Abstract
A filopodium is a cytoplasmic projection, exquisitely built and regulated, which extends from the leading edge of the migrating cell, exploring the cell's neighborhood. Commonly, filopodia grow and retract after their initiation, exhibiting rich dynamical behaviors. We model the growth of a filopodium based on a stochastic description which incorporates mechanical, physical, and biochemical components. Our model provides a full stochastic treatment of the actin monomer diffusion and polymerization of each individual actin filament under stress of the fluctuating membrane. We investigated the length distribution of individual filaments in a growing filopodium and studied how it depends on various physical parameters. The distribution of filament lengths turned out to be narrow, which we explained by the negative feedback created by the membrane load and monomeric G-actin gradient. We also discovered that filopodial growth is strongly diminished upon increasing retrograde flow, suggesting that regulating the retrograde flow rate would be a highly efficient way to control filopodial extension dynamics. The filopodial length increases as the membrane fluctuations decrease, which we attributed to the unequal loading of the membrane force among individual filaments, which, in turn, results in larger average polymerization rates. We also observed significant diffusional noise of G-actin monomers, which leads to smaller G-actin flux along the filopodial tube compared with the prediction using the diffusion equation. Overall, partial cancellation of these two fluctuation effects allows a simple mean field model to rationalize most of our simulation results. However, fast fluctuations significantly renormalize the mean field model parameters. The biological significance of our filopodial model and avenues for future development are also discussed.
INTRODUCTION
Cell migration, ubiquitous in many biological phenomena such as embryonic development, wound healing, and cancer metastasis, is a complex set of interacting mechanochemical processes (1). The leading edge of motile cells project μm-size finger-like protrusions based on parallel, bundled actin filaments called filopodia (2–4). Filopodia play an important role in guiding cell motility and participate in cell-cell communication. For example, the migration of tissue cells in embryonic development, under the guidance of external chemical cues, is facilitated by filopodia, which constantly grow and retract, exploring the complex extracellular matrix and directing the cell through the noisy environment (5–7). Filopodial misregulation results in developmental defects and diseases. In wound healing, fibroblast cells grow long filopodia that touch the other side of the cut, organizing the cells on the opposing sides and guiding smooth sealing of the opening (8,9). In cancer development, tumor cells may spread from their primary site to other places in the body through metastasis in which filopodia also play a role (10,11). Given the biological importance of filopodia, a number of recent works have investigated how the filopodial growth and retraction dynamics is regulated by various cellular structures and by internal and external signals (1–4,9,10,12–30).
A filopodium is a cytoplasmic projection, exquisitely built and regulated, which extends from the leading edge of the migrating cell, exploring the cell's neighborhood. Commonly, filopodia grow and retract after their initiation, exhibiting rich dynamical behaviors. The filopodial structure is supported by parallel actin filaments cross-linked into bundles by actin-binding proteins enclosed by the cell membrane. It has been suggested that a large protein complex at the filopodial tip plays an important role in regulating filopodial dynamics. Even though filopodial growth and retraction is largely driven by the polymerization and depolymerization of the actin filaments, the morphology and function of filopodia are also governed by the availability of monomeric actin and cross-linking proteins, the cell membrane, the tip complex, and other regulatory proteins (4,15,26,29,31–33).
The growth of filopodia involves complex mechanical, chemical, and biological processes that are intimately interwoven. Thus, filopodial growth modeling is both interesting and challenging. Though the overall structure and function of a filopodium has become quite clear, the comprehensive set of individual players and their detailed interactions remain to be elucidated (4,14,29). The complexity of the problem motivates building effective mathematical models that would allow one to gain deeper insights into filopodial dynamics and regulation. Prior modeling efforts were mainly concentrated on specific aspects of filopodial dynamics, considering a subset of participating proteins and a small part of the regulatory reaction network (34–38). In a pioneering work, Mogilner and Rubinstein proposed a simple deterministic model to study the filopodial initiation and growth (19). The mechanical properties of the actin filaments and bundles were studied from the point of view of elasticity theory. A one-dimensional reaction-diffusion equation was used to derive important length-force relations (19). However, the stochastic polymerization of each filament and the detailed membrane force on each filament were not considered. Later, the interaction of the filopodial tip and the membrane was investigated in detail by Sun and co-workers (23). In their elegant work they studied how the actin polymerization process depends on the membrane properties and external forces, but monomeric actin diffusion from the ventral part of the cell to the filopodial tip and the retrograde flow (4,19) were not considered in their model. More recently, a detailed modeling of the shape and size of stereocilia and microvilli was carried out based on mean field equations derived from the free energy function of various interactions and a master equation for the actin filament treadmilling (39). However, the effects of the membrane interaction with the tip on the polymerization were not considered, and the discrete noise due to the actin monomer was not discussed. These processes, however, play key roles in controlling filopodial growth and retraction, as discussed below.
A more realistic model of filopodial growth should simultaneously consider all the effects discussed above, since they are all important and together constitute the basis for the filopodial dynamical behaviors. Our model, reported in this work, provides a full stochastic treatment of the actin monomer diffusion and polymerization of each individual actin filament under stress of the fluctuating membrane. In principle, the randomness of the reaction and transport processes is expected to be important near the filopodial tip, since the actin monomer number density turns out to be very small at the tip (see below), necessitating that a continuous description be replaced by a discrete one (40–57). In this work, we investigated the length distribution of individual filaments in a growing filopodium and studied how it depends on various physical parameters. Our study identifies the key physical and chemical parameters that control filopodial growth. In particular, we discovered that retrograde flow rate plays a critical role in controlling filopodial extension. In addition, we found that membrane fluctuations significantly renormalize the polymerization rate, where stronger membrane fluctuations lead to shorter filopodia. Finally, our fully stochastic treatment of the filopodial growth process, which is based on only the minimal set of chemical and physical processes implicated in filopodial dynamics, produces only small amplitude steady-state fluctuations. In some experiments, filopodia undergo large amplitude fluctuations (1,14,26,58), suggesting that additional levels of regulation by signaling protein networks need to be incorporated into the computational models of filopodial dynamics.
The work is organized as follows. In the Model Development section, we discuss a number of determinants of filopodial growth dynamics: the monomeric actin diffusion, the actin filament polymerization and depolymerization, the membrane force, and the retrograde flow. Accordingly, we propose our physicochemical models for the corresponding processes. The model parameters and their typical values are also listed and discussed. Using mean field arguments, we provide in the Mean field solution at the steady state section a simple analytical solution of our filopodial model in the long time limit. The results of our more detailed stochastic simulations are discussed and rationalized in Stochastic simulations section. In the Discussion section, we point out the biological significance of our model and suggest avenues for future development and possible extensions.
MODEL DEVELOPMENT
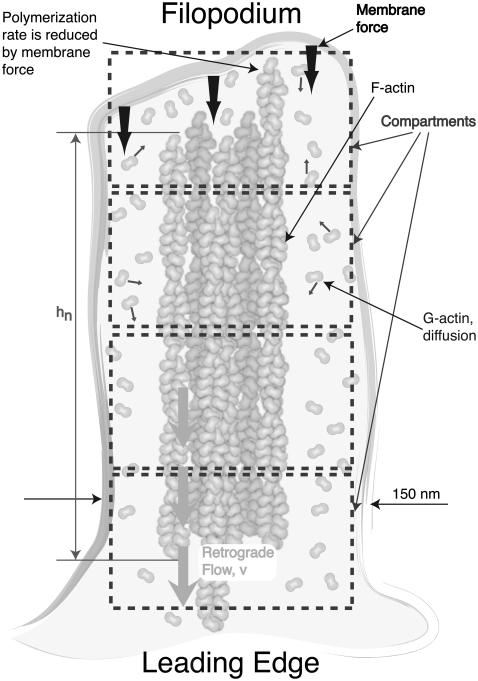
After initiation from its precursor, a filopodium is characterized by bundled actin filaments enclosed by the cell membrane with a protein complex at the tip, which forms a stable and universal structure (30). Fig. 1 shows a schematic cartoon of the filopodial structure and dynamics. Filopodia commonly grow out of the underlying dense web-like actin filament network (usually lamellipodia) (30). In addition to the rod-like stiff filaments and the floppy enclosing membrane, the filopodial structure includes cross-linking proteins which fasten the filament bundle and a possible surface protein complex which regulates the growth process (Fig. 1). However, these latter structures were not in our model here. Their role will be reported in future work.
FIGURE 1.
Schematic diagram of a matured filopodium. Only the most fundamental physicochemical components are shown; for example, the putative filopodial tip complex is not drawn. The following processes are included in our computational model: 1), monomeric G-actin diffusion along the filopodial tube; 2), polymerization and depolymerization of individual actin filaments; 3), fluctuating membrane under load which slows down the individual filament polymerization rate; and 4), a constant velocity retrograde flow, where actin filaments are continuously pulled into the cell body. Actin diffusion is modeled as a stochastic hopping process between compartments of size 50 nm along the filopodial tube. Rapid mixing is assumed in the transverse direction.
In cell migration or development, a filopodium usually grows or retracts at a speed of 0.1–0.2 μm/s up to lengths of 1–2 μm (1). In special cases, it may grow up to nearly 100 microns (6). Short filopodia are often straight, whereas long ones may become easily bent and tilted. Here, for simplicity, we assume straight filopodia. Filament bending in the context of our model is a subject of ongoing research. We use h to denote the average membrane position at the filopodial tip and hn to denote the length of the nth filament. It is reasonable to define h as
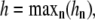 |
(1) |
since the membrane is supported by the filaments. The filopodial diameter varies between d = 100 nm and 300 nm depending on the number of filaments inside and the lateral interaction with the membrane. We take d = 150 nm in our model, which is derived from minimizing the membrane free energy and is reasonable if the number of filaments is not large (23). The number of actin filaments in one filopodium varies from 10 to 30 (19). For microvilli, it has been discussed in terms of the fluctuation of the membrane protein density (59). For filopodia, this number is determined in the initiation stage and will not change in a matured filopodium. Throughout the discussion, we considered a filopodium to consist of a fixed number N = 16 of filaments.
The following four major processes are the key components of our model: 1), the actin diffusion from the filopodial base to the tip; 2), the force applied by the membrane on individual filaments; 3), the actin filament polymerization and depolymerization at the barbed end; and 4), the depolymerization at the pointed end and the induced retrograde flow, vretr, of the filopodium as a whole. Below, we discuss all these aspects in detail and propose a modeling strategy for each of these processes.
Actin filaments and actin monomer diffusion to the filopodium tip
Filopodial protrusion and retraction is realized by the polymerization and depolymerization of the individual actin filaments. Because of dissimilar chemical affinities at the two ends of a filament, actin monomers usually add to the filament barbed end and dissociate from the pointed end. As a result, the filament as a whole marches in the direction of the barbed end, which is called “treadmilling” and constitutes the biochemical basis for the cytoskeletal dynamics (33,60). Free monomeric actin, called G-actin, is a globular protein that is 5.4 nm in diameter. When G-actin binds to an actin filament, the resulting polymeric species is called an F-actin. In our calculations, described below, each filament protrudes by δ = 2.7 nm upon a single G-actin addition, since two actin monomers are needed to protrude the filament by one step.
Each actin filament consists of two protofilaments, which form a right-handed double helix with a pitch distance of 37 nm. The resulting structure is very robust, with a persistence length, Lp ∼ 10 μm. Under a force, F, the critical length at which one filament buckles is
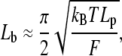 |
(2) |
where kB is the Boltzmann constant and kBT = 4.1 pN nm at room temperature (19). For a force F = 10 pN, the buckling length Lb = 101 nm. For weakly cross-linked bundles of N actin filaments, the buckling length of the bundle is 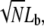 whereas it is
whereas it is 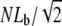 for tightly linked bundles (19).
for tightly linked bundles (19).
G-Actin monomers diffuse in cytoplasm with a diffusion constant of approximately D = 5 μm2/s (19). The crowded environment of filopodia possibly slows down the diffusion, but there are no experimental measurements to address this point. As the size of actin monomer is still much smaller than the filopodium size, we use the bulk diffusion constant. In the simulations, we examined the effect of the smaller values of the diffusion constant. As the diameter of a typical filopodium (150 nm) is much smaller than its length (several microns), we consider here only the diffusion of the actin monomer in the longitudinal direction (assuming quick mixing in the transverse direction). Although G-actin monomers are present at the high concentration of ρ0 = 10 μM in the bulk of a typical cell, the number of G-actin monomers at the filopodial tip may drop to essentially zero, due to the filament polymerization processes that consume G-actin. Therefore, we model G-actin diffusion along the filopodium as a random walk on a one-dimensional lattice. As shown in Fig. 1, the filopodium is divided into compartments, with a compartment height of ld = 50 nm, starting from the tip. We define the number of actin monomers in a compartment with index l as al. The transition rate for one particle moving between neighboring compartments is
 |
(3) |
where Cd is related to the diffusion constant, D, by 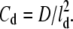 At the tip, we imposed the reflection boundary condition Ptrans(1 → 0) = 0, since the monomers cannot penetrate the cell membrane (the compartment index starts from the tip and runs down to the base). At the filopodial base, the G-actin concentration was kept at the constant bulk value, ρ0 = 10 μM.
At the tip, we imposed the reflection boundary condition Ptrans(1 → 0) = 0, since the monomers cannot penetrate the cell membrane (the compartment index starts from the tip and runs down to the base). At the filopodial base, the G-actin concentration was kept at the constant bulk value, ρ0 = 10 μM.
Our discretization of space assumes that the reaction processes are confined to a small enough spatial region, having a linear dimension of ζ (the so-called Kuramoto length (61)), such that particles diffuse across the region faster than the typical reaction times. We estimated ζ to be ∼150 nm at the tip of the filopodium. We chose a somewhat more conservative compartment size of 50 nm. The numerical results are not sensitive to small changes in the compartment size.
Polymerization and depolymerization
G-actin, when bound to ATP, exhibits a stronger propensity to polymerize than when bound to ADP. When G-actin turns into F-actin upon polymerization, an “aging” process starts (35), where ATP molecules are hydrolyzed into ADP. This, in turn, leads to enhanced dissociation rates at the pointed end. ADP in the resulting free G-actin monomer exchanges with ATP in the cytosol. Thus, the entire treadmilling process is energy driven, requiring participation and consumption of cells' universal energy currency, ATP.
In our model when a G-actin molecule diffuses to the filopodial tip, it has a certain probability, given by the rate k0, of being added to one of the filament ends. On the other hand, the probability of dissociation at the barbed end is small, determined by the dissociation rate of kd. These polymerization and depolymerization processes were stochastically modeled in our work. In prior experiments (62), it was shown that the polymerization rate sensitively depends on the membrane force, which was theoretically rationalized (19,36,37). In particular, in the Brownian ratchet model, the membrane fluctuations at the tip of the filopodium are considered (36,38). If the created space between the membrane and the filament tip is large enough for a G-actin monomer to fit sterically, a subsequent polymerization could happen with the rate k0. Thus, the effective polymerization rate, kn, on the nth filament equals the “bare” rate k0 times the probability of the gap opening at the tip of the nth filament. Because the membrane force suppresses the amplitude of the likely membrane fluctuations, a larger membrane force implies smaller kn. A convenient relation between the loading force fn and the polymerization rate was derived earlier (38),
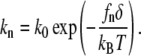 |
(4) |
Thus, to compute the effective polymerization rate, the membrane force on each individual filament has to be estimated, which is discussed next. In our model, the depolymerization rate is independent of the membrane fluctuations.
Membrane fluctuations and filament forces
Cell membranes consist of a double layer of phospholipids and may be thought of as a soft viscoelastic medium. Due to the osmotic pressure (63), electrostatic interactions (64), mechanical interactions, and exchange of lipids with internal reservoirs (65,66), the cells maintain a finite membrane tension, which in turn regulates the cytoskeletal dynamics in a variety of ways (67–69). Thus, enclosed by the cell membrane, filopodial filament growth is counteracted by the membrane force. In practice, depending on the membrane composition and mechanochemical conditions, the membrane force varies with the filopodium length and environmental cues. However, under quite general conditions and in a large range of parameter values, this variation is rather small (69,70). Therefore, for simplicity, the total membrane force acting on all filaments is a constant in our model. We used a typical value of f = 10 pN for some of our computations.
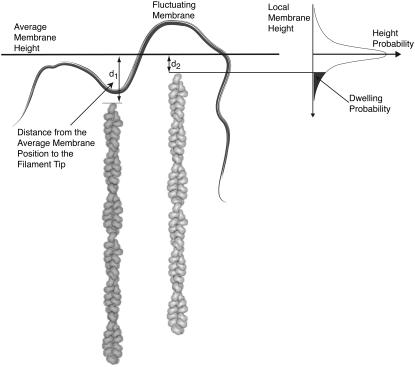
Prior studies indicated that submicron linear size membrane sheet fluctuations relax on the microsecond to millisecond timescale (71–74). Consequently, the membrane fluctuations may be assumed to be equilibrated on the timescale of the filament growth dynamics (subsecond). Each filament experiences an individual membrane force, fn, that is determined by the closeness of the filament end to the tip membrane average position. Thus, the total membrane force f is partitioned among the individual filaments, 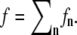 Here we propose a new scheme to calculate individual fns. The filopodial tip membrane fluctuates around some time-dependent average position and exerts a force when it makes contact with a specific filament (Fig. 2). It is reasonable to assume that on average the force on a filament is proportional to the membrane-filament contact dwelling probability, which is the probability that the membrane touches that filament. This interaction, in turn, critically depends on the amplitude of the membrane fluctuation near the filament and the filament length. Longer filaments are more likely to be in contact with a membrane and feel stronger membrane force. If the membrane fluctuations are assumed to be described by a Gaussian distribution around the membrane average position, the dwelling probability pn for the nth filament to be in contact with the membrane is proportional to the probability that the membrane height is found below the filament end (Fig. 2)
Here we propose a new scheme to calculate individual fns. The filopodial tip membrane fluctuates around some time-dependent average position and exerts a force when it makes contact with a specific filament (Fig. 2). It is reasonable to assume that on average the force on a filament is proportional to the membrane-filament contact dwelling probability, which is the probability that the membrane touches that filament. This interaction, in turn, critically depends on the amplitude of the membrane fluctuation near the filament and the filament length. Longer filaments are more likely to be in contact with a membrane and feel stronger membrane force. If the membrane fluctuations are assumed to be described by a Gaussian distribution around the membrane average position, the dwelling probability pn for the nth filament to be in contact with the membrane is proportional to the probability that the membrane height is found below the filament end (Fig. 2)
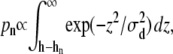 |
(5) |
where σd is the average membrane fluctuation amplitude (discussed next). Once pn is obtained, the force fn on each filament may be computed
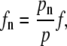 |
(6) |
where 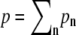 is a normalization factor. Substituting fn into Eq. 4, the time-dependent polymerization rate, kn, is computed for each filament under the membrane force.
is a normalization factor. Substituting fn into Eq. 4, the time-dependent polymerization rate, kn, is computed for each filament under the membrane force.
FIGURE 2.
Each filamental tip experiences some membrane force due to height fluctuations of the filopodial membrane. The sum of these individual filamental forces is equal to the overall average membrane load. The membrane fluctuations around an average height are modeled with a Gaussian distribution, having an average fluctuation amplitude of σd. The probability that a membrane is in contact with a particular filament (the dwelling probability, i.e., the probability of the local membrane height being found below the particular filament's tip) is proportional to the shaded area under the curve in the right-hand panel.
The biological membrane fluctuations could be driven both by thermal noise and by additional nonequilibrium processes (65,67,68,74–88). To roughly estimate the amplitude of thermal height fluctuations, the membrane may be modeled with the following free energy functional (89,90):
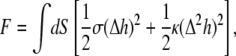 |
(7) |
where h indicates membrane height displacement at position x, y (Monge representation), σ represents membrane surface tension, and κ is the bending modulus. The fluctuation amplitude for a mode with wave vector q is (74,79)
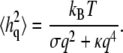 |
(8) |
Finally, the height fluctuations in real space may be computed using the definition of  Fourier transform (89)
Fourier transform (89)
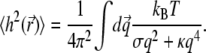 |
(9) |
For the surface tension coefficient in the physiological range of 10−6–10−4 N/m and the bending modulus between 10−20–10−19 J (65,74,86), the amplitude of thermal membrane fluctuations for a membrane sheet with submicron linear dimensions may be estimated to be a few nanometers using Eq. 9. However, a number of nonthermal, energy driven processes (73,82,85,87,91,92) may additionally increase severalfold the plasma membrane height fluctuations. Fluctuations on the order of tens of nanometers have been measured in red blood cell membranes (75,93). In this work, we found a noticeable dependence of the average filopodial length on the membrane fluctuation amplitude, when σd was varied between 0 nm and 60 nm (discussed below). For other calculations where the membrane fluctuation amplitude was kept constant, we take σd ∼ 10 nm, which is the likely upper bound to the filopodial membrane height fluctuations.
Retrograde flow and equation of motion
Prior experiments indicated that the depolymerization rate at the pointed end of a filament changes much less than the polymerization rate at the barbed end, whereas the depolymerization at the barbed end is almost negligible. The polymerization rate is very high when a filopodium is growing and near zero when shrinking. Thus, filopodial dynamics are strongly controlled by the polymerization at its tip (15,26). Both polymerization at the tip and depolymerization at the base result in a steady backward motion of the whole actin filament bundle, called the retrograde flow. In some cells, it is believed that specific myosin motors participate in creating the retrograde flow (17), which is then subject to the regulation by the signaling proteins. In our computations here, we neglect the fine regulation of the retrograde flow process and assume a constant average retrograde flow speed, vretr. The influence of retrograde flow rate fluctuations will be addressed in future work.
The above discussion of the barbed end dynamics and the retrograde flow process motivates us to write the following equation of motion for each filament length, hn,
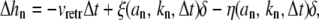 |
(10) |
where Δhn denotes the change of hn in a time interval Δt and ξ, η are two random variables that take discrete values {0, 1}. When there is one polymerization event on the nth filament during Δt, ξ = 1 and η = 0, and when there is one depolymerization event during Δt, η = 1 and ξ = 0. Otherwise, if some other stochastic events occur (for example, G-actin hopping between compartments), then η = 0 and ξ = 0. The probability that one polymerization event happens depends on the polymerization rate kn, the monomer number an in the compartment where the filament tip is located, and the time interval Δt itself. As discussed above, δ = 2.7 nm, which is half the actin monomer size.
In our computations, based on the Gillespie algorithm (94), the following steps are taken iteratively at each step: 1), Δt is determined by considering all possible chemical and diffusional events; and 2), one of the following events is chosen: a), G-actin monomer hopping between filopodial compartments, b), individual filament polymerization events, or c), depolymerization events. Subsequently, after learning Δt and the event type, we update the compartment monomer number am and the filament length hn. Then, upon incorporation of the retrograde flow (see Eq. 10), we compute hn, an, kn, where kn is modulated by membrane fluctuations and continue with the next iteration. The retrograde flow speed vretr is around 20 ∼ 200 nm/s (3,26,58,95,96), and we take vretr = 70 nm/s as the default value in our simulations.
Initial conditions
Our modeling starts with an already mature short filopodium. How this filopodium is initiated is a very interesting question, which is, however, outside the scope of this work. In a recent work, initiation of membrane protrusions was modeled based on membrane proteins with a convex spontaneous curvature that activates actin polymerization (97). We chose an initial filopodial length of 81 nm. This value only has some effect on the transient growth phase but does not influence the steady-state filopodial lengths. In addition, we used a bulk G-actin concentration of 10 μM as an initial concentration in the protrusion. Because of a relatively fast G-actin diffusion constant, this is a reasonable approximation for the short initial protrusion.
RESULTS
Mean field solution at the steady state
In the steady state, if we ignore all the fluctuations, take the deterministic continuum approximation, and use some reasonable assumptions, then an analytical approximation of the average filopodial length can be derived. Our stochastic simulations indicate that the individual filament lengths are narrowly distributed (discussed below). Thus, in the mean field approximation, we assume equal filamental lengths. This, in turn, implies that every filament is subject to the same force,
 |
(11) |
where N = 16 is the total number of actin filaments. Next, we compute the flux of G-actin monomers in the filopodial tube in the long time limit. One expects that the diffusion equation would adequately characterize how the spatial profile of the average actin concentration evolves in time (the accuracy of this assumption is examined below),
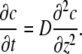 |
(12) |
In the steady state, 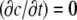 implies a linear G-actin concentration profile, c = az + b, where a and b are constants. Another way to represent this solution is to define the actin monomer concentration gradient, g,
implies a linear G-actin concentration profile, c = az + b, where a and b are constants. Another way to represent this solution is to define the actin monomer concentration gradient, g,
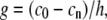 |
(13) |
where c0 and cn are the bulk and the tip actin monomer concentration, respectively, and h is the filopodial length.
In the steady state, the actin monomers consumed by the retrograde flow (the left-hand side in Eq. 14) are compensated by the net polymerization at the filopodial tip (the right-hand side in Eq. 14), which is sustained by the average actin diffusion flow (the right-hand side in Eq. 15), that is
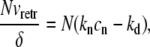 |
(14) |
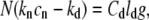 |
(15) |
where ld = 50 nm and Cd = 2000 s−1 are the size of our discretization compartments and the hopping rate between them, respectively. From the first equality, Eq. 14, we have
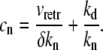 |
(16) |
Combining Eqs. 4 and 13–15, we obtain the steady-state filopodial length, h, as a function of the actin filament polymerization and depolymerization rates, k0 and kd, the retrograde flow rate, vretr, the monomeric actin concentration at the filopodial base, c0, and the membrane force, f,
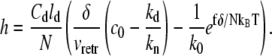 |
(17) |
We compare the mean field prediction of Eq. 17 with the numerical results from stochastic simulations, where from 100 to 1000 trajectories are averaged to produce 〈h〉 in the long time limit. In general, Eq. 17 agrees quite well with the stochastic simulation results; however, significant deviations show up in certain parameter regimes due to 1), the fluctuations considered in the numerical model but not in the mean field approximation; and 2), the discretization in the space coordinate along the filopodium in the numerical computation. We find that one of the main effects of fluctuations is to renormalize the parameters that enter the mean field solution (Eq. 17). More detailed discussion of the reasons for the observed discrepancies is given below.
Stochastic simulations
One of the main goals of this work is to observe and rationalize the temporal evolution of the filament lengths as physical parameters in the model are varied. We show below that the asymptotic average length of the filaments is a good observable to characterize the efficiency of the filopodial growth. All the calculations were carried out with the Gillespie algorithm (94). From 100 to 1000 trajectories were generated for each set of parameter values to achieve statistical convergence of trajectory analysis. The corresponding trajectory averages and variances are marked as circles in the reported plots. The parameter values and initial conditions are listed in Table 1. In the following calculations, when one of the parameters is varied, other parameters are kept at the values shown (Table 1). Spatially resolved stochastic simulations are computationally intensive—we ran our program in parallel on 128 processors (2.3 GHz Intel EM64T) for 10 days to generate all the necessary data, reported below.
TABLE 1.
Default parameter values used in this work
| Half actin monomer size | δ = 2.7 nm |
| Number of filaments | nf = 16 |
| Bulk concentration | 10 μM (five actin monomers/per compartment) |
| Diffusion rate | 5 μm2s−1(2000 s−1) |
| Thermal energy | kBT = 4.1 pN × nm |
| Membrane force | f = 10 pN |
| Polymerization rate | k0 = 10 μM−1s−1(21.8 s−1) |
| Depolymerization rate | 1.4 s−1 |
| Membrane fluctuation | σd = 10 nm |
| Retrograde flow speed | vretr = 70 nm/s |
| Initial length | 81 nm |
The grid size and the compartment volume were fixed in our computations; thus, the corresponding reaction rates and the protein diffusion rates are also given in “second” units in parenthesis.
Individual stochastic trajectories, along with the ensemble averaged trajectory, are shown in Fig. 3 a. A filopodium grows very quickly in the beginning then monotonically reaches a limiting value in a long time limit where the filament growth due to the polymerization at the barbed end is balanced against the retrograde flow. In a prior deterministic approach (19), filopodial average length was predicted to grow linearly with time at short times followed by 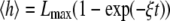 growth in the asymptotically long time limit. In this work, in contrast, we did not observe a linear time dependence at the initial phase or a subsequent intermediate growth phase. Instead, we find that the whole growth curve, from the initial segment onward, is nearly exactly described by the following expression:
growth in the asymptotically long time limit. In this work, in contrast, we did not observe a linear time dependence at the initial phase or a subsequent intermediate growth phase. Instead, we find that the whole growth curve, from the initial segment onward, is nearly exactly described by the following expression:
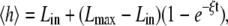 |
(18) |
where Lin = 112 nm, Lmax = 612 nm, and ξ = 0.08 s−1. Lin corresponds to our first recording of the stochastic trajectory at t = 0.5 s, which starts from an initial protrusion of 81 nm at t = 0 s. The filopodial growth speed, v ≈ e−ξt, which quickly diminishes at timescales above 12 s, may be thought of as having been derived from the following effective differential equation dv/dt ≈ −ξv. Thus, filopodial growth is analogous to the motion of a particle having a certain initial impulse in a viscous medium where the particle eventually comes to rest, indicating that ξ plays the role of an effective friction constant for the growth process.
FIGURE 3.
(a) Time evolution of the filopodial lengths obtained from individual trajectories. The trajectory average is shown with a thick solid line. The fit of Eq. 18 (solid line) to the ensemble-averaged filopodial growth curve (circles) is shown in the inset. (b) The probability distribution of filopodial lengths in the long time limit.
We next discuss the distribution of filament lengths in the long time limit, i.e., the asymptotic distribution. For the parameter values we used, at times longer than t = 300 s (600 s for longer filopodia), the system has already reached the steady state. If the filaments were to grow in the absence of the membrane and under constant G-actin concentration, the variance of the filament length distribution could grow large (35). However, membrane load preferentially diminishes the growth of longest filaments, decreasing the filament length variance. Similarly, tips of longer filaments are surrounded by a decreased number of G-actin monomers (Fig. 4 a), which also diminishes their polymerization rates compared with shorter filaments. Both of these effects act as negative feedback loops, resulting in a narrow filament length asymptotic distribution centered around 612 nm (Fig. 3 a). The distribution is slightly asymmetric; when the distribution mean and the variance are used to define a Gaussian, an appreciable deviation from the Gaussian behavior is observed (dashed line in Fig. 3 a). The asymmetry is attributed to the discrete noise and nonlinearity in the model.
FIGURE 4.
(a) G-actin monomer numbers along the filopodium at t = 300 s averaged over 1000 trajectories. Error bars indicate the amplitude of typical fluctuations. (b) 100 individual trajectories are shown. Filopodial tip is positioned at z = 0 nm, and the base is positioned near 625 nm (at t = 300 s).
Without the membrane force and the decreasing concentration of actin monomer along the filopodium, the length distribution of long filament is exponential (98), but the interfilament attractions could narrow the length distribution considerably (99). This is probably a mechanism for the filopodial precursor initiation in the cytosol (30). Although the prior deterministic approach, where only the average length of the filament bundle was considered (19), is consistent with the narrow filament length distribution obtained in this work, the exact values of the average filopodial length differ between the stochastic and the deterministic calculations because of coupling between model nonlinearity and fluctuations. In particular, the average number of G-action molecules at the filopodial tip compartment is found between 0.3 and 2; thus, discrete noise is fundamentally present at the microscopic level. The observed narrowness of the length distribution, on the other hand, motivates us to focus mainly on the filopodial average length as a function of various physical parameters.
Next, we examine the way the average steady-state length 〈h〉 depends on the G-actin diffusion constant (Fig. 5). As one would expect, 〈h〉 increases with the G-actin diffusion rate cdiff (Fig. 5 a). The dashed line in the figure is from the mean field approximation (Eq. 17). The major linear dependence of 〈h〉 on Cd is certainly captured. In addition, given the small copy number of actin molecules in filopodial compartments, one might expect significant fluctuations in monomeric G-actin in the filopodial tube. Indeed, whereas the G-actin ensemble average shows the expected behavior (an almost linear G-actin concentration profile, Fig. 4 a), individual simulation runs indicate significant diffusional noise (Fig. 4 b). For instance, near the filopodial tip, the number of G-actin monomers frequently drops to exactly zero when steady state is reached (Fig. 4 b). We anticipate that when significant stochastic noise is injected into filopodial dynamics by including important signaling regulators, such as capping proteins or formins, some nontrivial stochastic correlations may result between these noise sources and the monomeric G-actin diffusional noise.
FIGURE 5.
Dependence of the average filopodial equilibrium length, 〈h〉, on the monomeric G-actin diffusion constant, Cd. Circles represent the ensemble average obtained from 100 Gillespie trajectories, and the dashed line represents the mean field solution (Eq. 17).
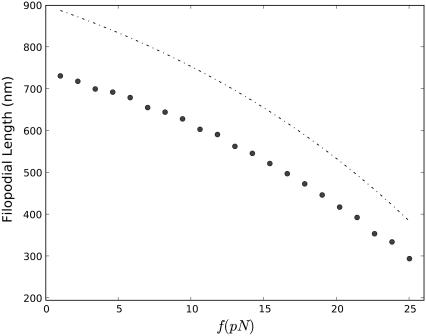
Generally, filopodial length decreases with increasing membrane force, as shown in Fig. 6. In the neighborhood of the zero force, however, the decrease is relatively slow, which indicates that within a certain range of small forces, 〈h〉 depends weakly on the membrane force. This is clearly seen from the mean field approximation (Eq. 17). Since the membrane load is shared among 16 filaments, small forces perturb filament polymerization rates only weakly. A similar point has also been made in prior works (19,23,62).
FIGURE 6.
Dependence of the average filopodial equilibrium length, 〈h〉, on the average membrane force, f, on the filament bundle tip. Circles represent the ensemble average obtained from 100 Gillespie trajectories, and the dashed line represents the mean field solution (Eq. 17).
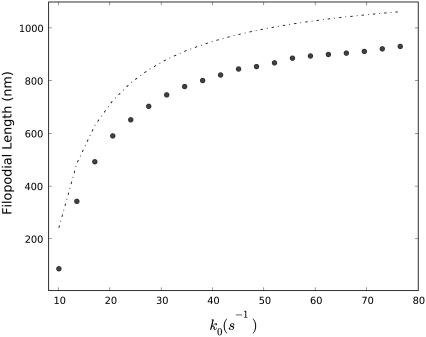
The dependence of the filopodial average length on the polymerization rate is shown in Fig. 7 a. 〈h〉 initially increases quickly with increasing bare polymerization rate k0 (see Eq. 4). However, when k0 reaches between 20 s−1 and 40 s−1, the 〈h〉 curve starts to approach a limiting value (Fig. 7 a). Thus, after this critical k0 value, the increase of k0 has little effect on 〈h〉. At this point, the polymerization at the tip becomes diffusion limited. Interestingly, the cell works near k0 ∼ 20 s−1, a value just below the point where the 〈h〉(k0) curve bends, perhaps to maximize the growth rate without sacrificing efficiency. The dashed line indicates the mean field prediction using Eq. 7. Although lifted as a whole from the Gillespie simulation, it correctly gives the general tendency.
FIGURE 7.
Dependence of the average filopodial equilibrium length, 〈h〉, on the bare polymerization rate, k0, at the barbed end. Circles represent the ensemble average obtained from 100 Gillespie trajectories, and the dashed line represents the mean field solution (Eq. 17).
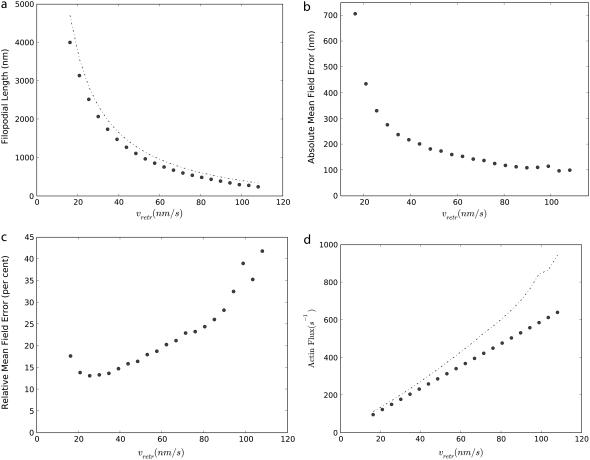
Next, we examine the effect of the retrograde flow. We found that 〈h〉 quickly increases with decreasing the speed of the retrograde flow rate (Fig. 8 a). This sensitive dependence suggests that regulating the retrograde flow rate may be a very effective strategy to control the filopodia growth and retraction. Again, the dashed line plots the analytical solution (Eq. 17). In terms of absolute error in 〈h〉, Eq. 17 matches well with the Gillespie trajectory averages for large vretrs but deviates significantly for small vretrs (Fig. 8 b). The absolute error in length is between 100 nm and 700 nm. When the relative error is plotted, Δh/h (Fig. 8 c), it is apparent that in shorter filopodia that the average lengths are overestimated from 30–50% when using the mean field (Eq. 17). Our detailed analysis of stochastic trajectories (data not shown) indicated that Eqs. 14 and 15 agree with stochastically obtained averages to high degree of accuracy, when cn and kn used in these equations were computed by ensemble averaging of stochastic trajectories. The main discrepancy arises because of the inadequacy of the diffusion equation (Eq. 12) to describe noisy G-actin transport from the filopodial base to the tip, in shorter filopodia. In particular, when actin flux is computed from the mean field (Eq. 13), it is significantly overestimated compared with the flux computed numerically (Fig. 8 d).
FIGURE 8.
(a) Dependence of the average filopodial equilibrium length, 〈h〉, on the speed of the retrograde flow, vretr, at the pointed end. Circles represent the ensemble average obtained from 1000 Gillespie trajectories, and the dashed line represents the mean field solution (Eq. 17). (b) The absolute mean field error defined as the difference between the mean field and exact solutions. (c) The relative difference between the mean field and exact solutions (absolute error divided by filopodial length). (d) Actin flux computed from stochastic simulations (circles) and the mean field (Eq. 13; dashed line).
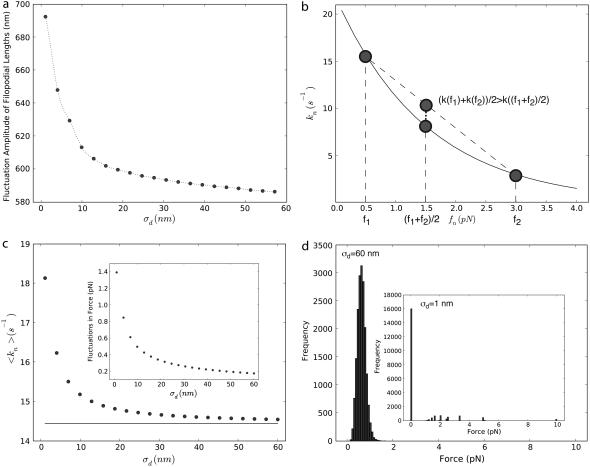
The effect of changing the membrane stiffness, σd, is shown in Fig. 9 a. In the whole computed parameter range, the filopodial length, 〈h〉, significantly increases as the membrane stiffness increases. The following line of reasoning explains this observation. When the amplitude of membrane fluctuations is small, forces acting on different filaments are unequal, resulting in unequal tip polymerization rates. On the other hand, when membrane fluctuations are large, the loading force is distributed nearly equally among filaments, resulting in very similar polymerization rates. Since the polymerization rate is a convex function of the force (Fig. 9, b and Eq. 4), the second scenario (large σd) leads to smaller average growth rates, resulting in shorter filopodia. To test this hypothesis, we computed the ensemble average of individual filament polymerization rates as a function of membrane stiffness from stochastic simulations. Indeed, 〈kn〉 decreases as σd increases (Fig. 9 c). The inset shows that force fluctuations among individual filaments also decrease with increasing σd, as expected (Fig. 9 c). When force distribution among filaments is compared between σd = 60 nm and σd = 1 nm simulations, it is apparent that in the former case filaments nearly equally bear the membrane load, and in the latter case only a few filaments are under stress; the large majority are stress free, and thus grow quickly (Fig. 9 d). Therefore, this analysis confirms that because of the convex nature of the polymerization rate curve as a function of membrane force, small amplitude fluctuations can significantly speed up filopodial growth rates compared with the mean field result.
FIGURE 9.
(a) Dependence of the filamental average length on the membrane fluctuation amplitude, σd. (b) Polymerization rate at the filament's barbed end as a function of force load. For the same average load, unequal distribution of forces results in faster average growth rates. (c) Ensemble average of individual polymerization rates, 〈kn〉, obtained from 1000 Gillespie trajectories. The solid line represents the mean field result, 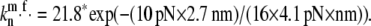 The inset indicates the force fluctuations among individual filaments,
The inset indicates the force fluctuations among individual filaments, 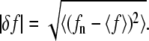 (d) Distribution of individual filament forces for σd = 60 nm and σd = 1 nm (inset).
(d) Distribution of individual filament forces for σd = 60 nm and σd = 1 nm (inset).
Here we considered only the growth of a matured filopodium. At the initiation stage, the membrane stiffness might play an even more important role (4,30,34). In addition to the filopodial length distribution average, we examined the dependence of the filopodial length distribution variance on σd (data not shown). Within the biologically relevant range of σd, the variance is rather small. Thus, it is unlikely that long single filaments grow without other filaments catching up. If this scenario was realized, at some point the filament would buckle, inducing abnormal conformational changes or, perhaps, collapse of the filopodium.
DISCUSSION
In this work, we proposed a computational model to explore the filopodium dynamics that combines complex mechanical, physical, and biochemical processes. In contrast to prior works, our model provides a full stochastic treatment of the polymerization and depolymerization of each individual actin filament under the stress of the fluctuating membrane, where actin monomer diffusion and retrograde flow are key model components. We examined the dependence of the filopodial length on the membrane force, polymerization rate, speed of the retrograde flow, and monomeric actin diffusion constant. We found that for the biologically relevant parameter values, the length distribution of actin filaments in one filopodium has a narrow width, as a consequence of the negative feedback created by the membrane load and monomeric G-actin gradient. Thus, the average filopodial length is a good parameter to characterize the filopodial growth under normal conditions. We also discovered that filopodial growth sensitively depends on the retrograde flow rate. The filopodial length significantly decreases as the membrane fluctuations increase, which we attributed to the convex shape of the rate of the filament tip polymerization as a function of loading force.
We derived a mean field approximation to filopodial length at steady state and compared the results of our extensive stochastic simulations with the mean field predictions. Overall, the mean field solution captured major trends, albeit with significant numerical discrepancies in some cases. Although the filament length fluctuations turned out to be small, they still significantly influenced the growth dynamics: 1), unequal loading of membrane force among individual filaments leads to faster filopodial tip polymerization rates, thus longer filopodia; and 2), very strong diffusional noise of G-actin monomer transport (only 0.3 G-actin molecules are present at the tip in some simulations) results in G-actin fluxes that are smaller than predicted from the diffusion equation. Since these two effects work in opposite directions, a partial cancellation of errors occurs that improves the accuracy of the mean field solution. However, the parameters of the mean field model become significantly renormalized by the fast fluctuations. With bare parameters, the error of the mean field filopodial length prediction can reach nearly 50%.
On the other hand, our model indicates that the large amplitude filopodial length fluctuations, observed in the in vivo experiments, cannot be accounted for by the minimally embedded stochastic processes that include actin polymerization and depolymerization and G-actin diffusion. Our work suggests that these fluctuations are induced by external or internal chemical cues through additional cell-signaling cascades. It is believed that the filopodial growth and retraction is controlled largely by the protein complex located at its tip. The role of an external cue can be played by chemical ligands or mechanical tension. The signaling molecules are transmitted along the filopodium by diffusion, retrograde flow, or motor proteins (3,100). Although much progress has been made in characterizing the filopodial machinery (14,15,26,29,31,101), the exact composition and function of the signaling pathways that regulate filopodial growth is still not entirely clear (4).
The model reported in this work does not include signaling by regulatory proteins or binding of cross-linking proteins, however, it provides a platform for further, more involved and realistic modeling. Our computations already hint what could be the key processes that should be regulated by signaling molecules. For example, we suggest that regulating the retrograde flow rate would be a highly efficient way to control filopodial extension dynamics. In an ongoing investigation, we are building into our model a number of additional regulatory proteins, such as CAPZ, Ena/Vasp, formin, profilin, and cofilin, considering their transportation along the filopodial protrusion and interaction with the G-actin monomers and actin filaments. For some of these large protein molecules, at low copy numbers, stochastic modeling is essential. The dynamics of the retrograde flow (17), the regulation of the membrane force (76), and their influence on the filopodial growth also need to be further investigated. Finally, close examination of physicochemical experiments on filopodia will point to additional important biochemical processes that need to be included in the model.
Acknowledgments
We are grateful to Dr. Steve Rogers and Pavel Zhuravlev for stimulating discussions. We thank Dr. Michael Rubinstein for critically reading the manuscript and providing helpful comments. All calculations were carried out using the UNC Topsail supercomputer.
This work was supported through National Science Foundation grant 0715225.
Editor: Alexander Mogilner.
References
- 1.Argiro, V., B. Bunge, and M. I. Johnson. 1985. A quantitative study of growth cone filopodial extension. J. Neurosci. Res. 13:149–162. [DOI] [PubMed] [Google Scholar]
- 2.Wicki, A., F. Lehembre, N. Wick, B. Hantusch, D. Kerjaschki, and G. Christofori. 2006. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of cytoskeleton. Cancer Cell. 9:261–272. [DOI] [PubMed] [Google Scholar]
- 3.Lidke, D. S., K. A. Lidke, B. Rieger, T. M. Jovin, and D. J. Arndt-Jovin. 2005. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J. Cell Biol. 170:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faix, J., and K. Rottner. 2006. The making of filopodia. Curr. Opin. Cell Biol. 18:18–25. [DOI] [PubMed] [Google Scholar]
- 5.Goodhill, G. J., M. Gu, and J. S. Urbach. 2004. Predicting axonal response to molecular gradients with a computational model of filopodial dynamics. Neural Comput. 16:2221–2243. [DOI] [PubMed] [Google Scholar]
- 6.Gustafson, T., and L. Wolpert. 1961. Studies on the cellular basis of morphogenesis in the sea urchin embryo: directed movements of primary mesenchyme cells in normal and vegetalized larvae. Exp. Cell Res. 24:64–79. [DOI] [PubMed] [Google Scholar]
- 7.Jacinto, A., W. Wood, T. Balayo, M. Turmaine, A. Martinez-Arias, and P. Martin. 2000. Dynamic actin-based epithelial adhesion and cell matching during Drosophila dorsal closure. Curr. Biol. 10:1420–1426. [DOI] [PubMed] [Google Scholar]
- 8.Wood, W., A. Jacinto, R. Grose, S. Woolner, J. Gale, C. Wilson, and P. Martin. 2002. Wound healing recapitulates the morphogenesis in Drosophila embryos. Nat. Cell Biol. 4:907–912. [DOI] [PubMed] [Google Scholar]
- 9.Wood, W., and P. Martin. 2002. Structures in focus-filopodia. Int. J. Biochem. Cell Biol. 34:726–730. [DOI] [PubMed] [Google Scholar]
- 10.Hsu, T., W. Liao, P. Yang, C. Wang, J. Xiao, and C. Lee. 2007. Dynamics of cancer cell filopodia characterized by super-resolution bright-field optical microscopy. Opt. Express. 15:76–82. [DOI] [PubMed] [Google Scholar]
- 11.Discher, D. E., P. Janmey, and Y.-L. Wang. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science. 310:1139–1143. [DOI] [PubMed] [Google Scholar]
- 12.Medeiros, N. A., D. T. Burnette, and P. Forscher. 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8:215–226. [DOI] [PubMed] [Google Scholar]
- 13.Schirenbeck, A., R. Arasada, T. Bretschneider, T. E. B. Stradal, M. Schleicher, and J. Faix. 2006. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc. Natl. Acad. Sci. USA. 103:7694–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steffen, A., J. Faix, G. P. Resch, J. Linkner, J. Wehland, J. V. Small, K. Rottner, and T. E. B. Stradal. 2006. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol. Biol. Cell. 17:2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mejillano, M. R., S. Kojima, D. A. Applewhite, F. B. Gertler, T. M. Svitkina, and G. G. Borisy. 2004. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 118:363–373. [DOI] [PubMed] [Google Scholar]
- 16.Tokuo, H., and M. Ikebe. 2004. Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 319:214–220. [DOI] [PubMed] [Google Scholar]
- 17.Heid, P. J., J. Geiger, D. Wessels, E. Voss, and D. R. Soll. 2005. Computer-assisted analysis of filopod formation and the role of myosin II heavy chain phosphorylation in Dictyostelium. J. Cell Sci. 118:2225–2237. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann, C., A. Finski, and T. Bonhoeffer. 2005. Local calcium transients regulate the spontaneous motility of dendritic filopodia. Nat. Neurosci. 8:305–312. [DOI] [PubMed] [Google Scholar]
- 19.Mogilner, A., and B. Rubinstein. 2005. The physics of filopodial protrusion. Biophys. J. 89:782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robles, E., S. Woo, and T. M. Gomez. 2005. Src-dependent tyrosine phosphorylation at the tips of growth cone filopodia promotes extension. J. Neurosci. 25:7669–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schirenbeck, A., R. Arasada, T. Bretschneider, M. Schleicher, and J. Faix. 2005. Formins and VASPs may co-operate in the formation of filopodia. Biochem. Soc. Trans. 33:1256–1259. [DOI] [PubMed] [Google Scholar]
- 22.Schirenbeck, A., T. Bretschneider, R. Arasada, M. Schleicher, and J. Faix. 2005. The diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 7:619–625. [DOI] [PubMed] [Google Scholar]
- 23.Atilgan, E., D. Wirtz, and S. X. Sun. 2006. Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys. J. 90:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connor, T. P., J. S. Duerr, and D. Bentley. 1990. Pioneer growth cone steering decisions mediated by single filopodium contacts in situ. J. Neurosci. 10:3935–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, D.-Y., and D. J. Goldberg. 1993. Regulated tyrosine phosphorylation at the tips of growth cone filopodia. J. Cell Biol. 123:653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallavarapu, A., and T. Mitchison. 1999. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 146:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krugmann, S., I. Jordens, K. Gevaert, M. Driessens, J. Vandekerckhove, and A. Hall. 2001. Cdc42 induces filopodia by promoting the formation of an IRSp53:Mena complex. Curr. Biol. 11:1645–1655. [DOI] [PubMed] [Google Scholar]
- 28.Berg, J. S., and R. E. Cheney. 2002. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat. Cell Biol. 4:246–250. [DOI] [PubMed] [Google Scholar]
- 29.Biyasheva, A., T. Svitkina, P. Kunda, B. Baum, and G. Borisy. 2003. Cascade pathway of filopodia formation downstream of SCAR. J. Cell Sci. 117:837–848. [DOI] [PubMed] [Google Scholar]
- 30.Svitkina, T. M., E. A. Bulanova, O. Y. Chaga, D. M. Vignjevic, S. Kojima, J. M. Vasiliev, and G. G. Borisy. 2003. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, T. Y., C. DerMardirossian, and G. M. Bokoch. 2006. Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 18:26–31. [DOI] [PubMed] [Google Scholar]
- 32.Kovar, D. R. 2006. Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 18:11–17. [DOI] [PubMed] [Google Scholar]
- 33.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- 34.Daniels, D. R., and M. S. Turner. 2005. Spicules and the effect of rigid rods on enclosing membrane tubes. Phys. Rev. Lett. 95:238101. [DOI] [PubMed] [Google Scholar]
- 35.Stukalin, E. B., and A. B. Kolomeisky. 2006. ATP hydrolysis stimulates large length fluctuations in single actin filaments. Biophys. J. 90:2673–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mogilner, A. 2006. On the edge: modeling protrusion. Curr. Opin. Cell Biol. 18:32–39. [DOI] [PubMed] [Google Scholar]
- 37.Mogilner, A., and G. Oster. 2003. Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys. J. 84:1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peskin, C. S., G. M. Odell, and G. F. Oster. 1993. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 65:316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prost, J., C. Barbetta, and J. F. Joanny. 2007. Dynamical control of the shape and size of stereocilia and microvilli. Biophys. J. 93:1124–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lan, Y., P. Wolynes, and G. A. Papoian. 2005. A variational approach to the stochastic aspects of cellular signal transduction. J. Chem. Phys. 125:124106. [DOI] [PubMed] [Google Scholar]
- 41.Lan, Y., and G. A. Papoian. 2006. The interplay between discrete noise and nonlinear chemical kinetics in a signal amplification cascade. J. Chem. Phys. 125:154901. [DOI] [PubMed] [Google Scholar]
- 42.Lan, Y., and G. A. Papoian. 2007. Stochastic resonant signaling in enzyme cascades. Phys. Rev. Lett. 98:228301. [DOI] [PubMed] [Google Scholar]
- 43.Kepler, T. B., and T. C. Elston. 2001. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys. J. 81:3116–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kærn, M., T. C. Elston, W. J. Blake, and J. J. Collins. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6:451–464. [DOI] [PubMed] [Google Scholar]
- 45.Sasai, M., and P. G. Wolynes. 2003. Stochastic gene expression as a many-body problem. Proc. Natl. Acad. Sci. USA. 100:2374–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walczak, A. M., J. N. Onuchic, and P. G. Wolynes. 2005. Absolute rate theories of epigenetic stability. Proc. Natl. Acad. Sci. USA. 102:18926–18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walczak, A. M., M. Sasai, and P. G. Wolynes. 2005. Self-consistent proteomic field theory of stochastic gene switches. Biophys. J. 88:828–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu, T., T. Shen, C. Zong, J. Hasty, and P. G. Wolynes. 2006. Statistics of cellular signal transduction as a race to the nucleus by multiple random walkers in compartment/phosphorylation space. Proc. Natl. Acad. Sci. USA. 103:16752–16757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao, C. V., D. M. Wolf, and A. P. Arkin. 2002. Control, exploitation and tolerance of intracellular noise. Nature. 420:231–237. [DOI] [PubMed] [Google Scholar]
- 50.Weinberger, L. S., J. C. Burnett, J. E. Toettcher, A. P. Arkin, and D. V. Schaffer. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: Hiv-1 tat fluctuation drive phenotypic diversity. Cell. 122:169–182. [DOI] [PubMed] [Google Scholar]
- 51.Thattai, M., and A. van Oudenaarden. 2001. Intrinsic noise in gene regulatory networks. Proc. Natl. Acad. Sci. USA. 98:8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozbudak, E. M., M. Thattai, I. Kurtser, A. D. Grossman, and A. van Oudenaarden. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31:69–73. [DOI] [PubMed] [Google Scholar]
- 53.Thattai, M., and A. van Oudenaarden. 2002. Attenuation of noise in ultrasensitive signaling cascades. Biophys. J. 82:2943–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thattai, M., and A. van Oudenaarden. 2004. Stochastic gene expressions in fluctuating environments. Genetics. 167:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hasty, J., M. Dolnik, V. Rottschäfer, and J. J. Collins. 2002. Synthetic gene network for entraining and amplifying cellular oscillations. Phys. Rev. Lett. 88:148101. [DOI] [PubMed] [Google Scholar]
- 56.Brastsun, D., D. Volfson, L. S. Tsimring, and J. Hasty. 2005. Delay-induced stochastic oscillations in gene regulation. Proc. Natl. Acad. Sci. USA. 102:14593–14598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Haastert, P. V., and M. Postma. 2007. Biased random walk by stochastic fluctuations of chemoattractant-receptor interactions at the lower limit of detection. Biophys J. 1:1787–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forscher, P., and S. J. Smith. 1988. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 107:1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gov, N. S. 2006. Dynamics and morphology of microvilli driven by actin polymerization. Phys. Rev. Lett. 97:018101. [DOI] [PubMed] [Google Scholar]
- 60.Pantaloni, D., C. Le Clainche, and M.-F. Carlier. 2001. Mechanism of actin-based motility. Science. 292:1502–1506. [DOI] [PubMed] [Google Scholar]
- 61.van Kampen, N. G. 1992. Stochastic Processes in Physics and Chemistry. North Holland Personal Library, Amsterdam.
- 62.Carlsson, A. E. 2001. Growth of branched actin networks against obstacles. Biophys. J. 81:1907–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charras, G. T., J. C. Yarrow, M. A. Horton, L. Mahadevan, and T. J. Mitchison. 2005. Non-equilibration of hydrostatic pressure in blebbing cells. Nature. 435:365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shoemaker, S. D., and T. K. Vanderlick. 2002. Intramembrane electrostatic interactions destabilize lipid vesicles. Biophys. J. 83:2007–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morris, C. E., and U. Homann. 2001. Cell surface area regulation and membrane tension. J. Membr. Biol. 179:79–102. [DOI] [PubMed] [Google Scholar]
- 66.Morris, C. E. 2002. How did cells get their size? Anat. Rec. 268:239–251. [DOI] [PubMed] [Google Scholar]
- 67.Raucher, D., and M. Sheetz. 2000. Cell spreading and lamellipodial extension rate is regulated by membrane tension. J. Cell Biol. 148:127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sheetz, M. P. 2001. Cell control by membrane-cytoskeleton adhesion. Nat. Rev. Mol. Cell Biol. 2:392–396. [DOI] [PubMed] [Google Scholar]
- 69.Sens, P., and M. S. Turner. 2006. Budded membrane microdomains as tension regulators. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 73:031918. [DOI] [PubMed] [Google Scholar]
- 70.Parton, R. G., and K. Simons. 2007. The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 8:185–194. [DOI] [PubMed] [Google Scholar]
- 71.Lin, L. C.-L., and F. L. H. Brown. 2004. Brownian dynamics in Fourier space: membrane simulations over long length and time scales. Phys. Rev. Lett. 93:256001. [DOI] [PubMed] [Google Scholar]
- 72.Pécréaux, J., H.-G. Döbereiner, J. Prost, J.-F. Joanny, and P. Bassereau. 2004. Refined contour analysis of giant unilamellar vesicles. Eur. Phys. J. E. 13:277–290. [DOI] [PubMed] [Google Scholar]
- 73.Gov, N. S., and S. A. Safran. 2005. Red blood cell membrane fluctuations and shape controlled by ATP-induced cytoskeletal defects. Biophys. J. 88:1859–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Safran, S., N. Gov, A. Nicolas, U. Schwarz, and T. Tlusty. 2005. Physics of cell elasticity, shape and adhesion. Physica A. 352:171–201. [Google Scholar]
- 75.Zeman, K., H. Engelhard, and E. Sackmann. 1990. Bending undulations and elasticity of the erythrocyte membrane: effects of cell shape and membrane organization. Eur. Biophys. J. 18:203–219. [DOI] [PubMed] [Google Scholar]
- 76.Khan, S., and M. P. Sheetz. 1997. Force effects on biochemical kinetics. Annu. Rev. Biochem. 66:785–805. [DOI] [PubMed] [Google Scholar]
- 77.Seifert, U. 1997. Configurations of fluid membranes and vesicles. Adv. Phys. 46:13–137. [Google Scholar]
- 78.Dai, J., M. P. Sheetz, X. Wan, and C. E. Morris. 1998. Membrane tension in swelling and shrinking molluscan neurons. J. Neurosci. 18:6681–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hirn, R., T. M. Bayerl, J. O. Rädler, and E. Sackmann. 1998. Collective membrane motions of high and low amplitude, studied by dynamic light scattering and micro-interferometry. Faraday Discuss. 111:17–30. [DOI] [PubMed] [Google Scholar]
- 80.Dai, J., and M. P. Sheetz. 1999. Membrane tether formation from blebbing cells. Biophys. J. 77:3363–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raucher, D., and M. P. Sheetz. 1999. Characteristics of a membrane reservoir buffering membrane tension. Biophys. J. 77:1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gov, N., A. G. Zilman, and S. Safran. 2003. Cytoskeleton confinement and tension of red blood cell membranes. Phys. Rev. Lett. 90:228101. [DOI] [PubMed] [Google Scholar]
- 83.Döbereiner, H.-G., B. Dubin-Thaler, G. Giannone, H. S. Xenias, and M. P. Sheetz. 2004. Dynamic phase transitions in cell spreading. Phys. Rev. Lett. 93:108105. [DOI] [PubMed] [Google Scholar]
- 84.Döbereiner, H.-G., B. J. Dubin-Thaler, G. Giannone, and M. P. Sheetz. 2005. Force sensing and generation in cell phases: analyses of complex functions. J. Appl. Physiol. 98:1542–1546. [DOI] [PubMed] [Google Scholar]
- 85.Girard, P., J. Prost, and P. Bassereau. 2005. Passive or active fluctuations in membranes containing proteins. Phys. Rev. Lett. 94:088102. [DOI] [PubMed] [Google Scholar]
- 86.Sheetz, M. P., J. E. Sable, and H.-G. Döbereiner. 2006. Continuous membrane-cytoskeleton adhesion requires continuous accommodation to lipid and cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct. 35:417–434. [DOI] [PubMed] [Google Scholar]
- 87.Solon, J., J. Pécréaux, P. Girard, M.-C. Fauré, J. Prost, and P. Bassereau. 2006. Negative tension induced by lipid uptake. Phys. Rev. Lett. 97:098103. [DOI] [PubMed] [Google Scholar]
- 88.Giannone, G., B. J. Dubin-Thaler, O. Rossier, Y. Cai, O. Chaga, G. Jiang, W. Beaver, H.-G. Döbereiner, Y. Freund, G. Borisy, and M. P. Sheetz. 2007. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 128:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Safran, S. A. 2003. Statistical Thermodynamics of Surfaces, Interfaces, and Membranes. Westview Press, Boulder, CO.
- 90.Farago, O., and P. Pincus. 2004. Statistical mechanics of bilayer membrane with a fixed projected area. J. Chem. Phys. 120:2934–2950. [DOI] [PubMed] [Google Scholar]
- 91.Gov, N. S., and A. Gopinathan. 2006. Dynamics of membranes driven by actin polymerization. Biophys. J. 90:454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin, L. C.-L., N. Gov, and F. L. H. Brown. 2006. Nonequilibrium membrane fluctuations driven by active proteins. J. Chem. Phys. 124:74903. [DOI] [PubMed] [Google Scholar]
- 93.Strey, H., M. Peterson, and E. Sackmann. 1995. Measurement of erythrocyte membrane elasticity by flicker eigenmode decomposition. Biophys. J. 69:478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gillespie, D. T. 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81:2340–2361. [Google Scholar]
- 95.Brown, M. E., and P. C. Bridgman. 2002. Retrograde flow rate is increased in growth cones from myosin IIB knockout mice. J. Cell Sci. 116:1087–1094. [DOI] [PubMed] [Google Scholar]
- 96.Huckaba, T. M., T. Lipkin, and L. A. Pon. 2006. Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. J. Cell Biol. 175:957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Veksler, A., and N. S. Gov. 2007. Phase transitions of the coupled membrane-cytoskeleton modify cellular shape. Biophys. J. 93:3798–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gov, N. S. 2007. Theory of the length distributions of tread-milling actin filaments inside bundles. Europhys. Lett. 77:68005. [Google Scholar]
- 99.Biron, D., E. Moses, I. Borukhov, and S. A. Safran. 2006. Inter-filament interactions narrow the length distribution of actin filaments. Europhys. Lett. 73:464–470. [Google Scholar]
- 100.Faretta, M. R., and B. Bassetti. 1998. Active filaments dynamics in motility assays for motor proteins: pure and anomalous diffusion. Europhys. Lett. 41:689–694. [Google Scholar]
- 101.Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]