Abstract
Flagellin is the subunit of the bacterial filament, the micrometer-long propeller of a bacterial flagellum. The protein is believed to undergo unfolding for transport through the channel of the filament and to refold in a chamber at the end of the channel before being assembled into the growing filament. We report a thermal unfolding simulation study of S. typhimurium flagellin in aqueous solution as an attempt to gain atomic-level insight into the refolding process. Each molecule comprises two filament-core domains {D0, D1} and two hypervariable-region domains {D2, D3}. D2 can be separated into subdomains D2a and D2b. We observed a similar unfolding order of the domains as reported in experimental thermal denaturation. D2a and D3 exhibited high thermal stability and contained persistent three-stranded β-sheets in the denatured state which could serve as folding cores to guide refolding. A recent mutagenesis study on flagellin stability seems to suggest the importance of the folding cores. Using crude size estimates, our data suggests that the chamber might be large enough for either denatured hypervariable-region domains or filament-core domains, but not whole flagellin; this implicates a two-staged refolding process.
INTRODUCTION
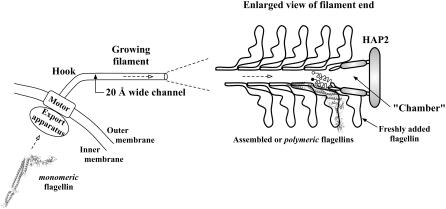
The flagellum is the motility device of bacterium, with the long tubelike filament used as a propeller constructed solely of one protein called “flagellin”. Flagellin of Salmonella typhimurium has four domains, with the terminal domain (D0) being highly conserved across bacterial strains (1) as it is involved in forming the inner tube of the filament (2). To reach the growing end of the filament, flagellin synthesized in the cytoplasm has to be exported though a narrow (20 Å diameter) channel that runs through the core of the filament (3). The conformation that flagellin adopts during transport remains a mystery. Fig. 1 shows a schematic diagram of flagellin export from the bacterium cytoplasm to the tip of the growing filament for assembly.
FIGURE 1.
Bacterial flagellin export. A flagellum with a few assembled flagellins near the tip of the filament is shown in this schematic diagram. The assembled or polymeric form of flagellin has been solved (PDB ID: 1UCU), from which the cytoplasmic or monomeric form with disordered termini helices has been obtained from our molecular dynamics simulation in solvent. The conformation of flagellin during transport through the channel is still unknown but is suggested to be highly unfolded due to the narrow channel cross section. Refolding then takes place in the chamber before assembly with the help of HAP2 chaperone. The newly added flagellin shown still has disordered termini helices, with solid circle representing the N-terminal.
Because the channel is too narrow for folded flagellin even if domain rearrangements are possible, it has been proposed that considerable unfolding is required before export can begin (3). The flagellar export system can be considered a variant of the Type-III secretion systems (4). In bacterial Type-III secretion systems, proteins are unfolded before export to the host cell through a needlelike conduit, with refolding occurring in the host cell (5). In many such translocons, the typically narrow (20–30 Å) channel requires secreted proteins to be in an unfolded state (6,7). For S. typhimurium flagellin, the small space below the filament cap measuring 70 Å deep by ∼40 Å across that is surrounded by domain D1 of the uppermost turn of assembled flagellin has been suggested to be where transported flagellin refolds before assembly into the filament (8).
The latest experimental investigation of flagellin-folding mechanism via thermal denaturation was carried out by Honda et al. (9) using differential scanning calorimetry (DSC) and circular dichroism (CD) melting measurements. They established that flagellin unfolds in stages in accord to its multidomain nature. By fitting a new theoretical model for multistate transitions to the DSC data, the authors obtained melting curves for each thermodynamic domain that is subsequently assigned to a unique structural domain via CD measurements. An unfolding order for the domains was suggested based on the melting temperatures. They have also characterized the interactions between domains.
In this article, we seek further understanding of the (re)folding pathway at the atomic level by performing in silico thermal unfolding. High temperature molecular dynamics (MD) simulations, when used in combination with experiments, affords a method to more fully describe the unfolding pathway (10–12). Relative stabilities of domains during unfolding have also been studied by such simulations (13). Use of elevated temperatures in simulations was shown not to grossly affect the unfolding pathway, as thermal denaturation is an activated process where lower energy barriers are surmounted first. The overall order of events is conserved across temperatures but their timescales do differ (14). Recent work from Day and Daggett showed that folding/unfolding from a structural nucleus obeys the principle of microscopic reversibility to a large extent, when performing simulations close to the melting temperature of the protein where folded and unfolded states are equally populated. Though the refolded protein at 200 ns is not identical to the crystal native state, it can be interpreted as the native state at the slightly elevated temperature (15). The effect of temperature on protein unfolding has been extensively discussed in the review article by Daggett (16). Thus, thermal unfolding at high temperatures affords a way to study the stability and kinetics involved in the folding process with reasonable computational effort despite possible bias/distortion to the pathway. Such bias can be reduced by running multiple simulations at a series of temperatures and taking an ensemble view of the process (17).
We have carried out MD simulation in aqueous solvent at 300 K (control) and five runs each at 400, 500, and 600 K to partially and fully denature the flagellin monomer. A similar order of unfolding as in the thermal denaturation experiment was found. We also found contacts within three-stranded β-sheets remaining for long periods of simulation times in the thermally denatured hypervariable-region (HVR) domains, suggestive of folding cores that would assist in the refolding of flagellin. The simulation results are used to interpret some of the findings in the thermal denaturation experiment (9) and in a mutagenesis study on flagellin mechanical stability (18).
MATERIALS AND METHODS
Simulation setup
The complete flagellin structure as adopted in the filament (PDB code: 1UCU) was taken as the starting coordinates and denoted as the polymeric form. We aligned the molecule with its longest extent along the z axis and immersed it in a periodic rectangular box of TIP3P waters with at least 120 Å between the protein atoms and box edge along the Z-direction and at least 8 Å in the X- and Y-directions. Such a large simulation box was initially meant for unfolding flagellin under applied force. AMBER 8 (19) with ff99 force field was used for all the simulations. Electrostatics was handled with the particle-mesh Ewald method (20) with a nonbonded real-space cutoff of 8 Å.
After energy minimization, the charge neutralized system was heated to 300 K, keeping restraints on the nonhydrogen atoms. Next, Berendsen temperature and pressure control was imposed (at 300 K and 1 atm) with restraints reduced in stages and turned off when equilibration has been reached. The system density approached the bulk solvent value after we activated SHAKE (21) to constrain motions of chemical bonds involving hydrogen. The flagellin conformation now obtained is denoted as the monomeric form.
For thermal denaturation studies, the equilibrated monomeric flagellin (with a 5 Å shell of surrounding solvent molecules) was resolvated in a wider rectangular box with at least 10 Å between the protein and box edges. The simulation box size becomes 216 Å × 105 Å × 84 Å and contains 159,162 atoms. The atoms were then reassigned velocities at 300 K and the system temperature increased to 600 K in stages, following the equilibration protocol as mentioned above. By setting different initial velocities, five independent sets of simulations were carried out at 400 K, 500 K, and 600 K for 8-ns, 6-ns, and-2 ns, respectively under the NVE condition (no temperature or pressure control) with a 1-fs time-step. It has been demonstrated that a small set of five to ten simulations are sufficient to capture the average properties and pathway during unfolding (17). A control simulation at 300 K with a 2-fs time-step was performed for 8 ns. We carried out these simulations with the PMEMD module of AMBER 8 on Itanium-2 computer clusters, taking 33 h/ns on 16 CPUs. Trajectory frames were saved every 0.5 ps, but resampled to 2-ps intervals for analysis.
RESULTS
Simulation reproduced monomeric flagellin with disordered termini
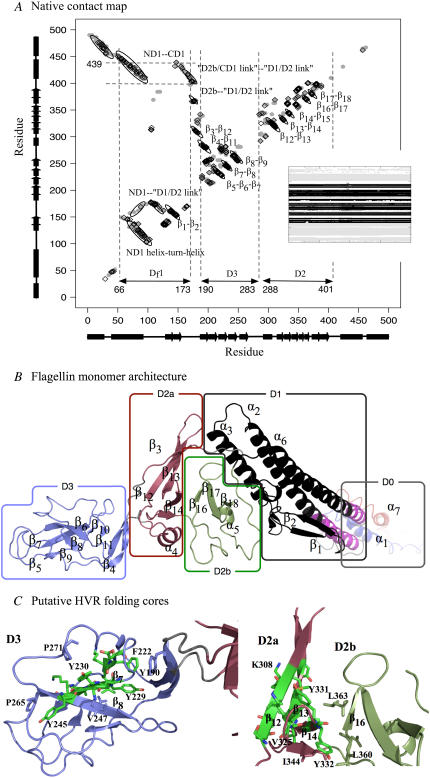
In Fig. 2 B, we show a model of the four-domain, 494-residue monomeric S. typhimurium flagellin with unstructured helices in domain D0, obtained from the polymeric form (1UCU) by running a short 1.4-ns equilibration MD simulation in solvent. Domains D0 and D1 are involved with formation of the inner and outer filament tubes respectively (filament-core domains). Domains D2 (composed of subdomains D2a and D2b) and D3 make up the HVR that is exposed on the filament exterior (3). The N-terminal part of D1 (ND1) contains α2, α3, β1, and β2. The C-terminal part (CD1) consists of α6 only. Similarly, ND0 and CD0 are made up of α1 and α7, respectively.
FIGURE 2.
(A) Residue contact map of native flagellin structure. Shaded circles indicate contacts in 1UCU. Black diamonds indicate persistent native contacts: contacts found in >70% of the snapshots taken from the last 1 ns of the control simulation as a monomer in solution. Dotted lines mark location of Df1 contact clusters (see Text). (B) The tertiary structure of flagellin monomer in solution obtained from MD simulation (starting from polymeric flagellin structure) showing DSSP-assigned α-helices and β-strands. Each (sub)domain is colored differently and labeled, with linkers colored gray. N-terminus (blue) and C-terminus (red) helices are disordered in the monomer. We have neglected the very small β-turn from residues 130 to 135 in our labeling scheme. Segment Df1 of D1 is in black, with the remaining portion in magenta. (C) The residues in the HVR domains (D2 and D3) forming persistent pairs of residue contacts in at least two of the five 500 K simulations are shown as sticks and colored by chemical element: carbon (green), oxygen (red), and nitrogen (blue). The β-sheets β6β7β8 and β12β13β14 might form folding cores. Molecular structures are rendered with PyMOL (52).
Limited proteolysis of monomeric flagellin showed the existence of a resistant central portion with disordered terminal region (22). The terminal helices are only marginally stable as determined from far-UV CD spectra (23) and NMR measurements (24), but became structured during filament assembly (25,26). The polymerization of monomeric flagellin into filaments made it difficult to obtain crystals of monomeric flagellin for x-ray structure determination. Hence, termini-truncated flagellin structure was first obtained by x-ray (PDB code: 1IO1) (27) and subsequently used as a guide to reconstruct the complete polymeric flagellin structure (PDB code: 1UCU) from cryo-EM density map of the filament (3). The contact map of 1UCU is shown in Fig. 2 A, computed using a program in the MMTSB Toolkit (28), which defines two residues to be in contact if their minimum heavy atom separation is <4.2 Å.
Persistent native monomer contacts from control simulation
The 8-ns MD simulation at 300 K served as a control for the higher temperature simulations. Using snapshots from the last 1-ns, we defined persistent native contacts as contacting residues that appeared in >70% of the snapshots. The fraction of such contacts will be used for monitoring the unfolding process. We have overlapped these persistent clusters on the 1UCU contact map where we can see a loss of intra- and interhelical contacts in D0 during the control simulation (Fig. 2 A). This observation is also reflected as a loss of α-helical content in the changes to DSSP (29) assigned secondary structures shown as an inset in the contact map. Contact clusters from β-stranded pairs dominate the contact map, but those between D0 and D1 helices and between unstructured domain linkers are also present.
D1-D2 interfacial H-bond network
Solvent molecules in the simulation could not penetrate deep enough into the D2a-ND1 interfacial space for most part of the 300 K 8-ns control simulation, resulting in strong bridging hydrogen bonds between side chains of D313 (in D2a) and S106/S104 (in ND1). The solvent-mediated H-bond network appeared only after 7.5 ns when interdomain arrangement between D1 and D2 changed. Under higher temperatures, however, solvent penetration occurred faster (results not shown).
Order of domain unfolding
For temperatures >300 K, unfolding of flagellin monomer was observed. ASAView (30) plots, which arrange residues on a spiral according to their relative solvent accessibility surface area (SASA), are an easy way to visualize the effect of thermal denaturation on a protein/domain (see Fig. S1 in the Supplementary Material, Data S1). For instance, a narrow thread of hydrophobic residues from the center of HVR domain D3 remains visible at 400 K but disappeared at 500 K, indicating loss of its hydrophobic core.
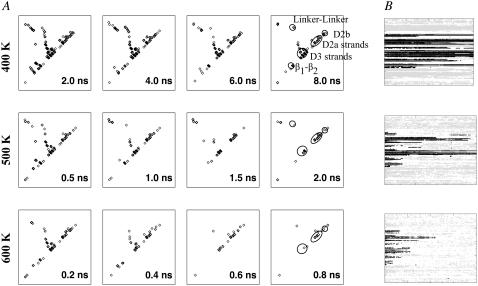
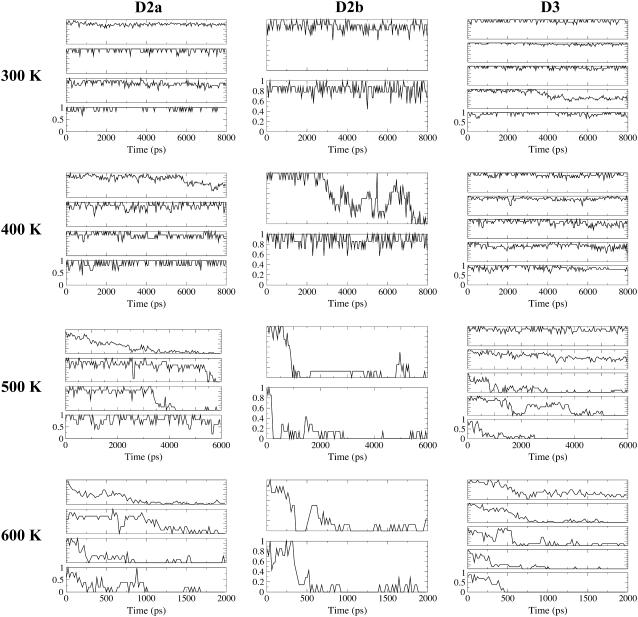
Time-averaged persistent contact maps from the first set of simulations indicated that pairs of β-strands in domains D2 and D3 remained for varying simulation lengths depending on the temperature (Fig. 3 A). Corresponding secondary structure changes in Fig. 3 B also revealed persistent β-strands in D2 and D3 while α-helices in D0 and D1 became denatured. To facilitate subsequent discussions, we need to introduce domain fragment Df1. This is the proteolysis-resistant portion of D1, which includes not just residues from the N-terminal side as originally defined (8) but also from the C-terminal side, as marked on the contact map (Fig. 2 A). Df1, colored black in Fig. 2 B, contains an elongated hydrophobic core that could account for its proteolytic resistance. The rigidity of Df1 hydrophobic core has been noted in a simulation of a 44-mer model of the filament (31). The remaining fragment of D1, colored magenta, is indeed found to be less structured during our simulations (Fig. 4).
FIGURE 3.
(A) Changes to persistent contact maps from the first set of simulations. Here, only persistent contacts within each time window of 2-ns (400 K), 0.5-ns (500 K), or 0.2-ns (600 K) are plotted. (B) Changes to DSSP assigned secondary structures from the same simulation set (black, β-strand; gray, α-helix), with plot for 300 K shown as inset to Fig. 2. Note that simulation lengths differ: 8-ns (400 K), 6-ns (500 K), and 2-ns (600 K).
FIGURE 4.
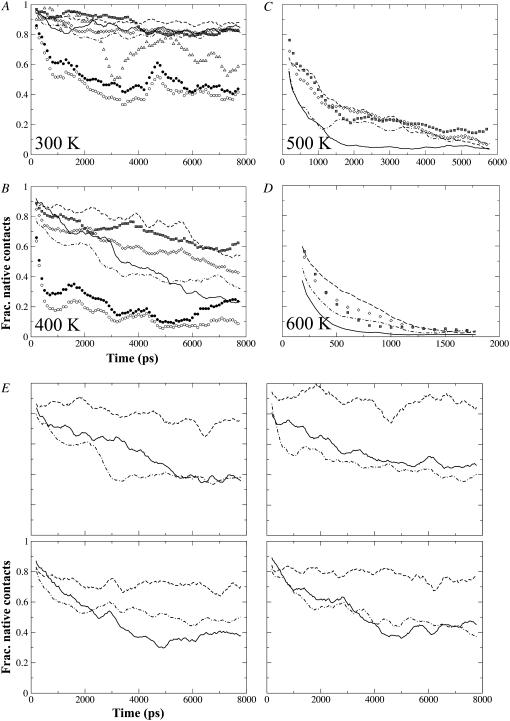
(A–D) Change of normalized fractional contacts of (sub)domains with simulation time, taken from the first set of simulations. Only contacts present in the starting structure were monitored and resulting fractional contacts normalized for better comparison between subdomains: Df1 (thick continuous line), D2 (open diamonds), D2a (dashed line), D2b (dot-dashed line), D3 (hatched squares), ND0 (solid circles), and CD0 (open circles). Note that D0 fractional contacts are shown only for 300 and 400 K. Fractional contacts for D1 excluding Df1 segment are shown only for 300 K (open triangles). (E) Plots from the remaining four 400 K trajectories of 8 ns each, showing only fractional contacts for Df1, D2a, and D2b. Note that abscissa and ordinate for all four plots are the same. In panels A–E, data points for Df1, D2a, and D2b have been resampled at 40-ps interval with a 20-ps window running-average applied for smoothing; for the rest, resampling interval is 100-ps with a 10-ps running-averaging.
The fraction of persistent native contacts that has been retained under thermal denaturing conditions is a crude indicator of the unfolding order. Fig. 4 shows data from the first set of simulations under each temperature, which indicated that D0 helices denatured very easily. Df1 and D2b were the first structured domains to be denatured. In contrast, for D2 (made up of D2a and D2b) and D3, the fraction remained >0.1 at <500 K (the value reached by CD0 at <400 K, taken to indicate complete unfolding). Despite a similar decay trend between D2 and D3, the D2a contacts were found to be generally better maintained than those of D3.
Although Df1 remained stable under the control simulation at physiological temperature of 300 K, it became unfolded under mild denaturation temperature of 400 K (Fig. 4 B). Looking at the fractional contacts between Df1 and D2a/b among the other four sets of 400 K simulation trajectories (Fig. 4 E), we noticed that D2a fractional contacts remained above or ∼0.7 while that for Df1 dropped to ∼0.4 and all plots seem to follow the profiles for D2b that is also marginally stable. Among these trajectories, hydrophobic side chains of residues on the D1-D2 linker loop remain well packed in the Df1 core and Df1 helices suffered minimal distortions. The D2a β12β13 hairpin and ND1a-ND1b helix-turn-helix segment are displaced relative to each other in some trajectories (thus breaking the D1-D2 interfacial H-bonds) but maintained in others. The further decrease of D2a fractional contacts after 6 ns during the first 400 K trajectory (Fig. 4 B) could have destabilized Df1 by further extracting the D1-D2 linker from the hydrophobic core: an increase in SASA for L167 and L169 occurred at ∼6 ns but L159 remained buried throughout (data not shown). There were also increased distortions to Df1 helices with loss of regular secondary structures corresponding to the drop of native fractional contacts to a lower value of 0.2.
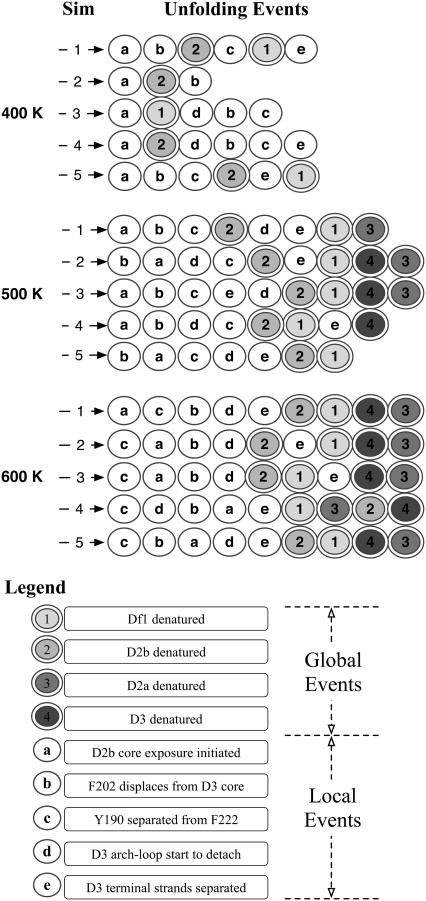
The sole use of fractional contacts to determine whether a (sub)domain is denatured does not work well for D2b, as the value can remain >0.1 while loss of secondary structures continues (data not shown). The situation is reversed for Df1. To capture both aspects of denaturation, we introduce a hybrid metric: the square-root of the product of fractional native contacts and fractional secondary structure content. When the 200-ps running average of this metric first dropped to <0.2, a value somewhat arbitrarily assigned, we say that the (sub)domain has denatured. Using these criteria to assign the global unfolding events and other criteria for the local unfolding events, we constructed a state-diagram for all the simulations as shown in Fig. 5. We noticed a general conservation of global unfolding events, such as D2b and D2a being the first and last to denature, respectively.
FIGURE 5.
A symbolic representation of major events in the thermal unfolding of flagellin from all simulations conducted in this study. The criteria used were as follows. States 1–4: when the 200-ps running average of the hybrid metric (see text for definition) first dropped below the 0.2 threshold. Fractional secondary structure content at time t in a trajectory is defined as the number of residues assigned as helix (Df1) or strand (D2a, D2b, D3) divided by the maximum number from the whole trajectory. State a: when the maximum instantaneous residue SASA among the D2b hydrophobic core residues first exceeded 30 Å2, which is the maximum observed during the control simulation. State b: similar to state a, but only one residue is considered and threshold is 25 Å2. State c: when minimum heavy atom separation of Y190 and F222 first exceeded the contact threshold without subsequent fluctuations about the threshold. States d and e: when the 200-ps running-averaged fractional native contact dropped below 0.2.
Partial unfolding/refolding of D2b
The D2b subdomain has a three-stranded β-sheet and only a weak hydrophobic core composed of residues V373, A385, and T347, according to ASAView (Fig. S1 in the Supplementary Material, Data S1). Threonine actually has a small hydropathy of −0.7 according to the Kyte and Doolittle scale, just below that of Ala and Gly. This weak core remains stable under control conditions. The surface-exposed aromatic cluster (Y349, Y380, and F390) was found to be even less stable against thermal denaturation than the weak hydrophobic core. The functional role of this cluster is as yet unknown.
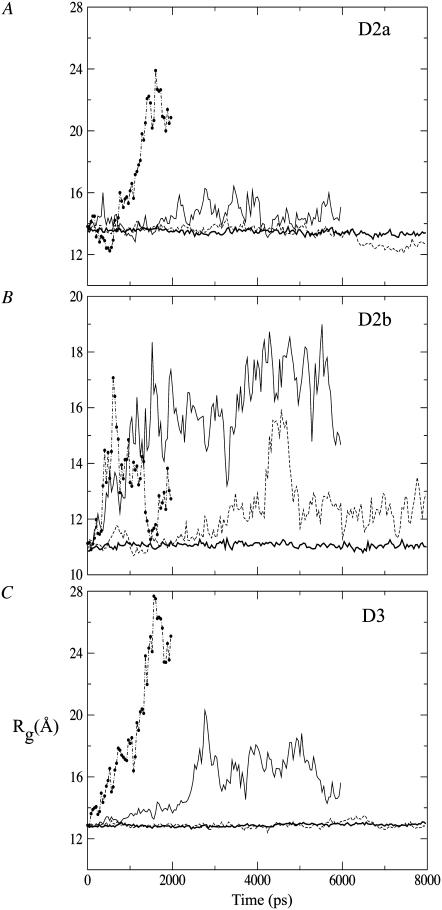
From the first 400 K simulation trajectory, we found that the Rg value increased gradually from 11 Å and suddenly jumped to 16 Å at ∼4 ns before dropping back at ∼12 Å (see Fig. 8 B). This spike could be due to the partial unfolding and refolding of the hydrophobic core region, with T347 separated from the A385-V373 pair. The opening-up of D2b is illustrated in Fig. S2 in the Supplementary Material, Data S1, via simulation snapshots. Combined SASAs of selected core residues from all simulations are shown in Fig. S3 in the Supplementary Material, Data S1. A threshold of 80 Å2 was adopted from the averaged SASA of polar residues reported in an early MD study (32). Fluctuations of D2b SASA at 400 K are large compared to those in the control or to those of D3 which remain folded. The above observations indicate that D2b has very low stability and would become folded only after D2a and D3.
FIGURE 8.
Radius of gyration of (A) subdomain D2a, (B) subdomain D2b, and (C) domain D3 under different temperatures from the first set of simulations: control (bold continuous), 400 K (dashed), 500 K (continuous), and 600 K (dot-dashed line with solid circles). Data points have been resampled at an interval of 20 ps.
Unfolding pathway of D3
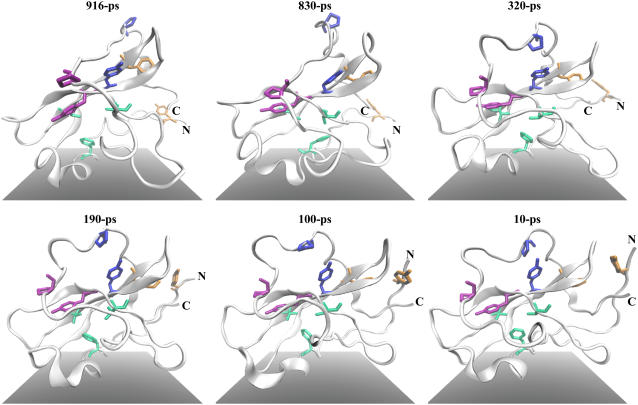
From the thermal unfolding simulations, we identified key events during the unfolding of domain D3. In the state-diagram (Fig. 5), we present the order of these events (local events b–d) as observed from each simulation. Conformation snapshots of D3 unfolding are presented in reversed time order in Fig. 6.
FIGURE 6.
Representative trajectory snapshots taken from the first 500 K thermal unfolding simulation (after root-mean-square fitting on D3 residues), presented in reversed time order to show folding of domain D3. The N- and C-termini are labeled. Folding starts from stabilization of the arch-loop conformation by proline-aromatic interactions between pairs P265-Y245 (magenta sticks) and P271-Y230 (blue sticks) across the loop and the β-folium sheet, denoting event d in Fig. 5. Aromatic-aromatic interaction between F222 and Y190 (on terminal β-sheet, which is also the D2-D3 linker) shown here as orange sticks helps to bind the terminal sheet against the rest of the domain (event c). The tight packing of F202 in the hydrophobic core (F202, V233, and V247 shown as light green sticks) takes the longest time (event b). Note the formation of the terminal β-sheet after F222-Y190 interaction becomes stable. Molecular scenes rendered by Tachyon ray tracer (53) within VMD.
Event b is the displacement of F202 from the hydrophobic core of D3, the precursor to solvent exposure of other core residues. Event c is the detachment of terminal anti-parallel β-sheet from the rest of D3 body, marked by separation of Y190 on the sheet from F222 on β6β7 hairpin. This separation allows extension of the β-sheet which then acts as a linker between D3 and D2, which allowed for relative rotations of the domains as observed from our simulations (data not shown). Y190 and F222 side chains form favorable CH···π interactions (33), which together with F229 makes up a surface aromatic cluster. Such clusters have been found to increase thermal stability of thermophiles (34). The Y190-F222-F229 aromatic cluster might have also protected the linker backbone hydrogen bonds from solvent attacks, which would weaken the linker, especially when mechanical stresses are applied to the linker region such as during rotation of the flagellum. Role of mechanical force has been attributed to destabilizing H-bonds to allow for replacement with residue-solvent ones (35). Lastly, event d marks the detachment of the arch-loop connecting β9 with β10 that wraps over the β-folium sheet when Proline (on the loop)-Tyrosine residue pairs separate (see Fig. 2 C). Subsequent unfolding reduces the size of the β-folium sheet. The consensus order of these events during folding might be: (d) to (c) to (b). This order suggests that completion of the hydrophobic core is the finishing touch and not the crucial phase in the folding of D3 (Fig. 6).
To further analyze the unfolding pathway, we performed property space, principal component analysis on all five trajectories at <400–600 K based on the following six properties: fraction of native contacts; native backbone H-bonds; β-sheet content; Rg; SASA; and SASA (hydrophobic) (36). This procedure allows us to assess which physical property changes the most during unfolding, judged by the size of the principal eigenvector components (Table S1 in the Supplementary Material, Data S1). Loss of native backbone H-bonds and increase in Rg dominates the second and third principal components under 500 K and 600 K. By projecting the property-space trajectories onto the first three principal eigenvectors, we observed divergence of the trajectories after 2 ns (500 K) and 0.5 ns (600 K) (Fig. S4 in the Supplementary Material, Data S1), indicative of a passage through the transition state. Hence, trajectories after these two time-points can be considered to be sampling the denatured ensemble. In some of the trajectories, native secondary structures are observed to be persistent.
Persistent structures in HVR domains
HVR domains in S. typhimurium contain high proportions of β-strands with a unique β-folium fold (27). D3 and D2a contained similar amounts of β-strands, found to be persistent during thermal denaturation (Fig. 3 B). The three β-strands in D2b, on the other hand, “dissolved” rapidly (data not shown).
Changes in fraction of persistent native contacts for D2a, D2b, and D3 under each denaturing temperature from the first set of simulations are presented in Fig. 7. For D2a at 500 K, although β14β16 is most persistent in this simulation, the top spot is taken by β13β14 among the other 500 K simulations and also in the first 600 K trajectory. The key stabilizing residue on β14 is Y332, which formed aromatic-aliphatic interactions with V325 on β13. The persistent contacts could have restricted the increase in Rg for D2a as compared to D2b at 500 K (Fig. 8). In the case of D3, though β6β7 is the most persistent at 500 K, β7β8 is also persistent at 600 K. Residues F222 and Y229 in β6β7 form favorable interaction with their side-chain aromatic rings tilted at a mean angle of 57° (from control simulation). Moreover, the lower Rg at 500 K relative to 600 K could be a result of compact region formation in the denatured state, involving hydrophobic residues 245, 247, and 229–231.
FIGURE 7.
Fractional contacts of β-stranded contact clusters, based on the first set of simulation data. There are four sets of multiplots under each of the columns for each flagellin subdomain, each representing a temperature. From the top of each multiplot, the change of fractional contacts with time for contact clusters are listed in the following order: β12β13, β13β14, β14β15, and β14β16 in D2a; β16β17 and β17β18 in D2b; β6β7, β7β8, β8β9, β5β7, and β6β10 in D3. Note that fractional contacts for 300 K were computed with respect to the equilibrated structure while values for 400 K and above were based on persistent native contacts obtained from the 300 K trajectory.
To identify which residues are involved in the thermal residual structures, lists of D2 and D3 residues in persistent contacts (>70% occurrence) during each of the 500 K simulations were compared and those residue pairs appearing in at least two of the five lists are selected and shown in Fig. 1 B. Of particular mention are the pairs V325-Y332 and D223-K228, which appeared in four and five lists, respectively. The latter pair formed a salt-bridge after ∼300 ps into two of the simulations, as determined by the VMD (37) Salt-Bridge plug-in. Although there is an abundance of Tyrosine residues in the persistent contacts, Tyr does not appear to be conserved among flagellin HVR sequences.
Volume of denatured flagellin
By calculating the principal moments of inertia of flagellin molecules and approximating the molecules as ellipsoids, we can get estimates for the dimensions (semi-major radius R1, and semi-minor radii R2 and R3) of the equivalent ellipsoids. From the radii, we can get rough estimates for the molecular volume. These estimates are presented in Table 1.
TABLE 1.
Estimates of refolding chamber and flagellin volumes
| Cylindrical approximation | CL1 (Å) | 2CR1 (Å) | CL2 (Å) | 2CR2 (Å) | Volume (× 1000 Å3) |
|---|---|---|---|---|---|
| Chamber* | 55 | 40 | 15 | 60 | 112 |
| Ellipsoidal approximation | 2R1 (Å) | 2R2 (Å) | 2R3 (Å) | Volume (× 1000 Å3) | |
| Polymeric-flagellin (1UCU) | 204 | 74 | 29 | 234 | |
| D0-D1 fragment | 172 | 28 | 25 | 61 | |
| D2-D3 fragment | 98 | 42 | 23 | 50 | |
| Monomeric-flagellin (Sim) | 200 | 70 | 38 | 280 | |
| D0-D1 fragment | 163 | 34 | 27 | 78 | |
| D2-D3 fragment | 96 | 41 | 26 | 54 | |
| D0-D1 (average 500 K Sim) | 74 (5) | 40 (6) | 53 (3) | 82 (7) | |
| D2-D3 (average 500 K Sim) | 77 (13) | 37 (5) | 49 (4) | 73 (12) |
Numbers in parentheses denote the standard deviations of the ellipsoidal radii and volume distributions obtained from the combined data set of five 500 K MD trajectories.
The refolding chamber volume is approximated by the sum of the volumes of two cylinders with length/radius given as CL1/CR1 and CL2/CR2.
The top panel of Table 1 shows our estimate of the refolding chamber volume by representing it as a combination of two cylinders. The main panel of the table presents estimated volumes for whole flagellin and its molecular fragments. Double the semi-major and semi-minor axes radii are reported for better comparison with chamber dimensions. Both the ellipsoidal approximations of polymeric (1UCU) and monomeric flagellin (starting structure for the denaturing simulations) are too large for the chamber, although their D0-D1 and D2-D3 fragments might fit. The volumes occupied by whole denatured flagellin are also computed. From conformation snapshots taken from the last 1-ns of 500 K and last 0.5 ns of 600 K simulations, we found broad and multimodal distributions for the volume with values ranging from 140,000 to 250,000 Å3, much larger than the estimated chamber volume. The ellipsoids representing denatured flagellin are ∼120 Å along the polar axis and 40–54 Å at the equator, too long for the chamber. Together, the results suggest that maybe half of flagellin might fit inside the chamber.
Physical extents and volumes of denatured flagellin fragments are estimated using 2-ps snapshots taken from the last 1-ns of the five 500 K trajectories. The D0-D1 and D2-D3 fragments are still separable despite being denatured. We present the average and standard deviation (in brackets) of the combined data sets in the lower panel of Table 1. The histograms of the fragment volume distributions from all simulations can be found in Fig. S5 in the Supplementary Material, Data S1. The results suggest that denatured D0-D1 and D2-D3 fragments could individually fit inside the refolding chamber but not whole flagellin.
DISCUSSION
Comparison with experimental denaturation study
The folding energetics of flagellin has been investigated by Honda et al. (9) by means of analyzing DSC experiments with a theoretical model formulated for multistate unfolding transitions. They determined from the melting temperatures that the unfolding of (sub)domains follows this order: Df1 to D2 to D3. From our thermal unfolding simulations of flagellin, we found a similar consensus order of unfolding where fragment Df1 and subdomain D2b unfolded before subdomain D2a and domain D3. The lower thermal stability of subdomain D2b (denaturing under 400 K) observed in our atomistic simulation corroborated that domain D2 should have a lower melting temperature as compared to D3. We have to employ much higher temperatures than used in experiments to observe significant changes in the protein over atomistic simulation timescales of nanoseconds.
The experimental study found an asymmetric DSC curve for Df1 with steeper slope after the melting temperature as compared to theoretical curve assuming no interdomain interactions. This steeper drop lead the authors to suggest that unfolding of D2 sped up that of Df1 (9). They also made the suggestion that Df1 is marginally stable even under physiological conditions (9). With the flagellin three-dimensional structure (PDB code 1IO1) obtained after the DSC study, we know that (N-terminal) D1-D2 linker is a long backbone segment devoid of any secondary structures that runs under the hydrophobic core of Df1 and residues L159 (most buried), I162, L167, L169, and L172 on the linker contribute side chains to the core. This linker is contiguous with β3 in D2a and has D2b packed beneath it (Fig. 2 B). There is almost no interface between D2b-Df1, and that of D2a-Df1 was found to be a weak one that is formed by solvent-mediated H-bonds between β12β13 hairpin turn in D2a and the turn across α2α3 in ND1, as found from the control simulation at 300 K. Hence, stabilization of Df1 by D2 does seem to be nonenthalpic as suggested (9).
From our thermal unfolding simulations at 400 K, we found that in cases where the Df1 fractional contacts dropped to and stabilized at ∼0.4 (Fig. 4 E), the maintenance of the native D2a-ND1 interface is not common to all. This finding supports the suggestion that the stabilization of Df1 by D2 is not enthalpic. An indication that unfolding of D2a might cause further unfolding of Df1 is shown in Fig. 4 B. However, D2b might have a larger influence on Df1 due to the seeming correlation in the fractional contact profiles, with those of the latter lagging behind the former (Fig. 4 E). If the correlations are real and assuming that the unfolding pathway is preserved (14), D2 unfolding might indeed hasten Df1 unfolding as suggested by experiment (9), through the D1-D2 linker. A possible way to assess the role of the linker is to constrain its conformational flexibility in further thermal unfolding simulations.
It was also demonstrated in the denaturation study that D2 and D3 interact both enthalpically and entropically to stabilize each other (9). The unfolding as monitored by fractional contacts do indicate similar profiles for the two domains among 400 K trajectories (data not shown) but the similarity alone might not be a convincing indicator of unfolding cooperativity.
To determine melting temperatures of individual domains and domain-domain folding cooperativity from simulation, we need to monitor changes in contacts over a wide range of temperatures and also increase the temperature within a simulation to mimic DSC experiments. The all-atom nature of our models limited temperature-scan types of equilibrium simulations to only a few selected temperatures due to the high computation cost. Temperature-ramp MD simulations might also be very time-consuming due to the need to increase the temperature in small steps and with sufficient equilibrium at each step to keep the system at quasi-equilibrium for proper simulation behavior. A simplified or coarse-grained model is thus desirable with the advantage of lower computation cost due to faster dynamics, which allows much longer simulations into microseconds, the realm of biological processes (38). Such an approach was employed by Chen and Dokholyan, who performed temperature-scan and temperature-ramp simulations to look at domain interactions and unfolding kinetics of vinculin (39).
HVR domains fold via nucleation?
Persistent secondary structures were detected in the denatured states of flagellin from our simulations at 500–600 K: β6 to β8 in D3 and β12 to β14 in D2a. These β-sheets might account for the relatively high melting temperatures of D2 and D3 found in experiment (9). Residue-residue contacts persistent under unfolding were identified using all five sets of 500 K trajectories and involved the following residues: 217, 220–223, 228–232, and 245 in D3 and 305–309, 321–326, 331–332, and 334 in D2a. These residues are located in the persistent β-sheets mentioned above (Fig. 2 C)
The K-Fold server, which uses a machine-learning algorithm trained on 63 proteins with known folding characteristics, predicts both folding rate and kinetics of proteins given the three-dimensional structure (40). The residues in the persistent secondary structures were found to be major contributors to the total contact order (data not shown). Demirel et al. (41) have suggested that strong dynamic coupling (high contact order) to all other residues implies that such residues are hubs in the intraprotein network. Such hubs have been associated with the folding nucleus in a theoretical study on the small-world nature of protein structures (42).
We thus propose that the persistent three-stranded anti-parallel β-sheets observed in D2 and D3 might form early in the (re)folding process, serving as spatially diffused folding nucleation sites or cores to promote folding of the rest of the domains, following the nucleation-condensation model proposed by Fersht (43). Folding cores are like the foundation pillars of a building, which needs to be present before the rest of the building can come together. They also define the shape of the building. Similar suggestions of persistent three-stranded sheets being folding cores have been made by other researchers via thermal unfolding simulations (11,13). Due to their local nature, such folding cores might even be formed during the flagellin transport phase. Sato and Raleigh reported the detection of significant backbone hydrogen bonds in the transition state of the ribosomal protein L9 N-terminal domain by means of amide H/D isotope exchange (44). Their work showed the existence of secondary structures in the transition/denatured state of proteins.
It has been deduced that new flagellin is added to the growing filament every second (45), implying that flagellin has 1 s to refold if the chamber is always occupied. According to a linear regression relationship between logarithm of experimental folding rate and absolute contact order (ACO, a measure of topological complexity defined for native structure) established for two- and multi-state folders (46), the ACO of a domain should be <∼20 for a folding time of <1 s. Based on the experimental structure of flagellin (1UCU.pdb), helical domains D0 and D1 have ACO at ∼3 while D2 has a value close to 10. The globular domain D3 has the largest value of ∼15, meaning that it could be the rate-limiting step in the refolding process. The numbers also suggests that D2-D3 might either fold cooperatively or fold independently but simultaneously, and then dock together. The presence of folding cores in HVR domains deduced from our simulations would enable fast folding of D2-D3 followed by docking of folded D1-D0 domains. Such a mechanism could help flagellin to complete refolding within 1 s, overcoming the limitation of high topological complexity.
A study comparing folding mechanisms of immunoglobulin-like β-sandwich proteins from diverse families found that interactions defining the structural topology also guided folding (47). Geierhaas et al. (48) have also found that the folding nucleus can be significantly deformed to preserve the same folding mechanism despite sequence variation in immunoglobulin-like domains. We suspect that HVR domains are under evolutionary pressure to adopt structurally stable folds despite sequence divergence. However, to date we only have the structures from S. typhimurium. It is interesting to see whether HVR domains from diverse flagellin species adopt similar folds. If so, HVR domains from other flagellin might also harbor folding cores.
Importance of folding cores to flagellin stability
In a mutagenesis study (18) screening for functional deletion flagellin mutants (retention of mobility), we found that the three deletion mutants reported to show the same mechanical stability as wild-type retained residues we have identified to be in the folding cores (except for Y245 that is absent in one of the mutants). Also, the folding and mechanical stability of mutants were not affected if residues 250 to ∼290 are missing: starting from the β-turn of β8β9, through the arching-loop to β10 and reaching the α-helix in D2a. These residues are not part of the D2 or D3 folding cores.
For mutants with more brittle filaments, they either have a reduced D2a or, in the case of C11 in this article, missing residues A231 and K232 from the putative folding cores and V233 from the native hydrophobic core (18). This mutant flagellin highlighted the greater importance of retaining folding core residues (especially the salt-bridge D223-K228) over residues in the hydrophobic core. This observation also indicated that the salt-bridge is not an artifact of simulating at elevated temperatures, as salt-bridges become more important at higher temperatures and confer thermostability upon thermophilic proteins (49). Interestingly, in the native state under physiological conditions, K228 forms a salt-bridge with E246 on β8 instead of D223 on β6, which suggests a switch in partner after the folding core has been formed and more extensive interactions are desired to form the rest of D3 (note: putative folding core ends at Y245). Lastly, functional mutants which could not swim but showed swarming after 12–16 h of growth were missing all of the folding core residues or all of D2 (18), affecting the formation of the native HVR domains. This mutagenesis study thus highlighted the existence and importance of the folding cores to flagellin mechanical stability.
With the identification of putative folding cores, amino-acid substitution experiments can now be directed toward replacing D223 (breaking only the folding core salt-bridge) or V233 to investigate their effects on D3 folding and stability. In the case of D2a, Valine pair 306–325 and the Tyrosine pair 331–332 in the folding core might be good candidates for stability studies. Destabilization of the D2a folding-core might be a way to test whether folding of Df1 is indeed dependent upon that of D2, as suggested by Honda et al. (9). The effect would be manifested as a slowdown or abolishment of filament growth since natively folded D1 domains are needed to build the outer tube of the filament.
Refolding of flagellin may happen in stages
Flagellin is likely to be substantially unfolded to be exported by the apparatus located at the entrance to the filament channel (Fig. 1). We can also assume that unfolded flagellin molecules travel in single-file along the filament channel, as in a mathematical model of filament growth (50). At the end of the channel under the filament cap (formed by HAP2 chaperone) is a cylindrical space surrounded by D1 domains from the last round of polymerized flagellin (8), in which refolding of flagellin occurs. The depth of the chamber at ∼70 Å is close to the span of the longest D1 helices. Because D1 of flagellin is conserved across bacterial species (1), the chamber size might also be conserved and independent of HVR domain sizes. Whether flagellin enters the chamber as a linear chain or as a loop (half-folded chain) remains unknown. Investigation into the mechanical forces needed to generate these two transport states have been performed in our group (C.-P. Chng and A. Kitao, unpublished).
Based on estimates of denatured flagellin volumes from our simulations and comparing with an estimated chamber volume, we suggest that fragments D2-D3 or D0-D1 can be accommodated, but not the whole molecule. Yonekura et al. (8) has commented that the chamber is large enough for—at most—one molecule. The refolding process may therefore be a two-stage process: chamber occupancy and folding of HVR (D2-D3) domains precedes the filament-core (D0-D1) domains (the more likely scenario). Furthermore, HVR domains have been suggested to fold cooperatively from DSC experiment (9) and might fold rapidly with the help of the folding cores we have identified, before being extruded to the filament exterior through the HAP2 gap (8,51). As D2b was found to fold late, it could still be rather fluid and makes flagellin extrusion easier. After filament-core domains become folded, docking of Df1 to folded D2 would complete the refolding process for flagellin. In bacterial species with short or negligible HVR segment, refolding of filament-core domains would constitute the major step.
CONCLUSIONS
Persistent native structures detected in the thermally denatured ensemble of multidomain S. typhimurium flagellin from simulation suggest that they might constitute folding initiation sites, which could speed up folding inside the refolding chamber before the assembly step. Similar folding cores, dictated by native topology, might exist in HVR domains from other flagellin species despite sequence variation. Volume estimates suggest that HVR and filament-core domains occupy the chamber at different times, suggesting a two-staged refolding mechanism for flagellin with significant HVR.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
A.K. thanks Dr. Keiichi Namba and Dr. Katsumi Imada for fruitful discussions. MD simulations were performed on supercomputers at the Research Center for Computational Science (Okazaki Research Facilities, National Institutes of Natural Sciences) and at the Japan Atomic Energy Agency.
This work was supported by the Next Generation Super Computing Project, Nanoscience Program, Grant-in-Aid for Scientific Research (B) and Grants-in-Aid for Scientific Research on Priority Areas to A.K. from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. C.-P.C. is supported by a MEXT graduate scholarship.
Editor: Ron Elber.
References
- 1.Beatson, S. A., T. Minamino, and M. J. Pallen. 2006. Variation in bacterial flagellins: from sequence to structure. Trends Microbiol. 14:151–155. [DOI] [PubMed] [Google Scholar]
- 2.Mimori-Kiyosue, Y., F. Vonderviszt, I. Yamashita, Y. Fujiyoshi, and K. Namba. 1996. Direct interaction of flagellin termini essential for polymorphic ability of flagellar filament. Proc. Natl. Acad. Sci. USA. 93:15108–15113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 424:643–650. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825. [DOI] [PubMed] [Google Scholar]
- 5.Galan, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 444:567–573. [DOI] [PubMed] [Google Scholar]
- 6.Holland, I. B. 2004. Translocation of bacterial proteins—an overview. Biochim. Biophys. Acta. 1694:5–16. [DOI] [PubMed] [Google Scholar]
- 7.Wickner, W., and R. Schekman. 2005. Protein translocation across biological membranes. Science. 310:1452–1456. [DOI] [PubMed] [Google Scholar]
- 8.Yonekura, K., S. Maki, D. G. Morgan, D. J. DeRosier, F. Vonderviszt, K. Imada, and K. Namba. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 290:2148–2152. [DOI] [PubMed] [Google Scholar]
- 9.Honda, S., H. Uedaira, F. Vonderviszt, S. Kidokoro, and K. Namba. 1999. Folding energetics of a multidomain protein, flagellin. J. Mol. Biol. 293:719–732. [DOI] [PubMed] [Google Scholar]
- 10.Oroguchi, T., M. Ikeguchi, K. Saeki, K. Kamagata, Y. Sawano, M. Tanokura, A. Kidera, and K. Kuwajima. 2005. Atomically detailed description of the unfolding of α-lactalbumin by the combined use of experiments and simulations. J. Mol. Biol. 354:164–172. [DOI] [PubMed] [Google Scholar]
- 11.Akanuma, S., and A. Yamagishi. 2005. Identification and characterization of key substructures involved in the early folding events of a (β/α)8-barrel protein as studied by experimental and computational methods. J. Mol. Biol. 353:1161–1170. [DOI] [PubMed] [Google Scholar]
- 12.Scott, K. A., L. G. Randles, S. J. Moran, V. Daggett, and J. Clarke. 2006. The folding pathway of spectrin R17 from experiment and simulation: using experimentally validated MD simulations to characterize states hinted at by experiment. J. Mol. Biol. 359:159–173. [DOI] [PubMed] [Google Scholar]
- 13.Sham, Y. Y., B. Ma, C. J. Tsai, and R. Nussinov. 2002. Thermal unfolding molecular dynamics simulation of Escherichia coli dihydrofolate reductase: thermal stability of protein domains and unfolding pathway. Proteins. 46:308–320. [DOI] [PubMed] [Google Scholar]
- 14.Day, R., B. J. Bennion, S. Ham, and V. Daggett. 2002. Increasing temperature accelerates protein unfolding without changing the pathway of unfolding. J. Mol. Biol. 322:189–203. [DOI] [PubMed] [Google Scholar]
- 15.Day, R., and V. Daggett. 2007. Direct observation of microscopic reversibility in single-molecule protein folding. J. Mol. Biol. 366:677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daggett, V. 2006. Protein folding—simulation. Chem. Rev. 106:1898–1916. [DOI] [PubMed] [Google Scholar]
- 17.Day, R., and V. Daggett. 2005. Ensemble versus single-molecule protein unfolding. Proc. Natl. Acad. Sci. USA. 102:13445–13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malapaka, R. R., L. O. Adebayo, and B. C. Tripp. 2007. A deletion variant study of the functional role of the salmonella flagellin hypervariable domain region in motility. J. Mol. Biol. 365:1102–1116. [DOI] [PubMed] [Google Scholar]
- 19.Case, D. D. T.A., T. I. Cheatham, C. Simmerling, J. Wang, R. Duke, R. Luo, K. Merz, B. Wang, D. Pearlman, M. Crowley, S. Brozell, V. Tsui, H. Gohlke, J. Mongan, V. Hornak, G. Cui, P. Beroza, C. Schafmeister, J. Caldwell, W. Ross, and P. Kollman. 2004. AMBER 8. University of California, San Francisco.
- 20.Essmann, U., L. Perera, M. Berkowitz, T. Darden, H. Lee, and L. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- 21.Ryckaert, J., G. Ciccotti, and H. Berendsen. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- 22.Kostyukova, A. S., M. G. Pyatibratov, V. V. Filimonov, and O. V. Fedorov. 1988. Flagellin parts acquiring a regular structure during polymerization are disposed on the molecule ends. FEBS Lett. 241:141–144. [DOI] [PubMed] [Google Scholar]
- 23.Vonderviszt, F., S. Kanto, S. Aizawa, and K. Namba. 1989. Terminal regions of flagellin are disordered in solution. J. Mol. Biol. 209:127–133. [DOI] [PubMed] [Google Scholar]
- 24.Ishima, R., K. Akasaka, S. Aizawa, and F. Vonderviszt. 1991. Mobility of the terminal regions of flagellin in solution. J. Biol. Chem. 266:23682–23688. [PubMed] [Google Scholar]
- 25.Aizawa, S.-I., F. Vonderviszt, R. Ishima, and K. Akasaka. 1990. Termini of salmonella flagellin are disordered and become organized upon polymerization into flagellar filament. J. Mol. Biol. 211:673–677. [DOI] [PubMed] [Google Scholar]
- 26.Tamura, Y., K. Gekko, K. Yoshioka, F. Vonderviszt, and K. Namba. 1997. Adiabatic compressibility of flagellin and flagellar filament of salmonella typhimurium. Biochim. Biophys. Acta. 1335:120–126. [DOI] [PubMed] [Google Scholar]
- 27.Samatey, F. A., K. Imada, S. Nagashima, F. Vonderviszt, T. Kumasaka, M. Yamamoto, and K. Namba. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 410:331–337. [DOI] [PubMed] [Google Scholar]
- 28.Feig, M., J. Karanicolas, and C. Brooks. 2004. MMTSB Tool Set: enhanced sampling and multiscale modeling methods for applications in structural biology. J. Mol. Graph. Model. 22:377–395.15099834 [Google Scholar]
- 29.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad, S., M. Gromiha, H. Fawareh, and A. Sarai. 2004. ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinformatics. 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitao, A., K. Yonekura, S. Maki-Yonekura, F. A. Samatey, K. Imada, K. Namba, and N. Gō. 2006. Switch interactions control energy frustration and multiple flagellar filament structures. Proc. Natl. Acad. Sci. USA. 103:4894–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng, C., C. F. Wong, and A. J. McCammon. 1990. Fluctuation of the solvent-accessible surface area of tuna ferrocytochrome c. Biopolymers. 29:1877–1883. [DOI] [PubMed] [Google Scholar]
- 33.Hunter, C., and J. Sanders. 1990. The nature of π-π interactions. J. Am. Chem. Soc. 112:5525–5534. [Google Scholar]
- 34.Kannan, N., and S. Vishveshwara. 2000. Aromatic clusters: a determinant of thermal stability of thermophilic proteins. Protein Eng. 13:753–761. [DOI] [PubMed] [Google Scholar]
- 35.Pabon, G., and L. M. Amzel. 2006. Mechanism of titin unfolding by force: insight from quasi-equilibrium molecular dynamics calculations. Biophys. J. 91:467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kazmirski, S. L., A. Li, and V. Daggett. 1999. Analysis methods for comparison of multiple molecular dynamics trajectories: applications to protein unfolding pathways and denatured ensembles. J. Mol. Biol. 290:283–304. [DOI] [PubMed] [Google Scholar]
- 37.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- 38.Ding, F., and N. V. Dokholyan. 2005. Simple but predictive protein models. Trends Biotechnol. 23:450–455. [DOI] [PubMed] [Google Scholar]
- 39.Chen, Y., and N. V. Dokholyan. 2006. Insights into allosteric control of vinculin function from its large scale conformational dynamics. J. Biol. Chem. 281:29148–29154. [DOI] [PubMed] [Google Scholar]
- 40.Capriotti, E., and R. Casadio. 2007. K-Fold: a tool for the prediction of the protein folding kinetic order and rate. Bioinformatics. 23:385–386. [DOI] [PubMed] [Google Scholar]
- 41.Demirel, M., A. Atilgan, R. Jernigan, B. Erman, and I. Bahar. 1998. Identification of kinetically hot residues in proteins. Protein Sci. 7:2522–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vendruscolo, M., N. V. Dokholyan, E. Paci, and M. Karplus. 2002. Small-world view of the amino acids that play a key role in protein folding. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 65: 061910. [DOI] [PubMed] [Google Scholar]
- 43.Fersht, A. R. 1997. Nucleation mechanisms in protein folding. Curr. Opin. Struct. Biol. 7:3–9. [DOI] [PubMed] [Google Scholar]
- 44.Sato, S., and D. P. Raleigh. 2007. Kinetic isotope effects reveal the presence of significant secondary structure in the transition state for the folding of the N-terminal domain of l9. J. Mol. Biol. 370:349–355. [DOI] [PubMed] [Google Scholar]
- 45.Ikeda, T., S. Yamaguchi, and H. Hotani. 1993. Flagellar growth in a filament-less salmonella fliD mutant supplemented with purified hook-associated protein 2. J. Biochem. (Tokyo). 114:39–44. [DOI] [PubMed] [Google Scholar]
- 46.Ivankov, D. N., S. O. Garbuzynskiy, E. Alm, K. W. Plaxco, D. Baker, and A. V. Finkelstein. 2003. Contact order revisited: influence of protein size on the folding rate. Protein Sci. 12:2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clarke, J., E. Cota, S. B. Fowler, and S. J. Hamill. 1999. Folding studies of immunoglobulin-like β-sandwich proteins suggest that they share a common folding pathway. Structure. 7:1145–1153. [DOI] [PubMed] [Google Scholar]
- 48.Geierhaas, C. D., E. Paci, M. Vendruscolo, and J. Clarke. 2004. Comparison of the transition states for folding of two Ig-like proteins from different superfamilies. J. Mol. Biol. 343:1111–1123. [DOI] [PubMed] [Google Scholar]
- 49.Missimer, J. H., M. O. Steinmetz, R. Baron, F. K. Winkler, R. A. Kammerer, X. Daura, and W. F. van Gunsteren. 2007. Configurational entropy elucidates the role of salt-bridge networks in protein thermostability. Protein Sci. 16:1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keener, J. P. 2006. How salmonella typhimurium measures the length of flagellar filaments. Bull. Math. Biol. 68:1761–1778. [DOI] [PubMed] [Google Scholar]
- 51.Maki-Yonekura, S., K. Yonekura, and K. Namba. 2003. Domain movements of hap2 in the cap-filament complex formation and growth process of the bacterial flagellum. Proc. Natl. Acad. Sci. USA. 100:15528–15533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano, W. L. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA.
- 53.Stone, J. 1998. An efficient library for parallel ray tracing and animation. Master's thesis, Computer Science Department, University of Missouri-Rolla.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.