability of the models by fitting perfectly within the zipper. In contrast to GNNQQNY zipper motifs, hydrogen bonding interactions provide a significant contribution to the overall stability of polyQ models. Molecular dynamics simulations carried out on monomeric polyQ forms (composed by 40–60 residues) show clearly that they can also assume structures stabilized by steric zipper motifs. Based on these findings, we build monomeric polyQ models that can explain recent data on the toxicity exerted by these species. In a more general context, our data suggests that polyQ models with interdigitated side chains can provide a structural rationale to several literature experiments on polyQ formation, stability, and toxicity.
INTRODUCTION
Protein organization in well-defined structural states is essential for all biological processes. Indeed, it is accepted commonly that protein function largely depends on its structure. Investigations carried out in the last decade(s) have highlighted that protein folding is a very delicate process because proteins may assume deleterious conformational states (1). These findings have led to the identification of a class of pathologies collectively designated as conformational diseases (2,3). Conformational diseases include widespread and often lethal neurodegenerative pathologies such as Alzheimer's disease, Parkinson's disease, Huntington's disease, and prion disease (3). Recent evidence also suggests that a much wider group of disorders, from liver cirrhosis to degenerative eye diseases, share the same general pathology (4). There is increasing evidence that these diseases have common molecular mechanisms, typically linked to protein misfolding and aggregation, although the proteins involved are radically different for function and native folding (5). Despite devoted efforts, the current understanding of the molecular basis of these disorders is rather limited. Consequently, their description is basically phenomenological.
An increasing number of human diseases have been linked to the pathological expansion of normal tracts of single amino acid repeats (6–8). Although recent reports have shown that polyA may lead to developmental malformations and/or to formation of insoluble protein complexes, most of these pathologies are primarily associated with expansions of polyglutamine (polyQ) repeats (9). These diseases share a number of similar characteristics (9), including formation of ubiquitinated inclusions, neural dysfunction, and type-specific cell death, although the precise mechanism of pathogenesis of the expanded polyQ tracts remains unknown.
From the molecular point of view, solution and solid state studies on polyQ peptide models have shown that, within the large class of conformational diseases, polyQ disorders show some distinctive features. Fiber diffraction analyses, although somewhat controversial (10), seem to indicate that polyQ peptides exhibit a high tendency to form crystallites and fibers that display, in addition to the typical meridional reflection at 4.8 Å resolution, a rather sharp equatorial reflection at 8.3 Å (11–13). In addition, kinetic studies (14,15) have indicated that the slow step in the nucleation dependent fiber growth process is the conversion of polyQ monomers into a conformational state, yet to be characterized, which is prone to form aggregates. Even more intriguing is the recent discovery that toxicity of polyQ fragment fused to thioredoxin is not linked to aggregation as monomeric species exert high toxicity (16).
Although several structural models for polyQ regions have been proposed over the years (10,17–23), this issue remains highly disputed (10,24). The cross-β-spine steric zipper motif, identified recently for several fiber-forming peptides (25–27), represents an attractive model for fibers formed by polyQ fragments. We report a detailed molecular dynamics investigation of polyQ models assembled by cross-β-spine steric zipper motifs. On the basis of our findings, we provide a structural rationale to several literature experiments on polyQ formation, stability, and toxicity.
MATERIALS AND METHODS
Systems and notation
PolyQ polymeric and monomeric forms endowed with a steric zipper motif were built using different approaches depending on the availability of related experimental structures.
In principle, for polymeric forms, three arrangements with interdigitated Gln side-chains are possible. Indeed, the strands in each sheet may be parallel (P) or antiparallel (A). Furthermore, the polypeptide chains of facing strands in opposite sheets can be oriented in a head-to-head (hh) or head-to-tail (ht) fashion. Taking into account the homopolymeric nature of polyQ peptides and the staggering of facing β-strands, the two options, hhA and htA, are equivalent. All three possible arrangements (htP, hhP, A) were considered.
Polymeric polyQ assemblies with parallel β-sheets, with an antiparallel relative orientation (htP), were generated using the structure of the peptide GNNQQNY (25) (Protein Data Bank code 1YJP) as a template. In particular, the fragments composed by six Gln were simply generated by replacing the side chains of the fragment NNQQNY. The structural parameters of these models (dihedral angles and geometry) were used to build larger chains.
The hhP arrangement was generated by preserving the orientation of a sheet of htP and reverting the direction of the other.
Finally, assemblies formed by A sheets were built manually by reverting the chain direction of selected strands in the two sheets of the htP model. Occasionally twisted fragments from the polyQ-containing ribonuclease A model proposed by Eisenberg et al. (23) were also considered.
A β-helix topology was assumed for single chain models. Compact polyQ β-helix models were generated either manually or by taking into account models from a previous molecular dynamics (MD) analysis (18). MD trials carried out on these compact models showed that they did not lead to the interdigitation of side chains in the timescale of the simulation (20 ns). β-helix models endowed with interdigitated side chains were generated by conducting restrained MD simulations (see later in the text for further details).
A general notation for the assemblies characterized in this study has been used. In particular, aggregates were identified by the following formula, SHx-STy-Qz, where x, y, and z are the number of sheets, the number of strands per sheet, and the number of Gln per strand, respectively. Finally, the initial letters (htP, hhP, A) denote parallel or antiparallel sheets and their relative orientations (see above). The monomeric forms have been denoted as Qx, where x identifies the number of Gln residues of the chain. A summary of the investigated models is reported in Table 1.
TABLE 1.
Summary of the simulations carried out
| Model | No. atoms peptide/atoms waters | Box dimensions (Å3) | Simulation time (ns) | T (K) |
|---|---|---|---|---|
| ASH2-ST4-Q15* | 1816/26538 | 99 × 52 × 56 | 20 | 300 |
| htPSH2-ST4-Q15* | 1816/25653 | 98 × 51 × 56 | 20 | 300 |
| hhPSH2-ST4-Q15* | 1816/24438 | 98 × 51 × 55 | 20 | 300 |
| ASH2-ST4-Q15* | 1816/26538 | 110 × 58 × 62 | 15 | 500 |
| htPSH2-ST4-Q15* | 1816/25653 | 109 × 57 × 62 | 10 | 500 |
| htPSH4-ST4-Q15* | 3632/28689 | 101 × 48 × 69 | 10 | 300 |
| htPSH4-ST4-Q15* | 3632/28689 | 110 × 53 × 76 | 15 | 500 |
| ASH2-ST10-Q6† | 1500/23943 | 60 × 57 × 78 | 10 | 300 |
| htPSH2-ST10-Q6 | 1500/18678 | 51 × 80 × 51 | 10 | 300 |
| htPSH2-ST4-Q6 | 600/18996 | 58 × 58 × 58 | 10 | 300 |
| Q57 | 687/19050 | 66 × 55 × 56 | 50 | 300 |
| Q41 | 495/18447 | 58 × 58 × 58 | 50 | 300 |
The notation of each model is reported in the Materials and Methods section.
Asp-Asp and Lys-Lys peptides were added as N- and C-terminal capping, respectively.
This starting model was extrapolated from the theoretical model proposed by Eisenberg et al. (26) for the construct RNaseA-Q10 (Protein Data Bank code 2APU).
Simulation procedure
MD simulations were carried out with the GROMACS software package.3.3 (28). The models were immersed in rectangular or cubic boxes filled with water molecules. The GROMOS43a1 force field and the SPC water model were used in the simulations. For all systems, to relax bond geometries, the potential energy of the system (peptides and water) was minimized by using the steepest-descent method until convergence was reached. The solvent was then relaxed by 50 ps of MD at 300 K, restraining protein atomic positions with a harmonic potential. The system temperature was brought to 300 K in a step-wise manner: 20 ps MD runs were carried out at 50, 100, 150, 200, 250, and 300 K. The timescale of the individual simulations is reported in Table 1. The simulations were run with periodic boundary conditions. Bond lengths were constrained by the LINCS algorithm (29). The electrostatic interactions were calculated using Part Mesh Ewald algorithm with a cutoff of 0.9 nm. The cutoff radius for the Lennard-Jones interactions was set to 1.2 nm. A dielectric constant of 1, and a time step of 2 fs were used. For 300 K simulations we used the NVT ensemble. The temperature was maintained constant using the Berendsen thermostat with a time constant of 0.1 ps. To investigate the influence of the force field on the results obtained we repeated the simulation for the system htPSH2-ST4-Q15 by using the OPLS force field (30). To check the stability of some of the assemblies, simulations were occasionally carried out at higher temperatures (Table 1). In these simulations we used the NPT ensemble. We used as starting structures the models obtained after 500 ps of the simulation carried out at 300 K. A stepwise procedure was used to increase the temperature to 500 K. The trajectory was checked to assess the quality of the simulation using GROMACS (28) and VMD routines (31). Pictures were generated using the programs VMD (31) and Molscript (32).
RESULTS
Structure and stability of polyQ aggregates with extended interfaces
The initial MD simulations were conducted on models with interdigitated Gln side chains endowed with an extended intersheet surface. Although it has been proposed that turns may be frequent in polyQ crystallites and fibers (10,11), these analyses were aimed at gaining insights into the structural features of polyQ interfaces in a wide and unstrained context. The basic unit of these extended models was a peptide containing 15 Gln residues. Because the termini of Q15 peptides were capped with Lys- and Glu-charged residues in literature experiments (33), these were also included in the simulated models (Table 1). Models composed by a pair of four stranded β-sheets, either parallel or antiparallel, were analyzed. We also considered two alternative relative orientations of the sheets within the pair (head-to-head or head-to-tail chain direction). As described in Materials and Methods, these options lead to three possible arrangements: head-to-tail sheets/parallel strands (htP), head-to-head sheets/parallel strands (hhP), and antiparallel strands (A).
Despite some irregularities present in the starting models, all models (htPSH2-ST4-Q15, hhPSH2-ST4-Q15, and ASH2-ST4-Q15) converged toward very stable and well-defined structures. Indeed, as shown by all indicators used commonly for analyzing MD simulations (secondary structure, root mean-square deviations (RMSD), total number of hydrogen bonds, gyration radius), the models reach a stable equilibrated state after ∼2000 ps (Figs. 1–3 in the Supplementary Material, Data S1). In both cases, the intersheet distance of the trajectory structures is ∼8.5 Å (Fig. 1), in close agreement with the value derived from fiber diffraction studies (10–12). The analysis of the average models derived from the plateau region of the trajectory clearly indicates that Gln residues of facing strands are regularly interdigitated (Fig. 2). This tight interdigitation is also highlighted by the analysis of the surface complementarity of the two facing sheets. For all systems, the value of surface complementarity (34) is very high (0.82–0.83). In contrast to GNNQQNY interface, which is essentially stabilized by Van der Waals interactions (25,35) and dipole-dipole interactions (36), the structure of these polyQ models is likely stabilized by hydrogen bonding interactions formed by the Nɛ2 atom of a Gln side chain with the oxygen backbone of a Gln located in the facing strand (Fig. 1). The impact of the force field on these hydrogen bonding interactions has been evaluated by repeating the simulation on htPSH2-ST4-Q15, carried out originally with the Gromos96 (see Methods), with the all-atom force field OPLS (30). As shown in Fig. 4 of the Supplementary Material (Data S1), this analysis fully confirms the data obtained using Gromos96.
FIGURE 1.
Intersheet distances of representative (A) htPSH2-ST4-Q15, (B) hhPSH2-ST4-Q15, and (C) ASH2-ST4-Q15 structures. The Cα-Cα distances and the hydrogen bonding interaction between the side-chain Nɛ2 atom and the main-chain O atom of two facing Gln residues are shown in black and gray, respectively.
FIGURE 2.
Average models for (A) htPSH2-ST4-Q15, (B) hhPSH2-ST4-Q15, and (C) ASH2-ST4-Q15.
FIGURE 3.
Root mean-square fluctuation of Gln side chains in (A) htPSH2-ST4-Q15, (B) hhPSH2-ST4-Q15, and (C) ASH2-ST4-Q15. In both cases, the equilibrated region of the trajectory (2000–20,000 ps) was considered.
FIGURE 4.
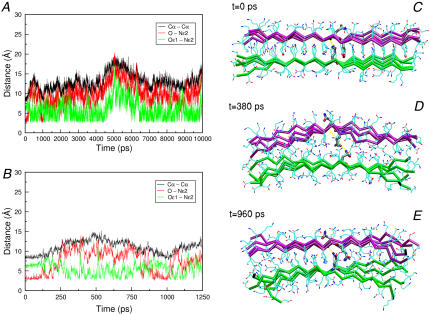
Transient interactions in the reversible local unfolding of htPSH2-ST4-Q15 at 500 K. (A) Reports the evolution of representative intersheet distances. The Cα-Cα distances and potential hydrogen bonding interactions between side chain–main chain atoms and side chain-side chain atoms of two facing Gln residues are shown in black, red, and green, respectively. (B) A snapshot on the first 1250 ps of simulation is presented. (C–E) Representative structures along the trajectory and hydrogen bonding interactions in yellow. For clarity, N, C, and O backbone atoms have been omitted.
The analysis of Gln side chain fluctuations indicate a very large difference between the mobility of solvent exposed residues and that exhibited by side chains inserted in the steric zipper motif (Fig. 3). Although this result is somewhat expected, it suggests that polar zipper interactions formed by Gln side chains in the solvent-exposed face of the sheet do not play a significant role in reducing their flexibility. This also indicates that there is no preorganization of Gln side chains embedded in single sheets for a steric zipper motif.
The overall stability of these models is confirmed by MD simulations carried out at high temperatures on ASH2-ST4-Q15 and on htPSH2-ST4-Q15, as representative of parallel sheets. As a general trend, these simulations indicate that these models exhibit very similar stabilities. Both models are stable in MD simulations carried out at 400 K. On the other hand, a significant destabilization of these structures is observed at 500 K. The analysis of ASH2-ST4-Q15/htPSH2-ST4-Q15 unfolding process at 500 K shows some interesting features on the stability of these aggregates. Indeed, it has been found that, in the early stage of the simulation within the first 1000 ps, the steric zipper motif is, in specific points, reversibly destroyed. As shown in Fig. 4, the H-bond formed between the side chain and the main chain of two facing Gln residues is replaced by side chain-side chain H-bonds. Although these bonds may be formed also in rather compact structures (intersheet separation of 9–10 Å), they occur mostly in structures where the two sheets are less packed against each other (distance = 12–13 Å). Although these side chain-side chain interactions are virtually absent in the trajectories obtained at 300 K, they may contribute to maintain the structure of the assembly in a compact state in destabilizing conditions.
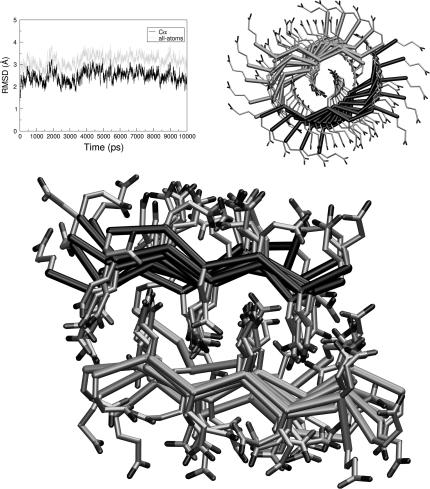
A distinctive characteristic of these models compared to those obtained from MD analyses on GNNQQNY aggregates (37) is the marginal level of twisting of the strands within the individual β-sheet. This feature, which is likely connected to the homopolymeric nature of the polyQ peptides, is suitable for lateral aggregation (crystallites) (10,11). Indeed, MD simulations carried out on the aggregate htPSH4-ST4-Q15 formed by four β-sheets show that that steric zipper interactions between nearly flat sheets may easily extend in direction orthogonal to the cross-β-spine axis (Fig. 5). This higher aggregate also exhibits higher stability. Indeed, in contrast to similar models formed by a single pair of sheet (htPSH2-ST4-Q15 and ASH2-ST4-Q15) the model htPSH4-ST4-Q15 is stable at 500 K in the timescale of the simulation (15 ns) (Fig. 5).
FIGURE 5.
Average model of htPSH4-ST4-Q15 derived from the MD trajectory (2000–10,000 ps). The RMSD of structures of the trajectory versus the starting model is reported in the inset.
Structure and stability of polyQ assemblies with limited intersheet interfaces
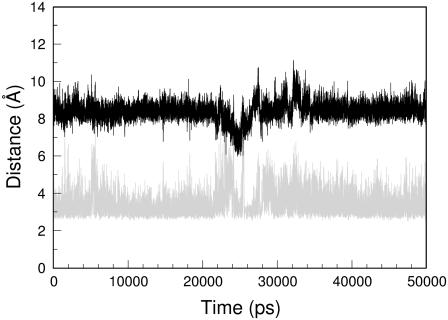
The high stability of the models with a large intersheet interface prompted us to check the stability of aggregates with a reduced interface. Initial analyses were conducted on peptides made of six Gln residues assembled in a pair of 10 stranded β-sheets (ASH2-ST10-Q6 and htPSH2-ST10-Q6). The model for htPSH2-ST10-Q6 was built manually using the structure of GNNQQNY as template. The MD simulation clearly indicates that this assembly is quite stable at 300 K (Fig. 5 in the Supplementary Material, Data S1). The final state closely resembles that obtained for larger aggregates. Similar results were obtained on the antiparallel model (ASH2-ST10-Q6) (data not shown). A plausible model for ASH2-ST10-Q6 is also embedded in the structure proposed by Eisenberg et al. for RNaseA.Q10 (Protein Data Bank entry 2APU) (23). Although the Q10 moiety of this model is characterized by a steric–zipper interface, the fine details of this structure are different from those that emerged in this analyses. In particular, Q10 moiety of RNaseA.Q10 exhibits a significant twisting of the strands within a single sheet and an intersheet separation of ∼9.5 Å, which is ∼1.0 Å higher that that found in the models obtained from the MD simulations. Although these differences may obviously be related to the different contexts of these polyQ assemblies, to verify the dependence of our results on the starting structure we also carried out MD analysis on a ASH2-ST10-Q6 model using the coordinates of the corresponding region of RNaseA.Q10. Our analysis indicates clearly that the model reaches a stable state in the simulation (Fig. 6). The average model emerged from the simulation is essentially formed by flat sheets with an intersheet distance of ∼8.5 Å (Fig. 6 in the Supplementary Material, Data S1). The formation of intersheet H-bonds is also observed (Fig. 6 in the Supplementary Material, Data S1). These findings indicate clearly that the features of polyQ models here elucidated are rather independent of the starting models.
FIGURE 6.
Average model of ASH2-ST10-Q6 derived from the MD trajectory (2000–10,000 ps). The left and right insets report the starting model and the RMSD values of the structure of the trajectory, respectively.
To obtain further insights into the structure and stability of small polyQ aggregates we also investigated the model htPSH2-ST4-Q6 by molecular dynamics. MD simulations show that also a model containing a limited number of glutamines (48 residues) is able to form stable steric zipper assemblies (Fig. 7 in the Supplementary Material, Data S1).
FIGURE 7.
Two different views of Q57 average model derived from the MD trajectory (5000–50,000 ps). The RMSD of structures of the trajectory versus the starting model (left) and the evolution of the secondary structure elements (right) are reported in the insets.
Structure and stability of polyQ monomeric forms
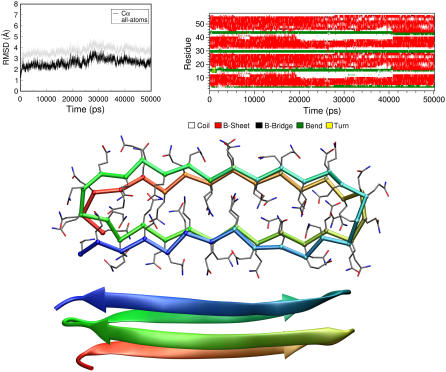
One of the most striking features of polyQ aggregation and toxicity is the key role played by monomeric species. It has been shown that the formation of a specific monomeric species is the slow step in the fiber formation process (14,15) and that monomeric species can exert toxic effects (16). In this framework, analyses aimed at identifying structural features of these forms are particularly valuable. Because, as illustrated in the previous section, even models containing a limited number of residues can form stable assemblies, we generated polyQ monomeric models by connecting within a single chain all strands of the small polyQ aggregates described above. We initially considered a polyQ composed of 57 residues, because a fragment of a comparable size fused to thioredoxin was shown to be toxic (16). Although, in principle, a large number of monomeric models may be built by using either parallel or antiparallel β-sheets and different covalent connections between the strands, we carried out our analysis on a representative model (Q57) made of parallel β-sheets linked as a collapsed β-helix motif, with interdigitated side chains (Fig. 7). The analysis of the MD trajectory shows that the model is very stable (Fig. 7). In particular, the evolution of Cα-Cα distance of facing Gln residues (Fig. 8) shows that the intersheet distance of the model in the equilibrated region is in line with that observed for the fibril-like assemblies and with the experimental values reported for polyQ aggregates. In addition to the intrasheet hydrogen bonding interactions that stabilize the β-sheet and the polar zipper motifs, rather stable H-bonds are also formed by Gln residues belonging to opposing sheets (Fig. 7 B). This network of H-bonds interactions makes even the exposed backbone groups (carbonyl and nitrogen atoms) of the model quite rigid. This is evident from the analysis of the root mean-square fluctuation values of the backbone atoms reported in Fig. 8 in the Supplementary Material (Data S1). The accumulation on Q57 surface (Fig. 9 in the Supplementary Material, Data S1) of rigid and fully exposed reactive groups is likely linked to toxic effects shown by these peptides.
FIGURE 8.
MD simulation of the model Q57: evolution of Cα–Cα distances (black) and main-chain side-chain H-bonding interactions (shaded) between representative facing Gln residues in the steric zipper interface.
FIGURE 9.
Two different views of Q41 average model derived from the MD trajectory (20,000–50,000 ps). The RMSD of structures of the trajectory versus the starting model (left) and the evolution of the secondary structure elements (right) are reported in the inset.
Along this line, we carried out additional simulations by further reducing the size of the monomeric form. In particular, we tried to generate models composed by ∼40 residues, a value comparable to the threshold observed for the disease onset (8). The manual generation of monomeric models endowed with regular steric zipper motifs and a β-structure proved to be a difficult task. A reasonable solution to this problem was achieved by resorting to the compact β-helix structure of Q41 obtained in a previous investigation (18). Although in this model there were direct contacts between Gln of opposing strands, they essentially involved H-bonds between side chains, with an intersheet distance of ∼12–13 Å. To achieve a state with interdigitated side chains, this model was subjected to a restrained MD simulation by imposing a 3.0 Å distance between atoms of opposite strands involved in main chain–side chain H-bond formation in models endowed with steric zipper. The stability of this model was checked by carrying out a standard MD simulation (50 ns), by removing the distance restraints. In contrast with the previous analysis, a rather long time (20 ns) was required to reach the equilibrium state (20–50 ns) (Fig. 9). However, the average structure of the equilibrated states retains the overall features (a collapsed β-structure) (Fig. 9). These results suggest that basic structural features of polyQ fibrils can be accommodated in rather small monomeric polyQ peptides.
DISCUSSION
Abnormal expansion of CAG triplets encoding Gln residues is linked to the insurgence of severe neurodegenerative diseases (3,24). Although the success rate of therapeutical approaches would likely increase with the availability of structural information on polyQ assemblies, current polyQ models are far from being generally accepted. The first goal of this investigation is the analysis of the compatibility of a steric zipper (25) with the structure of these assemblies. In addition, the involvement of the same basic structural element in the structure of monomeric and highly toxic forms has been evaluated.
The analysis conducted on a variety of polyQ aggregates with interdigitation of the side chain of opposing strands indicates clearly that they are very stable, even at high temperatures (400 K). The stability of these models relies heavily on hydrogen bonding interactions. Indeed, in addition to main chain–main chain and side chain-side chain (polar zipper) H-bonds within a single sheet (38), the structure of polyQ assemblies is also stabilized by HBs formed by Nɛ2 atom of a Gln residue with the oxygen backbone atom of Gln residue located in facing sheets. Because it has been shown that the primary amides in the side chains of glutamine residues are excellent hydrogen bond donors (39), it can be assumed that this latter interaction may significantly contribute to the overall stability of polyQ assembly. Interestingly, similar hydrogen bonding interactions are present in the structure of the peptide NNQQ (form 1, Protein Data Bank code 2ONX) solved recently at high resolution (27).
Notably, stable models can be generated by using either parallel or antiparallel β-sheets. This may be ascribed to the possibility of forming in both arrangements the three types of H-bonding interactions described above. This may suggest that, even in very similar experimental conditions, fibers with parallel or antiparallel β-sheets can be grown. Along this line, controversial indications on the chain direction of the strands within the cross-β-structure obtained from experimental studies (11) may, at least in part, reflect actual differences of the investigated samples. In line with the experimental data collected on polyQ model peptides (10–12), MD equilibrated structures show a conserved intersheet distance of ∼8.5 Å. In contrast to other simulations carried out on peptides assembled through a steric zipper interface (37,40,41) (A. De Simone, L. Esposito, C. Pedone, and L. Vitagliano, unpublished) the individual sheets are rather flat with a negligible twisting between consecutive strands. This feature, due likely to the homopolymeric nature of these peptides, strongly favors a higher level of aggregation through lateral association. This finding is in line with the observed tendency of polyQ peptides to frequently form crystallites rather than fibers (10,11). Our data are also in line with the sharpness of the equatorial reflection on the 8.3 Å exhibited usually by model peptides (10–12,17). Because our analyses indicates that different starting models converge to structures with intersheet distances of ∼8.5 Å, different attributions to the resolutions of the equatorial reflection (23) are due likely to the spread appearance of the spot, especially when the polyQ fragment is embedded in a protein context. In such conditions, it can be surmised that the presence of a protein matrix likely hampers a high level of lateral aggregation. This limited periodicity along the lateral direction may lead to rather broad equatorial reflections. Because, as shown here, even a single pair of interdigitated sheets can assume a stable cross-β-structure, limited lateral aggregation does not hamper fiber formation. In summary, our data, along with previous results and suggestions (10,23), clearly indicates that polyQ fiber structure is likely composed by facing nearly flat sheets with interdigitated Gln side chains. There is, however, an open question: is this structural motif relevant for the toxic polyQ species?
MD simulations carried out on small assemblies indicate that the strength of steric zipper interface makes stable models with a limited number of strands and of Gln per strand. This observation prompted us the analysis of plausible conformers of monomeric polyQ peptides. These polyQ forms have received great attention recently because it has been shown to be critical for fiber formation and polyQ toxicity (15,16). In particular, a polypeptide (Q57) with a size equivalent to that (62 Gln residues) fused to thioredoxin and endowed with toxic effects was investigated. The results show clearly that it can assume a β-structure stabilized by steric zipper interactions. The interdigitation of Gln side chains makes the strands of this structure very rigid, despite their exposure to the solvent. The accumulation of rigid and exposed groups on the Q57 surface, that may be also partially polarized (42), renders this assembly particularly reactive. Taking into account the intrinsic reactivity of edge β-strands (16,43,44) and the particular configuration (rigidity, concentration, solvent-accessibility, and polarization) of those located at the end of small and diffusible steric zipper assemblies, we suggested recently that the terminal strands of these objects can cross-react with a variety of cellular component giving rise to undesired and deleterious reactions (A. De Simone, L. Esposito, C. Pedone, and L. Vitagliano, unpublished). The data on Q57 perfectly fit into this scheme as the strength of Q57 interface is able to keep the edge strands in an “activated” state.
The structural motifs exhibited by Q57 may be also a feature of the stable monomeric intermediate that acts as the template for the quick fibril formation. In this scenario, the slow step in the fiber formation process is the organization of a native collapsed polyQ structure (45) into the well-defined assemblies presented here. The analyses of the peptide Q41 showed that conformers with rather stable β-structure and a steric zipper interface may be formed even with smaller sequences. However, we also found that the reduction of peptide size leads to increasing difficulties in building these models. Along this line, we suggest that the threshold of polyQ length for the disease insurgence and the dependence of its onset on polyQ lengths (8) are intimately linked to the need of a minimal number of residues to build a stable β-structure endowed with a stable interdigitated interface.
The occurrence in disease onset of a polyQ repeat threshold that resembles the one required to form stable monomeric species indirectly suggests that the process of protein aggregation follows the formation of “activated” monomeric species.
CONCLUSIONS
The extensive MD investigations here conducted on a variety of different polyQ models clearly indicate that side chain interdigitation, a motif found recently for several amyloid-like peptides, likely represents the basic element of polyQ fibers. Present analyses also indicate that a network of intrasheet and intersheet hydrogen bonding interactions contributes to the overall stability of these assemblies. More intriguingly, stable monomeric polyQ peptides with a size comparable to that endowed with toxicity could be generated using the same basic motif. Notably, the strength of the interface between the two β-sheets makes the overall structure of these monomeric species quite rigid. We propose that the reduced flexibility of the exposed backbone atoms, along with their partial polarization, further increase their intrinsic reactivity, thus leading to unwanted deleterious reactions with other cellular components. In addition, the structural correspondence between these monomeric species and the final fiber structure may also indicate that these models could be representative of the monomeric nuclei detected in the fiber formation process. Along this line, the increasing difficulties encountered in building this type of models for shorter peptides (<40 residues) is in line with the observed Gln repeat threshold in the onset and severity of polyQ-linked diseases. In conclusion, we show that models based on the same structural element can explain puzzling and diversified literature data on polyQ fibers and monomeric forms, thus providing a possible connection between structure and diseases.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
The authors thank the Centro Regionale di Competenza in Diagnostica e Farmaceutica Molecolari for providing some of the facilities used to carry out this work. The authors also thank Luca De Luca for technical assistance. CINECA Supercomputing (Project cne0fm4e) is acknowledged for computational support.
This work was supported by the Ministry of University and Research (FIRB Contract RBNE03PX83).
Editor: Feng Gai.
References
- 1.Chiti, F., and C. M. Dobson. 2006. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75:333–366. [DOI] [PubMed] [Google Scholar]
- 2.Buxbaum, J. N. 2003. Diseases of protein conformation: what do in vitro experiments tell us about in vivo diseases? Trends Biochem. Sci. 28:585–592. [DOI] [PubMed] [Google Scholar]
- 3.Ross, C. A., and M. A. Poirier. 2004. Protein aggregation and neurodegenerative disease. Nat. Med. 10(Suppl):S10–S17. [DOI] [PubMed] [Google Scholar]
- 4.Carrell, R. W. 2005. Cell toxicity and conformational disease. Trends Cell Biol. 15:574–580. [DOI] [PubMed] [Google Scholar]
- 5.Kayed, R., E. Head, J. L. Thompson, T. M. McIntire, S. C. Milton, C. W. Cotman, and C. G. Glabe. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 300:486–489. [DOI] [PubMed] [Google Scholar]
- 6.Gatchel, J. R., and H. Y. Zoghbi. 2005. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6:743–755. [DOI] [PubMed] [Google Scholar]
- 7.Brown, L. Y., and S. A. Brown. 2004. Alanine tracts: the expanding story of human illness and trinucleotide repeats. Trends Genet. 20:51–58. [DOI] [PubMed] [Google Scholar]
- 8.Perutz, M. F. 1999. Glutamine repeats and neurodegenerative diseases: molecular aspects. Trends Biochem. Sci. 24:58–63. [DOI] [PubMed] [Google Scholar]
- 9.Ross, C. A. 2002. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 35:819–822. [DOI] [PubMed] [Google Scholar]
- 10.Sikorski, P., and E. Atkins. 2005. New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules. 6:425–432. [DOI] [PubMed] [Google Scholar]
- 11.Sharma, D., L. M. Shinchuk, H. Inouye, R. Wetzel, and D. A. Kirschner. 2005. Polyglutamine homopolymers having 8–45 residues form slablike beta-crystallite assemblies. Proteins. 61:398–411. [DOI] [PubMed] [Google Scholar]
- 12.Bader, R., M. A. Seeliger, S. E. Kelly, L. L. Ilag, F. Meersman, A. Limones, B. F. Luisi, C. M. Dobson, and L. S. Itzhaki. 2006. Folding and fibril formation of the cell cycle protein Cks1. J. Biol. Chem. 281:18816–18824. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka, M., Y. Machida, Y. Nishikawa, T. Akagi, T. Hashikawa, T. Fujisawa, and N. Nukina. 2003. Expansion of polyglutamine induces the formation of quasi-aggregate in the early stage of protein fibrillization. J. Biol. Chem. 278:34717–34724. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharyya, A. M., A. K. Thakur, and R. Wetzel. 2005. polyglutamine aggregation nucleation: thermodynamics of a highly unfavorable protein folding reaction. Proc. Natl. Acad. Sci. USA. 102:15400–15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wetzel, R. 2006. Kinetics and thermodynamics of amyloid fibril assembly. Acc. Chem. Res. 39:671–679. [DOI] [PubMed] [Google Scholar]
- 16.Nagai, Y., T. Inui, H. A. Popiel, N. Fujikake, K. Hasegawa, Y. Urade, Y. Goto, H. Naiki, and T. Toda. 2007. A toxic monomeric conformer of the polyglutamine protein. Nat. Struct. Mol. Biol. 14:332–340. [DOI] [PubMed] [Google Scholar]
- 17.Perutz, M. F., B. J. Pope, D. Owen, E. E. Wanker, and E. Scherzinger. 2002. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagine-rich domains of Sup35 and of the amyloid beta-peptide of amyloid plaques. Proc. Natl. Acad. Sci. USA. 99:5596–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merlino, A., L. Esposito, and L. Vitagliano. 2006. Polyglutamine repeats and beta-helix structure: molecular dynamics study. Proteins. 63:918–927. [DOI] [PubMed] [Google Scholar]
- 19.Zanuy, D., K. Gunasekaran, A. M. Lesk, and R. Nussinov. 2006. Computational study of the fibril organization of polyglutamine repeats reveals a common motif identified in beta-helices. J. Mol. Biol. 358:330–345. [DOI] [PubMed] [Google Scholar]
- 20.Stork, M., A. Giese, H. A. Kretzschmar, and P. Tavan. 2005. Molecular dynamics simulations indicate a possible role of parallel beta-helices in seeded aggregation of poly-Gln. Biophys. J. 88:2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armen, R. S., B. M. Bernard, R. Day, D. O. Alonso, and V. Daggett. 2005. Characterization of a possible amyloidogenic precursor in glutamine-repeat neurodegenerative diseases. Proc. Natl. Acad. Sci. USA. 102:13433–13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchut, A. J., and C. K. Hall. 2007. Effects of chain length on the aggregation of model polyglutamine peptides: molecular dynamics simulations. Proteins. 66:96–109. [DOI] [PubMed] [Google Scholar]
- 23.Sambashivan, S., Y. Liu, M. R. Sawaya, M. Gingery, and D. Eisenberg. 2005. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature. 437:266–269. [DOI] [PubMed] [Google Scholar]
- 24.Temussi, P. A., L. Masino, and A. Pastore. 2003. From Alzheimer to Huntington: why is a structural understanding so difficult? EMBO J. 22:355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, R., M. R. Sawaya, M. Balbirnie, A. O. Madsen, C. Riekel, R. Grothe, and D. Eisenberg. 2005. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 435:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenberg, D., R. Nelson, M. R. Sawaya, M. Balbirnie, S. Sambashivan, M. I. Ivanova, A. O. Madsen, and C. Riekel. 2006. The structural biology of protein aggregation diseases: fundamental questions and some answers. Acc. Chem. Res. 39:568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawaya, M. R., S. Sambashivan, R. Nelson, M. I. Ivanova, S. A. Sievers, M. I. Apostol, M. J. Thompson, M. Balbirnie, J. J. Wiltzius, H. T. McFarlane, A. O. Madsen, C. Riekel, and D. Eisenberg. 2007. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 447:453–457. [DOI] [PubMed] [Google Scholar]
- 28.Van Der Spoel, D., E. Lindahl, B. Hess, G. Groenhof, A. E. Mark, and H. J. Berendsen. 2005. GROMACS: fast, flexible, and free. J. Comput. Chem. 26:1701–1718. [DOI] [PubMed] [Google Scholar]
- 29.Hess, B., H. Bekker, H. J. C. Berendsen, and J. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 30.Kaminski, G. A., R. A. Friesner, J. Tirado-Rives, and W. L. Jorgensen. 2001. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 105:6474–6487. [Google Scholar]
- 31.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph 14:33–38. [DOI] [PubMed] [Google Scholar]
- 32.Kraulis, P. J. 1991. MOLSCRIPT:a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24:946–950. [Google Scholar]
- 33.Perutz, M. F., T. Johnson, M. Suzuki, and J. T. Finch. 1994. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. USA. 91:5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence, M. C., and P. M. Colman. 1993. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234:946–950. [DOI] [PubMed] [Google Scholar]
- 35.Zheng, J., B. Ma, C. J. Tsai, and R. Nussinov. 2006. Structural stability and dynamics of an amyloid-forming peptide GNNQQNY from the yeast prion sup-35. Biophys. J. 91:824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez, A. 2005. What factor drives the fibrillogenic association of beta-sheets? FEBS Lett. 579:6635–6640. [DOI] [PubMed] [Google Scholar]
- 37.Esposito, L., C. Pedone, and L. Vitagliano. 2006. Molecular dynamics analyses of cross-beta-spine steric zipper models: beta-sheet twisting and aggregation. Proc. Natl. Acad. Sci. USA. 103:11533–11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perutz, M. 1994. Polar zippers: their role in human disease. Protein Sci. 3:1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberhardt, E. S., and R. T. Raines. 1994. Amide-amide and amide-water hydrogen bonds: implications for protein folding and stability. J. Am. Chem. Soc. 116:2149–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng, J., B. Ma, and R. Nussinov. 2006. Consensus features in amyloid fibrils: sheet-sheet recognition via a (polar or nonpolar) zipper structure. Phys. Biol. 3:1–4. [DOI] [PubMed] [Google Scholar]
- 41.Wu, C., Z. Wang, H. Lei, W. Zhang, and Y. Duan. 2007. Dual binding modes of Congo red to amyloid protofibril surface observed in molecular dynamics simulations. J. Am. Chem. Soc. 129:1225–1232. [DOI] [PubMed] [Google Scholar]
- 42.Tsemekhman, K., L. Goldschmidt, D. Eisenberg, and D. Baker. 2007. Cooperative hydrogen bonding in amyloid formation. Protein Sci. 16:761–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson, J. S., and D. C. Richardson. 2002. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. USA. 99:2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laidman, J., G. J. Forse, and T. O. Yeates. 2006. Conformational change and assembly through edge beta strands in transthyretin and other amyloid proteins. Acc. Chem. Res. 39:576–583. [DOI] [PubMed] [Google Scholar]
- 45.Crick, S. L., M. Jayaraman, C. Frieden, R. Wetzel, and R. V. Pappu. 2006. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc. Natl. Acad. Sci. USA. 103:16764–16769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.