Abstract
Enveloped viruses and nanosized biomimetic particles for drug and gene delivery enter target cells mainly through receptor-mediated endocytosis. A few models have been presented to elucidate the mechanics of particle engulfment by the cell membrane, showing how size and surface chemico-physical properties favor or oppose internalization. In this work, the effect of particle nonsphericity is addressed considering elliptical cylindrical particles with aspect ratio Γ. Using a continuum energetic approach, three different conditions have been identified: for sufficiently small Γ, the particle is not even wrapped by the cell membrane; for sufficiently large Γ, the particle is partially wrapped (“frustrated endocytosis”); and for intermediate values of Γ, the particle is fully wrapped and eventually internalized. Given the pleomorphism of viruses and the broad spectrum of shapes for nanosized biomimetic particles, the results presented may be of interest to virologists, pharmacologists, toxicologists, and nanotechnologists.
INTRODUCTION
Endocytosis is the general term used to define the process by which cells import and export selected extracellular molecules, viruses, microorganisms, and nanosized biomimetic particles. It includes a variety of pathways: clathrin-mediated and caveolae-mediated endocytosis; phagocytosis; and clathrin- and caveolae-independent endocytosis. The pathway chosen depends on the size and nature of the extracellular cargo (1): caveolae-mediated endocytosis is assisted by the activation of caveolae (plasma membrane invaginations) with a characteristic size of 50–60 nm. Clathrin-mediated endocytosis requires the concentration of transmembrane receptors and their bound ligands on the plasma membrane leading to the formation of vesicular cages with a characteristic size up to few hundreds microns (100–500 nm). Phagocytosis involves specific cell-surface receptors and signaling cascades with the formation of cell membrane protrusions that eventually envelop the external micrometer cargo (>1 μm). Clathrin- and caveolae-independent endocytosis is generally associated with the formation of invaginating vesicles smaller than 100 nm. Particle endocytosis is of fundamental importance in several fields as virology (2), drug and gene delivery (3), and in nanotoxicology (4).
For nanosized particles, both natural (as enveloped viruses) or artificial (as biomimetic particulates), the most effective internalization mechanism is the receptor-mediated endocytosis where molecules (ligands) distributed over the particle surface bind to counter-molecules (receptors) expressed over the cell membrane, which eventually bends to invaginate the foreign cargo (2,5). These receptors are generally collected at the site of invagination by surface diffusion, without which endocytosis either would not occur or would occur over a much longer timescale.
Recently a few theoretical analyses have been presented to elucidate the mechanics of the receptor-mediated endocytosis and how the chemical energy available upon formation of new ligand-receptor bonds is transformed into the elastic energy of the cell membrane, and employed to recruit membrane diffusion molecules at the internalization site. Gao et al. (6) have shown for cylindrical and spherical particles that a critical radius exists below which receptor-mediated endocytosis is not energetically favorable; this critical radius depends on the density of ligands decorating the particle, and receptors expressed at the cell membrane depend on the ligand-receptor affinity and cell-membrane-bending modulus. We have considered the contribution of nonspecific attractive and repulsive interactions at the particle/cell interface (7), demonstrating that these can be as important as the specific ligand-receptor binding. Sun and Wirtz (8) have emphasized the importance of the elastic deformation of the cytoskeleton for the final engulfment of the particle, showing how this may be even more important than membrane bending. All these analyses have considered circular cylindrical or spherical particles that are described through one geometrical parameter: the radius of the particle. What is the endocytic behavior of nonspherical particles?
A broad spectrum of nanoparticles to be used as drug carriers (drug delivery systems) and contrast agents (biomedical imaging systems) have been presented in the literature with different compositions, chemico-physical properties, sizes, and shapes. For instance, Mitragotri and co-workers have recently reported on a method to fabricate polystyrene particles with >20 different shapes and characteristic sizes ranging from 60 nm up to 30 μm (9) and has addressed the importance of the particle shape in phagocytic processes (10); DeSimone and co-workers have developed an extremely versatile and flexible method for the direct fabrication of shape-specific nano-biomaterials (11); and silicon microfabricated particles with an hemispherical shape have been produced by Cohen et al. (12). On the other side, Nature provides a variety of biological corpuscles with shapes substantially different from spherical, as virus particles (virions), which are commonly internalized through the receptor-mediated endocytosis pathway. In particular, an enveloped virus is composed of an external membrane, usually studded with proteins, containing the capsid with the virus genome. The International Committee on Taxonomy of Viruses has introduced a characteristic list for enveloped virus based on the virion shape identifying several different classes as filamentous, rod-shaped, bullet-shaped, ovoid, ellipsoid brick- and drop-shaped, and pleomorphic. It is not clear why viruses have developed such a large number of diverse shapes, but recently more attention is being given to possible correlations between the shape and molecular structure of virions and their infectivity and pathogenicity (13).
The objective of this study is to develop a model for analyzing the effect of particle shape and size on receptor-mediated endocytosis.
FORMULATION
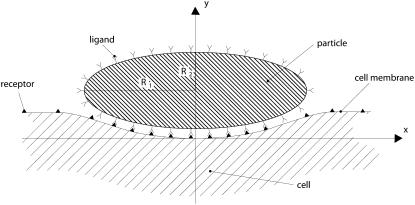
A cylindrical particle is considered to interact with a cell membrane through specific ligand-receptor bonds (Fig. 1). The surface of the particle is decorated with a fixed density ml of ligand molecules, whereas counter-molecules (receptors) are expressed over the cell membrane with a density mr. The cell receptors are free to diffuse over the membrane with a mobility M, and mr can change locally with time under proper stimuli. At time t = 0, the particle is in close proximity to the cell membrane so to nucleate a finite adhesive area and the density of the receptor molecules in the nonadhesive area is assumed to be equal to 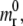 smaller than ml. The surface density of the cell receptors within the adhesive area is fixed and equal to the density of the ligands decorating the particle ml. As a consequence, the molecular bond surface density mb is equal to ml.
smaller than ml. The surface density of the cell receptors within the adhesive area is fixed and equal to the density of the ligands decorating the particle ml. As a consequence, the molecular bond surface density mb is equal to ml.
FIGURE 1.
A nonspherical particle sitting on a cell membrane during the internalization process.
The initially nucleated area can grow or recede over time depending on the biophysical and geometrical properties of the cell-particle system. A positive driving force for the growth of the adhesion area is given by the formation of new ligand-receptor bonds at the adhesive edge, whereas negative driving forces are associated with the deformation of the cell membrane and cytoskeleton complex required to make the cell conform to the particle surface, and the recruitment of receptor molecules at the adhesive front through membrane diffusion to provide fresh new receptors for molecular binding.
In the sequel, the Freund and Lin (14) formulation is recalled and readapted to analyze cell wrapping around a cylindrical particle with a sufficiently arbitrary cross section described by the function y(x) within the xy plane. The local curvature of the particle is given by
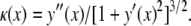 |
(1) |
A curvilinear abscissa u(x) is defined over the particle surface with 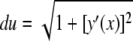 dx, and the length of the adhesive area Lw is given by
dx, and the length of the adhesive area Lw is given by
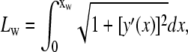 |
(2) |
where xw is the abscissa of the adhesive edge.
The power balance during cell wrapping
At the arbitrary time t, the length of the adhesive area is assumed to be a(t); and the surface density of the receptor molecules in the nonadhesive area is in general mr(x;t), different from 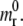 Starting from this configuration, it is assumed that the adhesive area increases by δa within the time δt, being δa sufficiently small to assume locally a fixed curvature equal to κ(a), and a fixed receptor density equal to mr(a+;t). The small increment δa is associated with a free energy variation δF obtained by summing up the contribution of the molecular bond formation (positive driving force), the bending of the membrane-cytoskeleton complex within the adhesive region (a, a + δa) where the curvature is κ(a), and the recruitment of new receptors through surface diffusion toward the adhesive front.
Starting from this configuration, it is assumed that the adhesive area increases by δa within the time δt, being δa sufficiently small to assume locally a fixed curvature equal to κ(a), and a fixed receptor density equal to mr(a+;t). The small increment δa is associated with a free energy variation δF obtained by summing up the contribution of the molecular bond formation (positive driving force), the bending of the membrane-cytoskeleton complex within the adhesive region (a, a + δa) where the curvature is κ(a), and the recruitment of new receptors through surface diffusion toward the adhesive front.
The chemical potential μr of a free receptor over the cell membrane is written as in Hill (15),
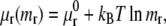 |
(3) |
where 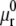 represents the chemical potential at a standard surface density of one molecule per unit area; the chemical potential of the ligand-receptor bond is written as
represents the chemical potential at a standard surface density of one molecule per unit area; the chemical potential of the ligand-receptor bond is written as
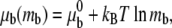 |
(4) |
where 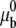 is related to
is related to 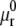 by
by 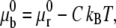 with C the binding energy factor. The free energy variation δFμ related to the formation of new bonds and to the redistribution of the receptor molecules over the cell membrane is given as
with C the binding energy factor. The free energy variation δFμ related to the formation of new bonds and to the redistribution of the receptor molecules over the cell membrane is given as
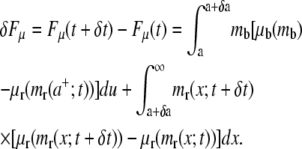 |
(5) |
Within the adhesive area (a, a + δa), the surface density of the receptors has increased from mr(a+;t) to form new bonds with a density mb = ml, as described by the first term on the right-hand side of Eq. 5. The recruitment of new receptors within the adhesive area induces their redistribution in the outer region (a + δa, ∞) changing m(x;t), as accounted for in the second term on the right-hand side of Eq. 5.
The free energy variation δFe related to the bending of the membrane-cytoskeleton complex over the portion δa of the particle surface is given as
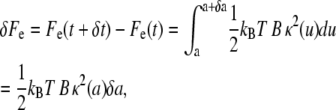 |
(6) |
with κ(a) fixed. Since this analysis is focused on particles with a characteristic size at least one order-of-magnitude smaller than that of the cell, the contribution of the cortical tension γ has been neglected (see Discussion).
Summing δFμ and δFe, and substituting for μr and μb as from Eqs. 3 and 4, the total free energy variation δF for the system is
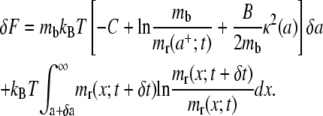 |
(7) |
Taking the derivative of δF with respect to time t, it follows
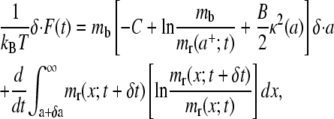 |
(8) |
and recalling the formulation proposed in Freund and Lin (14), it is finally derived
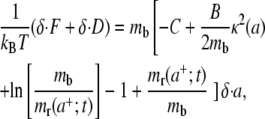 |
(9) |
where the quantity D is the energy dissipation as a result of the receptor transport over the cell membrane, and 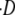 is the dissipation rate. As in a mechanical system, for a fixed temperature, the work available upon bond formation is in part used to make the cell conform to the particle (reversible bending) and in part is dissipated through irreversible receptor membrane diffusion. Therefore the relation in Eq. 9 gives the power balance for the cell/particle system which has to be zero at each time t during the system evolution. The first two terms on the right-hand side of Eq. 9 are related to the ligand-receptor binding and the membrane-cytoskeleton bending, whereas the following two terms are related to the recruitment of new receptors at the adhesive edge being their contribution null for mb = mr(a+;t). The only difference between Eq. 9 and a similar relation derived in Freund and Lin (14) is the bending energy contribution which is here related to a nonconstant curvature κ(a), varying with the size of the adhesive area a(t).
is the dissipation rate. As in a mechanical system, for a fixed temperature, the work available upon bond formation is in part used to make the cell conform to the particle (reversible bending) and in part is dissipated through irreversible receptor membrane diffusion. Therefore the relation in Eq. 9 gives the power balance for the cell/particle system which has to be zero at each time t during the system evolution. The first two terms on the right-hand side of Eq. 9 are related to the ligand-receptor binding and the membrane-cytoskeleton bending, whereas the following two terms are related to the recruitment of new receptors at the adhesive edge being their contribution null for mb = mr(a+;t). The only difference between Eq. 9 and a similar relation derived in Freund and Lin (14) is the bending energy contribution which is here related to a nonconstant curvature κ(a), varying with the size of the adhesive area a(t).
The surface density of receptor molecules
The receptor molecules are free to diffuse over the cell membrane with a mobility M and their distribution is governed by the classical dynamic diffusion equation (∂mr(x;t)/∂t + M, ∂2mr(x;t)/∂x2 = 0). Regardless of how the adhesive area a(t) evolves with time, the receptor density has to satisfy the same auxiliary conditions defined in Freund and Lin (14): m(x;0) = 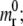 m(x;t) =
m(x;t) = 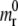 as x tends to infinity; and the flux condition across the adhesive front
as x tends to infinity; and the flux condition across the adhesive front 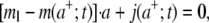 j(x;t) representing the number of receptor molecules passing a membrane section at x per unit time t. Therefore the function mr(x;t) has the same identical form of Freund and Lin (14), that is to say
j(x;t) representing the number of receptor molecules passing a membrane section at x per unit time t. Therefore the function mr(x;t) has the same identical form of Freund and Lin (14), that is to say
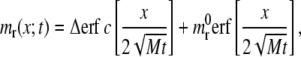 |
(10) |
where erf(z) and erf c(z) are, respectively, the error function and the complementary error function, with
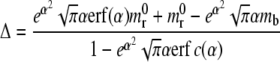 |
(11) |
and the adhesive area a having the form
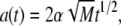 |
(12) |
with α a speed factor to be determined enforcing energy conservation at each time t.
The characteristic equation and incremental solution
The power balance presented in Eq. 9 and the receptor distribution given in the paragraph above hold for each time t during the system evolution. Then the speed factor α can be derived by substituting Eq. 12 and Eq. 10 in Eq. 9, leading to the characteristic equation
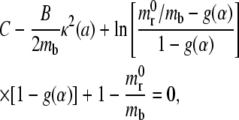 |
(13) |
with
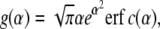 |
(14) |
which, for a fixed κ(a), would give the classical Freund and Lin equation (derived in (14)). For a particle with a nonfixed curvature, the parameter κ(a) changes as the adhesive area grows and consequently, the speed factor α changes over time and has to be estimated by solving Eq. 13 incrementally.
The particle profile y(x) is discretized in N nodes (N − 1 elements) with fixed unit length δa = Lw/(N−1). At time t = 0, the curvature to be considered in Eq. 13 is κ(0), and since all the biophysical parameters in Eq. 13 are known data (from Table 1), the corresponding speed factor can be readily estimated solving for α(0). If the speed factor α(0) is larger than zero, the adhesive front proceeds by δa over a time δt, which can be derived by discretizing Eq. 12 to obtain
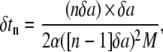 |
(15) |
where nδa is the actual length of the adhesion area (= δa for t = 0), and α([n−1]δa) is the actual speed factor (calculated at x = 0 for t = 0). At time t = δt1, the new curvature to be considered is κ(δa) leading to the speed factor α(δa) as from Eq. 13. Again from Eq. 15, the new time increment δt2 required is calculated as
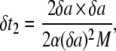 |
(16) |
considering that the new adhesive area length is 2δa (n = 2). The iteration over the N nodes continues as long as the speed factor α is larger than zero. When such condition is met the total length of the adhesive area is simply given by nδa, while the total time for wrapping is estimated by summing all the calculated 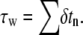 Note, in fact, that δa is fixed for each surface element, but δtn would change in that κ(a) would be different for each element.
Note, in fact, that δa is fixed for each surface element, but δtn would change in that κ(a) would be different for each element.
TABLE 1.
Biological and biophysical data for the problem
| B | M [m2/s] | C | 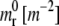 |
mt [m−2] | Rmin [nm] |
|---|---|---|---|---|---|
| 200 | 10−14 | 15 | 5 × 1014 | 5 × 1015 | 12.13 |
In this formulation, the contribution of nonspecific cell/particle interactions, as in our earlier work (7), has not been considered. However, following the same reasonings proposed in the paragraphs above, it can be shown that a more general expression for Eq. 13 including the nonspecific interactions is readily derived by introducing a nonconstant κ(a) in our characteristic equation from Decuzzi and Ferrari (7) and solving incrementally the corresponding system of equations.
RESULTS
The internalization process of cylindrical particles with circular and elliptical cross sections are analyzed in the sequel. The biophysical parameters of interest are listed in Table 1, where the initial surface density for the receptors is 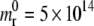 m−2; the fixed ligand density is ml = 5 × 1015 m−2(= mb); the mobility M of the cell receptors is 10−14 m2/s; the binding factor C is 15; and the bending factor B is 200. The bending factor used in this analysis considers the contribution of both the cell membrane and the internal cytoskeleton, as described in more detail in the Discussion.
m−2; the fixed ligand density is ml = 5 × 1015 m−2(= mb); the mobility M of the cell receptors is 10−14 m2/s; the binding factor C is 15; and the bending factor B is 200. The bending factor used in this analysis considers the contribution of both the cell membrane and the internal cytoskeleton, as described in more detail in the Discussion.
Circular cylindrical particle
For a circular particle, the critical radius Rmin below which internalization by receptor-mediated endocytosis is energetically unfavorable has been given explicitly in Eq. 6 as a function of B/ml and the ratio 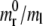 being
being
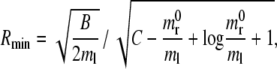 |
(17) |
which corresponds to a velocity parameter α = 0. Referring to the data given in Table 1, the minimum radius Rmin would be of ∼40 nm (strictly 38.35 nm), giving a critical curvature κmax = 1/Rmin = 0.026 nm−1, above which internalization would not be energetically favorable. Circular particles with R > 40 nm would be internalized (α > 0) with a wrapping time τw growing nonlinearly with the particle size as from τw = (πR)2/(4α2M), which has been derived by Eq. 12 with a = Lw = πR. On the other hand, particles with a radius <40 nm are predicted not to be internalized (Eq. 17 has no real solution).
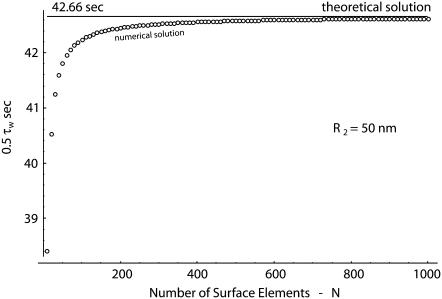
Following the discretization procedure described in the previous paragraph and the incremental solution of Eq. 13, the time 0.5τw to wrap half of the particle surface (Lw = πR/2) is estimated for a circular particle with a radius R = 50 nm, considering a growing number of nodes N. The variation of 0.5τw with N is plotted in Fig. 2 (dots) as the number of nodes increases, where the solid line represents the exact solution (0.5τw = 42.66 s). For N = 103, the difference between the theoretical and numerical results is of ∼0.1%. Similar errors have been observed for particles having a different size.
FIGURE 2.
The half-wrapping time for a circular particle with R = 50 nm: theoretical solution given by Eq. 12 (solid line) and numerical solution given by solving incrementally Eq. 13 (small open circles) with a growing number of elements N.
Elliptical cylindrical particle
What does happen when particles are not circular so that their curvature κ changes as wrapping proceeds? This is analyzed considering a particle with an elliptical cross-section described by the relation
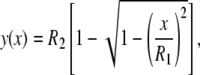 |
(18) |
where R1 is the semi-length of the major axis, and R2 is the semi-length of the minor axis (Fig. 1). The aspect ratio Γ of the ellipse is given by R1/R2, and the radii of curvature at the two rims are given respectively by R′eq = R2Γ2 (= R1Γ) at x = 0 and R″eq = R2Γ−1 (= R1/Γ2) at x = R1. The expression for the local curvature is given by κ(x) as defined in Eq. 1.
Two different conditions are considered in the sequel to assess the effect of Γ on the internalization process: elliptical particles with a fixed length R2 of the semi-minor axis and elliptical particles with a fixed volume V = πR1R2.
Fixed length R2 of the semi-minor axis
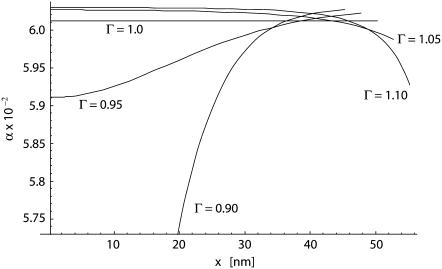
In Fig. 3, the variation of the speed factor α as a function of x, the projection of the adhesive edge along the x axis, is shown for different values of the aspect ratio Γ ranging between 0.9 and 1.1. The length R2 has been assumed to be of 50 nm. The speed factor is derived by solving Eq. 13 incrementally. For Γ = 1, the particle degenerates into a circular cylinder with a constant speed factor α ≃ 0.06 and xmax = 50 nm (= R1 = R2). As the shape deviates from circularity, the curvature of the particle is no more constant along the adhesive area and the speed factor α changes as wrapping proceeds.
FIGURE 3.
The variation of the speed factor α as wrapping proceeds for different values of Γ (= 0.9; 0.95; 1.0; 1.05, and 1.1) with a fixed R2 = 50 nm.
The geometry of the particle is fully defined by giving the values of Γ and R2. For particles having the major axis orthogonal to the cell membrane (Γ < 1), the first contact occurs at the rim x = 0 with a local curvature of κ(0) =1/(R2Γ2). If κ(0) is smaller than the limiting value κmax defined above, the wrapping of the cell around the particle is energetically favored. Otherwise, wrapping would not even start. This limiting condition leads to identify a first critical value for Γ as Γ′cr = (R2κmax)−1/2, defined as the aspect ratio below which wrapping cannot occur. As Γ increases above Γ′cr, κ(0) reduces and wrapping becomes more and more energetically favorable. This is shown in Fig. 3 by the curves Γ = 0.9 and 0.95 with a speed factor α growing steadily with the advancement of the adhesive front: the cell membrane encounters points on the particle profile having a decreasing curvature as proceeding from x = 0 to x = R1. Eventually, the particle is fully wrapped.
An opposite behavior is observed for aspect ratios Γ larger than unity, that is to say the major particle axis is parallel to the cell membrane. In this case, the limiting curvature is the one at the rim x = R1, where κ(R1) = Γ/R2. If κ(R1) is larger than κmax, the wrapping of the particle cannot be fully completed, leading to partial engulfment. This condition is associated with values of Γ larger than a second critical aspect ratio given by Γ″cr = R2κmax. For Γ smaller than Γ″cr, the particle is fully wrapped by the cell as shown in Fig. 3 for Γ = 1.05 and 1.1. For the data given in Table 1 and R2 = 50 nm, the critical values for the aspect ratio are Γ′cr ≃ 0.90 (= (R2κmax)−1/2) and Γ″cr ≃ 1.30 (= R2κmax).
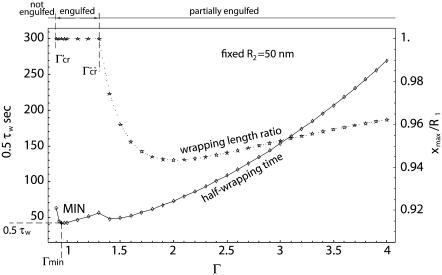
A more general picture of the evolution of the system is presented in Fig. 4, where the half-wrapping time 0.5τw and the wrapping length ratio xmax/R1 are presented as a function of Γ, ranging from 0.9 to 4, and for R2 = 50 nm. The parameter xmax is the projection of the wrapping length Lw over the x axis and for xmax/R1 = 1 complete wrapping is predicted for t = τw. For Γ < Γ′cr (∼0.9 in this case), wrapping cannot even start, leading to infinitely large τw and null ratios xmax/R1 (not shown in Fig. 4). For intermediate values of Γ, that is, Γ′cr < Γ < Γ″cr, the ratio xmax/R1 is always unity—meaning that complete engulfment of the particle occurs, leading eventually to particle internalization and 0.5τw grows with Γ almost linearly. For Γ > Γ″cr, xmax/R1 becomes smaller than unity but does not exhibit a monotonic behavior: the ratio xmax/R1 decreases, reaches a minimum, and then increases steadily, tending again to unity for infinitely large Γ. The half-wrapping time 0.5τw also decreases up to a minimum, to increase again steadily.
FIGURE 4.
The variation of the half-wrapping time and wrapping length ratio xmax/R1 as a function of the aspect ratio Γ, for a fixed R2 = 50 nm.
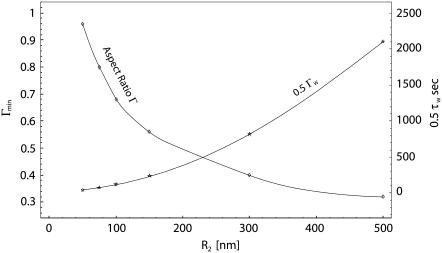
From Fig. 4, the existence of a minimum time for full wrapping (the point MIN) can be interestingly derived. And this minimum wrapping time is associated to a Γmin smaller then unity, as better shown by the curves given in Fig. 5, where the minimum half-time for internalization (0.5 τw) and the corresponding aspect ratio Γmin are plotted as a function of R2. The same data are then listed in Table 2, together with the different values of Γ′cr and Γ″cr. As the characteristic particle size R2 grows, the aspect ratio Γmin for which the internalization time is smaller reduces significantly, being 1 for R2 = Rmin and 0.32 for R2 = 500 nm. The comparison here is performed among particles having the same R2 and varying Γ, or R1. For fixed R2 as Γ reduces the total surface to be wrapped becomes smaller and smaller, favoring rapid internalization, and, on the other hand, the curvature at x = 0 increases, reducing the values of the speed factors during initial wrapping. A balance between these two opposite contributions leads to Γmin smaller than unity but evidently sufficiently larger than Γ′cr, as presented in Table 2. This is even more true as R2 grows.
FIGURE 5.
The variation of the minimum half-wrapping time and corresponding aspect ratio Γmin as a function of the semi-length of the ellipse minor axis (R2 = 50 nm).
TABLE 2.
The variation of the minimum wrapping half-time and corresponding aspect ratio Γmin as a function of R2
| R2 [nm] | Γmin | 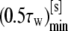 |
Γ′cr | Γ′cr |
|---|---|---|---|---|
| 38.35 (=Rmin) | 1 | ∞ | 1 | 1 |
| 50 | 0.96 | 41.60 | 0.87 | 1.30 |
| 75 | 0.80 | 78.33 | 0.71 | 1.95 |
| 100 | 0.68 | 123.4 | 0.62 | 2.61 |
| 150 | 0.56 | 243.7 | 0.50 | 3.91 |
| 300 | 0.40 | 823.6 | 0.36 | 7.82 |
| 500 | 0.32 | 2105.2 | 0.28 | 13.0 |
The corresponding critical values for Γ are also listed. Fixed R2 case.
Fixed volume
Differently from the case analyzed in the previous paragraph, in the sequel the comparison is performed between ellipse with the same cross-sectional area, that is to say with cylindrical particles having the same volume. As a reference, a circular particle with fixed radius Rs is considered, whose cross-sectional area is 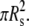 So that, considering the arbitrary elliptical particle with area
So that, considering the arbitrary elliptical particle with area 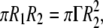 the condition is imposed through the relation
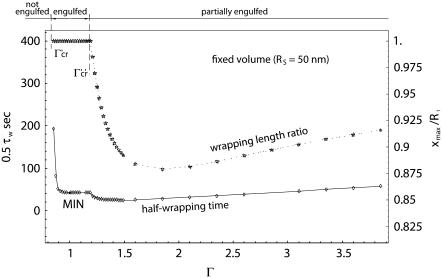
the condition is imposed through the relation 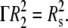 Therefore, the two new critical values of Γ are given by Γ′cr = (κmaxRs)−2/3 and Γ″cr = (κmaxRs)2/3. Fixing Rs = 50 nm, if Γ < Γ′cr (= 0.84), wrapping cannot even start; for Γ > Γ″cr (= 1.19), only partial wrapping occurs; but complete wrapping with particle internalization is predicted to occur for intermediate values of Γ.
Therefore, the two new critical values of Γ are given by Γ′cr = (κmaxRs)−2/3 and Γ″cr = (κmaxRs)2/3. Fixing Rs = 50 nm, if Γ < Γ′cr (= 0.84), wrapping cannot even start; for Γ > Γ″cr (= 1.19), only partial wrapping occurs; but complete wrapping with particle internalization is predicted to occur for intermediate values of Γ.
This is shown more clearly in Fig. 6, where the variation of the half-wrapping time 0.5τw and the wrapping length ratio xmax/R1 are presented as a function of Γ, and for Rs = 50 nm. The behavior of the curves is similar to that observed for the fixed R2 case. However, a significant difference is depicted: the minimum internalization time is associated with an almost-unity aspect ratio, as clearly reported by the data listed in Table 3, showing for each Rs, the Γmin and corresponding minimum time. Also, the minimum is pretty flat, implying that particles with the same volume and with small deviations from circularity are still characterized by a minimum wrapping and, eventually internalization, time. It should be recalled that circular cylindrical particles have the smaller perimeter for a fixed area, or smaller surface area for a fixed volume.
FIGURE 6.
The variation of the half-wrapping time and wrapping length ratio xmax/R1 as a function of the aspect ratio Γ, for a fixed volume (Rs = 50 nm).
TABLE 3.
The variation of the minimum wrapping half-time and corresponding aspect ratio Γmin as a function of Rs
| Rs [nm] | Γmin | 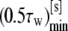 |
Γ′cr | Γ′cr |
|---|---|---|---|---|
| 38.35 (=Rmin) | 1 | ∞ | 1 | 1 |
| 50 | 1.055 | 42.48 | 0.84 | 1.19 |
| 75 | 1.001 | 95.30 | 0.64 | 1.56 |
| 100 | 1.003 | 169.45 | 0.53 | 1.89 |
| 150 | 1.000 | 381.15 | 0.40 | 2.48 |
| 300 | 0.996 | 1524.6 | 0.25 | 3.94 |
| 500 | 1.000 | 4235 | 0.18 | 5.54 |
The corresponding critical values for Γ are also listed. Fixed volume case.
DISCUSSION
Recently, the internalization of artificial particles with a nonspherical shape has been studied experimentally by Champion and Mitragotri (10), who considered nonopsonized and IgG-opsonized polystyrene particles with different shapes (oblate and prolate ellipsoids; elliptical and rectangular disks; as well as spherical) and with characteristic sizes ranging between 1 and 10 μm. The propensity to internalization and the internalization speed has been expressed as a function of a characteristic angle Ω. Particles with a small Ω (<45°) are elongated particles oriented normally to the cell membrane, corresponding to Γ smaller than unity within this work; particles with a large Ω (>45°) are elongated particles laying parallel to the cell membrane, corresponding to Γ larger then unity within this work; and spherical particles are always characterized by Ω = 45°. It was observed that the smaller the Ω, the larger the propensity for internalization and the internalization velocity; whereas for sufficiently large Ω, above 45°, the internalization velocity is zero, implying that phagocytosis cannot be completed, leading to what has been known for a long time as “frustrated phagocytosis”. These results are in qualitative agreement with those derived in this analysis, showing that elongated particles laying parallel to the cell membrane cannot be fully engulfed (Γ > Γ″cr). The smallest value of Ω considered is of ∼20°, possibly insufficiently small to depict the lower limit for internalization predicted by this analysis through the parameter Γ′cr.
Additional to this qualitative agreement, it should, however, be emphasized that the presented mathematical model has been developed for the receptor-mediated endocytosis (RME) of small particles (sub-micrometer) rather than the phagocytosis of large particles (1 μm up to the characteristic size of a phagocytic cell 10 μm). There are at least two significant differences between phagocytosis and RME for what concerns this analysis: the polymerization forces responsible for the formation of membrane protrusions (phagocytic cups) embracing the particle during phagocytosis, which are not present in RME; and the entity of cell deformation, which is much less dramatic in RME than during phagocytosis. For particle internalization, the endocytosis models in the literature (6,7) have considered the sole bending of the cell membrane, whereas Sun and Wirtz (8) have emphasized the importance of the actin cytoskeleton by modeling it as a linear elastic isotropic half-space with a 10 kPa Young's modulus. Evidently, the latest model predicts penetration depths for the particle smaller than those estimated through the other two approaches. Even if the role of the actin cytoskeleton during RME and phagocytosis and the mechanisms associated with the formation and transport of vesicles toward endosomes and lysosomes has not been fully clarified, it is sufficiently clear that the actin cytoskeleton participates in the internalization process, actively favoring or not favoring it, depending on the molecular recognition of the foreign object. For instance, the role of the actin regulation in clathrin-mediated endocytosis has been investigated in Herant et al. (16), showing that actin filaments are recruited at the site of vesicle formation during internalization and mediate the transport of the vesicle toward cell endosomes, thus favoring, rather than opposing, the process. Moreover, proteins involved in endocytosis have also been shown to be involved in the reorganization of the actin cytoskeleton to promote the entry of viral particles (17). Similarly in phagocytosis (18), the actin cytoskeleton tends to be considered as an active component of cells, with finely controlled rearrangement mechanisms regulating particle penetration and transport. Consequently, in this model, considering also that the focus is on sub-micrometric rather than large particles, the sole bending of the membrane-cytoskeleton complex has been considered introducing a combined modulus of the 200 kBT, as from Simson et al. (19), which is one order-of-magnitude larger than the bending modulus used generally for the sole cell membrane (O(10 kBT)). Therefore, this can be seen as an intermediate approach compared to the two extreme cases recalled above: cell membrane bending solely (6,7); and membrane bending and cytoskeleton indentation (8).
In addition to this, due again to the small size of the particle compared to the cell, the contribution of the membrane cortical tension γ in calculating the cell elastic energy can be neglected. This is reasonable if a spherical particle with a radius of 1 μm is considered to be internalized by a cell with a characteristic radius of 10 μm: the induced volume change in the cell would be ∼0.1%, with consequently no significant change in cell surface area. Also, the cortical tension has been experimentally proved to be constant and small (0.03 mN m−1), in the case of the phagocytosis by macrophages of latex beads as large as 3 μm (20).
CONCLUSIONS
It has been shown that the internalization of cylindrical particles with an elliptical cross section can be controlled through two geometrical parameters—the volume of the particle and its aspect ratio Γ. Three different behaviors of the particle-cell system have been observed, depending on the value of the aspect ratio: if Γ is smaller than a critical value Γ′cr, the particle is not wrapped at all; if Γ is larger than a critical value Γ″cr, the particle is partially wrapped, leading to what can be defined as frustrated endocytosis; if Γ ranges between Γ′cr and Γ″cr, the particle is fully wrapped and eventually internalized, giving the necessary time. Interestingly, the intermediate behavior with partial wrapping cannot be predicted for circular cylindrical particles. Comparing particles with the same volume, it has been also observed that circular particles and particles slightly deviating from circularity are associated with the minimum wrapping time, whereas particles laying parallel or orthogonally to the cell membrane may not be internalized if too elongated.
The internalization behavior can be significantly affected also by the nonuniform distribution of ligand molecules on the particle surface. If sufficiently large ligand densities are generated at rims with small curvature, the particle could be internalized regardless of its orientation with respect to the cell membrane.
The presented results not only may be of interest for the rational design of nanosized biomimetic particles for drug and gene delivery, but can be of help to virologists interested in correlating the shape and structure of virions with their infectivity and pathogenicity.
Acknowledgments
The authors thank the reviewers for their perceptive and constructive comments.
The authors were supported by the U.S. Department of Defense through grant No. W81XWH-04-2-0035 and by the Emerging Technology Fund by the State of Texas through the grant “Acquisition of Scientific Superiority in Biomedical Nanotechnology”. M.F. was also supported by NASA through grant No. SA23-06-017, “Nanotechnology for Space Medicine”.
Editor: Denis Wirtz.
References
- 1.Conner, S. D., and S. L. Schmid. 2003. Regulated portals of entry into the cell. Nature. 422:37–44. [DOI] [PubMed] [Google Scholar]
- 2.Marsh, M., and A. Helenius. 2006. Virus entry: open sesame. Cell. 124:729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasir, J. K., and V. Labhasetwar. 2006. Polymeric nanoparticles for gene delivery. Expert Opin. Drug Deliv. 3:325–344. [DOI] [PubMed] [Google Scholar]
- 4.Oberdorster, G., E. Oberdorster, and J. Oberdorster. 2005. Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 113:823–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith, A. E., and A. Helenius. 2004. How viruses enter animal cells. Science. 304:237–242. [DOI] [PubMed] [Google Scholar]
- 6.Gao, H., W. Shi, and L. B. Freund. 2005. Mechanics of receptor-mediated endocytosis. Proc. Natl. Acad. Sci. USA. 102:9469–9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decuzzi, P., and M. Ferrari. 2007. The role of specific and non-specific interactions in receptor-mediated endocytosis of nanoparticles. Biomaterials. 28:2915–2922. [DOI] [PubMed] [Google Scholar]
- 8.Sun, S. X., and D. Wirtz. 2006. Mechanics of enveloped virus entry into host cells. Biophys. J. 90:L10–L12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Champion, J. A., Y. K. Katare, and S. Mitragotri. 2007. Making polymeric micro- and nanoparticles of complex shapes. Proc. Natl. Acad. Sci. USA. 104:11901–11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champion, J. A., and S. Mitragotri. 2006. Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. USA. 103:4930–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolland, J. P., B. W. Maynor, L. E. Euliss, A. E. Exner, G. M. Denison, and J. M. DeSimone. 2005. Direct fabrication and harvesting of monodisperse, shape-specific nanobiomaterials. J. Am. Chem. Soc. 127:10096–10100. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, M. H., K. Melnik, A. A. Boiarski, M. Ferrari, and F. J. Martin. 2001. Microfabrication of silicon-based nanoporous particulates for medical applications. Biomed. Microdevices. 3:253–259. [Google Scholar]
- 13.Harris, A., G. Cardone, D. C. Winkler, J. B. Heymann, M. Brecher, J. M. White, and A. C. Steven. 2006. Influenza virus pleomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 103:19123–19127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freund, L. B., and Y. Lin. 2004. The role of binder mobility in spontaneous adhesive contact and implications for cell adhesion. J. Mech. Phys. Solids. 52:2455–2472. [Google Scholar]
- 15.Hill, T. L. 1960. An Introduction to Statistical Thermodynamics. Dover Publications, Mineola, New York.
- 16.Herant, M., V. Heinrich, and M. Dembo. 2006. Mechanics of neutrophil phagocytosis: experiments and quantitative models. J. Cell Sci. 119:1903–1913. [DOI] [PubMed] [Google Scholar]
- 17.Smythe, E., and K. R. Ayscough. 2006. Actin regulation in endocytosis. J. Cell Sci. 119:4589–4598. [DOI] [PubMed] [Google Scholar]
- 18.May, R. C., and L. M. Machesky. 2001. Phagocytosis and the actin cytoskeleton. J. Cell Sci. 114:1061–1077. [DOI] [PubMed] [Google Scholar]
- 19.Simson, R., E. Wallraff, J. Faix, J. Niewohner, G. Gerisch, and E. Sackmann. 1998. Membrane bending modulus and adhesion energy of wild-type and mutant cells of Dictyostelium lacking talin or cortexillins. Biophys. J. 74:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herant, M., V. Heinrich, and M. Dembo. 2005. Mechanics of neutrophil phagocytosis: behavior of the cortical tension. J. Cell Sci. 118:1789–1797. [DOI] [PubMed] [Google Scholar]