Summary
Insulin/IGF-1-like signaling (IIS) is central to growth and metabolism, and has a conserved role in aging. In C. elegans, reductions in IIS increase stress resistance and longevity, effects that require the IIS-inhibited FOXO protein DAF-16. The C. elegans transcription factor SKN-1 also defends against oxidative stress, by mobilizing the conserved Phase 2 detoxification response. Here we show that IIS not only opposes DAF-16, but also directly inhibits SKN-1 in parallel. The IIS kinases AKT-1,-2 and SGK-1 phosphorylate SKN-1, and reduced IIS leads to constitutive SKN-1 nuclear accumulation in the intestine and SKN-1 target gene activation. SKN-1 contributes to the increased stress tolerance and longevity resulting from reduced IIS, and delays aging when expressed transgenically. Furthermore, SKN-1 that is constitutively active increases lifespan independently of DAF-16. Our findings indicate that the transcription network regulated by SKN-1 promotes longevity, and is an important direct target of IIS.
Introduction
In many studies of aging, extended longevity correlates with enhanced resistance against oxidative damage and other stresses (Finkel and Holbrook, 2000; Lithgow and Walker, 2002). In C. elegans, both lifespan and stress resistance can be increased by the alteration of several processes, including Insulin/IGF-1-like signaling (IIS) (Friedman and Johnson, 1988; Gottlieb and Ruvkun, 1994; Kenyon et al., 1993; Larsen, 1993), caloric intake (Houthoofd et al., 2002), mitochondrial respiration (Dillin et al., 2002; Lakowski and Hekimi, 1996; Lee et al., 2003), and germline function (Arantes-Oliveira et al., 2002). It is a formidable but critical challenge to understand how modulation of these processes might inhibit or defend against stresses that contribute to aging.
Reductions in IIS have been associated with increased stress resistance and longevity in diverse species (Kenyon, 2005). Studies in worms and flies have established the paradigm that these effects derive from increased activity of the FOXO transcription factors, which are inhibited through IIS-induced phosphorylation (Figure 1A) (Antebi, 2007; Kenyon, 2005). FOXO proteins are also important in many of the biological effects of insulin in mammals (Accili and Arden, 2004; van der Horst and Burgering, 2007). In C. elegans, signaling through the IIS receptor DAF-2 ultimately directs the related kinases AKT-1, -2, and SGK-1 to phosphorylate the FOXO protein DAF-16, thereby inhibiting its accumulation in nuclei (Henderson and Johnson, 2001; Hertweck et al., 2004; Lin et al., 2001; Ogg et al., 1997). DAF-16 is required for phenotypes that are associated with decreased IIS, including increases in stress resistance, longevity, and the propensity to undergo diapause, the formation of long lived dauer larvae that can survive adverse environmental conditions (Kenyon, 2005; Kenyon et al., 1993; Kimura et al., 1997). DAF-16 regulates genes that represent diverse processes, including resistance to oxidative and other stresses (Antebi, 2007; Kenyon, 2005).
Figure 1. Accumulation of SKN-1 in intestinal nuclei is inhibited by DAF-2.
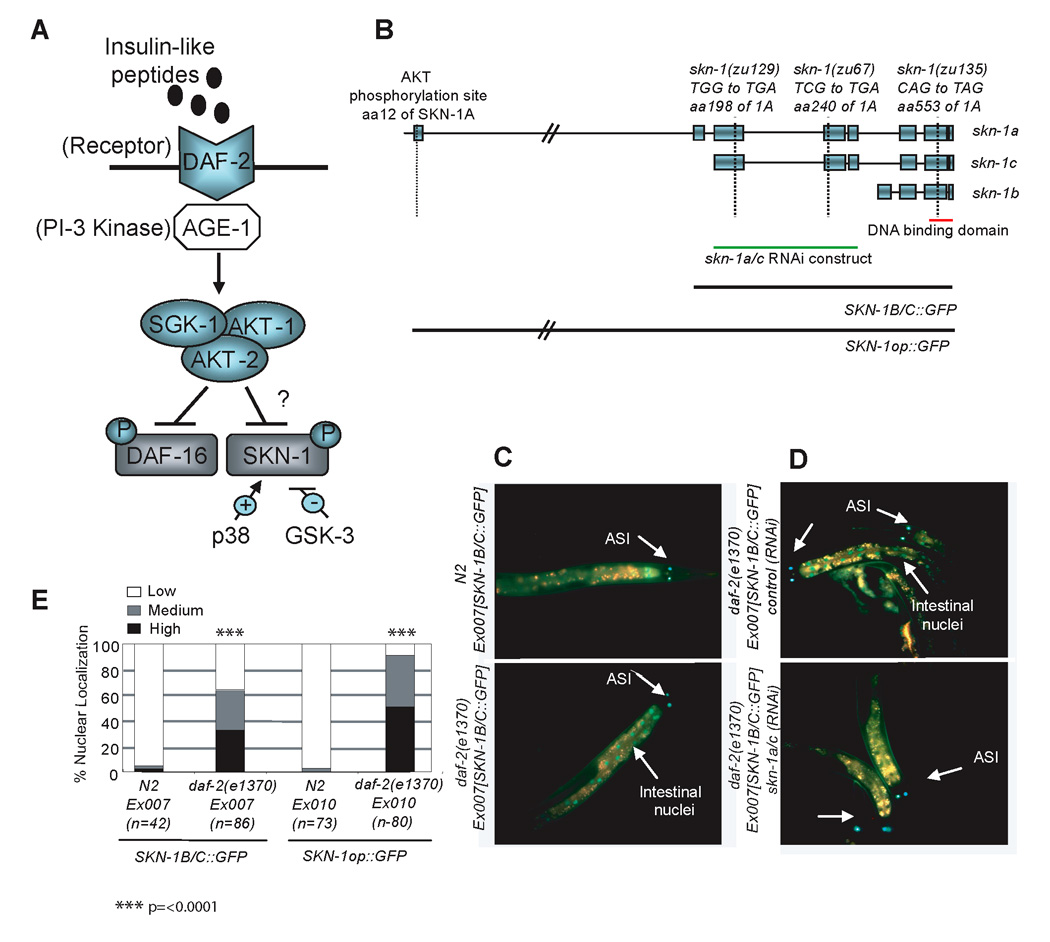
A) A partial schematic of C. elegans insulin-like signaling and our working model. B) skn-1 isoforms, alleles, and transgenes. The SKN-1A isoform is transcribed from an upstream operon promoter, and differs from SKN-1C by the addition of 90 amino acids at its N-terminus. SKN-1B and SKN-1C are each expressed from their own distinct upstream promoter (An and Blackwell, 2003; Bishop and Guarente, 2007). The skn-1(zu67), (zu129), and (zu135) mutations each create a premature stop codon (B. Bowerman, personal communication). Green Fluorescent Protein (GFP) is fused to the C-terminus of SKN-1 in the transgenes indicated at the bottom. Strains that carry these and other transgenes are described in Table S2. The skn-1a/c RNAi construct includes only coding sequence, and targets both SKN-1A and SKN-1C, but does not alter the levels of SKN-1B in the ASI neurons (Bishop and Guarente, 2007). C) SKN-1::GFP accumulates in intestinal nuclei when DAF-2 activity is reduced. D) Intestinal expression of SKN-1B/C::GFP in daf-2(e1370) derives from SKN-1C. E) Quantification of SKN-1 intestinal nuclear accumulation. Here and in Figure 2, results of experiments performed on L4 larvae have been combined and scored as described in the supplementary methods and in (An and Blackwell, 2003; An et al., 2005). p values were derived from a chi2 test.
The Phase 2 detoxification response also provides a conserved defense against oxidative stress, and consists of numerous enzymes that scavenge free radicals and other reactive compounds, and produce or transfer glutathione (McMahon et al., 2001). In C. elegans the Phase 2 response is orchestrated by the transcription factor SKN-1. SKN-1 initiates development of the feeding and digestive system during the earliest embryonic stages, then is required postembryonically for normal lifespan and stress resistance (An and Blackwell, 2003; Bowerman et al., 1992). SKN-1 is present in nuclei constitutively in the ASI neurons (putative hypothalamus) (An and Blackwell, 2003), where it is required for longevity to be extended by dietary restriction (DR), a condition that increases longevity in organisms as diverse as yeast and rodents (Bishop and Guarente, 2007). The stress resistance function of SKN-1 is mediated by its expression in the intestine (digestive system) (Bishop and Guarente, 2007), where SKN-1 accumulates in nuclei and activates Phase 2 gene expression inducibly in response to stress (An and Blackwell, 2003). In the intestine, phosphorylation of SKN-1 by p38/Mitogen Activated Protein Kinase (MAPK) signaling is required for its accumulation in nuclei, whilst negative regulation via Glycogen Synthase Kinase-3 (GSK-3) phosphorylation is needed to prevent this from occurring constitutively (Figure 1A) (An et al., 2005; Inoue et al., 2005).
As daf-16 is required for the increased stress resistance and longevity that are seen when IIS is reduced, it has seemed likely that these effects of decreased IIS could be accounted for by increased DAF-16 activity. However, we reasoned that if it is advantageous for IIS to inhibit stress response genes by acting on DAF-16, then IIS might also oppose SKN-1 (Figure 1A). Accordingly, here we show that reductions in IIS cause SKN-1 to accumulate constitutively in intestinal nuclei in the absence of stress, and to activate Phase 2 target genes. Importantly, these events do not require daf-16. AKT-1, -2 and SGK-1 phosphorylate SKN-1 at multiple sites, and mutation of an AKT phosphorylation site results in SKN-1 being present in intestinal nuclei constitutively. skn-1 mutations significantly suppress the oxidative stress resistance and longevity phenotypes associated with reduced IIS. Aging is delayed when SKN-1 is expressed transgenically, and a mutant SKN-1 form that localizes constitutively to intestinal nuclei increases lifespan in the absence of daf-16. In summary, our data indicate that IIS directly inhibits SKN-1 in parallel to DAF-16, that SKN-1 contributes to the stress and longevity phenotypes associated with reduced IIS, and that SKN-1 defines an important pro-longevity mechanism.
Results
DAF-2 signaling inhibits SKN-1 accumulation in intestinal nuclei
If IIS inhibits SKN-1 analogously to DAF-16 (Figure 1A), then reduced DAF-2 signaling should allow SKN-1 to accumulate in intestinal nuclei. To test this idea, we first investigated how reductions in DAF-2 activity affect the distribution of SKN-1 that is expressed from the previously described SKN-1B/C::GFP transgene, (Figure 1B). This transgene rescues the maternal lethality, stress sensitivity, and DR-associated longevity defects of skn-1 mutants (An and Blackwell, 2003; An et al., 2005; Bishop and Guarente, 2007). SKN-1B/C::GFP encodes two of three SKN-1 isoforms, SKN-1C and SKN-1B, which are expressed in the intestine and ASI neurons, respectively (Bishop and Guarente, 2007). In wild type (WT) animals that carry extrachromosomal or integrated SKN-1B/C::GFP, SKN-1::GFP is generally undetectable in intestinal nuclei except under stress conditions, or after inhibition of its phosphorylation by GSK-3 (An and Blackwell, 2003; An, et al., 2005). In contrast, SKN-1::GFP was constitutively present in intestinal nuclei of transgenic daf-2(e1370) and daf-2(e1368) animals, or in WT transgenics exposed to daf-2 RNA interference (RNAi) at either 20°C, or low temperature (16°C) Figure 1C and 1E; see also Figures S1A, S1B and S1C in the Supplemental Data available with this article online). This nuclear SKN-1 appeared to correspond to SKN- 1C, because RNAi against this isoform prevented SKN-1::GFP from accumulating in intestinal nuclei in response to daf-2 mutation (Figure 1D).
We also analyzed a transgene that encodes all three SKN-1 isoforms in the context of a large operon (SKN-1op::GFP; Figure 1B). In the WT background this transgene was expressed in a similar pattern to SKN-1B/C::GFP (not shown), and in daf-2(e1370) SKN-1 that was expressed from SKN-1op::GFP also accumulated in intestinal nuclei constitutively (Figure 1E). In contrast to these effects in the intestine, daf-2 mutation did not obviously alter the levels of nuclear SKN-1::GFP in the ASI neurons (Figure 1C and 1D; data not shown). We conclude that signaling through DAF-2 may be required to prevent SKN-1 from accumulating constitutively in intestinal nuclei.
DAF-2 signaling inhibits SKN-1 directly, through phosphorylation
Signaling through DAF-2 ultimately activates the AKT-1, -2 and SGK-1 kinases, which phosphorylate and inhibit DAF-16 (Figure 1A) (Cahill et al., 2001; Hertweck et al., 2004; Lin et al., 2001). RNAi knockdown of akt-1;akt-2 or sgk-1 dramatically increased the presence in intestinal nuclei of SKN-1::GFP that was expressed from either SKN- 1B/C::GFP or SKN-1op::GFP (Figure 2A and 2B), supporting the model that IIS inhibits SKN-1 directly. Importantly, daf-16 was not required for SKN-1 expression, or for the nuclear accumulation of SKN-1 that resulted from reductions in IIS. A constitutively nuclear SKN-1 substitution mutant in which a critical inhibitory GSK-3 phosphorylation site has been altered (SKN-1(S393A); An, et al., (2005)), was appropriately present in intestinal nuclei in null daf-16 animals (Figure S2). In addition, in a null daf-16 background, RNAi of the IIS kinases still increased the levels of intestinal nuclear SKN- 1::GFP expressed from SKN-1op::GFP (Figure 2A). Finally, in daf-2(e1370) animals, the nuclear accumulation of SKN-1 expressed from SKN-1B/C::GFP was not reduced by daf-16 RNAi (Figure S1D). We conclude that the IIS pathway inhibits SKN-1 in the intestine, and that this regulation occurs independently of DAF-16.
Figure 2. DAF-2 signaling inhibits nuclear accumulation of SKN-1 directly, and independently of DAF-16.
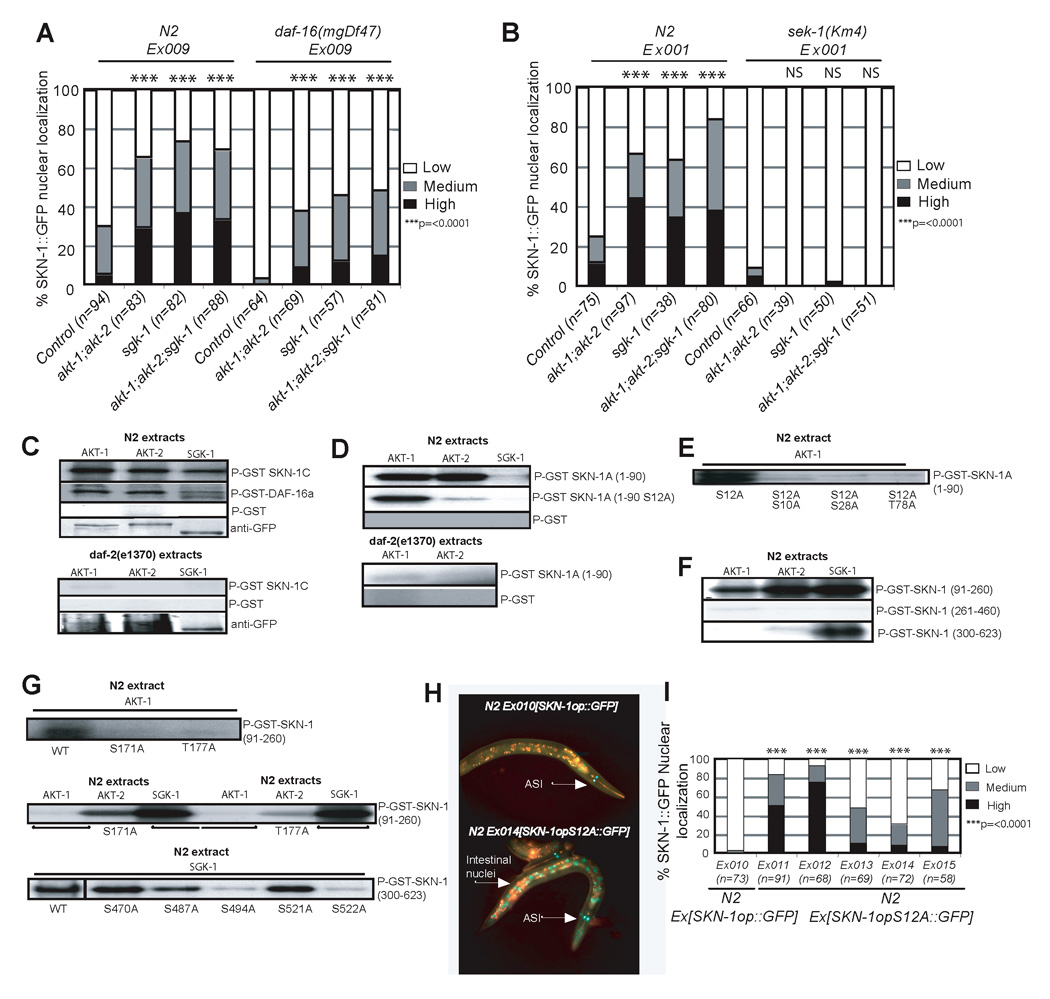
A) akt-1;akt-2 or sgk-1 RNAi allows daf-16-independent accumulation of SKN-1 in intestinal nuclei. Expression of SKN-1op::GFP was monitored. akt-1 or akt-2 RNAi had a similar effect to combined akt-1;akt-2 RNAi (data not shown). B) RNAi depletion of the downstream IIS kinases causes sek-1-dependent accumulation of SKN-1 in intestinal nuclei. Expression of SKN-1B/C::GFP was monitored. C) SKN-1C is phosphorylated by AKT-1, AKT-2, and SGK-1 comparably to DAF-16, dependent upon daf-2. Here and in panels (D–G), AKT-1::GFP, AKT-2::GFP and SGK-1::GFP were immunoprecipitated from WT or daf-2(e1370) C. elegans lysates with anti-GFP antibody, quantitated by Western blotting with anti-GFP, and used to phosphorylate purified GST-fused SKN-1 or DAF-16 proteins with [γP32]ATP. GST controls are shown in Figure S4B. These kinases appropriately required activation in C. elegans, as they did not phosphorylate SKN-1 after they had been expressed in E. coli (Figure S4A). D) The unique 90aa region at the SKN-A N-terminus (SKN-1A(1–90)) is phosphorylated by the AKT kinases but not SGK-1, dependent upon daf-2. E) Partial mapping of AKT-1 sites within SKN-1A(1–90). F) AKT and SGK phosphorylate particular regions of SKN-1C. Here and in panel (G), residues are numbered as in SKN-1A, so that the first residue of SKN-1C is numbered as 91. G) Partial mapping of AKT-1, AKT-2, and SGK-1 sites within SKN-1C fragments. H and I) Substitution of a strongly predicted AKT site within SKN-1A(1–90), Ser-12 (Table S1), results in nuclear accumulation of SKN-1A::GFP. In some lines low levels of SKN-1::GFP were observed in additional head neurons, hypodermis, and other tissues, particularly if Ser-12 was mutated (data not shown).
We previously observed that p38 MAPK signaling is required for SKN-1 to accumulate in intestinal nuclei not only in response to stress, but also when GSK-3 phosphorylation of SKN-1 is inhibited under normal conditions (An et al., 2005; Inoue et al., 2005). In the latter case, even though negative regulation of SKN-1 is impaired, the low-level background p38 signaling that is seen under normal conditions is still required. To test whether p38 signaling is also needed for SKN-1 to be present in intestinal nuclei when IIS is reduced, we performed akt-1;akt-2 and sgk-1 RNAi in animals that lack the p38 pathway MAPK kinase SEK-1 (Inoue et al., 2005). In sek-1(km4) mutants, SKN-1::GFP failed to accumulate in intestinal nuclei in response to IIS kinase RNAi (Figure 2B). Similarly, RNAi of the IIS kinases did not allow nuclear accumulation of a SKN-1 mutant in which two key p38 phosphorylation sites had been altered (Figure S3). We conclude that the SKN-1 nuclear accumulation that is allowed when IIS is reduced is appropriately dependent upon p38 MAPK signaling.
We next investigated whether SKN-1 is phosphorylated by the downstream IIS kinases AKT-1,-2 and SGK-1. Analysis using SCANSITE identified 7 potential AKT phosphorylation motifs within SKN-1C (Table S1). Four additional potential AKT phosphorylation motifs were detected within the unique N-terminal 90 amino acids of SKN-1A (SKN-1A(1–90); Table S1), which is predicted by expressed sequence tag analysis to be the most abundant SKN-1 isoform (www.wormbase.org). AKT-1::GFP, AKT-2::GFP and SGK-1::GFP that were purified from WT C. elegans (Hertweck et al., 2004) each phosphorylated full-length SKN-1C in vitro at least as robustly as they phosphorylated DAF-16 (Figure 2C). In contrast, SKN-1A(1–90) was phosphorylated by AKT-1 and AKT-2, but not SGK-1 (Figure 2D), consistent with the previous finding that AKT-1,-2 and SGK-1 differ with respect to their phosphorylation sites within the mammalian DAF-16 homolog FOXO3a (Brunet et al., 2001). Significantly, SKN-1 was not phosphorylated robustly by these same kinases if they were purified from daf- 2(e1370) animals (Figure 2C and 2D). We conclude that SKN-1 is a direct substrate of the downstream IIS kinases, and that their phosphorylation of SKN-1 depends upon signaling through DAF-2.
We then performed a partial mapping of IIS kinase phosphorylation sites in SKN-1. Within SKN-1A(1–90), Ala substitution at the strongly predicted AKT site Ser-12 reduced its phosphorylation by AKT-2 dramatically (Figure 2D) identifying Ser-12 as an AKT-2 site. AKT-1 phosphorylation was only slightly reduced by this mutation, but was substantially decreased by concurrent substitution of Ser-12 and each of three other sites within this region (Figure 2E). AKT and SGK also preferentially phosphorylated different regions or individual sites within the remainder of SKN-1, which corresponds to the SKN-1C isoform (Figure 2F and 2G; Table S1). Together, our data show that AKT- 1, -2 and SGK-1 phosphorylate SKN-1 at multiple overlapping and distinct sites.
To investigate whether SKN-1 is regulated directly by IIS kinase phosphorylation in vivo, we substituted Ala at its most strongly predicted AKT site (Ser-12 of SKN-1A) (Table S1), within the SKN-1op::GFP transgene. In each transgenic line examined, this substitution resulted in dramatic constitutive intestinal nuclear accumulation of SKN-1A (Figure 2H and 2I). This finding shows that SKN-1A is expressed in the intestine, and that the AKT phosphorylation site Ser-12 is essential for its appropriate regulation. This strongly supports the model that signaling through the DAF-2 pathway directly inhibits SKN-1 in vivo.
Inhibition of SKN-1 target genes by IIS
Given that IIS directly inhibits the SKN-1 protein in the intestine, it follows that IIS should also reduce expression of SKN-1 target genes. To test this idea, we first asked how reductions in IIS affect expression of two SKN-1 target reporter transgenes (Figure 3A and 3B). In the intestine, SKN-1 directly induces expression of gcs-1 (γ-glutamyl cysteine synthetase), which encodes an enzyme rate-limiting for glutathione biosynthesis (Figure 3A) (An and Blackwell, 2003). In the intestine a gcs-1::GFP transcriptional reporter is generally expressed at very low levels in the absence of stress, but was up-regulated by akt-1;akt-2 or sgk-1 RNAi (Figure 3C). This induction was abolished by mutation of an important SKN-1 binding site (gcs-1Δ2mut3; Figure 3A), but not by absence of daf-16 (Figure 3C), indicating that SKN-1 but not DAF-16 was required for IIS kinase RNAi to activate the gcs-1 promoter. Interestingly, gcs-1 was not induced by daf-2 RNAi, implying that its regulation by AKT and SGK-1 might involve an additional input (data not shown). In contrast, a translational fusion reporter for the SKN-1 target gene gst-7 (Glutathione S-Transferase; Figure S5) was comparably induced in the intestine by either daf-2 or IIS kinase RNAi (Figures 3B and 3D).
Figure 3. SKN-1 target genes downstream of IIS.
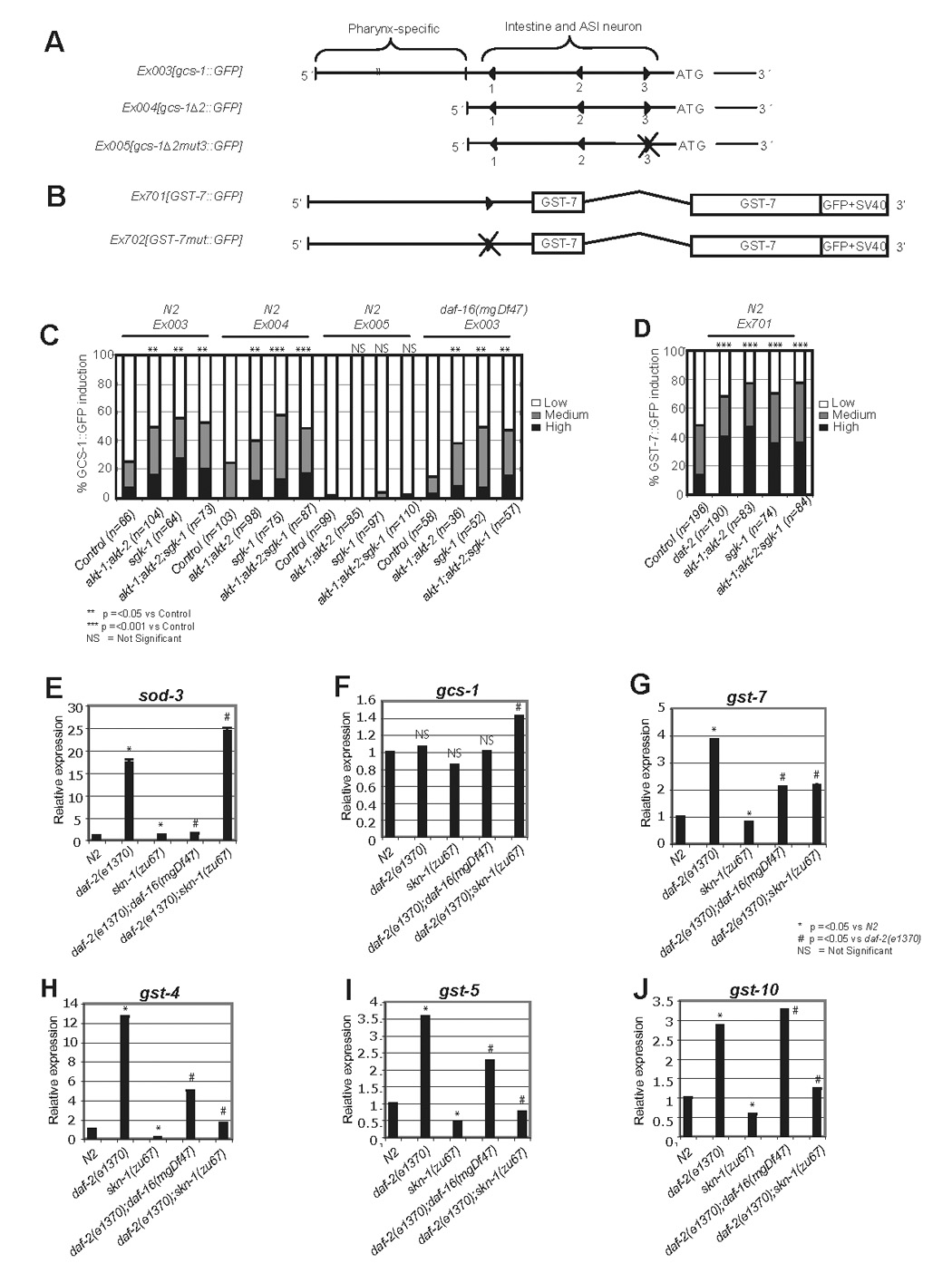
A and B) Schematics of the gcs-1 promoter (An and Blackwell, 2003), and the GST- 7::GFP translational fusion. Triangles mark SKN-1 binding sites. gcs-1Δ2::GFP lacks a promoter region required for SKN-1-independent pharyngeal expression, and gcs-1 Δ2mut3::GFP lacks a SKN-1 binding site that is important for skn-1-dependent gcs-1 expression in the intestine and ASI neurons (An and Blackwell, 2003). C) gcs-1::GFP is induced in the intestine independently of daf-16 in response to akt-1;akt-2 and sgk-1 RNAi. Here and in (D), results of multiple experiments performed on L4 larvae have been combined and scored as described in the supplementary methods and (An and Blackwell, 2003; An et al., 2005). p values were derived from a chi2 test. akt-1 and akt-2 RNAi were also performed separately, and shown to have a similar effect to combined akt-1;akt-2 RNAi (data not shown). D) GST-7::GFP is induced in response to daf-2, akt-1;akt-2, or sgk-1 RNAi. E–J) SKN-1- and DAF-16-responsive genes that are upregulated in daf-2(e1370) L4 larvae were identified by quantitative PCR, using actin (act-1) as a control. The level of skn-1 mRNA in these samples was also measured (Figure S6). One representative experiment is shown, error bars represent the SEM, and p values were derived from a Student’s t-test.
We next used quantitative (Q)-PCR to analyze how daf-2 mutation affected the levels of mRNA expressed from a set of known or predicted SKN-1 target Phase 2 genes (An and Blackwell, 2003; Kell et al., 2007). As would be expected, SKN-1 was not required for daf-2 mutation to induce expression of a negative control for our experiments, the DAF-16 target sod-3 (Figure 3E) (Antebi, 2007). The levels of gcs-1 mRNA were largely unaffected in this daf-2 mutant (Figure 3F), in agreement with our reporter data and the high levels of skn-1-independent gcs-1 expression seen in the pharynx (An and Blackwell, 2003). In contrast, four different gst genes were strongly upregulated by daf-2 mutation in a manner that was substantially or completely dependent upon skn-1. (Figure 3G–J). DAF-16 contributed to induction of some of these genes, but the SKN-1 target gst-10 was upregulated by daf-2 mutation independently of daf-16 (Figure 3G–J). Together, our results indicate that when DAF-2 pathway signaling is reduced, SKN-1 induces a gene expression program that overlaps only partially with that of DAF-16.
skn-1 contributes to the increased longevity and stress resistance associated with reduced IIS
Having established that DAF-2 and IIS inhibit SKN-1 directly, and thereby suppress SKN-1-dependent gene expression, we next asked whether SKN-1 contributes to phenotypes that are associated with decreased IIS. We engineered daf-2;skn-1 double mutants that each carried one of three skn-1 mutations, along with one of two daf-2 alleles. Each skn-1 mutation (Figure 1B) results in reduced stress resistance and longevity (Figure 4; Table 1) (An and Blackwell, 2003), failure of DR to extend lifespan (Bishop and Guarente, 2007), and maternally-derived embryonic lethality (Bowerman et al., 1992). The Class 2 daf-2 mutant e1370 is stress-resistant and lives much longer than WT at 20°C (Figure 4A and 4C), but this increased lifespan is accompanied by various dauer-like abnormalities (Gems et al., 1998; Kimura et al., 1997). The Class 1 daf-2 mutation e1368 increases lifespan to a lesser extent (Figure 4D), but these animals appear essentially normal. We also tested how skn-1 mutations affect similar phenotypes caused by RNAi of the IIS downstream kinases. At 25°C, sgk-1 RNAi increases both oxidative stress resistance and lifespan, dependent upon daf-16 (Hertweck et al., 2004). Previously, lifespan was not extended when akt-1 RNAi was performed in the akt-2(ok393) mutant (Hertweck et al., 2004), but we have now observed that akt-1;akt-2 RNAi significantly increases stress resistance and lifespan in an RNAi hypersensitive strain (Figure 4B and 4F; Table 1). This suggests that AKT-1,-2 and SGK-1 may contribute comparably to the effects of DAF-2 signaling on stress resistance and longevity.
Figure 4. skn-1 mutations suppress the longevity and stress resistance phenotypes conferred by reduced IIS.
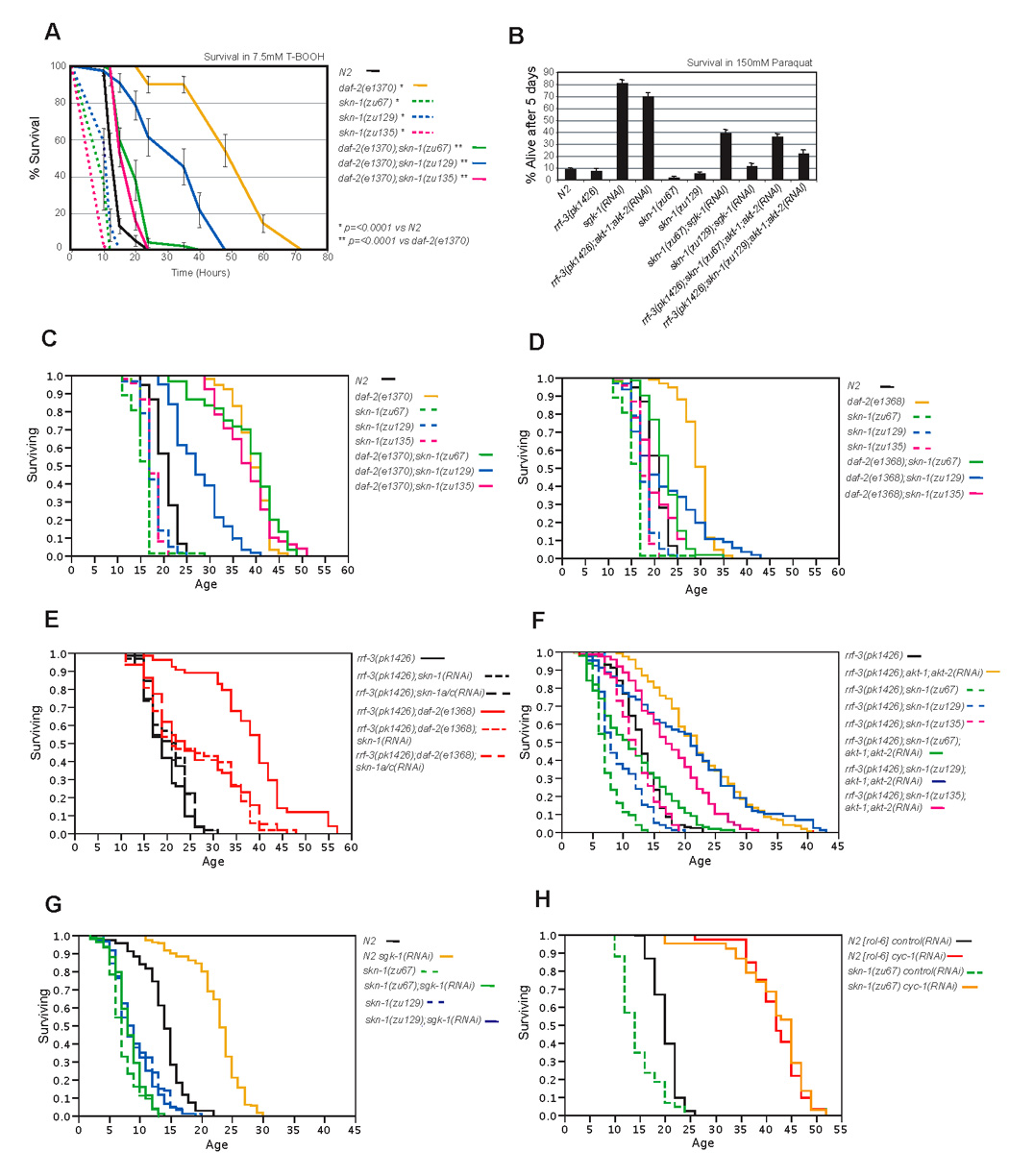
A) skn-1 contributes to the oxidative stress resistance phenotype of daf-2(e1370) mutants. A representative experiment is shown in which animals were exposed to 7.5 mM t-butyl hydrogen peroxide. B) skn-1 contributes to the stress resistance phenotypes conferred by sgk-1 or akt-1;akt-2 RNAi. Animals were scored after 5 days exposure to 150mM paraquat. Data were combined from at least three experiments with n>90 for each group. C) Lifespan analysis of daf-2(e1370); skn-1 mutants. D) skn-1 suppresses the longevity of daf-2(e1368) mutants. E) In the RNAi-sensitive rrf-3 background (Simmer et al., 2002), the lifespan increase seen in daf-2(e1368) is comparably suppressed by either skn-1 or skn-1a/c RNAi. RNAi bacteria were provided from the L1 stage. F, G) skn-1 mutations suppress the lifespan extension conferred by akt-1;akt-2 or sgk-1 RNAi. Survival assays were carried out at 25°C, are from adulthood, and represent the combined data from at least two experiments. H) skn-1 mutation does not suppress the lifespan extension conferred by cyc-1 RNAi. The lifespan experiments in C and D were carried out on OP50, but all others were performed on HT115 lawns. Lifespan data in this Figure are summarized in Table 1. Unless otherwise indicated, the analyses shown were performed at 20°C, and measured from hatching.
Table 1.
skn-1 contributes to the lifespan extension resulting from reduced IIS.
| Strain | mean life span ± SEM 20°C (days) | Median lifespan | 75th percentile 20°C (days) | 25th percentile 20°C (days) | p value against control | n | Trials |
|---|---|---|---|---|---|---|---|
| N2 | 20.44 ± 0.3 | 21 | 23 | 19 | - | 90/90 | 1 |
| daf-2 (e1368) | 29.42 ± 0.3 | 31 | 31 | 29 | <.0001a | 91/91 | 1 |
| daf-2 (e1370) | 39.39 ± 0.4 | 39 | 43 | 37 | <.0001a | 88/88 | 1 |
| daf-16 (mgDf47) | 16.80 ± 0.3 | 17 | 19 | 15 | <.0001a | 50/50 | 1 |
| daf-16 (mgDf47) ; daf-2 (e1370) | 20.01 ± 0.1 | 21 | 21 | 19 | <.0001b | 77/80 | 1 |
| skn-1(zu67) | 15.60 ± 0.3 | 17 | 17 | 15 | <.0001a | 73/73 | 1 |
| skn-1(zu129) | 17.67 ± 0.2 | 17 | 19 | 17 | <.0001a | 90/90 | 1 |
| skn-1(zu135) | 17.70 ± 0.3 | 17 | 19 | 17 | <.0001a | 57/57 | 1 |
| daf-2 (e1368); skn-1(zu67) | 22.63 ± 0.5 | 23 | 25 | 21 | <.0001c | 60/60 | 1 |
| daf-2 (e1368); skn-1(zu129) | 22.25 ± 1.1 | 19 | 29 | 15 | <.0001c | 58/60 | 1 |
| daf-2 (e1368); skn-1(zu135) | 19.83 ± 0.6 | 19 | 23 | 17 | <.0001c | 58/59 | 1 |
| daf-2 (e1370); skn-1(zu67) | 38.60 ± 1.0 | 41 | 43 | 35 | .0740b | 60/60 | 1 |
| daf-2 (e1370); skn-1(zu129) | 27.64 ± 0.7 | 27 | 31 | 23 | <.0001b | 62/62 | 1 |
| daf-2 (e1370); skn-1(zu135) | 33.16 ± 0.6 | 39 | 43 | 33 | .6016b | 25/25 | 1 |
| rrf-3(pk1426);control(RNAi) | 19.53 ± 0.5 | 19 | 24 | 17 | - | 57/60 | 1 |
| rrf-3(pk1426);skn-1(RNAi) | 20.43 ± 0.6 | 21 | 24 | 15 | .1531g | 60/60 | 1 |
| rrf-3(pk1426);skn-1a/c(RNAi) | 20.27 ± 0.6 | 21 | 24 | 15 | .2313g | 59/60 | 1 |
| rrf-3(pk1426);daf-2(e1368) control(RNAi) | 38.62 ± 1.3 | 40 | 44 | 34 | <.0001g | 52/55 | 1 |
| rrf-3(pk1426); daf-2(e1368) skn-1(RNAi) | 25.83 ± 1.3 | 21 | 36 | 17 | <.0001d | 56/56 | 1 |
| rrf-3(pk1426); daf-2(e1368) skn-1a/c (RNAi) | 26.32 ± 1.3 | 24 | 36 | 19 | <.0001d | 57/58 | 1 |
| N2 [rol-6]; control(RNAi) | 20.1 ± 0.3 | 20 | 22 | 18 | 84/84 | 1 | |
| skn-1(zu67); control(RNAi) | 14.6 ± 0.4 | 14 | 16 | 12 | <.0001e | 87/90 | 1 |
| N2 [rol-6]; cyc-1(RNAi) | 42.1 ± 0.9 | 42 | 45 | 40 | <.0001e | 32/34 | 1 |
| skn-1(zu67); cyc-1(RNAi) | 41.9 ± 1.2 | 45 | 47 | 38 | .5594f | 38/38 | 1 |
| N2 [rol-6]; cco-1(RNAi) | 31.52 ± 0.5 | 32 | 34 | 30 | <.0001e | 67/69 | 1 |
| skn-1(zu67); cco-1(RNAi) | 35.13 ± 0.5 | 36 | 38 | 34 | <.0001f | 46/46 | 1 |
| Strain | mean adult life span ± SEM 25°C (days) | Median lifespan | 75th Percentile 25°C (days) | 25th percentile 25°C (days) | p value against Control | n | No. Exp |
| rrf-3(pk1426); control(RNAi) | 12.90 ± 0.4 | 13 | 15 | 11 | - | 99/100 | 2 |
| daf-16(mgDf47) | 11.04 ± 0.2 | 11 | 13 | 9 | <.0001g | 138/138 | 3 |
| rrf-3(pk1426); akt1; akt-2(RNAi) | 22.50 ± 0.7 | 22 | 28 | 17 | <.0001g | 120/138 | 2 |
| daf-16(mgdf47); rrf-3(pk1426); akt1; akt-2(RNAi) | 11.03 ± 0.3 | 11 | 13 | 10 | <.0001h | 123/123 | 3 |
| rrf-3(pk1426);skn-1(zu67); control(RNAi) | 7.20 ± 0.2 | 7 | 8 | 6 | <.0001g | 106/106 | 2 |
| rrf-3(pk1426);skn-1(zu129);control(RNAi) | 9.30 ± 0.4 | 8 | 12 | 7 | <.0001g | 100/100 | 2 |
| rrf-3(pk1426);skn-1(zu135);control(RNAi) | 12.00 ± 0.3 | 12 | 15 | 9 | 0.1012g | 150/150 | 3 |
| rrf-3(pk1426);skn-1(zu67); akt-1; akt-2(RNAi) | 11.60 ± 0.6 | 11 | 17 | 6 | <.0001h | 104/104 | 2 |
| rrf-3(pk1426);skn-1(zu129); akt-1; akt-2(RNAi) | 20.40 ± 1.0 | 21 | 28 | 12 | 0.9400h | 101/104 | 2 |
| rrf-3(pk1426);skn-1(zu135); akt-1; akt-2(RNAi) | 17.69 ± 0.6 | 17 | 22 | 13 | <.0001h | 110/110 | 3 |
| N2; control(RNAi) | 13.8 ± 0.3 | 14 | 16 | 12 | - | 110/114 | 2 |
| N2; sgk-1(RNAi) | 22.6 ± 0.4 | 23 | 25 | 21 | <.0001e | 110/395 | 2 |
| skn-1(zu67); sgk-1(RNAi) | 8.2 ± 0.2 | 8 | 10 | 7 | <.0001i | 128/128 | 2 |
| skn-1(zu129); sgk-1(RNAi) | 9.1 ± 0.2 | 8 | 11 | 7 | <.0001i | 185/186 | 2 |
Corresponds to the data in Figure 4C–H. SEM: standard error of the mean. 75th and 25th percentiles refer to the day at which 75% or 25% of the population is dead. n represents total number of animals dying of old age vs those in total experiment (combined between trials where appropriate). In the case of sgk-1(RNAi) the majority of exclusions are due to Egl/bag of worms phenotypes. p values were calculated as follows:
N2,
daf-2(e1370),
daf-2(e1368),
rrf-3(pk1426);daf-2(e1368) control(RNAi),
N2 control(RNAi),
N2 with corresponding RNAi,
rrf-3(pk1426) control(RNAi),
rrf-3(pk1426) akt-1;akt-2(RNAi),
sgk-1(RNAi). Individual experiments, an additional analysis of daf-2(e1368) and daf-2(e1370), and other supporting data are in Table S5.
Initially, we tested whether the IIS-associated phenotypes of dauer formation and elevated stress resistance require skn-1. skn-1 RNAi did not prevent the constitutive dauer entry associated with the daf-2(e1370) mutation, or akt-1;akt-2 RNAi (supplemental data). In contrast, and consistent with its role in Phase 2 detoxification, mutations in skn-1 dramatically suppressed the increases in oxidative stress resistance seen in each of these contexts of reduced IIS (Figure 4A and 4B).
We then investigated how skn-1 contributes to the increased longevity deriving from decreased daf-2 function. Only skn-1(zu129) suppressed the increased longevity of the Class 2 daf-2 allele e1370 (Figure 4C; Table 1). In contrast, all three skn-1 mutations significantly reduced the lifespan of the Class 1 daf-2 mutant e1368 (Figure 4D; Table 1). These skn-1 mutations also altered the shape of the daf-2(e1368) survival plot, so that the longest-living double mutant animals appeared to be less dependent upon skn-1 than the remaining population (Figure 4D). Accordingly, when we excluded the 20% most long-lived animals from statistical analysis, two of three skn-1 mutations affected death rate more severely in daf-2(e1368) animals than WT (Figure S7; Tables S3 and S4). Importantly, an RNAi-sensitive genetic background skn-1 RNAi decreased the lifespan of daf-2(e1368) but not control animals (Figure 4E; Table 1 and Table S4), indicating that this daf-2 mutant is more susceptible than WT to a reduction in SKN-1 levels. daf-2(e1368) lifespan was similarly decreased by either skn-1 or skn-1a/c RNAi (Figure 4E; Table 1 and Table S4), indicating that the SKN-1 A and/or C isoforms are required for the longevity associated with daf-2(e1368). daf-2(e1368) lifespan was also decreased by feeding of skn-1 RNAi from the beginning of adulthood (Figure S8), indicating an effect on aging, not development.
Mutations in skn-1 also suppressed the lifespan extensions associated with reductions in IIS kinase levels. Each of two skn-1 alleles that we analyzed (zu67 and zu 129) completely eliminated the dramatic increase in lifespan deriving from sgk-1 RNAi (Figure 4G; Table 1 and Table S4). In addition, two of three skn-1 alleles significantly reduced the lifespan of akt-1;akt-2 RNAi animals (Figure 4F; Table 1). We conclude that SKN-1 makes important contributions to the increases in stress resistance and longevity that are associated with reductions in IIS activity.
We next asked whether skn-1 mutations might cause a general sickness that limits lifespan, by investigating whether skn-1 affects the long lifespan of animals with reduced mitochondrial electron transport chain activity (Dillin et al., 2002; Lee et al., 2003; Rea et al., 2007). In agreement with a recent report (Rea et al., 2007), a skn-1 mutation did not reduce the large increases in lifespan associated with RNAi against either cytochrome c reductase (cyc-1) or cytochrome c oxidase (cco-1) (Figure 4H and S9; Table 1). This indicates that while skn-1 is important for lifespan in long-lived animals in which IIS is reduced, lack of skn-1 does not limit longevity non-specifically.
SKN-1 promotes longevity
The involvement of skn-1 in the longevity effects of IIS led us to investigate whether SKN-1 has an independent pro-longevity function. Previously, lifespan was modestly increased by transgenic overexpression of either WT or constitutively nuclear forms of DAF-16 (Henderson and Johnson, 2001; Lin et al., 2001). To investigate whether modulation of SKN-1 expression can extend lifespan, we first analyzed otherwise WT strains in which either the SKN-1B/C::GFP or SKN-1B/C S393A::GFP transgenes were present. The SKN-1B/C S393A::GFP transgene, which expresses constitutively nuclear SKN-1 in the intestine, increased mean lifespan variably but consistently (by 5%–21%; Figure 5A; Table 2). A modest increase was also seen with SKN-1B/C::GFP (Ex001; Figure 5B; Table 2).
Figure 5. Transgenic expression of SKN-1 extends lifespan.
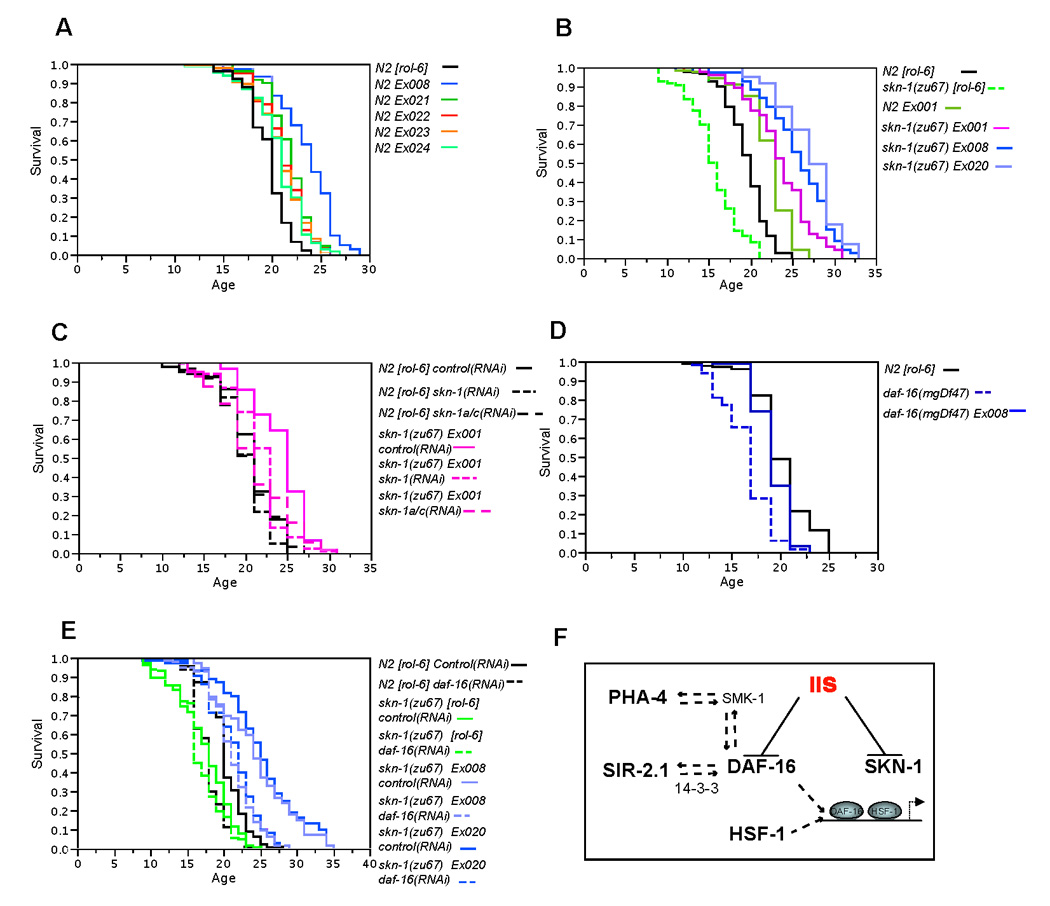
A) Lifespan extension in otherwise WT animals carrying the [SKN-1B/C S393A::GFP] transgene, which expresses constitutively nuclear SKN-1C in the intestine (see text). These independent extrachromosomal arrays were created by injecting 2.5ng/µl transgene DNA. B) Lifespan extension in skn-1(zu67) animals carrying skn-1 transgenes. The Ex001[SKN-1B/C::GFP], Ex008[SKN-1B/C S393A::GFP] and Ex020[SKN-1B/C S393A::GFP] extrachromosomal arrays were originally created in WT animals by injection of 2.5, 2.5, and 10 ng/µl transgene DNA, respectively, and then bred into skn-1(zu67). The presence of the transgene did not dramatically affect the developmental timing, brood size or age specific fecundity of either the WT or skn-1 mutants (Figure S14). The lifespan of WT animals carrying Ex001[SKN-1B/C::GFP] is shown for comparison. C) SKN-1C contributes to the lifespan of skn-1(zu67) Ex001[SKN-1B/C::GFP] animals. Lifespan is reduced comparably by skn-1 and skn-1a/c RNAi (Figure 1B). D) Presence of Ex008[SKN-1B/C S393A::GFP] increases the lifespan of daf-16(mgDf47) mutants. E) Transgene arrays that express constitutively nuclear SKN-1 (Ex008 or Ex020) increase skn-1(zu67) lifespan beyond WT when daf-16 is depleted by RNAi. Lifespan analyses were scored from hatching and performed at 20°C unless otherwise stated, and are summarized in Table 2. The experiments in panels A and B were carried out on OP50, and the others on HT115. Panels C and D are representative experiments, but otherwise data were combined from at least two experiments, within which all sample sets were analyzed in parallel. F) Cross talk among C. elegans transcriptional regulators that increase longevity when expressed transgenically. DAF-16 physically interacts with SIR-2.1 through 14-3-3 proteins, and is required for SIR-2.1 to extend lifespan (Berdichevsky et al., 2006; Tissenbaum and Guarente, 2001). HSF-1 and DAF-16 are predicted to bind to some of the same promoters, and regulate some pro-longevity target genes together (Hsu et al., 2003). PHA-4 shares a common cooperating cofactor with DAF-16 (SMK-1), extends lifespan when DAF-16 is absent, and is required for DR to extend lifespan (Panowski et al., 2007). SKN-1 and DAF-16 are inhibited directly and in parallel by IIS (this work).
Table 2.
Transgenic expression of SKN-1 extends lifespan.
| Strain | mean life span ± SEM 20°C (days) | 75th percentile 20°C (days) | p value against control | n | No.of Exp |
|---|---|---|---|---|---|
| N2 Ex[rol-6] | 19.57 ± 0.2 | 21 | - | 129/130 | 2 |
| N2 Ex008 [SKN-1 S393A:: GFP] 2.5ng/µl | 23.62 ± 0.3 | 26 | <.0001a | 78/78 | 2 |
| N2 Ex021 [SKN-1 S393A:: GFP] 2.5ng/µl | 21.80 ± 0.2 | 23 | <.0001a | 117/122 | 2 |
| N2 Ex022 [SKN-1 S393A:: GFP] 2.5ng/µl | 21.10 ± 0.3 | 23 | <.0001a | 62/62 | 2 |
| N2 Ex023 [SKN-1 S393A:: GFP] 2.5ng/µl | 20.89 ± 0.3 | 23 | <.0001a | 96/96 | 2 |
| N2 Ex024 [SKN-1 S393A:: GFP] 2.5ng/µl | 20.66 ± 0.4 | 23 | <.0001a | 63/63 | 2 |
| N2 Ex[rol-6] | 19.52 ± 0.2 | 21 | - | 236/243 | 3 |
| skn-1(zu67) Ex[rol-6] | 15.51 ± 0.3 | 18 | <0.0001a | 96/96 | 2 |
| N2 Ex001[SKN -1B/C:: GFP] 2.5ng/µl | 22.14 ± 0.3 | 25 | <.0001b,c | 146/148 | 2 |
| skn-1(zu67) Ex001[SKN -1B/C:: GFP] 2.5ng/µl | 23.53 ± 0.6 | 26 | <.0001b,c | 48/51 | 2 |
| skn-1(zu67) Ex008[SKN -1B/C S393A:: GFP] 2.5ng/µl | 25.74 ± 0.5 | 29 | <.0001b,c | 67/70 | 2 |
| skn-1(zu67) Ex020[SKN -1B/C S393A:: GFP] 10ng/µl | 27.16 ± 0.6 | 29 | <.0001b,c | 30/40 | 1 |
| N2 Ex[rol-6]; control(RNAi) | 20.60 ± 0.5 | 23 | - | 50/50 | 1 |
| N2 Ex[rol-6]; skn-1(RNAi) | 19.91 ± 0.4 | 21 | 0.0699f | 60/60 | 1 |
| N2 Ex[rol-6]; skn-1a/c(RNAi) | 20.37 ± 0.4 | 23 | 0.7880f | 59/60 | 1 |
| skn-1(zu67) Ex001[SKN -1B/C:: GFP] 2.5ng/µl ;control(RNAi) | 24.20 ± 0.4 | 27 | <.0001f | 62/64 | 1 |
| skn-1(zu67) Ex001[SKN -1B/C:: GFP] 2.5ng/µl;skn-1(RNAi) | 21.58 ± 0.4 | 23 | <.0001d | 84/84 | 1 |
| skn-1(zu67) Ex001[SKN -1B/C:: GFP] 2.5ng/µl ;skn-1a/c(RNAi) | 21.06 ± 0.5 | 25 | 0.0005d | 69/69 | 1 |
| N2 Ex[rol-6] | 20.60±0.5 | 23 | - | 50/50 | 1 |
| daf-16(mgDf47) | 15.87±0.3 | 17 | <.0001f | 58/61 | 1 |
| daf-16(mgDf47) Ex008[SKN-1 S393A:: GFP] 2.5ng/µl | 18.88±0.3 | 21 | <.0001e | 48/48 | 1 |
| N2 Ex[rol-6]; control(RNAi) | 20.02 ± 0.2 | 22 | - | 256/259 | 4 |
| N2 Ex[rol-6]; daf-16(RNAi) | 17.77 ± 0.1 | 19 | <.0001f | 266/266 | 4 |
| skn-1(zu67) Ex[rol-6]; control(RNAi) | 17.38 ± 0.3 | 20 | <.0001f | 145/146 | 3 |
| skn-1(zu67) Ex[rol-6]; daf-16(RNAi) | 16.59 ± 0.3 | 19 | 0.0240g | 142/145 | 3 |
| skn-1(zu67)Ex001[SKN-1b/c:: GFP] 2.5ng/µl; control(RNAi) | 22.95 ± 0.5 | 26 | <.0001g | 62/62 | 2 |
| skn-1(zu67) Ex001[SKN-1b/c:: GFP] 2.5ng/µl; daf-16(RNAi) | 16.53 ± 0.3 | 18 | 0.1655h | 59/59 | 2 |
| skn-1(zu67) Ex008[SKN-1 S393A:: GFP] 2.5ng/µl; control(RNAi) | 24.28 ± 0.6 | 28 | <.0001g | 64/76 | 3 |
| skn-1(zu67) Ex008[SKN-1 S393A:: GFP] 2.5ng/µl; daf-16(RNAi) | 21.12 ± 0.4 | 23 | <.0001h,i | 76/87 | 3 |
| skn-1(zu67) Ex020[SKN-1 S393A:: GFP] 10ng/µl; control(RNAi) | 25.10 ± 0.6 | 28 | <.0001g | 121/132 | 3 |
| skn-1(zu67) Ex020[SKN-1 S393A:: GFP] 10ng/µl; daf-16(RNAi) | 21.39 ± 0.4 | 24 | <.0001h,i | 124/140 | 3 |
Corresponds to the data in Figure 5A–E. p values were calculated as follows:
N2 Ex[rol-6],
skn-1(zu67)[rol-6],
significantly enhanced compared to N2 [rol-6] (p=<0.0001 in each case),
skn-1(zu67) Ex001 control(RNAi),
daf-16(mgDf47) control(RNAi),
N2 Ex[rol-6] control(RNAi),
skn-1(zu67)[rol-6] control(RNAi),
skn-1(zu67)[rol-6] daf-16(RNAi),
Interestingly, we had previously observed that skn-1 transgenes not only rescue the oxidative stress sensitivity of skn-1(zu67) mutants, but also increase their stress resistance to well beyond that of WT (An et al., 2005). Similarly, strains in which skn-1(zu67) was rescued with these transgenes also lived longer than wild type animals that carried the same transgene arrays (arrays Ex001 and Ex008, Figures 5A and 5B; Table 2). This difference was corroborated in independent strains that were generated by breeding the Ex001[SKN-1B/C::GFP] array from the WT into skn-1(zu67) and skn-1(zu129) (Figures S10A, S10B, and S11), and in an additional skn-1(zu67) strain carrying SKN-1B/C S393A::GFP (Ex020, Figure 5B; Table 2). Perhaps an optimal window of SKN-1 expression is important for longevity, because we found that expression of SKN-1 from high-copy arrays can be toxic (data not shown). Significantly, rescued skn-1(zu67) Ex001[SKN-1B/C::GFP] animals maintained the ability to move longer than WT (Figure S10C), indicating that transgenic skn-1 expression delayed the aging process. RNAi that was targeted to transgenic SKN-1C (skn-1a/c RNAi, Figure 1B) eliminated expression of intestinal nuclear SKN-1 from SKN-1B/C S393A::GFP (Figure S12), and suppressed the extended lifespan of a transgenically rescued skn-1(zu67) strain (Figure 5C). Taken together, our results indicate that SKN-1 promotes longevity, an effect that appears to require the SKN-1C isoform.
We next investigated whether daf-16 is required for transgenic SKN-1 to increase lifespan, because of the general importance of daf-16 for C. elegans longevity, and because SKN-1 and DAF-16 overlap in regulating some target genes (Figure 3E–J). When we bred a SKN-1 S393A transgenic array (Ex008) into the null mutant daf-16(mgDf47), lifespan was modestly but significantly increased in 2 of 3 experiments (Figure 5D; Table 2 and Table S6). Similarly, after daf-16 RNAi the lifespan of skn-1(zu67) animals was reduced, but presence of an array that expresses constitutively nuclear SKN- 1(S393A) (Ex008 or 020) still increased skn-1(zu67) lifespan to beyond that of WT (Figure 5E; Table 2). Interestingly, daf-16 RNAi completely suppressed the lifespan extension associated with transgenic WT SKN-1 (Figure S13; Table 2), suggesting that reductions in DAF-16 levels not only reduce lifespan, but also might free up negative regulatory mechanisms that act on SKN-1. Taken together, our data suggest that SKN-1 can promote longevity independently of DAF-16, provided that SKN-1 is released from negative regulation in the intestine.
Discussion
It is well established that FOXO proteins are inhibited directly by the IIS pathway, and that the FOXO protein DAF-16 is required for the increased longevity and stress resistance that result from decreased IIS in C. elegans (Kenyon, 2005). This has made it difficult to imagine that another transcription factor might be regulated similarly by IIS in parallel to FOXO, and could play a major role in IIS-regulated processes. However, here we have identified SKN-1 as the second transcription factor that is known to be inhibited directly by IIS. We have also shown that IIS inhibits SKN-1 target genes, and that SKN-1 contributes to the stress resistance and longevity phenotypes of reduced IIS, and functions as a pro-longevity factor.
Direct regulation of SKN-1 by IIS
Multiple lines of evidence indicate that IIS inhibits SKN-1 in the intestine directly (Figure 1A). Reductions in IIS activity resulted in SKN-1A and SKN-1C being present in intestinal nuclei in the absence of either stress or DAF-16, and in induction of SKN-1 target genes (Figure 1E, Figures 2A, 2B, and Figure 3). SKN-1 isoforms were phosphorylated by AKT-1,-2 and SGK-1 at multiple sites, and mutation of an AKT site (Ser-12) led to SKN-1A being present in intestinal nuclei constitutively (Figure 2H and 2I). The AKT and SGK kinases differed in their preferences for sites within SKN-1, consistent with previous analyses of their phosphorylation of FOXO (Brunet et al., 2001). The complexity of this regulation could potentially allow SKN-1 to integrate inputs from multiple signals or molecular interactions.
We also observed that SKN-1 contributes significantly to some IIS associated phenotypes. skn-1 mutations dramatically suppressed the increases in stress resistance deriving from sgk-1 RNAi, akt-1;akt-2 RNAi, and the daf-2(e1370) mutation (Figure 4A and 4B). Lack of skn-1 completely suppressed the increased lifespan deriving from sgk-1 RNAi, reduced the extended longevity of akt-1;akt-2 RNAi animals (Figures 4F and 4G) and suppressed the increased lifespan of the Class 1 daf-2 allele e1368 (Figure 4D). Importantly, skn-1 RNAi reduced the lifespan of daf-2(e1368) but not WT animals (Figure 4E), indicating that daf-2(e1368) was more severely affected than WT by partial loss of skn-1 function. In contrast, skn-1 mutation did not suppress the long life associated with RNAi of mitochondrial respiratory chain components (Figure 4H and Figure S9), indicating that lack of skn-1 does not reduce lifespan non-specifically. Interestingly, skn-1 mutations also did not consistently suppress the lifespan extension of daf-2(e1370) mutants (Figure 4C). However, dauer-like phenotypes are associated with this Class 2 allele (Gems et al., 1998), suggesting that this difference might reflect the apparent dispensability of SKN-1 for dauer formation (Supplementary data).
The simplest model to explain the contributions of SKN-1 to IIS-associated longevity is that they derive from Phase 2 detoxification gene expression in the intestine. Our finding that skn-1a/c RNAi suppresses daf-2(e1368) longevity (Figure 4E) is consistent with this idea, although we cannot exclude that other tissues or SKN-1 functions are involved. We conclude that daf-16 is necessary but not sufficient to confer the increases in stress resistance and longevity that are associated with reduced IIS, and that these increases also involve skn-1.
In contrast to the regulation of SKN-1 by IIS in the intestine, daf-2 mutation did not obviously affect SKN-1 levels in the ASI neurons (Figure 1C; not shown), in which SKN-1B is required for DR to extend lifespan (Bishop and Guarente, 2007). However, we cannot exclude that IIS might subtly influence the levels or activity of endogenous SKN-1 in ASI. This possibility could help resolve the apparent paradox that in C. elegans most DR protocols extend lifespan independently of daf-16, and therefore seemingly without input from IIS (Antebi, 2007; Bishop and Guarente, 2007; Greer et al., 2007; Houthoofd et al., 2003; Kenyon, 2005; Panowski et al., 2007).
SKN-1 is the sequence and functional counterpart to the mammalian NF-E2-related factor (Nrf) proteins (An and Blackwell, 2003), which orchestrate the Phase 2 detoxification response (Hayes and McMahon, 2001). Our results predict that the conserved IIS pathway might also regulate mammalian Nrf proteins, which include potential AKT sites (Table S1). Why would it be advantageous for IIS to decrease stress resistance? Many signaling cascades are influenced by redox status (Tonks, 2005). In particular, IIS is attenuated by Protein Tyrosine Phosphatase (PTP)1B, which is inhibited by reactive oxygen species (ROS) via the oxidation of conserved cysteine residues (Goldstein et al., 2005; Tonks, 2005). Thus, by inhibiting DAF-16/FOXO and SKN-1/Nrf, IIS could suppress antioxidant defenses that would lead to a reducing environment, and that in turn might decrease IIS effectiveness. This is consistent with evidence that insulin induces starved cells to produce a ‘burst’ of ROS, and that IIS functions more effectively under oxidative conditions (Goldstein et al., 2005).
Pro-longevity functions of SKN-1
Our finding that transgenic SKN-1 expression extends lifespan (Figures 5A and 5B) demonstrates that skn-1 has an independent pro-longevity function. This activity appears to require the action of the intestinal SKN-1C isoform, because it is suppressed by skn-1a/c RNAi (Figure 5C). This pro-longevity activity of SKN-1 thus appears to be distinct from its ASI-mediated role in DR-induced longevity (Bishop and Guarente, 2007). The observation that transgenic SKN-1 increased lifespan more robustly in skn-1 mutants than in the WT (Figures 5A and 5B) suggests that the systemic effects of SKN-1 expression may be complex. However, taken together, our data demonstrate that SKN-1 levels can be adjusted to extend lifespan and delay aging. We propose that the ancestral function of SKN-1 in Phase 2 detoxification can be modulated by either reductions in IIS or adjustment of SKN-1 expression to extend lifespan, raising the exciting possibility that this detoxification system might be of general and conserved importance for longevity.
In the context of reduced IIS, SKN-1 and DAF-16 appeared to have both distinct and overlapping functions (Figure 3). daf-16 contributed to expression of some skn-1-dependent Phase 2 detoxification genes (Figure 3E–I), but other SKN-1 targets were daf-16-independent (Figure 3J), consistent with our finding that transgenic expression of SKN-1 S393A increased longevity independently of daf-16 (Figure 5D and 5E). Our data indicate that SKN-1 has important activities that are largely independent of DAF-16, making it of intense interest to dissect the extent to which these two transcription factors cooperate or act independently under normal, stress, and reduced IIS conditions. It is interesting that transgenic overexpression of neither DAF-16 or SKN-1 recapitulates the dramatic increases in longevity seen with reductions in DAF-2 function ((Lin et al., 2001); Figure 5). One possibility is that simultaneous overexpression of both transcription factors is necessary, although modifications of these proteins or other signals associated with decreased IIS might also be required. Our intriguing observation that daf-16 RNAi suppressed lifespan extension by the WT SKN-1B/C::GFP transgene (Figure S13) suggests that DAF-16 may influence processes that regulate SKN-1. This could involve DAF-16 regulating genes that affect SKN-1, but a simpler model is that SKN-1 and DAF-16 compete for binding to negative regulators that include AKT-1, -2 and SGK-1 (Figure 1A). Thus, decreasing DAF-16 levels would increase the ratio of negative regulators available to sequester SKN-1 in the cytoplasm.
Our results add SKN-1 to the small group of C. elegans transcription regulators that extend longevity when expressed transgenically (Figure 5F). SIR-2.1, PHA-4, DAF-16, and SKN-1 or their orthologs have all been shown to influence aspects of metabolism, and thereby presumably may affect the rates of damage that leads to aging (Accili and Arden, 2004; Bishop and Guarente, 2007; Kenyon, 2005; Panowski et al., 2007; van der Horst and Burgering, 2007). Importantly, SKN-1, HSF-1, and each of these other proteins has also been implicated in resistance to free radical or other stresses (Figure 5F; see text), supporting the idea that stress defenses and detoxification are fundamentally important for longevity. A remarkable web of functional interactions among these factors has been uncovered, including our demonstration here that DAF-16 and SKN-1 are regulated in parallel by IIS (Figure 5F). These interactions could be important for coordinating how these proteins respond in different tissues to various scenarios of nutrient availability and stress. As these regulators are also evolutionarily conserved, it seems likely that a deeper understanding of interactions among them will be broadly relevant to elucidating how these proteins influence metabolism, stress resistance, and possibly longevity across species.
Experimental procedures
All supplemental data and experimental procedures, including details of cloning, strains, RNAi, GFP scoring system, and assays for lifespan, movement, and reproduction are available online.
In vitro kinase assay
Kinases were isolated from sonicated worm lysates of N2 Ex[AKT-1::GFP], N2 Ex[AKT-2::GFP] and N2 Ex[AKT-1::GFP] (Hertweck et al., 2004), using a GFP antibody bound to protein A/G agarose beads. These purified kinases were used to phosphorylate 6µg of bacterially expressed GST-SKN-1 fusion proteins in the presence of 6 µCi [γP32]ATP. After phosphorylation, samples were washed in 50 mM Tris/HCl (pH 8.0), 100 mM NaCl, 10% glycerol, and 1% Triton X-100, and analyzed by SDS-PAGE and autoradiography.
RNAi
Feeding RNAi was carried out as previously described, with empty pL4440 as the control ((Kamath et al., 2001); supplementary methods). To avoid maternal lethality in experiments using skn-1 or skn-1a/c RNAi, animals were hatched on HT115 bacteria expressing control RNAi, and were transferred to skn-1 RNAi at the L1 stage or on the first day of adulthood. For lifespan experiments that required feeding of other RNAi, animals were exposed to bacteria expressing the appropriate construct continually starting from hatching.
Lifespan analysis
Prior to experiments, all animals were maintained at the permissive temperature and grown for at least two generations in the presence of food to assure health. Lifespan assays were performed essentially as described (Hsin and Kenyon, 1999)(Supplementary methods). Survival plots, p values (Log-Rank) and proportional hazards were determined using JMP software, version 5.1.
Oxidative stress resistance assay
To assess stress resistance, young adults were transferred to plates that contained 7.5 mM t-butyl hydrogen peroxide (Sigma) or 150 mM paraquat (Sigma) in nematode growth media. These plates had been seeded with OP50 or HT115 expressing the appropriate dsRNAs. Animals were incubated on these plates at 20°C, and periodically scored for survival.
RNA isolation and quantitative PCR
Animals were picked to clean plates to minimize contamination, then total RNA was extracted from approximately 200 animals suspended in 50µl M9. RNA was extracted using Trizol (Sigma) and cDNA was synthesized using the Invitrogen SuperscriptTM Reverse Transcriptase Kit. SYBR Green Real Time PCR was carried out using the ABI 7700 and normalized to act-1. Primer sequences are available on request.
Supplementary Material
Acknowledgements
We thank Siu Sylvia Lee, Gary Ruvkun, and Laura Mitic for generously providing advice and strains, Stacey Robida, Anne Oelmann, Jeeyong Lee and Michael Lucke for invaluable technical support, and members of the Blackwell lab, Javier Apfeld, and Rohit Kulkarni for critically reading this manuscript. Work was supported by funding from the NIH and Iacocca Foundation (TKB), the Fonds der Chemischen Industrie, BMBF NGFN2, Qualitaetsoffensive BW and Deutsche Forschungsgemeinschaft CRC746 (RB), and EC Network of Excellence LifeSpan (MH), as well as an NRSA (RO), an NIH training grant (JB), KRF (2006-353-C00035) and MOST/KOSEF(R112000078010010) (JHA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, Matsumoto K, Blackwell TK. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C. Regulation of lifespan by germ-line stem cells in Caenorhabditis elegans. Science. 2002;295:502–505. doi: 10.1126/science.1065768. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill CM, Tzivion G, Nasrin N, Ogg S, Dore J, Ruvkun G, Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem. 2001;276:13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein BJ, Mahadev K, Wu X. Redox paradox: insulin action is facilitated by insulin-stimulated reactive oxygen species with multiple potential signaling targets. Diabetes. 2005;54:311–321. doi: 10.2337/diabetes.54.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 2001;174:103–113. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol. 2002;37:1371–1378. doi: 10.1016/s0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, Matsumoto K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell A, Ventura N, Kahn N, Johnson TE. Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Radic Biol Med. 2007;43:1560–1566. doi: 10.1016/j.freeradbiomed.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, Walker GA. Stress resistance as a determinate of C. elegans lifespan. Mech Ageing Dev. 2002;123:765–771. doi: 10.1016/s0047-6374(01)00422-5. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- van der Horst A, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


