Abstract
ATP-sensitive K+ (KATP) channels are known to play important roles in various cellular functions, but the direct consequences of disruption of KATP channel function are largely unknown. We have generated transgenic mice expressing a dominant-negative form of the KATP channel subunit Kir6.2 (Kir6.2G132S, substitution of glycine with serine at position 132) in pancreatic beta cells. Kir6.2G132S transgenic mice develop hypoglycemia with hyperinsulinemia in neonates and hyperglycemia with hypoinsulinemia and decreased beta cell population in adults. KATP channel function is found to be impaired in the beta cells of transgenic mice with hyperglycemia. In addition, both resting membrane potential and basal calcium concentrations are shown to be significantly elevated in the beta cells of transgenic mice. We also found a high frequency of apoptotic beta cells before the appearance of hyperglycemia in the transgenic mice, suggesting that the KATP channel might play a significant role in beta cell survival in addition to its role in the regulation of insulin secretion.
Keywords: sulfonylurea receptor, inward rectifier, apoptosis
ATP-sensitive K+ (KATP) channels are thought to regulate various cellular functions such as secretion and muscle and neural excitability by linking the cell’s metabolic state to its membrane potential (1–7). We have shown previously that KATP channels comprise at least two subunits, a sulfonylurea receptor (SUR), which belongs to the ATP-binding cassette superfamily (8), and the inward rectifier K+ channel member Kir6.2 (9). SUR is thought to confer the ATP and sulfonylurea sensitivities of KATP channels, and Kir6.2 is thought to form the K+ ion-selective pore (9–12). However, a recent study (13)suggests that Kir6.2 alone might confer ATP-sensitivity. Although mutations of SUR1, the other subunit of pancreatic beta cell KATP (SUR1/Kir6.2) channels, is known to cause familial persistent hyperinsulinemic hypoglycemia of infancy (PHHI) due to a loss of functional KATP channels (14–18), the consequences of inhibition of Kir6.2 function are unknown.
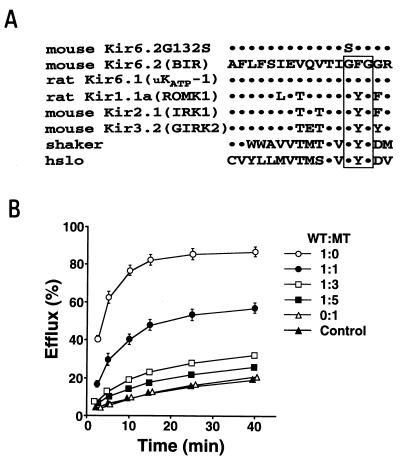
The putative K+ ion permeable domain (H5) is highly conserved in K+ channels (19), and the motif Gly-Tyr (or Phe)-Gly in H5 is thought to be critical for K+ ion selectivity (19–21). A substitution of the first residue of the Gly-Tyr-Gly motif with serine (residue 156) is found in the G-protein-gated inward rectifier GIRK2 (Kir3.2) of weaver mice (22). This mutation is responsible for their abnormalities of neuronal differentiation and development (23–26). In the present study, we have replaced the Gly-312 with Ser at the corresponding position of Kir6.2 (Kir6.2G132S) by in vitro mutagenesis. We found by 86Rb+ flux measurements that Kir6.2G132S functions as a dominant-negative mutant. We then generated transgenic mice expressing Kir6.2G132S in pancreatic beta cells and studied the morphological and functional changes in the pancreatic islets.
MATERIALS AND METHODS
Construction of Mutant Kir6.2.
Mouse Kir6.2 cDNA (9) was subcloned into pALTER (Promega). A single strand template of pALTER Kir6.2 was prepared using helper phage R408. Gly-132 (GGT) in Kir6.2 was mutated to Ser (AGT) by replacing guanine with adenine (Kir6.2 G132S) by site-directed mutagenesis (Fig. 1A), according to the manufacturer’s instructions (in vitro mutagenesis systems; Promega). For 86Rb+ efflux measurement, Kir6.2G132S cDNA was subcloned into the mammalian expression vector pCMV6b. Plasmid constructions were confirmed by DNA sequencing.
Figure 1.
(A) Alignment of amino acid sequences of the H5 regions of K+ channel members. The residues identical to mouse Kir6.2 are shown by dots. The Gly (G)-Phe (F) [or Tyr (Y)]-Gly (G) motifs are boxed. The Gly-132 (G) of mouse Kir6.2 was mutated to Ser (S) to generate the mutant Kir6.2 (Kir6.2G132S). The sequences are mouse Kir6.2 (GenBank accession no. D50581), rat Kir6.1(GenBank accession no. D42145), rat Kir1.1a (GenBank accession no. X72341), mouse Kir2.1(GenBank accession no. X73052), mouse Kir3.2 (GenBank accession no. U11859), shaker (GenBank accession no. M17211), and hslo (GenBank accession no. U11717). (B) 86Rb+ efflux in COS-1 cells transfected with wild-type Kir6.2 (WT) and the mutant Kir6.2G132S (MT) at various ratios together with SUR1. 86Rb+ efflux in COS-1 cells transfected with SUR1 alone is used as the control efflux (▴). The ratio of WT to MT is 1:0 (○), 1:1 (•), 1:3 (□), 1:5 (▪), and 0:1 (▵). Some of the error bars are not seen because of very small standard errors.
Cell Culture and Transfection.
COS-1 cells were plated at a density of 1 × 105 cells per dish (35 mm in diameter) for single-channel recordings, and 3 × 105 cells per dish for 86Rb+ efflux measurements. They were cultured in DMEM supplemented with 10% fetal bovine serum (9). For single-channel recordings, pCMV6c carrying wild-type SUR1 (1.5 μg) (9) and pCMV6b carrying Kir6.2G132S (1.5 μg) were transfected into COS-1 cells with Lipofectamine and Opti-MEM I reagents (Life Technologies, Gaithersburg, MD), according to the manufacturer’s instructions. For single-channel recordings, the expression plasmid vector for green fluorescence protein (pSRαGFP, 0.05 μg) as a reporter gene for transfection was cotransfected (10). For 86Rb+ efflux measurements, COS-1 cells were transfected with wild-type Kir6.2 (WT) cDNA and Kir6.2G132S (MT) cDNA in various ratios together with hamster SUR1 cDNA (0.5 μg) (8). The ratios of WT to MT used were 1:0 (WT = 0.5 μg, MT = 0 μg), 1:1 (WT = 0.5 μg, MT = 0.5 μg), 1:3 (WT = 0.5 μg, MT = 1.5 μg), 1:5 (WT = 0.5 μg, MT = 2.5 μg), and 0:1 (WT = 0 μg, MT = 0.5 μg). Total DNA amounts for transfection were adjusted to 3.5 μg by supplementation of pCMV6b vector.
86Rb+ Efflux Measurement.
Two days after transfection, 86RbCl (1 mCi/ml; 1 Ci = 37 GBq) (Amersham) was added into fresh culture medium and incubated for 12–24 hours. The cells were further incubated for 30 min at 37°C in Krebs–Ringer solution containing 1 mCi 86RbCl/ml in the presence of the metabolic inhibitors oligomycin (2.5 mg/ml) and 2-deoxy-d-glucose (1 mM). After washing the cells once in 86Rb+-free Krebs-Ringer solution containing the metabolic inhibitors, 86Rb+ efflux was measured at 37°C, as described (9). Portions of medium from each time point were evaluated and the values were summed to determine efflux. The data are expressed as the percentage of 86Rb+ content at the beginning of the incubation.
Generation of Kir6.2G132S Transgenic Mice.
For the transgene vector, Kir6.2G132S cDNA was inserted downstream of the human insulin promoter region (27). The expression unit was excised from the resulting pINSKir6.2G132S plasmid by restriction enzyme digestion, purified, and microinjected into fertilized eggs of (C57BL/6 × DBA2)F2 mice by standard procedures (28). Transgenic mice were selected from possible founders by Southern blot analysis and were backcrossed to C57BL/6. F2 mice were used for the experiments.
Electrophysiology.
For single-channel recordings of COS-1 cells, the cells were transfected with Kir6.2G132S cDNA and SUR1 cDNA and cultured for 48–72 hours before electrophysiological recordings. Single-channel recordings were done in the excised inside–out membrane patch configuration 48–72 hours after transfection, as described (9). The intracellular solution contained either 110 mM potassium aspartate or 110 mM sodium aspartate, and 30 mM KCl, 2 mM MgSO4, 1 mM EGTA, 0.084 mM CaCl2, 1 μM K2ATP, and 10 mM Mops (pH 7.4). The pipette solution contained either 140 mM NaCl or KCl, 2 mM CaCl2, and 5 mM Mops (pH 7.4). For whole-cell recordings of single pancreatic beta cells, the cells were prepared by collagenase digestion method (29). Pancreatic beta cells isolated from transgenic (lines M4 and M10) and control mice were cultured in DMEM supplemented with 10% fetal bovine serum, plated into 3.5-cm dishes containing CELLocate Coverslips (Eppendorf, Hamburg, Germany), and incubated at 37°C for 24–72 hours before the experiments. Whole-cell current recordings were made with a patch-clamp amplifier EPC-7 (List Electronics, Darmstadt, Germany), according to the method described by Ribar et al. (30). The extracellular solution contained 135 mM NaCl, 5 mM KCl, 5 mM CaCl2, 2 mM MgSO4, and 5 mM Hepes (pH 7.4). Pipette solution contained 107 mM KCl, 11 mM EGTA, 2 mM MgSO4, 1 mM CaCl2, and 11 mM Hepes (pH 7.2, adjusted with KOH). The membrane potential of the beta cells was measured by the perforated patch-clamp method (31) in the current clamp mode. Extracellular solution contained 125 mM NaCl, 5 mM KCl, 1.2 mM KH2PO4, 2 mM CaCl2, 1 mM MgSO4, 10 mM Hepes, and 2.8 mM glucose (pH 7.4). The pipette solution contained 130 mM potassium aspartate, 10 mM KCl, 10 mM EGTA, 10 mM Mops (pH 7.2), and 100 μg/ml nystatine. After recordings, the beta cells were identified by immunostaining with the anti-insulin antibody.
Measurement of Intracellular Calcium Concentrations ([Ca2+]i).
Dispersed islet cells were cultured for 1–2 days in DMEM containing 10% fetal bovine serum as described above, loaded with 1 μM fura-2 acetoxymethyl ester (Dojindo, Kumamoto, Japan) for 30 min in perifusion buffer containing 130 mM NaCl, 5 mM KCl, 2.5 mM CaCl2, 0.6 mM NaHCO3, 0.8 mM MgCl2, 2.8 mM glucose, 20 mM Hepes (pH 7.4), and 0.2% BSA, and mounted on the stage of the microscope. The perifusion rate was approximately 1.0 ml/min at 37°C. [Ca2+]i was measured by a dual-excitation wavelength method (340/380 nm) as described (31). The beta cells were identified as described above.
Histological Analysis.
The pancreata were fixed in 10% formalin and embedded in paraffin, and serial paraffin sections (4 μm thick) were prepared and processed by the indirect immunoperoxidase method and the avidin-biotinylated peroxidase complex method for staining pancreatic beta cells and alpha cells, using guinea pig anti-insulin and rabbit anti-glucagon antibodies, respectively. The number of beta cells in the pancreatic islets of transgenic and control mice was counted under a light microscope. The beta cell population is expressed as a percent of total islet cells (mean ± SE), by averaging the results from five different pancreatic islets in each mouse. Apoptotic cells were detected in serial sections of pancreatic islets of transgenic mice and control littermates using TUNEL (terminal deoxynucleotideyltransferase-mediated dUTP nick end labeling) assay (32).
Measurements of Blood Glucose and Insulin Levels.
Serum insulin levels were determined by ELISA kit (Morinaga, Yokohama, Japan). Glucose measurements were performed on whole blood with Antosense Glucose (Sankyo). An i.p. glucose tolerance test was performed in male F2 mice fasted for 16 h at 8 weeks of age. Glucose (1 g/kg) was injected into the i.p. space after anesthesia with sodium pentobarbital (75 mg/kg). Blood samples were taken from the tail vein.
RESULTS AND DISCUSSION
Since inward rectifier K+ channels are thought to function as hetero- or homomultimers (33–36), the mutant Kir6.2G132S if coexpressed with wild-type Kir6.2 could decrease the activity of KATP channels by a dominant-negative mechanism. To ascertain this, we measured the 86Rb+ efflux from COS-1 cells transfected with Kir6.2 and Kir6.2G132S at various ratios with SUR1 (Fig. 1B). Transfection of COS-1 cells with Kir6.2G132S and SUR1 had no effect on endogenous efflux and produced no significant 86Rb+ efflux upon metabolic poisoning. Metabolic poisoning of COS-1 cells transfected with Kir6.2 (Kir6.2:Kir6.2G132S = 1:0) and SUR1 remarkably increased 86Rb+ efflux above endogenous efflux (control). This increased efflux decreased as the relative amounts of Kir6.2G132S increased. No significant Na+, K+, or Cl− currents were detected in the COS-1 cells transfected with Kir6.2G132S (Kir6.2: Kir6.2G132S = 0:1) and SUR1 by patch-clamp recordings. These results indicate that Kir6.2G132S acts as a dominant-negative inhibitor of KATP channels.
To investigate directly the role of KATP channels in pancreatic beta cells, we generated transgenic mice expressing Kir6.2G132S in beta cells under the regulation of the human insulin promoter. Southern blot analysis identified 13 founders from a total of 55 mice. Breeding of these founders with C57BL/6 established seven transgenic lines showing moderate to severe hyperglycemia at 4–6 weeks of age. Northern blot and reverse transcription-PCR analyses confirmed that the Kir6.2G132S transgene was expressed only in pancreas among the tissues examined in these transgenic mice, which included liver, brain, heart, and kidney (data not shown). We next examined functional and morphological changes in the endocrine pancreas of transgenic mice during development after birth.
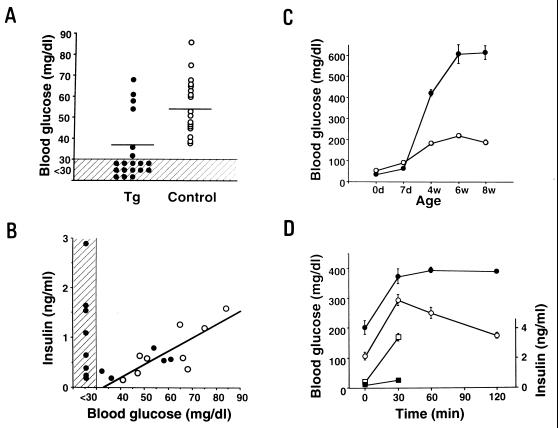
Blood glucose levels in neonate transgenic mice (line M45) were significantly lower than those of controls [transgenic mice, 37.0 ± 3.3 (mean ± SE) mg/dl; control mice, 63.7 ± 5.3 mg/dl, P < 0.001] (Fig. 2A). Spearman rank analysis shows a positive correlation between blood glucose levels and insulin levels (r = 0.70, P < 0.05, n = 9) in controls. By contrast, there is no correlation in transgenic mice (r = −0.24, p = 0.41, n = 13): despite low glucose levels, insulin levels remained relatively high (Fig. 2B). This indicates unregulated insulin secretion in transgenic mice, and suggests the phenotype of PHHI in humans, which is caused by mutations of SUR1 (14–18).
Figure 2.
(A) Blood glucose levels in Kir6.2G132S transgenic mice (line M45) (Tg, •, n = 19) and control mice (control, ○, n = 18) in neonates (day 0). The values below 30 mg/dl (minimum detectable value) were calculated as 30 mg/dl for statistical analysis. The bars indicate the mean values. (B) Relationship between blood glucose levels and serum insulin levels in transgenic mice (•, n = 13) and control mice (○, n = 9) in neonates. The correlation between blood glucose and serum insulin levels in control mice is indicated. The shaded area indicates values below the sensitivity of assay in A and B. (C) Blood glucose levels in the nonfasted state in transgenic (•) and control mice (○) at the indicated ages. Each point was determined by 10–19 mice. (D) Blood glucose and insulin responses to i.p. glucose load in transgenic (n = 4) and control (n = 4) mice at 8 weeks of age. Filled and open circles indicate blood glucose levels in transgenic mice and control mice, respectively. Filled and open squares indicate serum insulin levels in transgenic and control mice, respectively.
Blood glucose levels in transgenic mice at 4 weeks became markedly elevated [transgenic mice, 424 ± 45.1 mg/dl (n = 9); controls, 185 ± 15.5 mg/dl (n = 10)], and increased further to 610 ± 45.5 mg/dl (n = 9) at 6 weeks of age (Fig. 2C). The blood glucose levels after i.p. glucose loading in transgenic mice remained significantly higher than those in controls, and the insulin secretion in transgenic mice was markedly reduced (Fig. 2D).
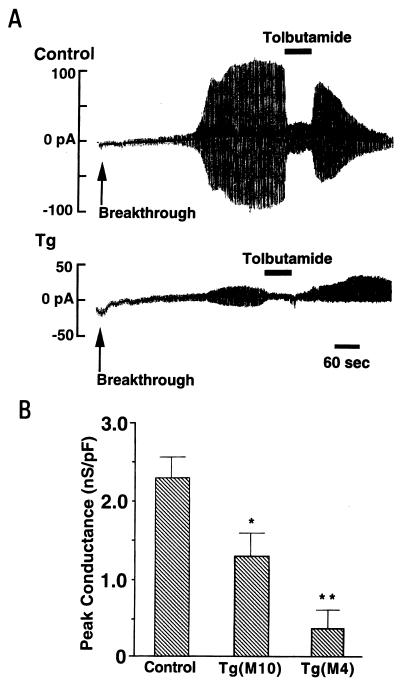
Electrophysiological analysis by whole-cell configuration showed that in the absence of ATP in the pipette solution, a progressive increase in K+ conductance in response to ±10 mV amplitude pulses was observed in beta cells of control mice, and that the increased currents were inhibited by addition of the sulfonylurea tolbutamide (Fig. 3A Upper) (17). In contrast, there was only a slight or moderate increase of K+ conductance in beta cells of transgenic mice (line M4) (Fig. 3A Lower). The ATP-sensitive K+ conductance, normalized by dividing by the membrane capacitance, was significantly reduced in the beta cells of transgenic mice (lines M4 and M10) (Fig. 3B), confirming the KATP channel function to be impaired in the beta cells.
Figure 3.
(A) Representative traces of whole-cell recordings of pancreatic beta cells from control and Kir6.2G132S transgenic mice (line M4). The holding potential was −70 mV and alternate voltage pulses of ±10 mV and 200-ms duration every 2 s were applied. Removal of intracellular ATP beginning at the time indicated (breakthrough) caused a progressive increase in K+ conductance in response to ±10 mV amplitude pulses, and addition of 0.5 mM tolbutamide promptly inhibited the conductance in a beta cell of a control mouse (control). There was only a slight increase in K+ conductance in a beta cell of a transgenic mouse (Tg) under the same conditions. (B) Normalized ATP-sensitive K+ conductance of beta cells in control and transgenic mice (lines M4 and M10). Because the membrane area of each beta cell varied, the K+ conductance was normalized by dividing by the membrane capacitance measured for each cell. Control mice, 2.29 ± 0.27 nS/pF (mean ± SE, n = 20); line M10 transgenic mice, 1.27 ± 0.29 nS/pF (n = 20); line M4 transgenic mice, 0.37 ± 0.25 nS/pF (n = 10). ∗, P < 0.02, ∗∗, P < 0.001 (comparing with control).
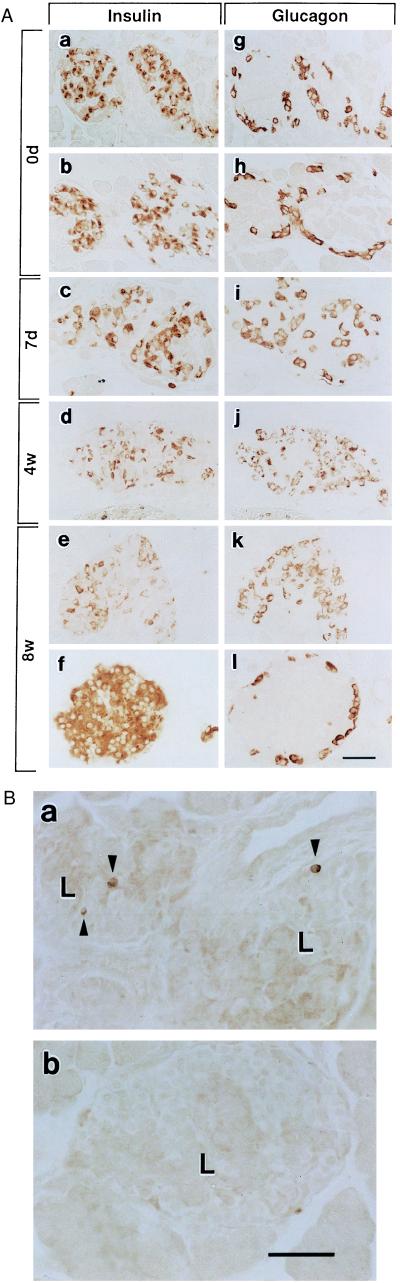
Histological analysis revealed that the beta cell population of transgenic mice (line M45) in neonates (day 0) was not significantly different from that of control mice, and that the morphological configuration of the islet cells in transgenic mice was normal (Fig. 4A). At postnatal day 7, the beta cell population in transgenic mice was slightly decreased, and glucagon-positive alpha cells, which are present normally in the periphery of pancreatic islets, began to also appear in the central region. This abnormality in the morphological configuration of the beta cells and alpha cells of transgenic mice became more pronounced as the beta cell population decreased with age. The severity of hyperglycemia increased with the decrease in beta cell population. The islets of transgenic mice were decreased in size and irregular in shape due to the loss of beta cells.
Figure 4.
(A) Histological changes of pancreatic islets from Kir6.2G132S transgenic and control mice. (a-l) Immunostaining of islet cells at indicated ages: 0d, transgenic and control mice in neonates; 7d, 7-day transgenic mice; 4w, 4-week transgenic mice; 8w, 8-week transgenic and control mice. The insulin column shows staining with anti-insulin antibody (a-f); the glucagon column shows staining with anti-glucagon antibody (g-l). Control mice are shown in b, f, h, and l. Three independent transgenic lines (M7, M8, and M45) have been analyzed. Histological changes of these three lines are similar. Representative examples of M45 mice are presented. The average beta cell population (per islet) of transgenic and control mice are 60.8 ± 4.6% and 63.8 ± 4.7% at day 0, 44.8 ± 2.5% and 62.7 ± 4.9% at 1 week, 34.7 ± 3.6% and 75.2 ± 5.3% at 4 weeks, and 32.5 ± 3.7% and 72.7 ± 4.4% at 8 weeks, respectively. (B) Detection of apoptotic cells in the islets by TUNEL method. (a) Apoptotic beta cells (indicated by arrowheads) are present in the islets (indicated by L) of a transgenic mouse. (b) No apoptotic cells are detected in the islets of a control mouse. (Bar = 100 μm).
Interestingly, apoptotic cell death, as determined by TUNEL method, was found in most of the islets examined in 2-week-old transgenic mice (1–3 apoptotic cells per islet section). Apoptotic beta cells, estimated from observation of serial sections, were found at the rate of 7–20 per islet in transgenic mice (Fig. 4Ba), but were rarely detected in the islets of control mice (Fig. 4Bb), suggesting that the loss of beta cells in these transgenic mice is due to acceleration of apoptosis. We also measured resting membrane potential and basal [Ca2+]i in the beta cells of transgenic and control mice. The resting membrane potential of the beta cells of transgenic mice (line M4) (−46.7 ± 3.9 mV, n = 30) was significantly higher than that of control mice (−63.5 ± 2.0 mV, n = 22) (P < 0.001). Basal [Ca2+]i also was significantly elevated in the beta cells of transgenic mice: transgenic mice (line M4), 137.1 ± 15.9 nM, n = 12; control mice, 90.6 ± 3.5, n = 23 (P < 0.001). In Kir6.2G132S transgenic mice, accordingly, it is likely that the loss of KATP channel function causes membrane depolarization and high basal [Ca2+]i in the beta cells, which results in hypoglycemia from unregulated insulin secretion in neonates, and that chronic membrane depolarization and Ca2+ overloading finally promote apoptotic cell death, which contributes to the hyperglycemia in adults.
In weaver mice, the chronic membrane depolarization due to the loss of K+ ion selectivity or K+ channel function could cause the excitotoxicity that ultimately results in granule cell death in the cerebellum (26). In addition, it has been suggested that KATP channels are involved in protection from cell death in brain and heart during ischemia and hypoxia (37, 38).
Considering these findings together, we propose that some inward rectifier K+ channels are important for cell survival. The targeted expression of a dominant-negative mutation in transgenic mice should be useful in the analysis of the physiological roles of the various inward rectifier K+ channels.
Acknowledgments
We thank K. Kotake, C.-Z. Wang, N. Ozaki, J. Kawaki, and J. Kawagishi for their involvement with part of the animal experiments. We also thank T. Tokuhisa and M. Hatano for helpful advice during the course of the study. This work was supported by scientific research grants from the Ministry of Education, Science, and Culture and from the Ministry of Health and Welfare, Japan; by a grant from Uehara Memorial Foundation; by a grant from the Naito Foundation; a grant for studies on the pathophysiology and complications of diabetes from Tsumura Pharma Ltd.; by a grant from Yamanouchi Foundation for Research on Metabolic Disorders; and the Juvenile Diabetes Foundation International.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: KATP channel, ATP-senstive K+ channel; SUR, sulfonylurea receptor; PHHI, persistent hyperinsulinemic hypoglycemia of infancy; WT, wild-type Kir6.2; MT, Kir6.2G132S; TUNEL, deoxynucleotidyltransferase-mediated dUTP nick end labeling; [Ca2+]i, intracellular calcium concentration.
References
- 1.Noma A. Nature (London) 1983;305:147–148. doi: 10.1038/305147a0. [DOI] [PubMed] [Google Scholar]
- 2.Cook D L, Hales C N. Nature (London) 1984;310:271–273. doi: 10.1038/311271a0. [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft F M, Harrison D E, Ashcroft S J H. Nature (London) 1984;312:446–448. doi: 10.1038/312446a0. [DOI] [PubMed] [Google Scholar]
- 4.Rorsman P, Trube G. Pflügers Arch. 1985;405:305–309. doi: 10.1007/BF00595682. [DOI] [PubMed] [Google Scholar]
- 5.Spruce A E, Standen N B, Stanfield P R. J Physiol (London) 1987;382:213–236. doi: 10.1113/jphysiol.1987.sp016364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashford M L, Sturgess N C, Trout N J, Gardner N J, Hales C N. Pflügers Arch. 1988;412:297–304. doi: 10.1007/BF00582512. [DOI] [PubMed] [Google Scholar]
- 7.Bernardi H, DeWeille J R, Epelbaum J, Mourre C, Amoroso S, Slama A, Fosset M, Lazdunski M. Proc Natl Acad Sci USA. 1993;90:1340–1344. doi: 10.1073/pnas.90.4.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilar-Bryan L, Nichols C G, Wechsler S W, Clement J P, IV, Boyd A E, III, Gonález G, Herrera-Sosa H, Nguy K, Bryan J, Nelson D A. Science. 1995;268:423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- 9.Inagaki N, Gonoi T, Clement J P, IV, Namba N, Inazawa J, Gonález G, Aguilar-Bryan L, Seino S, Bryan J. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 10.Inagaki N, Gonoi T, Clement J P, IV, Wang C-Z, Aguilar-Bryan L, Bryan J, Seino S. Neuron. 1996;16:1011–1017. doi: 10.1016/s0896-6273(00)80124-5. [DOI] [PubMed] [Google Scholar]
- 11.Sakura H, Ämmälä C, Gribble P A, Ashcroft F M. FEBS Lett. 1995;377:338–344. doi: 10.1016/0014-5793(95)01369-5. [DOI] [PubMed] [Google Scholar]
- 12.Isomoto S, Kondo C, Yamada M, Matsumoto S, Higashiguchi O, Hoyio Y, Matsuzawa Y, Kurachi Y. J Biol Chem. 1996;271:24321–24324. doi: 10.1074/jbc.271.40.24321. [DOI] [PubMed] [Google Scholar]
- 13.Tucker S J, Gribble F M, Zhao C, Trapp S, Ashcroft F M. Nature (London) 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- 14.Thomas P M, Cote G J, Wohllk N, Haddad B, Mathew P M, Rabl W, Aguilar-Bryan L, Bryan J. Science. 1995;268:426–429. doi: 10.1126/science.7716548. [DOI] [PubMed] [Google Scholar]
- 15.Aguilar-Bryan L, Bryan J. Diabetes Rev. 1996;4:336–346. [Google Scholar]
- 16.Permutt M A, Nestorowicz A, Glaser B. Diabetes Rev. 1996;4:347–355. [Google Scholar]
- 17.Kane C, Shepherd R M, Squires P E, Johnson P R V, James R F L, Milla P J, Aynsley-Green A, Lindley K J, Dunne M J. Nat Med. 1996;2:1344–1347. doi: 10.1038/nm1296-1344. [DOI] [PubMed] [Google Scholar]
- 18.Dunne M J, Kane C, Shepherd R M, Sanchez J A, James R F L, Johnson P R V, Aynsley-Green A, Lu S, Clement J P, IV, Lindley K J, Seino S, Aguilar-Bryan L. N Engl J Med. 1997;336:703–706. doi: 10.1056/NEJM199703063361005. [DOI] [PubMed] [Google Scholar]
- 19.Jan L Y, Jan Y N. Nature (London) 1994;371:119–122. doi: 10.1038/371119a0. [DOI] [PubMed] [Google Scholar]
- 20.Heginbotham L, Abramson T, MacKinnon R. Science. 1992;258:1152–1155. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- 21.Kerr I D, Sansom M S P. Nature (London) 1995;373:112. doi: 10.1038/373112a0. [DOI] [PubMed] [Google Scholar]
- 22.Patil N, Cox D R, Bhat D, Faham M, Myers R M, Peterson A S. Nat Genet. 1995;11:126–129. doi: 10.1038/ng1095-126. [DOI] [PubMed] [Google Scholar]
- 23.Slesinger P A, Patil N, Liao Y J, Jan Y N, Jan L Y, Cox D R. Neuron. 1996;16:321–331. doi: 10.1016/s0896-6273(00)80050-1. [DOI] [PubMed] [Google Scholar]
- 24.Kofuji P, Hofer M, Millen K J, Millonig J M, Davidson N, Lester H A, Hatten M E. Neuron. 1996;16:941–952. doi: 10.1016/s0896-6273(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 25.Navarro B, Kennedy M E, Velimirovic B, Bhat D, Peterson A S, Clapham D E. Science. 1996;272:1950–1953. doi: 10.1126/science.272.5270.1950. [DOI] [PubMed] [Google Scholar]
- 26.Hess E J. Neuron. 1996;16:1073–1076. doi: 10.1016/s0896-6273(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 27.Ishihara H, Tashiro F, Ikuta K, Asano T, Katagiri H, Inukai K, Kikuchi M, Yazaki Y, Oka Y, Miyazaki J-I. FEBS Lett. 1995;371:329–332. doi: 10.1016/0014-5793(95)00932-y. [DOI] [PubMed] [Google Scholar]
- 28.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 29.Wollheim C, Meda P, Halban P A. Methods Enzymol. 1990;192:188–223. doi: 10.1016/0076-6879(90)92071-k. [DOI] [PubMed] [Google Scholar]
- 30.Ribar T J, Jan C R, Augustine G A, Means A R. J Biol Chem. 1995;270:28688–28695. doi: 10.1074/jbc.270.48.28688. [DOI] [PubMed] [Google Scholar]
- 31.Gonoi T, Mizuno N, Inagaki N, Kuromi H, Seino Y, Miyazaki J-I, Seino S. J Biol Chem. 1994;269:16989–16992. [PubMed] [Google Scholar]
- 32.Gerrieli Y, Sherman Y, Ben-Sasson S A. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krapivinsky G, Gordon E A, Wickman K, Velimirovic B, Krapivinsky L, Clapham D E. Nature (London) 1995;374:135–141. doi: 10.1038/374135a0. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Jan Y N, Jan L Y. Neuron. 1995;15:1441–1447. doi: 10.1016/0896-6273(95)90021-7. [DOI] [PubMed] [Google Scholar]
- 35.Kofuji P, Davidson N, Lester H A. Proc Natl Acad Sci USA. 1995;92:6542–6546. doi: 10.1073/pnas.92.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tinker A, Jan Y N, Jan L Y. Cell. 1996;87:857–868. doi: 10.1016/s0092-8674(00)81993-5. [DOI] [PubMed] [Google Scholar]
- 37.Amoroso S, Schmid-Antomarchi H, Fosset M, Lazdunski M. Science. 1990;247:852–854. doi: 10.1126/science.2305257. [DOI] [PubMed] [Google Scholar]
- 38.Terzic A, Jahangir A, Kurachi Y. Am J Physiol. 1995;269:C525–C545. doi: 10.1152/ajpcell.1995.269.3.C525. [DOI] [PubMed] [Google Scholar]