Abstract
Previous studies on the functional analysis of the human vascular endothelial growth factor (VEGF) promoter using the full-length VEGF promoter reporter revealed that the proximal 36-bp region (−85 to −50 relative to transcription initiation site) is essential for basal or inducible VEGF promoter activity in several human cancer cells. This region consists of a polypurine (guanine) tract that contains four runs of at least three contiguous guanines separated by one or more bases, thus conforming to a general motif capable of forming an intramolecular G-quadruplex. Here, we demonstrated that the G-rich strand in this region is able to form an intramolecular propeller-type parallel-stranded G-quadruplex structure in vitro by using the electrophoretic mobility shift assay (EMSA), dimethyl sulfate (DMS) footprinting technique, DNA polymerase stop assay, CD spectroscopy, and computer-aided molecular modeling. Two well-known G-quadruplex interactive agents, TMPyP4 and Se2SAP, stabilize G-quadruplex structures formed by this sequence in the presence of potassium ion, although Se2SAP is at least 10 fold more effective in binding to the G-quadruplex than TMPyP4. Between these two agents, Se2SAP better suppresses VEGF transcription in different cancer cell lines, including HEC-A1 and MDA-MB-231. Collectively, our results provide evidence that specific G-quadruplex structures can be formed in the VEGF promoter region, and that the transcription of this gene can be controlled by ligand-mediated G-quadruplex stabilization. Our results also provide further support for the idea that G-quadruplex structures may play structural roles in vivo and therefore might provide insight into novel methodologies for rational drug design.
Keywords: G-quadruplex, VEGF, angiogenesis, G-quadruplex-interactive agents, Transcription, Drug design
INTRODUCTION
The formation of new blood vessels, angiogenesis, is essential for tumor growth and metastasis since it provides oxygen and nutrients to proliferating tumor cells, thus favoring tumor progression (1). The switch to an angiogenic phenotype is mediated by a number of key regulators, such as fibroblast growth factors (FGFs), vascular endothelial growth factors (VEGFs), and angiopoietins (2, 3). Among them, VEGF (or VEGF-A) has been considered to be the key mediator of tumor angiogenesis by stimulating the proliferation, migration, survival, and permeability of endothelial cells (4, 5). Recently, a number of agents designed specifically for targeting VEGF and/or its receptors, including antagonistic antibodies, ribozymes, immunotoxins, and synthetic small molecular inhibitors, are being evaluated in various clinical trials in cancer patients (6–15). While a strategy for inhibiting angiogenesis had been proposed more than three decades ago by Folkman (16), basic and translational studies aimed at cultivating a mechanistic understanding of the central role of VEGF in tumor angiogenesis have only recently led to important proof-of-concept clinical trials that convincingly demonstrate the benefits of anti-VEGF therapy in patients with advanced cancer (15).
The molecular basis of VEGF gene expression has been extensively studied by characterizing the cis-acting elements and transcription factors involved in constitutive VEGF expression in human cancer cells (17–21). VEGF expression is known to be induced by a variety of factors, including hypoxia, pH, activated oncogenes, inactivated tumor suppressor genes, and growth factors (reviewed in ref 21). These studies revealed that the proximal 36-bp region (−85 to −50 relative to transcription initiation site) is essential for basal or inducible VEGF promoter activity in several human cancer cells (21). These cis-regulatory elements contain at least three Sp1 binding sites, which consist of guanine/cytosine-rich sequences (Fig. 1A). Although there are binding sites for other factors, such as two Egr-1 elements and one AP-2 element, all Sp1 binding sites were found to be functionally significant in constitutive VEGF promoter activity (22). The existence of polyguanine/polycytosine tracks in the proximal promoters throughout the human genome has been reported in a number of mammalian genes that are mainly growth-related (reviewed in ref 23). These elements in the promoter regions of mammalian genes are known to be very dynamic in their conformation, easily adopting non-B-DNA conformations such as melted DNA, hairpin structures, slipped helices, or others under physiological conditions, sometimes even in the absence of conformational or torsional stress (23–32). In particular, G-rich sequences from a number of TATA-less mammalian genes, including c-MYC, HIF1-α, VEGF, BCL-2, PDGF-A, KRAS, RET and c-KIT have been reported to form parallel or antiparallel G-quadruplex structures consisting of two or more G-tetrads in the presence of monovalent cations such as Na+ and K+ (24–31). The polypurine tract in the VEGF promoter contains five runs of three or more contiguous guanines separated by one or more bases, conforming to a general motif capable of forming an intramolecular G-quadruplex: G4 (C) G3 (CC) G5 (C) G4 (TCCCGGC) G4 (CGG) (21). In our previous study, we demonstrated that the polypurine/polypyrimidine tract of the VEGF promoter region is very dynamic in its structure and can easily adopt a non-B-DNA conformation under physiological conditions (26). In addition, we predicted that the G-rich strand of this tract could form specific G-quadruplex structures in the presence of K+ or G-quadruplex-interactive agents. In this report, we demonstrated that the G-rich strand in the proximal promoter region of the VEGF gene is capable of forming stable intramolecular G-quadruplex structures in vitro, and G-quadruplex interactive agents such as TMPyP4 and Se2SAP further stabilize these G-quadruplex structures.
Figure 1.
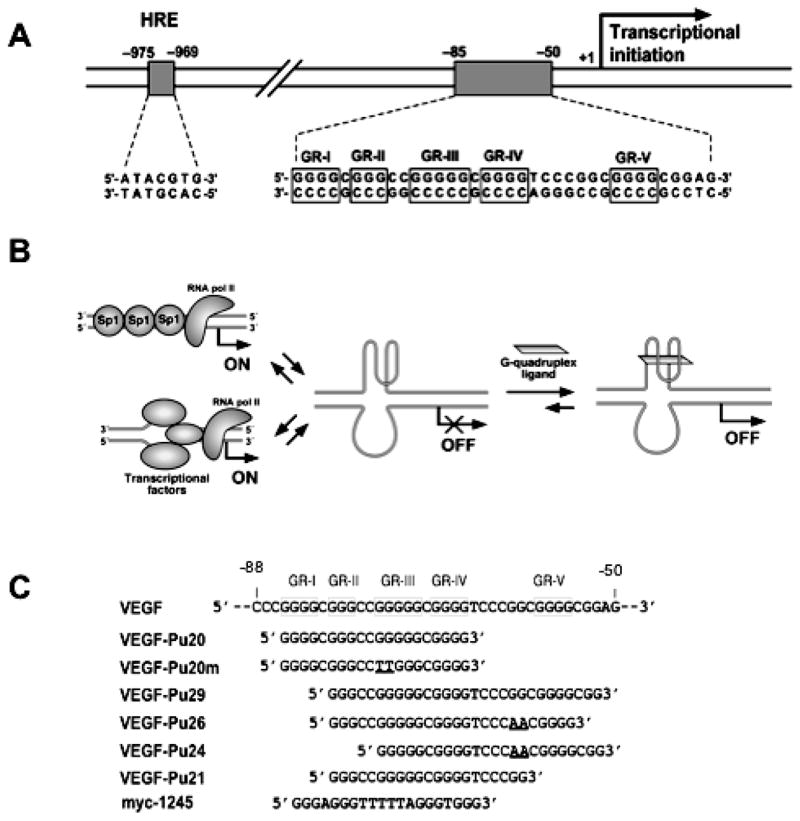
(A) Schematic diagram showing the location of HRE and the binding sites for transcription factor Sp1 in the proximal promoter region of the VEGF gene. Runs of guanines are underlined (enclosed in boxes). (B) Models for the repression of VEGF gene transcription by G-quadruplex-interactive agents. (C) Oligonucleotides used in this study.
Moreover, the suppression of VEGF transcription was observed in various human cancer cells cultured in the presence of Se2SAP. These results provide support for the idea that G-quadruplex structures may play structural roles in vivo and therefore provide insight into novel methodologies for rational drug design by targeting the G-quadruplex structures formed in the VEGF promoter (Fig. 1B).
MATERIALS AND METHODS
Drugs, Enzymes and Oligonucleotide DNA
TMPyP4, TMPyP2, and Se2SAP were synthesized in this laboratory. Drug molecules were dissolved in dimethyl sulfoxide (DMSO). T4 polymerase kinase and Taq DNA polymerase were from Promega. PAGE purified oligonucleotides were obtained from Sigma Genosys.
Preparation and End-Labeling of Oligonucleotides
The 5′-end-labeled single-strand oligonucleotides were obtained by incubating the oligonucleotides with T4 polynucleotide kinase and [γ–32P]ATP for 1 h at 37°C. Labeled oligonucleotides were purified with a Bio-Spin 6 chromatography column (BioRad) after inactivation of the kinase by heating at 95°C for 3 min.
EMSA and dimethyl sulfate (DMS) Footprinting
The 32P-labeled DNA oligomers (100 nM) were denatured by heating at 90°C for 5 min and then cooled slowly to room temperature over 2 to 3 hours in 20 mM Tris-HCl buffer with or without 100 mM KCl. Each annealed DNA was treated with DMS (1%) for 2 min to methylate DNA. Modification reactions were stopped by adding the same volume of stop buffer containing 50% glycerol and 3 μg calf thymus DNA, and the modification products were electrophoresed in 1X TBE buffer on an 16% native polyacryamide gel to separate single stranded DNA and intramolecular G-quadruplexes from other intermolecular quadruplexes by difference in the electrophoretic mobility. Each species of DNA was isolated from the gel, ethanol-precipitated twice, and treated with piperidine (10%). After ethanol precipitation, the cleaved products were resolved on a 16% denaturing polyacrylamide gel.
DNA Polymerase Stop Assay
The template DNAs containing various G-quadruplex forming regions were annealed with primers (P28) labeled with [γ]-32P, and the primer-annealed DNA templates were separated from excess labeled primer or remaining template DNA by electrophoresis through an 8% nondenaturing polyacrylamide gel. The resulting primer-annealed template DNAs were gel-purified and used in a primer extension assay by Taq DNA polymerase as described previously (33).
Circular Dichroism (CD) Spectroscopy
CD spectra were recorded on a Jasco-810 spectropolarimeter (Jasco, Easton, MD) using a quartz cell of 1 mm optical path length and an instrument scanning speed of 100 nm/min, with a response time of 1 s and over a wavelength range of 200–330 or 200–600 nm for the titration experiments with drugs. All DNA samples were dissolved in Tris-HCl buffer (20 mM, pH 7.6) to a strand concentration of 5 μM. Where appropriate, the samples also contained 100 mM KCl. The CD spectra herein are representations of four averaged scans taken at 25°C and are baseline corrected for signal contributions due to the buffer.
Cell culture, drug treatment and RT-PCR analysis of the VEGF mRNA synthesis in human cancer cells
Human breast cancer cell line MDA-MB231, human endometrial cancer cell line HEC-A1, and human kidney cancer cell lines A-498 and 786-0 were obtained from American Type Culture Collection (Rockvile, MD). These cell lines were cultured at 37°C in RPMI 1640 (Cellgro) with 10% fetal bovine serum. For the drug treatment of cell lines, exponentially growing cells were plated at approximately 105 cells /ml in T-75cm2 flasks, and drug molecules were added to the media and incubated for the desired times. After drug treatment, the medium in the culture flask was aspirated, and the remaining monolayer cells were thoroughly washed once with PBS. The washed cells were trypsinized and harvested for a cell count and subsequently, for the preparation of total RNA. Total RNA was extracted from cultured cells using RNAwiz™ (Ambion) according to the manufacturer’s protocol. Extracted RNA was pretreated with DNase I (Promega) and reverse transcription was performed using oligo(dT)18 primer with RETROscript™ (Ambion) to generate single-stranded cDNA. Primer sequences for VEGF gene were designed against a commom region to all isoforms of the VEGF mRNA: the forward primer 5′-TGCATTGGAGCCTTGCCTTG-3′ (nucleotides 1054–1073 from NM 003376.4) and the reverse primer 5′-CGGCTCACCGCCTCGGCTTG-3′ (nucleotides 1664–1683 from NM 003376.4). In parallel, the amplification of beta-actin cDNA was performed as an internal standard according to the manufacturer’s protocol (Ambion). All of the reactions involved initial denaturation at 95°C for 3 min followed by 40 cycles for VEGF or 30 cycles for beta-actin at 94°C for 30s, 55°C for 30s and 72°C for 40s on GeneAmp PCR system 9600 (Perkin Elmer).
RESULTS
Formation of intramolecular G-quadruplex structures by the G-rich strand of the VEGF proximal promoter region
The G-rich strand (−85 to −50) of the VEGF promoter is characterized by the presence of five runs of at least three adjacent guanines, which we designated GR-I through GR-V (Fig. 1A). To determine which guanine repeats were required for folding into intramolecular G-quadruplex structures, we prepared a series of DNA oligonucleotides spanning various portions of the G-rich sequence (Fig. 1C). Each oligonucleotide (100 nM) was 5′ end-labeled with 32P, denatured by heating at 90°C for 5 min, slow cooled to room temperature with or without 100 mM KCl, and then treated with DMS (0.5%) for 2 min to methylate the N-7 position of the guanine base within given oligonucleotides. It is well known that the guanine bases involved in the formation of G-quadruplex structures are inaccessible to methylation, since the N7 position of those guanines are involved in Hoogsteen bonding to form the G-tetrad (24). The methylated oligos were then subjected to a native polyacryamide gel electrophoresis (PAGE) to separate intramolecular G-quadruplexes from other intermolecular quadruplexes by differences in electrophoretic mobility. As shown in Figure 2A, a native gel electrophoresis of the 5′-end-labeled VEGF-Pu20 incubated in the presence of 100 mM KCl resulted in the formation of one major band (band “5” in Fig. 2A), while two major bands were formed in the absence of KCl (bands “3” and “4” Fig. 2A). Each DNA was excised from the gel and treated with piperidine, and the cleavage products were resolved on a sequencing gel. The pattern of N7 guanine methylation produced by the band “5” was consistent with the intramolecular quadruplex, containing three stacked G-tetrads (lane 5 in Fig. 2A). However, DMS cleavage pattern of the band “3” corresponds to that of the single-stranded form (lane 3 in Fig. 2A), while the band “4” (lane 4 in Fig. 2A), is believed to be an intermolecular G-quadruplex due to its much slower electrophoretic mobility compared with the intramolecular form of G-quadruplex structure (band “5”). Initially, the formation of intermolecular G-quadruplex structures under salt-deficient conditions in VEGF-Pu20 appears to be intriguing. However, we speculate that intermolecular G-quadruplex formation in VEGF-Pu20 under salt-deficient conditions during gel electrophoresis could still be facilitated by small amounts of sodium ion (2–3 mM) present in TBE buffer. DMS cleavage of the band “5” showed that three stacked G-tetrads formed by four G-stretches were protected from the methylation by DMS except G14, while guanine residues within the central loop (in sequence to right of lane 5 in Fig. 2A) showed hypersensitivity to DMS. To probe the effect of mutation of G11 and G12 which show hypersensitivity to DMS a mutant oligomer (VEGF-Pu20m in Fig. 2B) was investigated. The DMS cleavage at G14 residue of VEGF-Pu20 disappeared when G10 and G11 within the central loop were replaced with thymine residue in VEGF-Pu20m (lane 3 in Fig. 2B), indicating a possible interaction of this guanine residue with bases within the central 4 base loop. Overall, the results from this study suggest the strong tendency of VEGF-Pu20 to form an intramolecular G-quadruplex structure in the presence of KCl, while forming either single stranded structures or intermolecular G-quadruplex structures in the absence of KCl. The results of DMS footprinting for VEGF-Pu20 and VEGF-Pu20m in the presence 100 mM KCl are summarized in Figure 2C.
Figure 2.
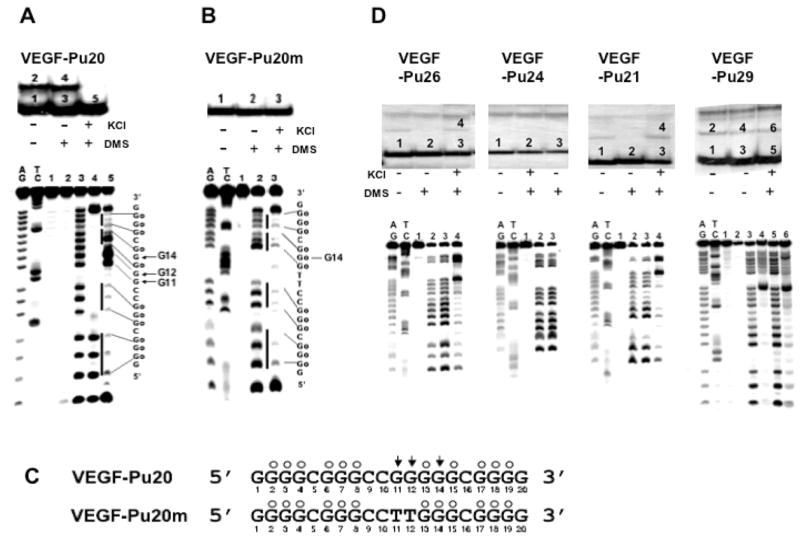
Determination of the intramolecular G-quadruplex structures formed by the G-rich sequence of the VEGF promoter region in the presence of 100 mM KCl. (A) Top, EMSA of VEGF-Pu20 pre-incubated under the conditions specified in the figure using 16% native polyacrylamide gel. Bottom, DMS footprinting of each band (1–5) from EMSA. AG and TC are the sequencing lanes, and lanes 1–5 correspond to the bands from EMSA. (B) Top, EMSA of VEGF-Pu20m pre-incubated under the conditions specified in the figure using 16% native polyacrylamide gel. Bottom, DMS footprinting of each band (1–3) from EMSA. AG and TC are the sequencing lanes, and lanes 1–3 correspond to the bands from EMSA. The vertical bars to the left of lane 5 (Fig. 2A) and lane 3 (Fig. 2B) correspond to DMS protected guanine repeats, and the sequence to the right shows the protected guanines (Go). (C) Summary of DMS footprinting of VEGF-Pu20 and VEGF-Pu20m in the presence of 100 mM KCl. The protected guanines from DMS are indicated by open circles, and guanine residues hypermethylated by DMS are indicated by arrowheads. (D) Determination of the structures of G-quadruplexes formed from VEGF-Pu29, Pu26, Pu24, and Pu21 in the presence of 100 mM KCl. Top, EMSA of VEGF-Pu26, Pu24, Pu21, and Pu29 (left to right) pre-incubated under the conditions specified in the figure using 16% native polyacrylamide gel. Bottom, DMS footprinting of each band from EMSA. AG and TC are the sequencing lanes, and each lane corresponds to the bands from corresponding EMSA.
In contrast with VEGF-Pu20, the major band of all other oligonucleotides (VEGF-Pu29, Pu26, and Pu21) incubated in the presence of 100 mM KCl failed to show the N7 guanine protection from methylation, indicating that these oligonucleotides do not form intramolecular G-quadruplexes (see lane 5 for VEGF-Pu29; lane 3 for VEGF-Pu26, VEGF-Pu24, and VEGF-Pu21 in Fig. 2D). Only the minor band with lower electrophoretic mobility (bands “4” or “6” in Fig. 2D) representing intermolecular G-quadruplexes exhibited the protection of guanine residues from DMS (see lanes 4 and 6 for VEGF-Pu29; lane 4 for VEGF-Pu26 and VEGF-Pu21 in Fig. 2D), suggesting these structures form only intermolecular G-quadruplexes. Collectively, this result indicates that the intramolecular G-quadruplex formed within the G-rich sequence of the VEGF promoter requires four G blocks (GR-I to GR-IV), consisting of twelve guanines, while other G blocks such as GR-V may be required for the formation of intermolecular G-quadruplex, which is less likely to be biologically relevant.
Sequence VEGF-Pu20 forms an intramolecular propeller-type parallel-stranded G-quadruplex
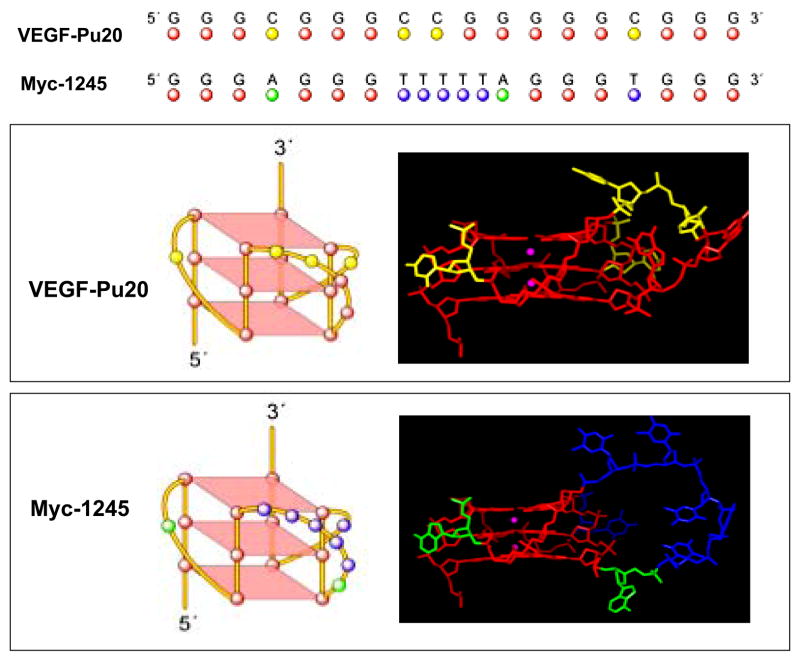
Like the G-quadruplex formed from four consecutive human telomeric DNA repeats (34), the sequence myc-1245, a mutant type of the Pu27-mer from the NHE III1 of the c-MYC (see Fig. 3A), was shown by NMR to form a stable intramolecular propeller-type parallel-stranded G-quadruplex in K+-containing solution (35). To compare the structure of VEGF-Pu20 with that of myc-1245, we first determined the number of stacked G-tetrads and the guanines involved in the G-quadruplex structure formed by myc-1245 by utilizing EMSA and DMS footprinting method. As shown in Figure 3B and C, the DMS footprinting experiment revealed that this structure involves a core of three stacked G-tetrads formed by four G-stretches and three loops bridging three G-tetrad layers, conforming to the folding pattern determined by the NMR study (35). The central loop contains six residues, while the two other loops contain only one residue. Next, the CD spectra of the VEGF-Pu20 and the myc-1245 were compared to determine the direction of the backbone (e.g., antiparallel or parallel orientation) in the G-quadruplex structure formed by the VEGF-Pu20, since CD spectroscopy has been widely used to infer the presence of G-quadruplexes and is particularly useful for differentiating parallel and antiparallel G-quadruplexes (36–37). All guanosines in the structure of a parallel quadruplex have an anti-conformation of the glycosyl bonds, resulting in a positive band at ~265 nm and a small negative band at ~245 nm in this CD spectrum (36–37). In contrast, the guanosines in an antiparallel quadruplex have alternating syn- and anti- glycosyl conformations along each strand, exhibiting a positive band at ~295 nm that is associated with an antiparallel strand arrangement. As shown Figure 3D, the CD spectra of both sequences showed a positive band at ~265 nm and a small negative band at ~245 nm in the CD spectrum in K+-containing solution, which is consistent with a parallel structure (36–37).
Figure 3.
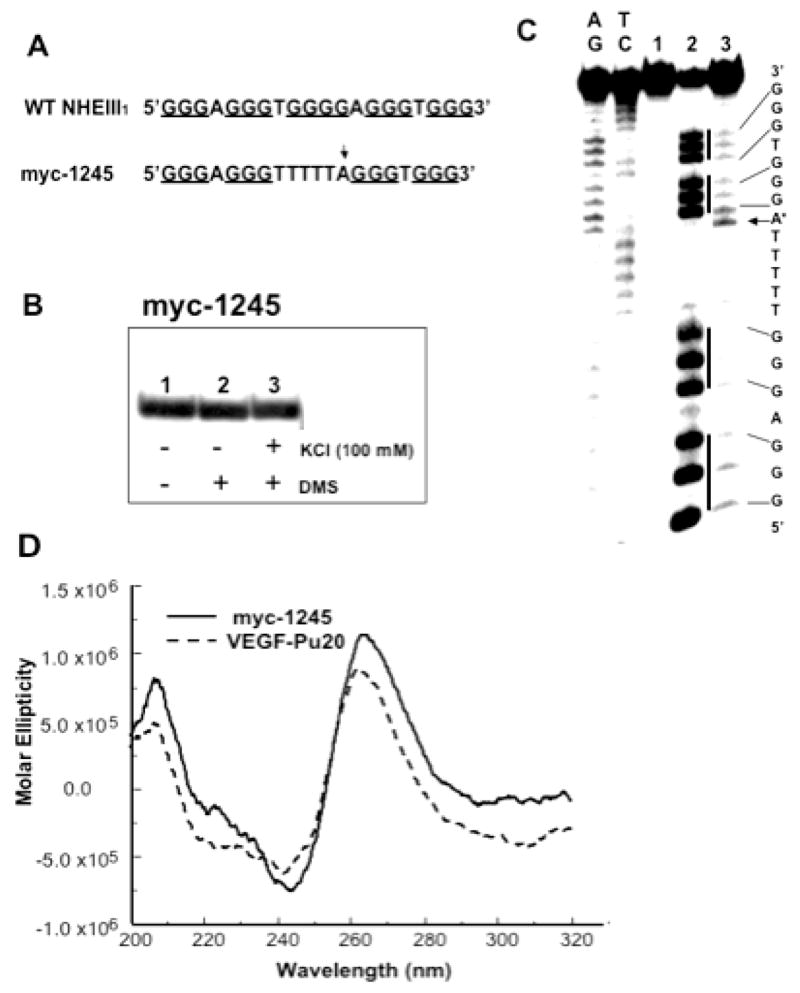
EMSA, DMS footprinting and CD spectrum of the sequence myc-1245. (A) Sequence of the wild type (WT) and mutant (myc-1245) NHEIII1 in the C-MYC promoter. (B) EMSA of the sequence myc-1245 pre-incubated under the conditions specified in the figure using a 16% native polyacrylamide gel. (C) DMS footprinting of each band from EMSA. (D) CD spectra of the myc-1245 (solid line) in comparison with that of VEGF-Pu20 (dashed line). The CD data were obtained with a 5 mM strand concentration in the presence of 100 mM KCl at 25°C.
Models for G-Quadruplex Structures Formed in the Guanine-Rich Strands of the VEGF Promoter
Based on information obtained from our study utilizing biochemical and biophysical methods, including EMSA, DMS footprinting, and CD spectroscopy, we have determined the number of stacked G-tetrads, the guanines involved in the G-quadruplex, and the probable direction of the backbone (e.g., antiparallel or parallel orientation) in the major G-quadruplex structures formed by the G-rich sequence in the promoter region of the VEGF gene. In the sequence myc-1245 examined by the Patel group (35), there are two one-base loops analogous to those found in the VEGF sequences flanking a six-thymine base loop analogous to the 4- base loops in the VEGF (Fig. 4). Therefore, the principal differences between VEGF and the Patel sequence is the size (4 versus 6) and identity of the bases (purines versus pyrimidines) in the central loop. This analogous sequence provides some insight into the probable folding pattern of the VEGF G-quadruplex structures. This Patel structure involves a core of three stacked G-tetrads formed by four parallel G-stretches with all anti conformations in the nucleosides and three double-chain-reversal loops bridging three G-tetrad layers (Fig. 4). Although VEGF-Pu20 and the Patel sequence contain two different sets of G-stretches, their overall folding topologies look strikingly similar, with a parallel-stranded G-tetrad core and three double-chain-reversal loops, two of which are one-residue loops. These G-quadruplexes were found to be stable during molecular minimization and dynamics calculations (Figure 4). All of the nucleotides of the tetrads exist in an anti conformation. Single base loops connecting parallel guanine strands, as well as all guanine bases of the strands connected by single base loops in the tetrad arrangement, were found to be stable. The main difference between the three structures lies in the size and sequence of the central loop, which is a four- and six-residue loop in VEGF-Pu20 and myc1245, respectively (see Figure 4).
Figure 4.
Molecular modeling structures for G-quadruplexes formed by VEGF-Pu20 in comparison to the known folding pattern of myc-1245. Top, alignment of both sequences. Middle, schematic illustration (left panel) and molecular modeling structure (right panel) of the VEGF parallel quadruplex. Bottom, schematic illustration (left panel) and molecular modeling structure (right panel) of the myc-1245 parallel quadruplex. The structure of the human c-myc parallel quadruplex was used as a starting structure (35). Necessary replacements and deletions were done. Loop geometries were obtained as in the case of the human telomeric DNA parallel quadruplex (PDB code 1KF1) (34). Modeling was performed using the Biopolymer module with Insight II modeling software and charges, and potential types were assigned using Amber forcefield within Insight II. Structures were minimized using 2500 steps of Discover 3 minimization as described previously (42).
G-quadruplex interactive agents stabilize an intramolecular G-quadruplex structure formed by the G-rich strand of the VEGF proximal promoter regions
In order to test whether G-quadruplex interactive agents stabilize an intramolecular G-quadruplex structure formed by the G-rich strand of the VEGF proximal promoter region, a DNA polymerase stop assay was utilized with a DNA template (MT-1 in Fig. 5A) containing the VEGF-Pu20 {d(G4CG3C2G5CG4)} annealed with 32P labeled primers. Figure 5B shows a potassium dependent pause of DNA polymerase extension through this DNA template at 37°C. In the absence of KCl, the DNA polymerase bypasses the G-repeat region; however, in the presence of KCl, pausing is observed immediately 3′ to the fourth guanine run (GR-IV), presumably by stabilizing the intramolecular G-quadruplex structure, confirming that this region is indeed capable of forming intramolecular G-quadruplex structures. We next examined two well-known G-quadruplex interactive agents (TMPyP4, and Se2SAP) for their ability to interact with the G-quadruplexes using the polymerase stop assay (Fig. 5C). TMPyP4 is a cationic porphyrin that has been shown to interact specifically with G-quadruplexes; Se2SAP is a core-modified expanded porphyrine analogs with significantly reduced photoreactivity compared with to TMPyP4 and TMPyP2. These G-quadruplex interactive agents used in this experiment have been described in our previous studies (30, 33, 34, 37–39). These agents are capable of interacting with the G-quadruplex in the VEGF promoter as shown by a concentration dependent increase in the amount of arrest compared to the amount of arrest seen with potassium alone, suggesting that these agents stabilize the G-quadruplex formed by this sequence (Fig. 5D). Interestingly, Se2SAP also turned out be at least 10-fold more effective in binding to the VEGF G-quadruplex than TMPyP4. In contrast, TMPyP2 (structure shown in Fig. 5C), a structural isomer of TMPyP4 that is known to have low affinity for G-quadruplexes due to its inability to enter or stack into G-tetrad structures (40), had little effect on the formation of the arrest products (see Fig. 5D).
Figure 5.
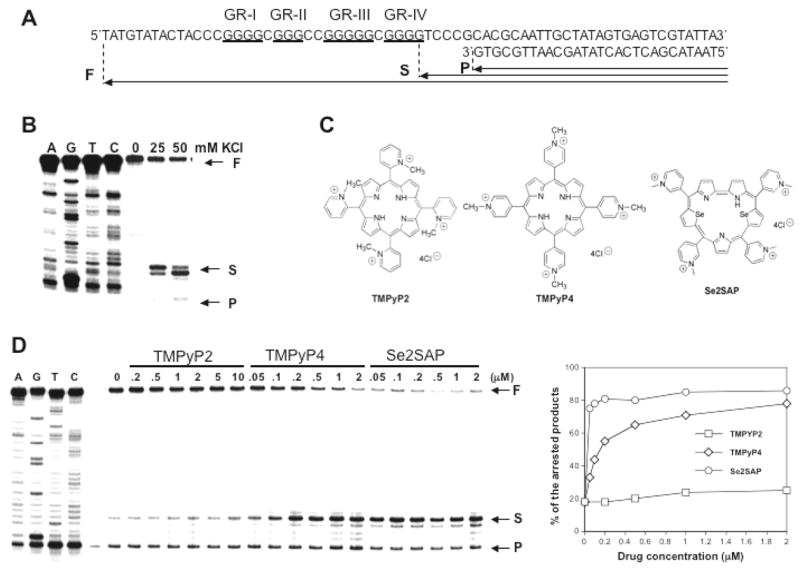
Taq DNA polymerase stop assay showing the stabilization of G-quadruplex structures by K+ and TMPyP2, TMPyP4, and Se2SAP. (A) Sequence of the single-stranded DNA template annealed with primer used in DNA polymerase stop assay. (B) KCl-dependent pausing of DNA polymerase DNA synthesis at the 3′ side of the fourth guanine run in the DNA template containing the VEGF-Pu20 sequence. (C) Structures of TMPyP2, TMPyP4, and Se2SAP. (D) Stabilization of G-quadruplex structure formed by the VEGF-Pu20 with the addition of increasing concentrations of TMPyP2, TMPyP4, and Se2SAP (left panel). Arrows indicate the positions of the full-length product (“F”) of DNA synthesis, the G-quadruplex pause sites (“S”), and the free primer (“P”). Lanes A, G, T, and C represent dideoxysequencing reactions with the same template as a size marker for the precise arrest sites. The percentage of the major arrest products of each sample to the total product was plotted against drug concentrations (right panel)
G-quadruplex-interactive agent Se2SAP causes a decrease in the expression of VEGF in human tumor cells
Since we demonstrated that the G-rich strand of Sp1 binding sites within the VEGF promoter region is capable of forming intramolecular G-quadruplex structures in vitro and that G-quadruplex interactive agents further stabilize these G-quadruplex structures, we next determined whether the expression of the VEGF gene is affected by G-quadruplex interactive agents in various human cancer cell lines, including kidney (A498 and 786-0), endometrial (HEC-A1), and breast (MDA-MB231) cancers. As shown in Fig. 6A-B, Se2SAP decreased VEGF gene expression by 50 and 80% in MDA-MB231 and HEC-A1 cells, respectively, at 48 h post-treatment. This compound also decreased VEGF gene expression in the two kidney cell lines A498 and 786-O by 50–60% within 24h after treatment (see Fig. 6C-D). In contrast, TMPyP2 has little effect on the expression of the VEGF gene at the range of concentrations which resulted in similar growth inhibition of these cell lines by TMPyP4 and Se2SAP (data not shown), suggesting that the transcriptional repression of VEGF gene by G-quadruplex interactive agents is not simply a result of decreased proliferation of HEC-A1 cells. TMPyP4, which is less effective in binding to the VEGF G-quadruplex than Se2SAP, only has a marginal effect on the inhibition of VEGF expression in HEC-A1 cell lines, although it is capable of interacting with VEGF intramolecular G-quadruplex structures. Therefore, a substantial difference between different G-quadruplex interactive agents in binding affinity to G-quadruplexes could be attributed to their varying degree of potency in downregulating VEGF expression.
Figure 6.
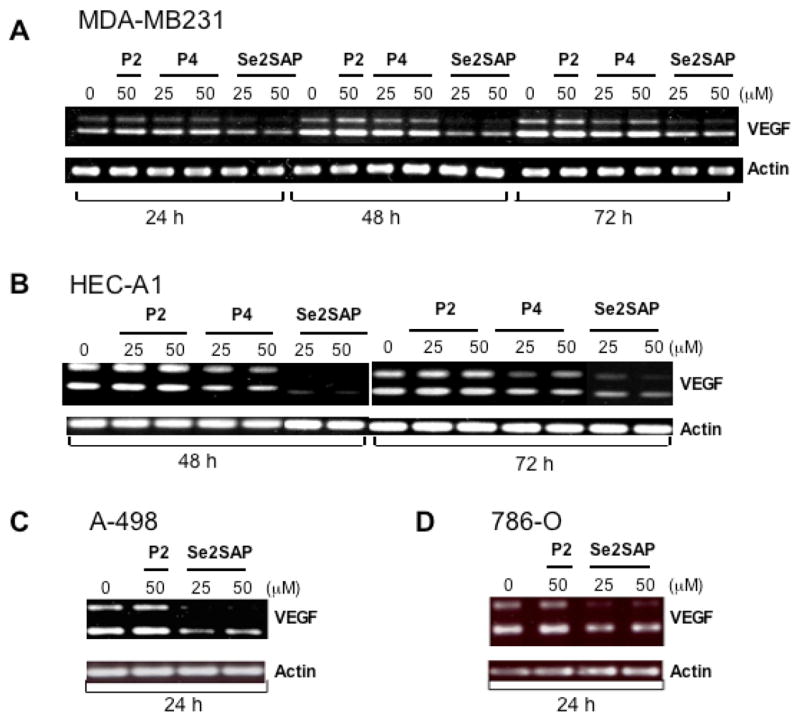
Effect of G-quadruplex interactive agents on VEGF mRNA synthesis in various human cancer cells, including human breast cancer cell line MDA-MB231 (A), human endometrial cancer cell line HEC-A1 (B), and human renal cancer cell lines A498 and 786-O (C, left to right). Human cancer cells were treated with TMPyP2, TMPyP4, and Se2SAP at various concentrations for indicated times, and VEGF mRNA and beta-actin mRNA were measured by RT-PCR. Less than 30% reduction in the number of cells occurred over 3 days after treatment with 50 μM TMPyP4, TMPyP2 and Se2SAP (data not shown). Two products were obtained after RT-PCR, which correspond to c-DNAs of VEGF121, and VEGF165.
DISCUSSION
In an expanding tumor mass, VEGF expression is physiologically induced in many types of tumor by hypoxia and hypoglycemia, particularly in regions surrounding necrosis (4, 5). In addition, tumor cells may produce VEGF in the absence of physiological stimuli due to the loss of tumor suppressor genes, such as VHL and p53, as the result of the activation of certain growth factors signaling cascades, such as EGFR, and the stimulation of the tumor cells by cytokines, growth factors, gonadotropins, and several other extracellular molecules from normal or malignant cells, which act in an endocrine, paracrine, or autocrine fashion. (17–26). Thus, VEGF and its receptors are regarded as potential targets for therapeutic intervention (6–8).
In this study, we explored a new potential strategy to inhibit the production of VEGF in tumor cells by targeting the G-quadruplex structure formed by the G-rich sequences in the promoter region of the VEGF gene. Direct evidence for the existence of G-quadruplexes in vivo is beginning to emerge, and the ability of some of the G-rich sequences to form very stable G-quadruplex structures in vitro suggests that G-quadruplex DNA may play an important role in several biological events (29–34). For instance, a recent study provided compelling evidence that a specific G-quadruplex structure formed in the c-Myc promoter functions as a transcriptional repressor element, establishing the principle that c-Myc transcription can be controlled by ligand-mediated G-quadruplex stabilization (34). This principle has now been independently confirmed by a second group using structurally distinct molecules (41). More recently, the folding pattern (35) and solution structure (42) of the parallel G-quadruplex in the c-Myc promoter resulting from the four 3′ runs of guanine in the NHEIII1 has been determined. Significantly, this G-quadruplex has a melting point of more than 85°C and is a mixture of four loop isomers.
The promoter element encompassing the three Sp1 binding sites contains five runs of three or more contiguous guanines (Fig. 1A) and has a potential to form two alternative intramolecular G-quadruplex structures. Of these alternative G-quadruplex structures, that involving the four 5′ runs of guanine appears to be most stable. In the absence of the terminal 5′ four guanines, neither of the G-quadruplex structures is able exist in a stable form. Within the four 5′ runs of guanine the second has only three guanines limiting the G-quadruplex to three tetrads. DMS footprinting confirms the formation of a G-quadruplex containing three tetrads, although there is one anomaly in the second run of three guanines we shall discuss later. The result from CD spectroscopy suggests that the G-quadruplex is a parallel G-quadruplex and two single nucleotides (C) which separate contiguously DMS-protected runs of three guanines suggests two double chain reversal loops similar to those found in the C-MYC G4 structures. In this mutant sequence derived from the NHEIII1 in the c-MYC promoter, the central run of guanines is substituted in the four thymines (Figure 3A). However, in the case of VEGF, the third loop contains four nucleotides (CCGG) in which the two guanines show hypersensitivity to DMS. Overall, the DMS (Fig. 2A) and CD (Fig. 3D) characteristics of this sequence closely align to those associated with the folding pattern of a mutant C-MYC G-quadruplex sequence (myc-1245) in which there are 3 double chain reversal loops containing 1, 6 and 1 nucleotides in each loop. In comparison the VEGF core sequence has 1, 4, and 1 nucleotide size loops. An unexplained enigma of the DMS cleavage pattern is the cleavage of a guanine in the central tetrad that disappears upon substitution of the two guanines in the four base loop with thymine bases (compare Figure 2A and B). This is suggestive of a central modified tetrad structure of as yet undetermined structure, which could be determined by NMR in a separate study.
The second important goal of this study was to provide proof of the principle that G-quadruplex interactive agents could be potentially used as a new class of anti-angiogenic agents by inhibiting the transcriptional activity of the VEGF gene. Therefore, we utilized the well-characterized G-quadruplex-interactive agents TMPyP4 and Se2SAP in our initial studies, since previous studies clearly demonstrated that these compounds showed varying degrees of selectivity for G-quadruplexes over duplex or other forms of secondary structures (30, 33, 34, 37–39). As described in the results, compared to TMPyP4, the expanded porphyrin Se2SAP was proven highly effective in binding to the VEGF G-quadruplex. Significantly, Se2SAP was also very effective in inhibiting VEGF transcription in various human cancer cell lines, including HEC-A1, MDA-MB231, A-498 and 786-O, although some variation in the degree of the inhibition of the VEGF gene expression with Se2SAP was observed between the cell lines. In order to avoid general cytotoxic effects, TMPyP2 was dosed at the same range of concentrations that resulted in similar growth inhibition of these cell lines by TMPyP4 and Se2SAP. At these concentrations, TMPyP2 has little effect on the expression of on VEGF transcription. The spatial distribution of expressed VEGF isoforms is known to vary both within and between tumor types, although VEGF121 and VEGF165 appear to predominate in human cancer (43). We observed that the expression of VEGF121 was the most predominant in all cell lines, while the expression of VEGF165 varied between cell lines (low in MDA-MB231, A-498, and 786-O, higher in HEC-A1 cell line). Since Se2SAP suppresses the expression of both VEGF mRNA isoforms in these cell lines, our data suggests that G-quadruplex interactive agents might have little influence on the splicing process of VEGF mRNA isoforms. While it is attractive to suggest a mechanism in which the G-quadruplex interactive compounds such as Se2SAP downregulate VEGF by direct interaction with the G-quadruplex in the promoter region, thereby inhibiting transcriptional activation, we cannot at this time rule out alternative mechanisms.
In summary, in order to develop a new potential strategy to inhibit the production of VEGF in tumor cells, we characterized specific G-quadruplex structures formed in the VEGF promoter region and determined whether the transcription of this gene can be controlled with G-quadruplex-interactive agents targeting the G-quadruplex structure formed by the G-rich sequences in the promoter region of the VEGF gene. We demonstrated that the G-rich strands in this region are able to form intramolecular G-quadruplex structures in vitro, which are further stabilized by G-quadruplex interactive agents. These agents are able to suppress VEGF transcription in human tumor cells, raising the possibility that a new antiangiogenic therapy can be evolved by targeting specific G-quadruplex structures formed in the promoter region of the VEGF gene and inhibiting the transcription of this gene with G-quadruplex-interactive agents. Thus, the identification of novel anti-angiogenic compounds based on G-quadruplex-interactive agents could have important applications in the treatment of human cancers. Furthermore, the combination of certain G-quadruplex-interactive agents with a number of recently developed agents for targeting VEGF and/or its receptors (6–15) with different mechanisms of action is likely to produce an enhanced antiangiogenic effect in human cancer.
Acknowledgments
This research was supported by the National Institutes of Health (CA109069 and CA94166). We are grateful to Dr. Sridevi Bashyam for synthesizing Se2SAP for this study and to David Bishop for preparing, proofreading, and editing the final version of the manuscript and figures.
Grant support: National Cancer Institute/NIH grants CA109069 and CA94166.
Abbreviations
- CD
Circular dichroism
- DMS
Dimethyl sulfate
- EMSA
Electrophoretic mobility shift assay
- HRE
Hypoxia responsive element
- NHEIII1
Nuclease hypersensitive element III1
- PAGE
Polyacryamide gel electrophoresis
- VEGF
Vascular endothelial growth factor
References
- 1.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–8. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan DC, Bicknell R. New molecular pathways in angiogenesis. Br J Cancer. 2003;89:228–31. doi: 10.1038/sj.bjc.6601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giordano FJ, Johnson RS. Angiogenesis: the role of the microenvironment in flipping the switch. Curr Opin Genet Dev. 2001;11:35–40. doi: 10.1016/s0959-437x(00)00153-2. [DOI] [PubMed] [Google Scholar]
- 4.Goodsell DS. The molecular perspective: VEGF and angiogenesis. Stem Cells. 2003;21:118–9. doi: 10.1634/stemcells.21-1-118. [DOI] [PubMed] [Google Scholar]
- 5.Jain RK. Tumor angiogenesis and accessibility: role of vascular endothelial growth factor. Semin Oncol. 2002;29:3–9. doi: 10.1053/sonc.2002.37265. [DOI] [PubMed] [Google Scholar]
- 6.Sato Y. Molecular diagnosis of tumor angiogenesis and anti-angiogenic cancer therapy. Int J Clin Oncol. 2003;8:200–6. doi: 10.1007/s10147-003-0342-8. [DOI] [PubMed] [Google Scholar]
- 7.Martiny-Baron G, Marme D. VEGF-mediated tumour angiogenesis: a new target for cancer therapy. Curr Opin Biotechnol. 1995;6:675–80. doi: 10.1016/0958-1669(95)80111-1. [DOI] [PubMed] [Google Scholar]
- 8.Bikfalvi A, Bicknell R. Recent advances in angiogenesis, anti-angiogenesis and vascular targeting. Trends Pharmacol Sci. 2002;23:576–82. doi: 10.1016/s0165-6147(02)02109-0. [DOI] [PubMed] [Google Scholar]
- 9.Sepp-Lorenzino L, Thomas KA. Antiangiogenic agents targeting vascular endothelial growth factor and its receptors in clinical development. Expert Opin Investig Drugs. 2002;11:1447–65. doi: 10.1517/13543784.11.10.1447. [DOI] [PubMed] [Google Scholar]
- 10.Niederman TM, Ghogawala Z, Carter BS, et al. Antitumor activity of cytotoxic T lymphocytes engineered to target vascular endothelial growth factor receptors. Proc Natl Acad Sci USA. 2002;99:7009–14. doi: 10.1073/pnas.092562399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin N, Chen W, Blazar BR, et al. Gene therapy of murine solid tumors with T cells transduced with a retroviral vascular endothelial growth factor--immunotoxin target gene. Hum Gene Ther. 2002;13:497–508. doi: 10.1089/10430340252809793. [DOI] [PubMed] [Google Scholar]
- 12.Stefanik DF, Fellows WK, Rizkalla LR, et al. Monoclonal antibodies to vascular endothelial growth factor (VEGF) and the VEGF receptor, FLT-1, inhibit the growth of C6 glioma in a mouse xenograft. J Neurooncol. 2001;55:91–100. doi: 10.1023/a:1013329832067. [DOI] [PubMed] [Google Scholar]
- 13.Hess C, Vuong V, Hegyi I, et al. Effect of VEGF receptor inhibitor PTK787/ZK222584 [correction of ZK222548] combined with ionizing radiation on endothelial cells and tumour growth. Br J Cancer. 2001;85:2010–16. doi: 10.1054/bjoc.2001.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holash J, Davis S, Papadopoulos N, et al. VEGF-Trap: a VEGF blocker with potent antitumor effects. Proc Natl Acad Sci USA. 2002;99:11393–8. doi: 10.1073/pnas.172398299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JC, Haworth L, Sherry RM, et al. A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N Engl J Med. 2003;349:427–34. doi: 10.1056/NEJMoa021491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–86. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 17.Schafer G, Cramer T, Suske G, et al. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J Biol Chem. 2003;278:8190–8. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- 18.Maeno T, Tanaka T, Sando Y, et al. Stimulation of vascular endothelial growth factor gene transcription by all trans retinoic acid through Sp1 and Sp3 sites in human bronchioloalveolar carcinoma cells. Am J Respir Cell Mol Biol. 2002;26:246–53. doi: 10.1165/ajrcmb.26.2.4509. [DOI] [PubMed] [Google Scholar]
- 19.Pal S, Datta K, Mukhopadhyay D. Central role of p53 on regulation of vascular permeability factor/vascular endothelial growth factor (VPF/VEGF) expression in mammary carcinoma. Cancer Res. 2001;61:6952–57. [PubMed] [Google Scholar]
- 20.Pal S, Datta K, Khosravi-Far R, et al. Role of protein kinase Czeta in Ras-mediated transcriptional activation of vascular permeability factor/vascular endothelial growth factor expression. J Biol Chem. 2001;276:2395–403. doi: 10.1074/jbc.M007818200. [DOI] [PubMed] [Google Scholar]
- 21.Loureiro RM, D’Amore PA. Transcriptional regulation of vascular endothelial growth factor in cancer. Cytokine Growth Factor Rev. 2005;16:77–89. doi: 10.1016/j.cytogfr.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Finkenzeller G, Sparacio A, Technau A. Sp1 recognition sites in the proximal promoter of the human vascular endothelial growth factor gene are essential for platelet-derived growth factor-induced gene expression. Oncogene. 1997;15:669–76. doi: 10.1038/sj.onc.1201219. [DOI] [PubMed] [Google Scholar]
- 23.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–13. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddiqui-Jain A, Grand CL, Bearss DJ, et al. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99:11593–8. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Armond R, Wood S, Sun D, et al. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44:16341–50. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 26.Sun D, Guo K, Rusche JJ, et al. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–80. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J Am Chem Soc. 2006;128:5404–15. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–49. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirude PS, Okumus B, Ying L, et al. Single-molecule conformational analysis of G-quadruplex formation in the promoter DNA duplex of the proto-oncogene c-kit. J Am Chem Soc. 2007;129:7484–5. doi: 10.1021/ja070497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quin Y, Rezler EM, Gokhale V, et al. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkm538. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo K, Pourpak A, Beetz-Rogers K, et al. Formation of pseudo-symmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J Am Chem Soc. 2007;129:10220–8. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai J, Chen D, Jones RA, et al. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–44. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han H, Hurley LH, Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27:537–42. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–80. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 35.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004;126:8710–16. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray DM, Gray CW, Mou TC, et al. CD of single-stranded, double-stranded, and G-quartet nucleic acids in complexes with a single-stranded DNA-binding protein. Enantiomer. 2002;7:49–58. doi: 10.1080/10242430212192. [DOI] [PubMed] [Google Scholar]
- 37.Seenisamy J, Rezler EM, Powell TJ, et al. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J Am Chem Soc. 2004;126:8702–9. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 38.Seenisamy J, Bashyam S, Gokhale V, et al. Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J Am Chem Soc. 2005;127:2944–59. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 39.Kim MY, Vankayalapati H, Shin-Ya K, et al. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. J Am Chem Soc. 2002;124:2098–9. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 40.Han H, Langley DR, Rangan A, et al. Selective interactions of cationic porphyrins with G-quadruplex structures. J Am Chem Soc. 2001;123:8902–13. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 41.Ou TM, Lu YJ, Zhang C, et al. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J Med Chem. 2007;50:1465–74. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 42.Ambrus A, Chen D, Dai J, et al. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–58. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 43.Yu JL, Rak JW, Klement G, Kerbel RS. Vascular Endothelial Growth Factor Isoform Expression as a Determinant of Blood Vessel Patterning in Human Melanoma Xenografts. Cancer Res. 2002;62:1838–46. [PubMed] [Google Scholar]