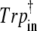Abstract
The recently discovered photoreceptor proteins containing BLUF (sensor of blue light using FAD) domains mediate physiological responses to blue light in bacteria and euglena. In BLUF domains, blue light activates the flavin chromophore yielding a signaling state characterized by a ∼10 nm red-shifted absorption. We developed molecular models for the dark and light states of the BLUF domain of the Rhodobacter sphaeroides AppA protein, which are based on the crystal structures and quantum-mechanical simulations. According to these models, photon absorption by the flavin results in a tautomerization and 180° rotation of the Gln side chain that interacts with the flavin cofactor. This chemical modification of the Gln residue induces alterations in the hydrogen bond network in the core of the photoreceptor domain, which were observed in numerous spectroscopic experiments. The calculated electronic transition energies and vibrational frequencies of the proposed dark and light states are consistent with the optical and IR spectral changes observed during the photocycle. Light-induced isomerization of an amino acid residue instead of a chromophore represents a feature that has not been described previously in photoreceptors.
INTRODUCTION
Light is an essential environmental factor, and many organisms have evolved the capability to respond to light intensity and spectrum. There are three distinct flavin-containing photoreceptor families that respond to blue light: cryptochromes, light oxygen voltage (LOV), and sensor of blue light using FAD (BLUF) domains containing proteins (1–3). There is no significant relationship in sequence, structure, or photoactivation mechanism between the families. BLUF domains (4) are present in proteins from various Bacteria and lower Eukarya, where they control a range of physiological responses, including photosynthesis gene expression and negative phototaxis (3,5,6). BLUF domains consist of ∼100 amino acids and contain a noncovalently bound FAD chromophore. Typically, the BLUF domain serves as an N-terminal sensor that communicates a blue light signal to various C-terminally attached effector domains or to downstream proteins. In AppA, an anti-repressor involved in photosynthesis gene expression in Rhodobacter sphaeroides (5,7–10), the BLUF domain controls light-dependent protein-protein interactions (10). In the Escherichia coli YcgF/BlrP protein (9,11), an N-terminal BLUF domain is linked to an EAL domain that exhibits cyclic-di-GMP-specific phosphodiesterase activity (4,12). The BLUF domain controls adenylyl cyclase activity of the photoactivated adenylyl cyclase, PAC, involved in photophobic response in unicellular flagellate Euglena gracilis (13). In Synechocystis PCC6803, the BLUF domain of the Slr1694 protein controls activity of the downstream partner involved in phototactic avoidance (6). It is noteworthy that BLUF domains function modularly, as it has been shown that the BLUF domain of AppA can be replaced by the BLUF domain of PAC without abolishing the light-dependent antirepressor activity of AppA (14).
Light-absorption by a chromophore yields a signaling state of the photosensory domain, which transmits the light changes to the effector domain or protein, thereby activating it. In BLUF domains, absorption of a blue light photon by the isoalloxazine ring of flavin results in the transient formation of anionic and neutral flavin radical species (15) that produce the light state on a 100 ps time-scale (16–19). The long-lived light state is characterized by a ∼10 nm red-shift of the flavin absorption spectrum that has been attributed to alterations in the hydrogen bonds that the isoalloxazine ring forms with the protein (20–27). The light state decays in the dark to the initial resting state on the timescale ranging from seconds for BlrB, Tll0078, and Slr1694 proteins (18,22,28) to almost 30 min for the BLUF domain of AppA (16,17,19).
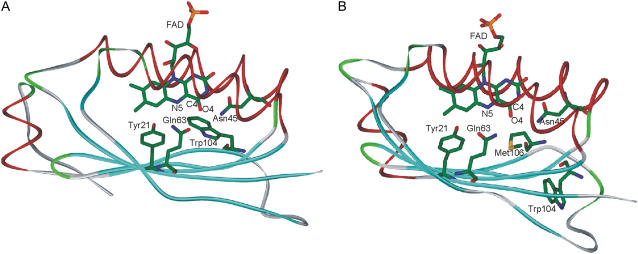
Recently, structures of several BLUF domains have been determined (29–34). They show a ferredoxin-like fold, consisting of a five-stranded β-sheet that serves as a platform for two α-helices located atop as shown in Fig. 1. The isoalloxazine ring of the FAD chromophore is sandwiched between the helices with the ribityl chain pointing toward the solvent. Most residues interacting with the isoalloxazine ring are highly conserved in the BLUF domains, and it has been shown by site-mutagenesis studies that Tyr21 and Gln63 (AppA numbering) are essential for the photoreaction (17,27,35–37). Two incompatible models of the BLUF active site have been suggested. In the AppA BLUF structure determined by Anderson et al. (30), the side chain amide group of Gln63 donates hydrogen bonds to the flavin N5 and Tyr21 hydroxyl whereas the OE1 oxygen atom of Gln63 accepts a hydrogen bond from the indolyl nitrogen of the conserved Trp104 (Fig. 1 A). Anderson et al. (30) proposed that light induces a 180° rotation of the Gln side chain that results in a change of the hydrogen bonds such that Gln63 becomes a hydrogen bond acceptor for the Tyr21 hydroxyl rather than a donor and a new hydrogen bond by Gln63 is formed with the flavin O4 atom instead of with flavin N5 atom. This new interaction was proposed to underlie the observed spectral changes of the light state. However, the majority of the available dark state crystal structures of BLUF domains (29,31–33) show interactions of the Gln63 side chain proposed by Anderson et al. (30) for the light state (Fig. 1 B). Moreover, in these structures, the position of the Trp next to the Gln side chain is taken up by the fully conserved Met residue whereas the Trp side chain points toward the solvent (Trpout conformation, Fig. 1 B) in lieu of being located in the flavin binding pocket (Trpin conformation, Fig. 1 A).
FIGURE 1.
Schematic representation of the crystal structures of the AppA BLUF domain. (A) In the structure determined by Anderson et al. (pdb 1YRX, (30)), the side chain of Trp104 is located in the flavin binding pocket (conformation Trpin), forming a hydrogen bond with the Gln63 side chain. (B) In the structure determined by Jung et al. (pdb 2IYG, (32)), Met106 is located in the flavin binding pocket instead of Trp104 (conformation Trpout).
Identification of the dark-adapted and light-induced states is of crucial importance to any photoreaction mechanism. Currently, there is considerable debate on the assignment of the Trpin and Trpout conformations to the functional states of the BLUF photocycle. Jung et al. (32) assigned the Trpout and Trpin conformations to the dark and light states of the BLUF domain, respectively, and proposed a structural pathway for the transition between the two states, which is initiated by a light-induced rotation of Gln63. On the other hand, in several recent studies (25,27,34,38), the dark state of the BLUF domain was proposed not to have a hydrogen bond between Gln63 and the flavin C4O4 carbonyl group and, therefore, was suggested to correspond to the Trpin conformation (30). However, this assignment is inconsistent with the short (2.7 Å) distance between the flavin oxygen atom O4 and the side chain carbonyl oxygen in Gln63. Such short distance indicates a hydrogen bond between these oxygen atoms. Moreover, it is difficult to rationalize why the Trpin conformation would represent the energetically preferred dark state, given that the structures of the BLUF domains of AppA, BlrB, Tll0078, and Slr1694 (9 of 10 monomers in the asymmetric unit) display the Trpout conformation (29,31–33).
To assign the dark and light states of the AppA BLUF domain we applied modern molecular modeling tools that allowed us to analyze hydrogen bonding interactions of the isoalloxazine ring with the protein. To this end, we constructed all-atom molecular models of the photoreceptor by combined quantum mechanical/molecular mechanical (QM/MM) (39) calculations using the coordinates of the two available crystal structures of the BLUF domain of AppA. Our results support the assignment of the Trpout conformation to the dark state of the photoreceptor. Further, the Trpin structure can only be realized if one assumes a tautomerized Gln63 side chain. The calculated molecular properties of the complex of the isoalloxazine and the imidic acid Gln63 side chain agree with the experimental characterization of the light state of the photoreceptor. We propose that the side chain of Gln63 interacting with the isoalloxazine switches from the amide form in the dark state to the imidic acid form in the light state. Therefore, the mechanism of BLUF domain photoreception involves a chemical reaction occurring between the photoexcited chromophore and amino acid side chains resulting in isomerization of the Gln side chain but not of the chromophore itself.
METHODS
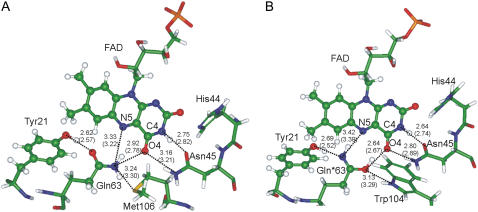
For calculations of the all-atom models (Fig. 2) we used a QM/MM method based on the theory of effective fragment potentials (39–41). This approach allows calculations close to an ab initio treatment of the entire molecular system. Molecular groups assigned to the MM part are represented by effective fragments that contribute their electrostatic potentials expanded up to octupoles to the quantum Hamiltonian. These one-electron electrostatic potentials, as well as contributions from interactions of effective fragments with the QM region, are obtained in preliminary quantum chemical calculations by using ab initio electron densities. The exchange-repulsion potentials to be combined with the electrostatic terms are also created in preliminary ab initio calculations. The computer program used in the simulations was based on the GAMESS(US) quantum chemistry package (42) (more specifically on its Intel-specific version, PC GAMESS (43)).
FIGURE 2.
Flavin binding pocket in the optimized QM/MM models in the Trpout (A) and Trpin (B) conformation of the AppA BLUF domain. The quantum part is shown in ball and stick representation. Optimized distances between heavy atoms are given in angstroms; the values in parentheses correspond to the distances in the crystal structures. The short OE1(Gln63)-O4 bond distance reported for the Trpin conformation pdb 1YRX (30) could be reproduced only by assuming an imidic tautomer of the glutamine, denoted as Gln*63.
The initial positions of heavy atoms were obtained from the crystallographic coordinates of the A molecules in the asymmetric units of the structures Protein Data Bank (pdb) 1YRX (30) and 2IYG (32), comprising residue Val17 to residue Cys110. When hydrogen atoms were introduced, all Lys and Arg residues were assumed positively charged (protonated), whereas Asp and Glu residues were considered negatively charged (deprotonated). A total of 115 solvent water molecules were added to the system. The quantum subsystem included the isoalloxazine part of the chromophore and the side chains of Tyr21 and Gln63. The remaining atoms of the model systems assigned to the MM part were represented by 368 small effective fragments interacting with the QM part and among themselves. The Hartree-Fock approach and the 6–31G basis sets were used in the QM part. The side chains of His44, Asn45, Trp104, and Met106 along with other residues in the vicinity of the reaction center were represented by effective fragment and did contribute to the QM/MM energies and forces in our model. The QM/MM optimizations of all internal coordinates in the QM part and positions of effective fragments in the MM part were carried out to locate the stationary points. The coordinates of outer molecular groups, farthest from the chromophore unit, were kept frozen as in the crystal structures.
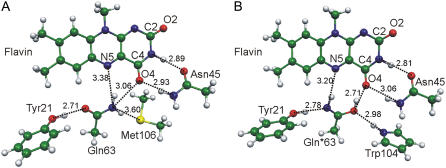
Geometry configurations of the model molecular clusters (Fig. 3) were optimized using the density functional theory B3LYP/6-31G(d) approximation. The harmonic vibrational analysis was carried out for the computed equilibrium structures. The estimated harmonic frequencies were scaled by 0.96 (44) to correct for the systematic errors of the computational method. The excited electronic states of the model clusters were treated using conventional multiconfigurational quantum chemistry approaches, including the configuration interaction method with single and double excitations (CI-SD), the complete active space self-consistent field (CASSCF) method and the second order multiconfigurational quasidegenerate perturbation theory (MCQDPT2) (45). Energies and the wave functions of the five lowest singlet states were calculated by the CI-SD method by considering electronic excitations within a window of the 32 highest occupied and 32 lowest virtual Hartree-Fock orbitals. The vertical electron transition energies were obtained in the CASSCF approximation with the energy functional averaged over the five lowest roots with the choice of the active space based on the obtained CI-SD solutions. The active space consisted of three bonding π, one nonbonding np, and one antibonding π* MOs localized on the lumiflavin and phenol molecules. The obtained CASSCF energies were corrected for dynamic correlation using the MCQDPT2 method. The CASSCF wave functions were used to calculate molecular properties in the ground and excited electronic states, including dipole moment and Loewdin atomic charges. All calculations were carried out with the PC GAMESS quantum-chemistry package (43).
FIGURE 3.
Optimized geometry configurations of the model clusters representing the flavin interactions in the conformations Trpout (A) and Trpin (B) of the BLUF photoreceptor. The internuclear distances are given in angstroms.
RESULTS
All-atom molecular models of the two conformations of the BLUF domain
All-atom models of the AppA BLUF domain were constructed by the combined quantum mechanical- molecular mechanical (QM/MM) method in the flexible effective fragment version (41,46), which has proven its efficiency in simulations of properties and reactivity of biomolecules (47,48). QM/MM geometry optimization of the Trpout conformation of AppA BLUF converged to a structure very similar to the starting crystal structure reported by Jung et al. (32). In contrast, when starting from the heavy atom coordinates of the Trpin conformation reported by Anderson et al. (30), the QM/MM geometry optimization resulted in significant structural relaxation caused by the energetically unfavorable short distance of 2.7 Å between the O4 atom of flavin and the OE1 carbonyl oxygen atom of Gln63. To explain such a short OE1(Gln63)-O4 distance, a hydrogen-bonding interaction is required between the two oxygen atoms, which can only be achieved in a tautomerized Gln side chain (imidic acid form, HN=CR-OH). Fig. 2 illustrates the flavin binding pockets in the computed all-atom models. The QM/MM optimized OE1(Gln63)-O4 distance between the imidic acid (denoted as Gln*63 in Fig. 2 B) and C4O4 as well as other geometry parameters are in good agreement with the crystal structure, prompting us to suggest that the Trpin conformation of the BLUF domain contains the tautomeric imidic acid form of the Gln63 side chain.
The light state of BLUF exhibits a ∼10 nm red-shifted absorption maximum compared to the dark state. To characterize the flavin absorption, the first two ππ* transitions of the flavin were estimated for both optimized structures shown in Fig. 2. The excitation energies of the chromophore were calculated using the multiconfigurational complete active space self-consistent field (CASSCF) approximation in the quantum part with the conventional state-averaging technique. Absorption of the flavin interacting with the tautomer Gln*63 in the Trpin model (Fig. 2 B) was found to be 10 nm red-shifted compared to the flavin interacting with the amide side chain of Gln63 in the Trpout model (Fig. 2 A). Although quantitative conclusions cannot be formulated using the results of these CASSCF simulations, the qualitative agreement with the observations is remarkable. Thus, the higher energy form containing the imidic acid side chain Gln*63 is predicted to have an absorption that is similar to the absorption of the light-induced state of BLUF.
In the nonphotocycling Gln63Leu mutant of AppA BLUF, a 10 nm blue shift was observed compared to the wild-type dark state (27). To verify the structure of the dark state we qualitatively simulate the effect of the Gln63Leu mutation by replacing the Gln63 side chain with Ala in the Trpout QM/MM model. The calculated wavelength of the first optical transition in flavin for the mutant model was 20 nm blue shifted compared to the initial model containing Gln63. Apparently, the blue shift of the Gln63 mutant absorption spectrum is caused by the loss of the hydrogen bond in which the amide group of Gln63 is a hydrogen donor whereas C4O4 and N5 of flavin serve as a bifurcated acceptor. The reproduced blue shift of the mutant absorption suggests that the bifurcated hydrogen bond between the Gln63 side chain and flavin is present in the dark state of the BLUF domain, supporting the assignment of the Trpout conformation to the dark state.
Accurate characterization of hydrogen bonding of flavin in model clusters
To refine conclusions drawn from the QM/MM simulations, we re-computed the energetic and electronic properties of two forms of the BLUF domain using more accurate quantum approaches for a series of molecular clusters. The model clusters mimicking the interactions of the isoalloxazine in the Trpin and Trpout conformations are shown in Fig. 3. The chromophore is represented by a lumiflavin molecule and the Tyr, Gln, and Met side chains are modeled by phenol, acetamide, and dimethylthioether molecules, respectively. To account for the hydrogen bond between the side chains of Gln63 and Trp104, we included a pyrrole molecule in the Trpin cluster model (Fig. 3 B). According to the QM/MM simulations, the Trpin cluster must contain the imidic acid tautomer of acetamide. We used the same designations of the molecular fragments as those used in the QM/MM models. The atomic coordinates of the model clusters were fully optimized (starting from the reference positions in the QM/MM models) by the B3LYP/6-31G(d) method. The optimized structures were confirmed as true minima on the potential energy surfaces by harmonic vibrational analysis.
The optimized internuclear distances specifying the positions of the molecular fragments relative to each other in the cluster models (Fig. 3) agreed with the distances in the crystal structures and the QM/MM models (Fig. 2), although they tended to be somewhat longer in the clusters in the absence of the protein molecule. On Gln63 tautomerization, the entire hydrogen-bonding network changes. There is a slightly shorter hydrogen bond if the imidic acid Gln*63 interacts with the C4O4 group of lumiflavin instead of the amide Gln63 form. The hydrogen bonds between the Asn45 analog and lumiflavin are also affected by the presence of the imidic acid. This rearrangement of hydrogen bonds is likely to cause the spectral shifts of the light state observed experimentally (24–27).
FTIR and Raman studies on BLUF domains showed characteristic light-induced changes in the C=O vibrations of the chromophore and the protein (20,21,23,24,28). In Table 1, we compare the calculated harmonic frequencies of the C=O groups in the two clusters models. The C4O4 frequency of the flavin appears at 1705 cm−1 in the complex with the amide Gln63, whereas it downshifts to 1662 cm−1 in the complex with the imidic acid Gln*63. Upshifts are predicted for the C=O vibration of Asn45 in the presence of Gln*63 (1685 cm−1) instead of Gln63 (1700 cm−1) and for the C=N vibration (1685 cm−1) of Gln*63 compared to the C=O vibration of Gln63 (1655 cm−1). Both the downshift of the flavin C4O4 frequency and the upshift of the protein C=O frequency were observed in the vibration spectra of the light state of the AppA BLUF domain (24,26).
TABLE 1.
Frequencies (cm−1) and tentative assignment of the CO stretching vibrations according to B3LYP/6-31G(d) calculations in harmonic approximation with scaling factor 0.96
| Model
|
||
|---|---|---|
| Tentative assignment |  |
 |
| C2=O2 in flavin | 1742 | 1748 |
| C4=O4 in flavin | 1705 | 1662 |
| C=O in Asn45 | 1685 | 1700 |
| C=O in Gln63 or C=N in Gln*63 | 1655 | 1685 |
In the light-induced difference FTIR spectrum of AppA BLUF, a negative band at 1709 cm−1 and a positive shoulder at 1679 cm−1 have been assigned to the C4O4 stretching vibration of the chromophore in the dark and light states, respectively (26). Raman spectra show a ∼20 cm−1 downshift of the C4O4 vibration on formation of the photoproduct, which has been attributed to a new hydrogen bond between C4O4 and Gln63 when adapting the orientation observed in the Trpout crystal structure (24,27). In both QM/MM and cluster models representing the Trpout structure, the NE2-H(Gln63)…O4 angle in the bifurcated hydrogen bond is rather small (131° in the Trpout cluster model), indicating that this is not a strong interaction. In computational models previously published (models 1 and 3A in Unno et al. (27)), the interaction of the Gln63 amide with the flavin produced only a 5 cm−1 downshift of the C4O4 frequency. This effect is in agreement with the small downshift of the C4O4 stretching frequency in the wild-type dark state compared to the Gln63Leu mutant (27), but not with the 20 cm−1 downshifted C4O4 vibration in the photoproduct compared to the dark state (24). Therefore, the bifurcated hydrogen bond, implicating a rather weak interaction of the amide Gln63 with C4O4 in the Trpout structure, can be attributed to the dark state of the BLUF photosensor. According to the data presented in Table 1, replacement of the weak interaction between the Gln63 and C4O4 in the Trpout model by a strong hydrogen bond that the imidic acid Gln*63 forms with C4O4 in model Trpin results in a significant downshift of the C4O4 frequency.
In a recent time-resolved IR study of wild-type AppA BLUF and the Gln63Leu mutant, a transient absorption at 1666 cm−1, typical for the photoactive state of the protein, has been attributed to the carbonyl vibration of Gln63 (49). This transient absorption disappeared in the spectrum of the light state of the BLUF domain concomitant with the appearance of another band at 1690 cm−1. Our results, shown in Table 1, indicate that these spectral changes can be accounted for by the light-induced tautomerization of Gln63. Indeed, a keto-enol tautomerism of the Gln63 amide group was proposed by these authors (49).
The calculated energies of the low-lying singlet states are listed in Table 2. In both model clusters, the calculated four lowest excited singlet states correspond to single-electron excitations to the lowest unoccupied molecular orbital of the cluster, which belongs to flavin. The first singlet excited state represents the npπ* transition in the flavin followed by two optically active flavin ππ* transitions and a ππ* type electron transfer from the phenol molecule (representing Tyr21) to the flavin. In the cluster containing amide Gln63, the calculated energies of the two ππ* transitions in the flavin, 2.80 and 3.43 eV, are in good agreement with the experimental absorption bands of the dark state AppA BLUF, 2.78 eV at 446 nm and 3.26 eV at 380 nm (16). In the cluster containing the tautomerized form Gln*63, the energy of the first ππ* (flavin) state is predicted to be lower that corresponds to a slightly red-shifted absorption of this complex compared to that of the cluster model containing the amide Gln63. We attribute the characteristic red shift of the BLUF photoproduct to the enhanced interactions of the tautomerized side chain Gln*63 with the N5 and O4 atoms of flavin. Thus, the cluster model Trpin reproduces both optical and IR spectral shifts observed for the light state of the BLUF domain. It is therefore suggestive to assign the Trpin conformation to the light state of BLUF.
TABLE 2.
Energies of the excited singlet electronic states (eV) in cluster models calculated in the MCQDPT2-CASSCF//B3LYP/6-31G(d) approximation
Reaction mechanism
Ultra-fast spectroscopy experiments have established that the BLUF photoreaction starts from the singlet excited state of the flavin chromophore and proceeds via an electron transfer from Tyr21 to flavin (15,17). We identified a charge transfer ππ* excited state corresponding to an electron transfer from Tyr21 to the flavin with energies of 4.09 and 3.78 eV for the clusters Trpout and Trpin, respectively. Vertical excitation to this state yields the anionic semiquinone and cationic phenyl radicals with calculated net charges of −0.96e and +0.93e in the Trpin and −0.99e and +0.92e in the Trpout clusters. This biradical state is characterized by a large dipole moment that is likely to be compensated by proton transfer from the phenyl cation radical to the semiquinone anion radical via the Gln63 amide group. A possible pathway resulting in the tautomerization of the Gln63 side chain is presented in Fig. 4. The formed imidic acid tautomer rotates by 180° to adopt a position allowing radical recombination and favorable interactions with the flavin.
FIGURE 4.
Proposed scheme of the photoreaction of the BLUF photoreceptor.
DISCUSSION
Our analysis of the Trpout and Trpin structures of the AppA BLUF domain by quantum-chemical calculations suggests a tautomeric imidic acid form of Gln63 when it is located next to Trp104 in the flavin binding pocket (Trpin conformation). This finding allowed us to re-assign the hydrogen bonds around the isoalloxazine ring compared to those reported by Anderson et al. (30). Remarkably, our assignment was consistent with all crystallographically deduced hydrogen-bonded distances between the isoalloxazine and the conserved residues forming the binding pocket. The conserved Gln residue in the form of imidic acid is incorporated into an intricate hydrogen bonding network interacting with the isoalloxazine ring and the Tyr21 and Trp104 side chains (Fig. 2 B). We showed by quantum mechanical calculations that interactions of the flavin with the tautomeric Gln side chain account for the red shift of the flavin absorption and the downshift of the C=O vibration observed experimentally in the light state (24,26). The good agreement between the predicted and experimental properties suggests that the glutamine tautomer is a chemical product of the primary photoreaction in BLUF.
Ultra-fast spectroscopy on BLUF domains has shown that the light-driven reaction proceeds within the flavin-glutamine–tyrosine reactive core via a radical pair mechanism involving electron and proton transfers (15). The phenyl group of the conserved tyrosine was suggested to serve as an electron and proton donor to the flavin in the primary photoreaction (15,17,50). Based on the assignment of hydrogen bonds proposed by Anderson et al. (30) and the NMR spectra of the dark and light state of AppA BLUF (25,34,50), the Tyr21 hydroxyl group was proposed to be a proton acceptor in the dark state and a proton donor in the light state. Our hydrogen bond assignment contradicts this model and suggests a proton donor role for the phenyl ring incorporated in the hydrogen bond network of the flavin chromophore in both functional states (Fig. 4). Recent FTIR studies have confirmed the tyrosine side chain to be a proton donor in both dark and light states of the Tll0078 BLUF (51).
In the Trpin conformation of AppA BLUF (30), the region from residue Trp104 to residue Ser109 takes up a regular β-strand structure, β5-strand, which requires positioning of the Trp104 side chain inside the flavin binding pocket (Fig. 1 A). Analysis of the all-atom QM/MM models showed that the orientation of Gln63 proposed by Anderson et al. (30) involves an energetically unfavorable contact between the two carbonyl groups causing significant structural relaxation and an optimized geometry different from the geometry observed in the crystal structure. On the other hand, the Gln*63 tautomer is compatible with the structure of the β5-strand in the Trpin conformation. The Trpout conformation of AppA BLUF contains the Gln63 side chain in the amide form that “pushes” the Trp104 side chain outside the flavin binding pocket resulting in the shortened β5-strand (Fig. 1 B). Thus, there is a direct connection between the chemical structure of the conserved Gln side chain and the secondary structure of the BLUF domain. We propose the following model of photoactivation in AppA BLUF. The dark state corresponds to the crystal structure Trpout and contains the Gln63 side chain in the amide form resulting in the shortened β5-strand. The light state corresponds to the crystal structure Trpin and contains the tautomerized Gln63 side chain interacting with the Trp104 side chain inside the flavin binding pocket allowing an elongated β5-strand (32). Thus, light activation of BLUF results in a change in secondary structure that has also been reported by FTIR measurements (52).
The important role of the Trp104 residue in the AppA protein has been realized and analyzed by mutagenesis experiments (38,50,52,53). The Trp104Ala mutant exhibits a normal photocycle with accelerated dark relaxation (52,53) but functionally acts as the light state even without light activation (38). A similar functional behavior was reported for the nonphotocycling Gln63Leu mutant (27,38). This finding was interpreted as evidence for a hydrogen bond between the Gln63 and Trp104 side chains in the dark state, which was therefore assigned to the Trpin crystal structure (38). Our model offers a different interpretation for these observations: on mutation of either Gln63 or Trp104, the β5-strand adapts the elongated conformation without light-activation. This may also be the case for the Tyr21 mutants, for which light-induced transient reduction of the flavin by Trp104 has been reported, indicating that the Trp104 side chain is located inside of the flavin binding pocket (50). However, it remains to be seen whether the Tyr21 mutants structurally and functionally represent the dark state.
Based on the data reported here, we propose that tautomerization of the highly conserved Gln63 represents the light switch in AppA BLUF. The structural and functional similarities of different BLUF domains suggest that this mechanism of photoactivation is shared by all members of the photoreceptor family. Isomerization reactions are often used in light receptors, although, only isomerization of the chromophore has been identified to date (3). In contrast to the chromophores with linear structure in the rhodopsin, phytochrome, and PYP light-receptors, the condensed ring system of the FAD chromophore is not prone to isomerization. Instead, the flavin containing photoreceptors rely on photoreactions of the excited chromophore with neighboring amino acids using flavin redox properties. In LOV domains, photo-induced electron and proton transfers from the active site Cys to the flavin takes place, yielding a covalent Cys-flavin adduct (54). In cryptochromes, a flavin semiquinone radical results from electron transfer assisted by Trp residues (55,56). We propose that in BLUF domains, electron and proton transfers from the phenyl group of the conserved Tyr to flavin result in the isomerization of the active site Gln residue. Formation of specific hydrogen-bonding network in the presence of the tautomerized Gln allows fast recombination of the radical products yielding the light-induced state of BLUF, containing the imidic acid form of Gln. Thus, the two chemical strategies used by the photoreceptors, photoreduction and isomerization, are combined in light sensing by the BLUF domains.
Acknowledgments
We are very grateful to Mark Gomelsky and Max Cryle for helpful comments on the manuscript.
This work is supported in part by grants from the Deutsche Forschungsgemeinschaft (FOR 526) and from the Russian Foundation for Basic Researches (project No. 07-03-00059).
Editor: Janos K. Lanyi.
References
- 1.van der Horst, M. A., W. Laan, S. Yeremenko, A. Wende, P. Palm, D. Oesterhelt, and K. J. Hellingwerf. 2005. From primary photochemistry to biological function in the blue-light photoreceptors PYP and AppA. Photochem. Photobiol. Sci. 4:688–693. [DOI] [PubMed] [Google Scholar]
- 2.Christie, J. M., and W. R. Briggs. 2001. Blue light sensing in higher plants. J. Biol. Chem. 276:11457–11460. [DOI] [PubMed] [Google Scholar]
- 3.van der Horst, M. A., and K. J. Hellingwerf. 2004. Photoreceptor proteins, “star actors of modern times”: a review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc. Chem. Res. 37:13–20. [DOI] [PubMed] [Google Scholar]
- 4.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497–500. [DOI] [PubMed] [Google Scholar]
- 5.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45:827–836. [DOI] [PubMed] [Google Scholar]
- 6.Okajima, K., S. Yoshihara, Y. Fukushima, X. X. Geng, M. Katayama, S. Higashi, M. Watanabe, S. Sato, S. Tabata, Y. Shibata, S. Itoh, and M. Ikeuchi. 2005. Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J. Biochem. 137:741–750. [DOI] [PubMed] [Google Scholar]
- 7.Shimada, H., K. Iba, and K. Takamiya. 1992. Blue-light irradiation reduces the expression of Puf and Puc operons of Rhodobacter sphaeroides under semi-aerobic conditions. Plant Cell Physiol. 33:471–475. [Google Scholar]
- 8.Gomelsky, M., and S. Kaplan. 1995. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 177:4609–4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomelsky, M., and S. Kaplan. 1998. AppA, a redox regulator of photo system formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel FAD binding domain. J. Biol. Chem. 273:35319–35325. [DOI] [PubMed] [Google Scholar]
- 10.Masuda, S., and C. E. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 110:613–623. [DOI] [PubMed] [Google Scholar]
- 11.Rajagopal, S., J. M. Key, E. B. Purcell, D. J. Boerema, and K. Moffat. 2004. Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochem. Photobiol. 80:542–547. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iseki, M., S. Matsunaga, A. Murakami, K. Ohno, K. Shiga, K. Yoshida, M. Sugai, T. Takahashi, T. Hori, and M. Watanabe. 2002. A blue-light-activated adenylyl cyclase mediates photo avoidance in Euglena gracilis. Nature. 415:1047–1051. [DOI] [PubMed] [Google Scholar]
- 14.Han, Y., S. Braatsch, L. Osterloh, and G. Klug. 2004. A eukaryotic BLUF domain mediates light-dependent gene expression in the purple bacterium Rhodobacter sphaeroides 2.4.1. Proc. Natl. Acad. Sci. USA. 101:12306–12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gauden, M., I. H. M. van Stokkum, J. M. Key, D. C. Luhrs, R. van Grondelle, P. Hegemann, and J. T. M. Kennis. 2006. Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc. Natl. Acad. Sci. USA. 103:10895–10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauden, M., S. Yeremenko, W. Laan, I. H. van Stokkum, J. A. Ihalainen, R. van Grondelle, K. J. Hellingwerf, and J. T. Kennis. 2005. Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry. 44:3653–3662. [DOI] [PubMed] [Google Scholar]
- 17.Dragnea, V., M. Waegele, S. Balascuta, C. Bauer, and B. Dragnea. 2005. Time-resolved spectroscopic studies of the AppA blue-light receptor BLUF domain from Rhodobacter sphaeroides. Biochemistry. 44:15978–15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zirak, P., A. Penzkofer, T. Schiereis, P. Hegemann, A. Jung, and I. Schlichting. 2006. Photodynamics of the small BLUF protein BlrB from Rhodobacter sphaeroides. J. Photochem. Photobiol. B. 83:180–194. [DOI] [PubMed] [Google Scholar]
- 19.Zirak, P., A. Penzkofer, T. Schiereis, P. Hegemann, A. Jung, and I. Schlichting. 2005. Absorption and fluorescence spectroscopic characterization of BLUF domain of AppA from Rhodobacter sphaeroides. Chem. Phys. 315:142–154. [Google Scholar]
- 20.Hasegawa, K., S. Masuda, and T. A. Ono. 2004. Structural intermediate in the photocycle of a BLUF (Sensor of Blue Light Using FAD) protein Slr1694 in a Cyanobacterium Synechocystis sp. PCC6803. Biochemistry. 43:14979–14986. [DOI] [PubMed] [Google Scholar]
- 21.Masuda, S., K. Hasegawa, A. Ishii, and T. A. Ono. 2004. Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr1694 of synechocystis sp. PCC6803. Biochemistry. 43:5304–5313. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa, K., S. Masuda, and T. A. Ono. 2005. Spectroscopic analysis of the dark relaxation process of a photocycle in a sensor of blue light using FAD (BLUF) protein Slr1694 of the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 46:136–146. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa, K., S. Masuda, and T. Ono. 2006. Light induced structural changes of a full-length protein and its BLUF domain in YcgF(Blrp), a blue-light sensing protein that uses FAD (BLUF). Biochemistry. 45:3785–3793. [DOI] [PubMed] [Google Scholar]
- 24.Unno, M., R. Sano, S. Masuda, T. A. Ono, and S. Yamauchi. 2005. Light-induced structural changes in the active site of the BLUF domain in AppA by Raman spectroscopy. J. Phys. Chem. B. 109:12620–12626. [DOI] [PubMed] [Google Scholar]
- 25.Grinstead, J. S., M. Avila-Perez, K. J. Hellingwerf, R. Boelens, and R. Kaptein. 2006. Light-induced flipping of a conserved glutamine sidechain and its orientation in the AppA BLUF domain. J. Am. Chem. Soc. 128:15066–15067. [DOI] [PubMed] [Google Scholar]
- 26.Masuda, S., K. Hasegawa, and T. Ono. 2005. Light-induced structural changes of apoprotein and chromophore in the sensor of blue light using FAD (BLUF) domain of AppA for a signaling state. Biochemistry. 44:1215–1224. [DOI] [PubMed] [Google Scholar]
- 27.Unno, M., S. Masuda, T. A. Ono, and S. Yamauchi. 2006. Orientation of a key glutamine residue in the BLUF domain from AppA revealed by mutagenesis, spectroscopy, and quantum chemical calculations. J. Am. Chem. Soc. 128:5638–5639. [DOI] [PubMed] [Google Scholar]
- 28.Fukushima, Y., K. Okajima, Y. Shibata, M. Ikeuchi, and S. Itoh. 2005. Primary intermediate in the photocycle of a blue-light sensory BLUF FAD-Protein, T110078, of Thermosynechococcus elongatus BP-I. Biochemistry. 44:5149–5158. [DOI] [PubMed] [Google Scholar]
- 29.Kita, A., K. Okajima, Y. Morimoto, M. Ikeuchi, and K. Miki. 2005. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J. Mol. Biol. 349:1–9. [DOI] [PubMed] [Google Scholar]
- 30.Anderson, S., V. Dragnea, S. Masuda, J. Ybe, K. Moffat, and C. Bauer. 2005. Structure of a Novel Photoreceptor, the BLUF Domain of AppA from Rhodobacter sphaeroides. Biochemistry. 44:7998–8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung, A., T. Domratcheva, M. Tarutina, Q. Wu, W. H. Ko, R. L. Shoeman, M. Gomelsky, K. H. Gardner, and I. Schlichting. 2005. Structure of a bacterial BLUF photoreceptor: insights into blue light-mediated signal transduction. Proc. Natl. Acad. Sci. USA. 102:12350–12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung, A., J. Reinstein, T. Domratcheva, R. L. Shoeman, and I. Schlichting. 2006. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J. Mol. Biol. 362:717–732. [DOI] [PubMed] [Google Scholar]
- 33.Yuan, H., S. Anderson, S. Masuda, V. Dragnea, K. Moffat, and C. Bauer. 2006. Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry. 45:12687–12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grinstead, J. S., S. T. Hsu, W. Laan, A. M. Bonvin, K. J. Hellingwerf, R. Boelens, and R. Kaptein. 2006. The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling. ChemBioChem. 7:187–193. [DOI] [PubMed] [Google Scholar]
- 35.Kraft, B. J., S. Masuda, J. Kikuchi, V. Dragnea, G. Tollin, J. M. Zaleski, and C. E. Bauer. 2003. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: a novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry. 42:6726–6734. [DOI] [PubMed] [Google Scholar]
- 36.Laan, W., M. A. van der Horst, I. H. van Stokkum, and K. J. Hellingwerf. 2003. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine dinucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: a key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochem. Photobiol. 78:290–297. [DOI] [PubMed] [Google Scholar]
- 37.Okajima, K., Y. Fukushima, H. Suzuki, A. Kita, Y. Ochiai, M. Katayama, Y. Shibata, K. Miki, T. Noguchi, S. Itoh, and M. Ikeuchi. 2006. Fate determination of the flavin photoreceptions in the cyanobacterial blue light receptor TePixD (T110078). J. Mol. Biol. 363:10–18. [DOI] [PubMed] [Google Scholar]
- 38.Masuda, S., Y. Tomida, H. Ohta, and K. Takamiya. 2007. The critical role of a hydrogen bond between Gln63 and Trp104 in the blue-light sensing BLUF domain that controls AppA activity. J. Mol. Biol. 368:1223–1230. [DOI] [PubMed] [Google Scholar]
- 39.Warshel, A., and M. Levitt. 1976. Theoretical studies of enzymic reactions - dielectric, electrostatic and steric stabilization of carbonium-ion in reaction of lysozyme. J. Mol. Biol. 103:227–249. [DOI] [PubMed] [Google Scholar]
- 40.Gordon, M. S., M. A. Freitag, P. Bandyopadhyay, J. H. Jensen, V. Kairys, and W. J. Stevens. 2001. The effective fragment potential method: A QM-based MM approach to modeling environmental effects in chemistry. J. Phys. Chem. A. 105:293–307. [Google Scholar]
- 41.Nemukhin, A. V., B. L. Grigorenko, I. A. Topol, and S. K. Burt. 2003. Flexible effective fragment QM/MM method: validation through the challenging tests. J. Comput. Chem. 24:1410–1420. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, M. W., K. K. Baldridge, J. A. Boatz, S. T. Elbert, M. S. Gordon, J. H. Jensen, S. Koseki, N. Matsunaga, S. Su. Nguyen, T. L. Windus, M. Dupuis, and J. A. Montgomery. 1993. The general atomic and molecular electronic structure system. J. Comput. Chem. 14:1347–1363. [Google Scholar]
- 43.Nemukhin, A. V., B. L. Grigorenko, and A. A. Granovsky. 2004. Molecular modeling by using the PC GAMESS program: From diatomic molecules to enzymes. Moscow State Univ. Res. Bull. Khimia. 45:75–102. [Google Scholar]
- 44.Takahashi, M., Y. Ishikawa, J. Nishizawa, and H. Ito. 2005. Low-frequency vibrational modes of riboflavin and related compounds. Chem. Phys. Lett. 401:475–482. [Google Scholar]
- 45.Schmidt, M. W., and M. S. Gordon. 1998. The construction and interpretation of MCSCF wave functions. Annu. Rev. Phys. Chem. 49:233–266. [DOI] [PubMed] [Google Scholar]
- 46.Grigorenko, B. L., A. V. Nemukhin, I. A. Topol, and S. K. Burt. 2002. Modeling of biomolecular systems with the quantum mechanical and molecular mechanical method based on the effective fragment potential technique: Proposal of flexible fragments. J. Phys. Chem. A. 106:10663–10672. [Google Scholar]
- 47.Grigorenko, B. L., A. V. Rogov, I. A. Topol, S. K. Burt, H. M. Martinez, and A. V. Nemukhin. 2007. Mechanism of the myosin catalyzed hydrolysis of ATP as rationalized by molecular modeling. Proc. Natl. Acad. Sci. USA. 104:7057–7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grigorenko, B. L., A. V. Nemukhin, M. S. Shadrina, I. A. Topol, and S. K. Burt. 2007. Mechanisms of guanosine triphosphate hydrolysis by Ras and Ras-GAP proteins as rationalized by ab initio QM/MM simulations. Proteins. 66:456–466. [DOI] [PubMed] [Google Scholar]
- 49.Stelling, A. L., K. L. Ronayne, J. Nappa, P. J. Tonge, and S. R. Meech. 2007. Ultrafast structural dynamics in BLUF domains: transient infrared spectroscopy of AppA and its mutants. J. Am. Chem. Soc. 129:15556–15564. [DOI] [PubMed] [Google Scholar]
- 50.Gauden, M., J. S. Grinstead, W. Laan, H. M. van Stokkum, M. Avila-Perez, K. C. Toh, R. Boelens, R. Kaptein, R. van Grondelle, K. J. Hellingwerf, and J. T. M. Kennis. 2007. On the role of aromatic side chains in the photoactivation of BLUF domains. Biochemistry. 46:7405–7415. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi, R., K. Okajima, H. Suzuki, H. Nakamura, M. Ikeuchi, and T. Noguchi. 2007. FTIR study on the hydrogen bond structure of a key tyrosine residue in the flavin-binding blue light sensor TePixD from Thermosynechococcus elongatus. Biochemistry. 46:6459–6467. [DOI] [PubMed] [Google Scholar]
- 52.Masuda, S., K. Hasegawa, and T. A. Ono. 2005. Tryptophan at position 104 is involved in transforming light signal into changes of β-sheet structure for the signaling state in the BLUF domain of AppA. Plant Cell Physiol. 46:1894–1901. [DOI] [PubMed] [Google Scholar]
- 53.Laan, W., M. Gauden, S. Yeremenko, R. van Grondelle, J. T. Kennis, and K. J. Hellingwerf. 2006. On the mechanism of activation of the BLUF domain of AppA. Biochemistry. 45:51–60. [DOI] [PubMed] [Google Scholar]
- 54.Salomon, M., J. M. Christie, E. Knieb, U. Lempert, and W. R. Briggs. 2000. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 39:9401–9410. [DOI] [PubMed] [Google Scholar]
- 55.Bouly, J. P., E. Schleicher, M. Dionisio-Sese, F. Vandenbussche, D. Van Der Straeten, N. Bakrim, S. Meier, A. Batschauer, P. Galland, R. Bittl, and M. Ahmad. 2007. Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J. Biol. Chem. 282:9383–9391. [DOI] [PubMed] [Google Scholar]
- 56.Berndt, A., T. Kottke, H. Breitkreuz, R. Dvorsky, S. Hennig, M. Alexander, and E. Wolf. 2007. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J. Biol. Chem. 282:13011–13021. [DOI] [PubMed] [Google Scholar]