Abstract
We have developed a high-resolution scanning surface confocal microscopy technique capable of imaging single virus-like particles (VLPs) on the surfaces of cells topographically and by fluorescence. The technique combines recently published single-molecule-resolution ion-conductance microscopy that acquires topographical data with confocal microscopy providing simultaneous fluorescent imaging. In our experiments we have demonstrated that the cell membrane exhibits numerous submicrometer-sized surface structures that could be topographically confused with virus particles. However, simultaneous acquisition of confocal images allows the positions of fluorescently tagged particles to be identified. Using this technique, we have, for the first time, visualized single polyoma VLPs adsorbed onto the cell membrane. Observed VLPs had a mean width of 108 ± 16 nm. The particles were randomly distributed across the cell membrane, and no specific interactions were seen with cell membrane structures such as microvilli. These experiments demonstrate the utility of this new microscope for imaging the interactions of nanoparticles with the cell surface to provide novel insights into the earliest interactions of viruses and other nanoparticles such as gene therapy vectors with the cell.
INTRODUCTION
Understanding the details of the passage of viruses, bacteria, or other nanoparticles across the cell membrane is essential for developing new antiviral/antibacterial agents and gene therapy vectors. Current methods for nanoparticle visualization include confocal microscopy and total internal reflection (TIRF) microscopy coupled with fluorescent labeling of particles, scanning and transmission electron microscopy, and atomic force microscopy (AFM) (1). Although fluorescence confocal microscopy is one of the most commonly used microscopy techniques in the virology field, it provides very limited information about single particles crossing the membrane because of its limited lateral and vertical resolution. TIRF has better vertical resolution and signal/noise ratio and has been used for single particle tracking (2). However, TIRF is limited to following events that occur on the basal cell membrane, because it relies on the imaging to a depth of ∼100 nm from the glass surface and is not applicable to studying apical cell surfaces or more complex cell cultures/tissues.
Electron microscopy, by contrast, has single-particle resolution and provides visual information about surface topography or cross-sectional data that are more positionally informative than optical images because the cell membrane can be defined. However, this requires fixation and specialized treatments of the cells, which can create structural aberrations and are incompatible with live studies.
Scanning probe microscopy techniques may address a number of these issues by providing topographical data on surfaces. AFM has been used to study cell surfaces, but because of the direct interaction of its probe with the sample, imaging soft and mobile cell surfaces is difficult, limiting most studies to fixed cells, which are more rigid (3). On the other hand, scanning ion conductance microscopy (SICM), which uses a glass micropipette as an imaging probe (4), allows noncontact visualization of the topography of living cells (5,6). This methodology has recently been combined with simultaneous confocal fluorescence imaging, resulting in a new technique named scanning surface confocal microscopy (SSCM) (7). SSCM allows the relation between cell surface membrane structures and fluorescently labeled nanoparticles to be studied in both fixed and live samples, opening up the possibility of following viral entry in real time. However, the topographical resolution of the first-generation SSCM was not sufficient to detect single virus-like particles (VLPs), making it difficult to study their internalization. Although the resolution of confocal fluorescence images remains limited by the diffraction limit of light, the resolution of SSCM topography can be increased by using smaller-diameter pipettes and improved mechanical stability of the microscope. We have recently developed a modified SICM capable of scanning at much higher resolution that uses nanopipettes with inner diameters as small as 12.5 nm, which is 20 times smaller than the average pipette normally used for SICM imaging. For example, high-resolution SICM can visualize single proteins in a monolayer of S-layer (cell surface layer) proteins from Bacillus sphaericus CCM 2177 on a mica surface 13.1 nm pitch with 3- to 6-nm resolution (8). We therefore combined these two techniques into a single new high-resolution SSCM instrument to explore if we could directly image nanoparticles on the cell surface. As a step toward this ultimate goal, we have exploited the improvement in the lateral resolution of SSCM to directly visualize single polyoma VLPs on the apical membrane of cells using the simultaneously recorded fluorescence image to distinguish VLPs on the cell surface from other structures of similar dimensions.
MATERIALS AND METHODS
SSCM
SSCM is based on a combination of SCM and SICM. SICM is a scanning probe microscopy technique (6,9) in which the ion current flowing into a nanopipette is used to control the vertical (z axis) position of the cell relative to the pipette tip. As shown diagrammatically in Fig. 1 A (not to scale), in SSCM the cell is moved up and down in the z direction while scanning is done in the x and y directions, so its surface is always the same distance from the nanopipette. A laser is passed up a high numerical aperture objective so that it is focused just at the tip of the nanopipette, and a pinhole is positioned at the image plane so that the confocal volume is just below the pipette, as described (9). Thus, a fluorescence image of the cell surface is obtained in a single scan, as is a simultaneously captured image of the cell topography.
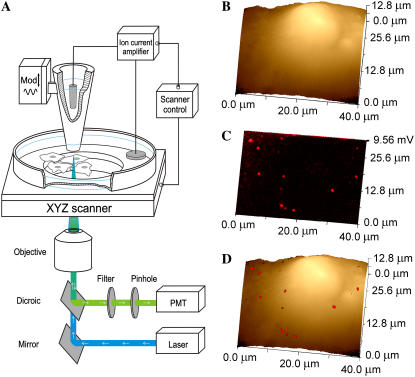
FIGURE 1.
SSCM of VLPs on the surface of a cell membrane. (A) Schematic diagram of the SSCM (not to scale). A conical glass nanopipette is placed over the surface of the cells and aligned with the laser beam and photomultiplier detector for simultaneous topographical and fluorescence measurements while the sample stage scans in XYZ directions. (B) A low-resolution topographical image of a COS7 cell formaldehyde fixed after addition of fluorescently labeled VLPs. (C) Corresponding fluorescence surface confocal image of VLPs obtained simultaneously with topography shown in B. (D) Fluorescent on topography overlay image showing the distribution of VLPs on the surface of the cell detected by fluorescence.
The SCIM scanning head was developed in collaboration with Ionscope Limited, UK, and mounted on a Nikon TE2000-U Inverted Microscope (Nikon, Tokyo, Japan). The sample holder was attached to a 100-μm HERA XY Nanopositioning System (Physik Instrumente, Karlsruhe, Germany) used for lateral scanning. Vertical measurement and modulation were provided by 12-μm LISA XY Nanopositioning System (Physik Instrumente). Both piezo stages were mounted on 25-mm translation stage DC motors (Physik Instrumente) to provide coarse lateral and vertical approach. The setups were controlled via a computer with a SBC6711 DSP board equipped with A4D4 ADC/DAC modules (Innovative Integration, Simi Valley, CA) using SICM software v. 1.2.00 (Ionscope Limited, London, UK). The time to acquire a 512 × 512 pixel image was ∼10 min.
Two types of nanopipettes were used for the experiments. For low-resolution images, nanopipettes with internal diameters ∼150 nm were pulled from borosilicate glass capillaries. High-resolution imaging was made using quartz nanopipettes with internal diameters ∼70 nm. The nanopipettes were made from 1.00-mm outer diameter, 0.5-mm inner diameter capillaries with inner filament (Sutter Instrument, San Rafael, CA) using a laser-based Brown-Flaming puller (model P-2000, Sutter Instrument).
The nanopipettes, backfilled with phosphate-buffered saline (PBS) and lowered into PBS produced a resistance of ∼300 MΩ for quartz and 100 MΩ for borosilicate pipettes. The maximum ion current measured using an Axopatch 200B (Axon Instruments, Sunnyvale, CA), was ∼0.7 nA for quartz and ∼1.5 nA for borosilicate pipettes. The set point for imaging was 1% of the maximum of modulated ion current.
The excitation light source was provided by a GPNT-02 laser diode (532-nm wavelength, IQ1A 635 nm laser (Power Technology, Little Rock, AR). The optical recording system consisted of a Nikon TE2000-U Inverted Microscope equipped with a 100× 1.3 N.A. oil-immersion objective. The excitation light was fed through an epifluorescent filter block, and emitted light collected by a photomultiplier with a pinhole (model D-104-814; Photon Technology International, West Sussex, UK).
Image processing and data analysis
Matching VLP topographical structure to its corresponding fluorescent signal was done as follows. Fluorescent confocal images were used as threshold to subtract the background, and positions of individual fluorescent spots were marked by arrowheads and multiple spots (where individual signals could not be resolved) circled. All positional markers were then grouped into one template and placed over the simultaneously recorded topographical image. As a result of this procedure, those topographical features having corresponding fluorescent signal were indicated.
Image contrast enhancement included slope correction and high-pass filtration performed similarly to previously described procedures (10,11). The slope was calculated and subtracted from images by least-squares algorithm (12), resulting in image flattening. A high-pass filter is a filter that passes high frequencies well but reduces frequencies lower than a certain specified (cutoff) frequency. When applied to a three-dimensional image, high-pass filtration results in finer (i.e., high-frequency) details standing out because larger features have been eliminated.
Cells and viruses
COS-7 cells (ECACC No.: 87021302) were grown in DMEM supplemented with 5% fetal calf serum at 37°C. Before scanning, cells were plated on a glass coverslip at a density of 5 × 104 cells for 16 h.
VLPs derived from self-assembling of VP1 of the murine polyomavirus were purified by sucrose and CsCl gradient centrifugation from Hi5 insect cells infected with recombinant baculovirus, as described (13), and resuspended in sterile 20 mM Hepes, pH 7.5, at 3–6 mg/ml.
For labeling, Cy5-succinimidyl ester kit (Amersham, Little Chalfont, UK) was reconstituted with 80 μl DMSO, and 1 μl dye was added to 30 μg of VLPs for 45 min at room temperature.
For scanning VLPs directly on glass, to improve VLP adhesion, the coverslips were incubated for 2 h with DMEM containing 5% FCS at 37°C. Coverslips plated with cells, or prepared as described above, were incubated with Cy5-labeled VLPs at 25 μg/ml in 300 μl Optimem (Gibco, Grand Island, NY) for 30 min on ice. Coverslips were washed with ice-cold PBS, fixed with 3% formaldehyde in PBS supplemented with 5% sucrose for 15 min on ice, and then washed with PBS.
RESULTS
The entry of VLPs into COS7 cells has been studied in detail previously, making this a good model system on which to validate our method (14). Previous work on this system has shown that VLPs may enter the cell by two routes, only one of which is productive for gene transfer. Cleaving of cell surface sialic acid by neuraminidase blocks VLP-mediated gene expression. However, the bulk of the VLPs are still bound to the cell at 0°C and internalized in the presence of neuraminidase, suggesting entry via a different route. Other research has reported that experiments with and without neutralizing antibody showed no difference in the efficiency or kinetics of uptake (15). We imaged the cells after addition of the VLPs for 30 min, where they are adsorbed on the cell surface but the majority have not yet entered the cell. Then to establish whether we could identify VLPs on the cell membrane, we performed a series of experiments on fixed cells so that there were no changes in viral particle position with time and so that the cells could be scanned repeatedly.
Fig. 1, B and C, shows typical low-resolution topographical and corresponding fluorescence images of a COS7 cell fixed immediately after addition of fluorescently labeled polyoma VLPs using the new SSCM instrument with a ∼150-nm-diameter pipette. An overlay of the fluorescence and topographical images is presented in Fig. 1 D and visualizes the distribution of VLPs on the surface of the cell. At this resolution, only large aggregates of VLPs can be detected on the optical image, and they could not be resolved topographically. We next subjected cells to higher-resolution imaging using finer pipettes made of quartz glass with inner diameters of ∼70 nm.
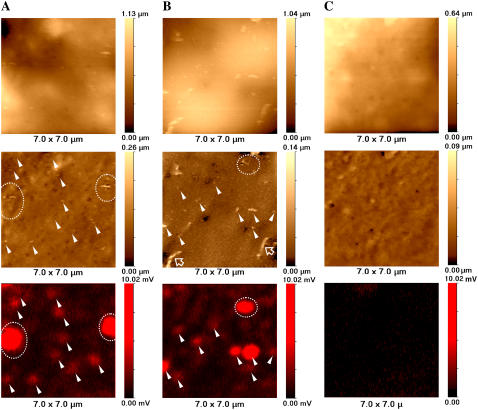
Fig. 2, A and B, represents high-resolution topographical unprocessed (top row) and flattened (slope-corrected and high-pass filtered, see Materials and Methods) (middle row) and confocal (bottom row) images of COS7 cells fixed with formaldehyde after Cy5-labeled VLPs were adsorbed to the outer surface by incubation at 4°C. For comparison, Fig. 2 C shows control images of fixed COS7 cell (no VLPs added). Based on electron microscopy (16) and other data (17,18), polyoma viruses and VLPs comprise 40- to 60-nm-diameter spheres. As can be seen from unprocessed images (top row), the relatively high slope gradient and large undulations of the cell membrane make it impossible to detect VLPs on the cell surface without additional image processing. In contrast, processed images (middle row) exhibit many nanometer-scale structures not only on cells to which VLPs were added but also in the control sample. Some of these structures are seen as protrusions, whereas others are indentations. Not all the protruding features represent VLPs, as they are also present in the control cells and are likely to be cellular structures such as elongated ridge-like structures (Fig. 2 B, hollow arrows) that may represent microvilli similar to those described in our previous articles (7). To identify which protrusions could represent VLPs, we analyzed the corresponding fluorescence data (bottom row).
FIGURE 2.
High-resolution SSCM imaging of single VLPs on COS7 cell membranes. (A and B) SSCM images of COS 7 cells fixed immediately after addition of fluorescently labeled polyoma VLPs. (C) Control SSCM images of fixed COS 7 cells (no VLPs added). (Top row) Unprocessed topographical images. (Middle row) Processed (slope-corrected and high-pass-filtered, see Materials and Methods) topographical images. (Bottom row) Fluorescent images of Cy5-labeled VLPs. Matching topographical and fluorescent representations of VLPs are indicated with arrowheads and dashed circles. Hollow arrows point to microvilli.
To determine which topographical features correspond to fluorescently labeled VLPs, the positions of fluorescent spots were mapped, and their locations were superimposed on the topographical image as described in Materials and Methods. For each pair of images (topography and fluorescence), VLP-related structures were seen on the cell membrane as topographical protrusions corresponding to fluorescence signals (arrowheads). Places where the VLPs were aggregated or their density was too high to distinguish singles by fluorescence corresponded with larger fluorescent spots (circled with dashed line).
All three confocal images presented in Fig. 2 (bottom row)—A and B of infected cells and C of a control cell—are equally scaled raw data fluorescent images. As can be seen, there is almost no fluorescence detected in the image of a control cell (Fig. 2 C, bottom row), although its topographic counterpart exhibits protrusions of sizes comparable to VLPs (Fig. 2 C, middle row). Therefore, we can conclude that only those protrusions associated with corresponding fluorescent signals can be considered to be real VLPs (marked with arrowheads). The control experiment in Fig. 2 C was repeated five times and gave a fluorescence background of 0.35 ± 0.11 arbitrary units compared with the mean intensity of a single VLP of 1.01 ± 0.27 arbitrary units. This shows that fluorescent spots are detected only when VLPs are present.
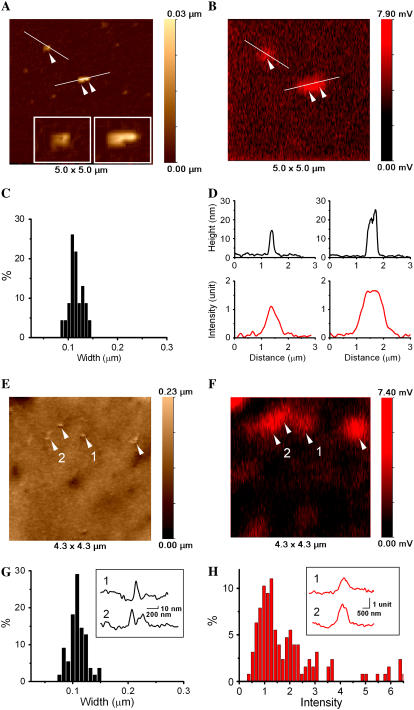
The size of the VLP-related features on the cell surface could correspond to single particles or small aggregates of particles. The majority of these structures were larger than 50–60 nm. Therefore, to determine whether it was likely that a single VLP could be topographically detected by SSCM, we imaged Cy5-labeled VLPs on a flat surface (glass coverslip) and compared these results with images obtained on the cell surface. Fig. 3 compares SSCM high-resolution images of protruding features on glass and on the cell. Topographical and corresponding fluorescence images of VLPs on glass showing one single and one aggregate of two VLPs are shown in Fig. 3, A and B. The insets in Fig. 3 A demonstrate clearly that high-resolution topographical data of nanometer structures can be obtained by SSCM. Fig. 3 C shows the distribution of the VLP width calculated from topographical images of VLPs on glass (N = 23). The corresponding topographical and fluorescence intensity profiles of VLPs on glass are shown in Fig. 3 D. The mean width of the VLPs was calculated as 115 ± 14 nm. This is larger than measurements obtained by electron microscopy (16) but similar to values acquired by AFM measurements (7,14). Such a difference in feature size estimation between scanning probe and electron microscopy has been previously observed. The above increase in feature size is common for scanning probe microscopy technique and is known as “convolution” caused by the finite dimensions of the imaging probe. The mean full width at half-maximum of the fluorescence images of individual VLPs gave a value of 640 ± 50 nm, which is limited by the resolution of the confocal microscope.
FIGURE 3.
High-resolution SSCM imaging of single VLPs on the surface of glass and on the cell membrane. (A) Topographical image showing single VLPs on glass. Insets show zoom of the corresponding particles (single and pair). (B) Fluorescence image of VLPs on glass acquired simultaneously with topographical image shown in A. (C) VLP width distribution calculated from the topographical images of VLPs on glass (N = 23). (D) Topographical (top row) and fluorescence intensity (bottom row) profiles plotted from A and B, respectively. (E) Topographical image of single VLPs on the cell membrane. (F) Fluorescence images of VLPs on the cell membrane acquired simultaneously with the topographical image shown in E. (G) VLP width distribution calculated from the topographical images of VLPs on cell membrane (N = 57). Inset shows topographical profiles of the corresponding VLPs obtained from E. (H) Fluorescence intensity distribution calculated from SSCM images of VLPs on cell membrane (N = 127). Inset shows fluorescence intensity profiles of the corresponding VLPs obtained from E.
Note that the topographical image of the surface of the coverslip also reveals structures that do not have corresponding fluorescence. Some of these structures may be unlabeled VLPs, and others may be impurities in the preparation. The fact that unlabeled features were detected clearly illustrates that topographical data on their own are not enough to identify VLPs even in such simplified artificial conditions and highlights the potential difficulties of imaging VLPs on the surface of the cell membranes.
Fig. 3 E shows a topographical image of VLPs on the surface of the cell. Most of the observed topographical features on the cell surface had corresponding fluorescence signals (Fig. 3 F). To determine the likelihood that protrusions on the cell surface detected by topography and correlated with fluorescent signal were VLPs, their diameters were measured and compared with a similar analysis of VLPs on glass. The results showed VLP features on the cell surface of 108 ± 16 nm (N = 57) (Fig. 3 G). The inset shows topographical profiles of VLPs on the cell surface from Fig. 3 E. Fluorescence intensity distribution of VLPs on the cell membrane revealed three peaks that correspond to single, double, and triple VLP aggregates (Fig. 3 H). A similar distribution of VLP fluorescence intensity acquired on glass was published previously (7). The inset shows the fluorescence intensity profiles of single (1) and double (2) VLPs plotted from the fluorescent image shown in Fig. 3 E. Thus, taking the exact correspondence with the fluorescence signals and the size measurements together, we conclude that SSCM can visualize single VLPs on the surface of the cells.
In formaldehyde-fixed cells, this technique has revealed a cell surface with numerous nanometer-scale protusions and indentations. VLPs were distributed across this surface apparently at random, and no specific interactions were seen with structures such as microvilli.
DISCUSSION
Here we demonstrate that the recent improvement of topographical resolution made for SICM (8) can be applied to the SSCM to allow direct imaging of VLPs on the cell surface. In these experiments, the mean diameter of the structures associated with VLP-linked fluorescence was 115 ± 14 nm. Although this is greater by more than twofold for the size estimated by techniques such as electron microscopy and x-ray crystallography, an increase in apparent feature size has been previously described for scanning probe microscopies and is also in agreement with our previous observations of analysis of single particles by AFM (7,19). It is also noteworthy that both techniques appear to foreshorten the height measurement of the VLPs (Fig. 3) (7,14). Nonetheless, using the size of the features and colocalization with fluorescence allows us now to identify topographical features that correspond to single 50-nm-diameter VLPs.
This ability to simultaneously image both structural features and fluorescence allows the assignment of individual VLPs on the cell surface with considerable certainty. It is important to note that distinguishing between various cell surface features and other nano objects is problematic for microscopy techniques such as scanning probe microscopy and scanning electron microscopy because of their lack of molecular specificity. That is why we believe that the combination of SICM and confocal microscopy, where fluorescence can be used to identify the virus particles, is highly advantageous. With the combination of these techniques, it becomes possible to detect the precise position of virus particle with regard to the cell membrane by topography and then to prove that the topographical feature observed is a real virus particle by using its fluorescent signal.
We are now investigating the possibility of carrying out experiments on live cells. However, there are some limitations in the system that need to be overcome relating to the fact that scan speeds are still slow (∼20 min/scan) and large (micrometer) features on the cell surface or rapid cell movements can disrupt high-resolution scanning. Despite these limitations, the advances we describe here are of particular importance as, for the first time, nanoparticle interactions with the apical surface of cells under conditions adaptable to live cell studies can be characterized. Further, unequivocal differentiation can be made between particles that are still on the outer surface of the cell and those that have recently been endocytosed, permitting analysis of surface interactions preceding receptor docking and entry. Previous imaging of the cell surface by SICM and SSCM (7) has revealed an undulating surface with protrusions and indentations distributed across it. With high-resolution SSCM, cell membrane structures such as endocytic pits have been revealed (20), and, therefore, the position and relations of VLPs to these structures can also now be studied.
Although we describe imaging of VLPs in this article, the technique can be applied to any virus or other nanoparticle, such as gene delivery agents, that can be labeled with a fluorescent moiety. Thus, it is anticipated that this procedure will be useful in characterizing the earliest interactions of these particles with the cell and contribute to identifying novel drugs to prevent viral infection as well as in the development of gene therapy reagents.
Acknowledgments
This work was funded by the Biotechnology and Biological Sciences Research Council.
Editor: Michael Edidin.
References
- 1.Kuznetsov, Y. G., J. G. Victoria, W. E. Robinson, Jr., and A. McPherson. 2003. Atomic force microscopy investigation of human immunodeficiency virus (HIV) and HIV-infected lymphocytes. J. Virol. 77:11896–11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewers, H., A. E. Smith, I. F. Sbalzarini, H. Lilie, P. Koumoutsakos, and A. Helenius. 2005. Single-particle tracking of murine polyoma virus-like particles on live cells and artificial membranes. Proc. Natl. Acad. Sci. USA. 102:15110–15115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuznetsov, Y. G., S. Datta, N. H. Kothari, A. Greenwood, H. Fan, and A. McPherson. 2002. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV). Biophys. J. 83:3665–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansma, P. K., B. Drake, O. Marti, S. A. Gould, and C. B. Prater. 1989. The scanning ion-conductance microscope. Science. 243:641–643. [DOI] [PubMed] [Google Scholar]
- 5.Korchev, Y. E., C. L. Bashford, M. Milovanovic, I. Vodyanoy, and M. J. Lab. 1997. Scanning ion conductance microscopy of living cells. Biophys. J. 73:653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korchev, Y. E., M. Milovanovic, C. L. Bashford, D. C. Bennett, E. V. Sviderskaya, I. Vodyanoy, and M. J. Lab. 1997. Specialized scanning ion-conductance microscope for imaging of living cells. J. Microsc. 188:17–23. [DOI] [PubMed] [Google Scholar]
- 7.Gorelik, J., A. Shevchuk, M. Ramalho, M. Elliott, C. Lei, C. F. Higgins, M. J. Lab, D. Klenerman, N. Krauzewicz, and Y. Korchev. 2002. Scanning surface confocal microscopy for simultaneous topographical and fluorescence imaging: application to single virus-like particle entry into a cell. Proc. Natl. Acad. Sci. USA. 99:16018–16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shevchuk, A. I., G. I. Frolenkov, D. Sanchez, P. S. James, N. Freedman, M. J. Lab, R. Jones, D. Klenerman, and Y. E. Korchev. 2006. Imaging proteins in membranes of living cells by high-resolution scanning ion conductance microscopy. Angew. Chem. Int. Ed. Engl. 45:2212–2216. [DOI] [PubMed] [Google Scholar]
- 9.Shevchuk, A. I., J. Gorelik, S. E. Harding, M. J. Lab, D. Klenerman, and Y. E. Korchev. 2001. Simultaneous measurement of Ca2+ and cellular dynamics: combined scanning ion conductance and optical microscopy to study contracting cardiac myocytes. Biophys. J. 81:1759–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kienberger, F., V. P. Pastushenko, G. Kada, T. Puntheeranurak, L. Chtcheglova, C. Riethmueller, C. Rankl, A. Ebner, and P. Hinterdorfer. 2006. Improving the contrast of topographical AFM images by a simple averaging filter. Ultramicroscopy. 106:822–828. [DOI] [PubMed] [Google Scholar]
- 11.Olin, H. 1994. Design of a scanning probe microscope. Meas. Sci. Technol. 5:976–984. [Google Scholar]
- 12.Savitzkiy, A., and M. J. B. Golay. 1964. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36:1627–1629. [DOI] [PubMed] [Google Scholar]
- 13.Krauzewicz, N., C. Cox, E. Soeda, B. Clark, S. Rayner, and B. E. Griffin. 2000. Sustained ex vivo and in vivo transfer of a reporter gene using polyoma virus pseudocapsids. Gene Ther. 7:1094–1102. [DOI] [PubMed] [Google Scholar]
- 14.Krauzewicz, N., J. Stokrova, C. Jenkins, M. Elliott, C. F. Higgins, and B. E. Griffin. 2000. Virus-like gene transfer into cells mediated by polyoma virus pseudocapsids. Gene Ther. 7:2122–2131. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, J. M., I. G. Goldberg, and T. L. Benjamin. 2003. Cell penetration and trafficking of polyomavirus. J. Virol. 77:2615–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsson, J., N. Miyazaki, L. Xing, B. Wu, L. Hammar, T. C. Li, N. Takeda, T. Miyamura, and R. H. Cheng. 2005. Structure and assembly of a T = 1 virus-like particle in BK polyomavirus. J. Virol. 79:5337–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stehle, T., Y. Yan, T. L. Benjamin, and S. C. Harrison. 1994. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 369:160–163. [DOI] [PubMed] [Google Scholar]
- 18.Yan, Y., T. Stehle, R. C. Liddington, H. Zhao, and S. C. Harrison. 1996. Structure determination of simian virus 40 and murine polyomavirus by a combination of 30-fold and 5-fold electron-density averaging. Structure. 4:157–164. [DOI] [PubMed] [Google Scholar]
- 19.Bishop, C. L., M. Ramalho, N. Nadkarni, K. W. May, C. F. Higgins, and N. Krauzewicz. 2006. Role for centromeric heterochromatin and PML nuclear bodies in the cellular response to foreign DNA. Mol. Cell. Biol. 26:2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchuk, A. I., P. Hobson, M. J. Lab, D. Klenerman, N. Krauzewicz, and Y. E. Korchev. 2008. Endocytic pathways: combined scanning ion conductance and surface confocal microscopy study. Pflugers Arch. 456:227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]