Abstract
In the yeast Saccharomyces cerevisiae, the CKI1-encoded choline kinase catalyzes the committed step in the synthesis of phosphatidylcholine via the CDP-choline branch of the Kennedy pathway. Analysis of a PCKI1-lacZ reporter gene revealed that CKI1 expression was regulated by intracellular levels of the essential mineral zinc. Zinc depletion resulted in a concentration-dependent induction of CKI1 expression. This regulation was mediated by the zinc-sensing and zinc-inducible transcriptional activator Zap1p. A purified Zap1p probe interacted with two putative UASZRE sequences (ZRE1 and ZRE2) in the CKI1 promoter. Mutations of ZRE1 and ZRE2 to a nonconsensus UASZRE attenuated the induction of CKI1 expression in response to zinc depletion. A UASINO element in the CKI1 promoter was responsible for stimulating CKI1 expression, but this element was not involved with the regulation by zinc depletion. The induction of CKI1 expression in zinc-depleted cells translated into increased choline kinase activity in vitro and in vivo, and an increase in phosphatidylcholine synthesis via the Kennedy pathway.
Recent studies have uncovered a novel relationship between the control of zinc homeostasis and the regulation of membrane phospholipid synthesis in the yeast Saccharomyces cerevisiae (1). Zinc is an essential mineral for the growth and metabolism of S. cerevisiae; it serves as a cofactor for hundreds of enzymes and it is a structural component of several transcription factors (2-4). The membrane function of zinc transport plays a central role in controlling the cellular levels of zinc (5). Zinc transporters are found in the plasma membrane (e.g. Zrt1p, Zrt2p, and Fet4p) (6-8), and membranes of the endoplasmic reticulum (Msc2p, Zrg17p) (4, 9), mitochondria (Mrs3p, Mrs4p) (10), and vacuole (e.g. Zrt3p, Cot1p, Zrc1p) (11-14). In controlling the cytoplasmic levels of zinc, some transporters mediate zinc influx (e.g. Zrt1p, Zrt2p, Fet4p, Cot1p, and Zrc1p), whereas some transporters mediate zinc efflux (e.g. Zrt3p) (5, 15).
The zinc transporters are regulated at the level of gene expression, and indeed, the most highly regulated transporter is Zrt1p (5, 6, 16). The expression of Zrt1p is induced for increased zinc uptake when the cytoplasmic levels of zinc are limiting (6). The increase in Zrt1p expression is dependent on the zinc-sensing and zinc-inducible transcriptional activator Zap1p, which interacts with a UASZRE sequence in the ZRT1 promoter to activate transcription (6, 17, 18). Conversely, when the cytoplasmic levels of zinc are high, the expression of Zrt1p is reduced to attenuate zinc uptake (6). Reduced Zrt1p expression is due to the loss of Zap1p activation of ZRT1 expression (18). Moreover, when the cytoplasmic levels of zinc are limiting, Zrt1p is a stable protein, and when the levels of zinc are high, Zrt1p is removed from the plasma membrane by endocytosis and is proteolytically degraded in the vacuole (19). Mechanisms to attenuate zinc uptake are important because an excess amount of cellular zinc is toxic (2).
Phospholipids are major components of cellular membranes. Because of their amphipathic nature, they are responsible for the membrane bilayer structure (20). In addition, phospholipids serve as precursors for the synthesis of complex membrane macromolecules (21-25), they are reservoirs of second messengers (26), and they regulate the structure, topography, and function of membrane transporters (27-34). Whereas little is known about the role of phospholipids in the structure and/or function of zinc transporters, it is known that the synthesis of membrane phospholipids in S. cerevisiae is coordinately regulated with the expression of zinc transporters in response to zinc depletion (1).
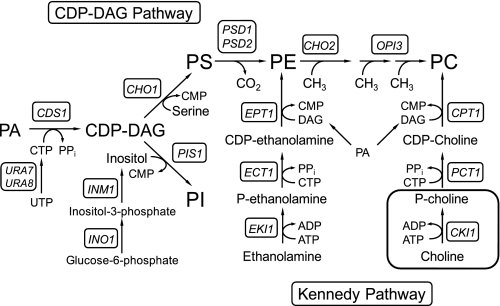
In S. cerevisiae, the major membrane phospholipid PC2 is synthesized (de novo) from the phospholipid precursor phosphatidate by the complementary CDP-DAG and Kennedy (CDP-choline branch) pathways (Fig. 1) (35, 36). When wild type cells are depleted for zinc, the activities of the CDP-DAG pathway enzymes PS synthase, PS decarboxylase, and the PE and phospholipid methyltransferases are repressed (37). On the other hand, zinc depletion causes the induction of PI synthase activity (37, 38). The regulation of the PS synthase and PI synthase enzymes in response to zinc depletion occurs at the level of transcription. The repression of CHO1, which encodes PS synthase, is mediated by the phospholipid synthesis repressor Opi1p (37), whereas the induction of PIS1, which encodes PI synthase, is mediated by Zap1p (38). Opi1p attenuates transcription of CHO1 (and other UASINO-containing genes in the pathway) by interaction with Ino2p, a component of the transcriptional activator Ino2p-Ino4p complex that interacts with a UASINO in the promoter (1, 36). Zap1p induces transcription of PIS1 by interaction with UASZRE sequences in the PIS1 promoter (38). The transcriptional regulation of the PS synthase and PI synthase enzymes translates into a decrease in the cellular PE content3 and an increase in the cellular PI content, respectively (37). The reciprocal regulation of these CDP-DAG branch point enzymes is part of an overall mechanism by which the synthesis of PI is coordinately regulated with the synthesis of phospholipids by way of the CDP-DAG pathway (1, 39, 40).
FIGURE 1.
Phospholipid synthesis in S. cerevisiae. The pathways shown for the synthesis of phospholipids include the relevant steps discussed throughout the article. The genes encoding enzymes responsible for the reactions in the pathways are indicated in the figure. The reaction catalyzed by the CKI1-encoded choline kinase enzyme is highlighted in the box. P-choline, phosphocholine; P-ethanolamine, phosphoethanolamine.
Despite the fact that the CDP-DAG pathway activities are repressed in response to zinc depletion, this growth condition does not have a significant effect on the cellular PC content (37). Because of the complementary nature of the phospholipid synthesis pathways in S. cerevisiae, we hypothesized that the decrease in CDP-DAG pathway enzyme activities is compensated by an increase in PC synthesis via the CDP-choline branch of the Kennedy pathway (Fig. 1). We focused our studies on the regulation of CKI1-encoded choline kinase, the enzyme that catalyzes the committed step in the pathway (Fig. 1). Indeed, choline kinase is a highly regulated enzyme that is controlled by transcriptional (41-43) and post-translational (44, 45) mechanisms to regulate PC content in S. cerevisiae. We found that the expression of CKI1 was induced by zinc depletion, and that the mechanism for this regulation involved the zinc-regulated transcriptional activator Zap1p. Moreover, this transcriptional regulation translated into increased choline kinase activity and PC synthesis via the Kennedy pathway.
EXPERIMENTAL PROCEDURES
Materials—All chemicals were reagent grade. Growth medium supplies and yeast nitrogen base lacking zinc sulfate were purchased from Difco and BIO 101 Inc., respectively. Clontech was the supplier of the Yeastmaker™ transformation kit. The QuikChange site-directed mutagenesis kit was purchased from Stratagene. Sigma was the source of ampicillin, aprotinin, benzamidine, bovine serum albumin, choline, phosphocholine, CDP-choline, leupeptin, O-nitrophenyl β-d-galactopyranoside, pepstatin, phenylmethylsulfonyl fluoride, and IGEPAL CA-630. Avanti Polar Lipids was the source of phosphatidylcholine. Silica Gel 60 thin layer chromatography plates were from EM Science. Protein assay reagents, electrophoretic reagents, and acrylamide solutions were purchased from Bio-Rad. Radiochemicals and scintillation counting supplies were purchased from PerkinElmer Life Sciences and National Diagnostics, respectively. Genosys Biotechnology, Inc. prepared oligonucleotides for PCR and electrophoretic mobility shift assays. ProbeQuant G-50 columns were purchased from Amersham Biosciences. Liqui-Nox detergent was from Alconox, Inc.
Strains and Growth Conditions—The strains used in this work are listed in Table 1. Yeast cultures were grown in YEPD medium (1% yeast extract, 2% peptone, 2% glucose) or in synthetic complete medium (46) containing 2% glucose at 30 °C. The appropriate amino acids of synthetic complete medium were omitted for selection purposes. Zinc-free medium was synthetic complete medium (46) prepared with yeast nitrogen base lacking zinc sulfate. To deplete internal stores of zinc (47), the following routine was followed. Cells were first grown for 24 h in synthetic complete medium containing 1.5 μm zinc sulfate. Saturated cultures were diluted into zinc-free medium at an initial concentration of 1 × 106 cells/ml, and grown for 24 h. Cultures were then diluted to 1 × 106 cells/ml and grown in zinc-free medium containing 0 or 1.5 μm zinc sulfate. The growth medium for the inositol auxotrophic ino2Δ and ino4Δ mutants (48) was supplemented with 75 μm inositol. Plasmid maintenance and amplification were performed in Escherichia coli strain DH5α. E. coli cells were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.4) at 37 °C. Ampicillin (100 μg/ml) was added to bacterial cultures that carried plasmids. For growth on plates, yeast and bacterial media were supplemented with 2 and 1.5% agar, respectively. Yeast cell numbers in liquid medium were determined spectrophotometrically at an absorbance of 600 nm. Exponential phase cells were harvested at a density of 1.5 × 107 cells/ml. Glassware were washed with Liqui-Nox, rinsed with 0.1 mm EDTA, and then rinsed several times with deionized distilled water to remove zinc contamination.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype or relevant characteristics | Source or Ref. |
|---|---|---|
| E. coli | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hdR17(rk−mk+) phoA supE44 l−thi-1 gyrA96 relA1 | 49 |
| S. cerevisiae | ||
| W303-1A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 77 |
| DY1457 | MATα ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-52 | 78 |
| ZHY6 | MATaade6 can1-100oc his3 leu2 ura3 zap1Δ::TRP1 | 78 |
| ZHY3 | MATα ade6 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-52 zrt1Δ::LEU2 zrt2Δ::HIS3 | 79 |
| SH304 | MATahis3Δ200 leu2Δ1 trp1Δ63 ura3-52 opi1Δ::LEU2 | S. A. Henry |
| SH303 | MATahis3Δ200 leu2Δ1 trp1Δ63 ura3-52 ino2Δ::TRP1 | S. A. Henry |
| SH307 | MATα his3Δ200 leu2Δ1 trp1Δ63 ura3-52 ino4Δ::LEU2 | S. A. Henry |
| Plasmid | ||
| pSD90 | PCRD1-lacZ reporter gene containing the CRD1 promoter with URA3 | W. Dowhan |
| pKSK11 | PCKI1-lacZ reporter gene containing the CKI1 promoter with URA3 | This work |
| pCK-ZRE1 | Derivative of pKSK11 with mutations in ZRE1 | This work |
| pCK-ZRE2 | Derivative of pKSK11 with mutations in ZRE2 | This work |
| pCK-ZRE1,2 | Derivative of pKSK11 with mutations in ZRE1 and ZRE2 | This work |
| pCK-UASino | Derivative of pKSK11 with mutations in the UASINO element | This work |
| pDg2 | PCYC1-ZRE-lacZ reporter gene containing the UASZRE element with URA3 | 17 |
DNA Manipulations, PCR, and Site-directed Mutagenesis—Standard methods were used for isolation and manipulation of DNA (49). PCR were optimized as described previously (50). Site-specific mutations in plasmids were generated with the QuikChange site-directed mutagenesis kit. All mutations were confirmed by DNA sequencing.
Plasmid Constructions—The plasmids used in this work are listed in Table 1. The pKSK11 plasmid contains the CKI1 gene promoter fused to the coding sequence of the lacZ gene of E. coli. This plasmid was constructed by replacing the CRD1 gene promoter in pSD90 (a plasmid based on YEp357R) with the CKI1 gene promoter sequence at the SphI/KpnI sites. The CKI1 promoter was obtained by PCR (primers, 5′-TCAGCATGCCTGCAGATATGAATTCCATAGG-3′ and 5′-CGAGGTACCCCTGGACGTGATTCTTGTAC-3′) using strain W303-1A genomic DNA as the template. The PCR primer used in the forward direction corresponds to -308 bp to the start codon, and the primer used in the reverse direction corresponds to +23 bp to the start codon. The correct orientation of the CKI1 promoter was confirmed by restriction enzyme digestion.
Plasmid pCK-UASino is a derivative of pKSK11, in which the UASINO element (39) sequence (5′-TATTCACAT-3′) in the CKI1 promoter was mutated to a nonconsensus sequence 5′-TATTTTTTTT-3′. Mutagenesis was performed with the QuikChange site-directed mutagenesis kit using plasmid pKSK11 as the template and the mutagenic primers 5′-CTTGTTCTTTGTTCTTTATGGTATAATATTTTTTTTGTGCTCTACCGTTTTTCTTGTCGGCCCAGC-3′ and 5′-GCTGGGCCGACAAGAAAAACGGTAGAGCACAAAAAAAATATTATACCATAAAGAACAAAGAACAAG-3′. Plasmids pCK-ZRE1 (primers, 5′-CTAAGCGATTGGTAACCAAAAAAAAAAAAGAACACCACCAAC-3′ and 5′-GTTGGTGGTGTTCTTTTTTTTTTTTGGTTACCAATCGCTTAC-3′) and pCK-ZRE2 (primers, 5′-CAGATCGTTCTCTTGTTCTTTGAAAAAAAAAAAATAATATTCACATGGTGCTCTAC-3′ and 5′-GTAGAGCACCATGTGAATATTATTTTTTTTTTTTCAAAGAACAAGAGAACGATCTG-3′) were also derivatives of pKSK11, in which the putative UASZRE element sequences ZRE1 and ZRE2 in the CKI1 promoter were mutated, respectively, to a nonconsensus sequence 5′-AAAAAAAAAAA-3′. Plasmid pCK-ZRE1, 2 is a derivative of pKSK11 where both ZRE1 and ZRE2 were mutated to the nonconsensus sequence. pCK-ZRE1, 2 was constructed by PCR amplification of plasmid pCK-ZRE1 using the primers for the construction of pCK-ZRE2. After PCR amplifications, all samples were digested with DpnI to eliminate the template DNA. The plasmids were amplified in E. coli, purified, and sequenced to confirm the mutations in the CKI1 promoter. Transformation of yeast (51, 52) and bacteria (49) with plasmids were performed as described previously.
Electrophoretic Mobility Shift Assays—Double-stranded oligonucleotides (Table 2) were prepared by annealing 25 μm complementary single-stranded oligonucleotides in a reaction mixture (0.1 ml) containing 10 mm Tris-HCl (pH 7.5), 100 mm NaCl, and 1 mm EDTA. The annealing reactions were incubated for 5 min at 100 °C in a heat block, and then kept in the heat block for another 2 h after it had been turned off. The annealed oligonucleotides (100 pmol), which had a 5′ over-hanging end, were labeled with [α-32P]dATP (400-800 Ci/nmol) and Klenow fragment (5 units) for 30 min at room temperature. The labeled oligonucleotides were separated from unincorporated nucleotides by gel filtration using ProbeQuant G-50 spin columns. Purified recombinant GST-Zap1p687-880 (38) was incubated with 1 pmol of radiolabeled DNA probe (2.0 × 105 cpm/pmol) for 15 min at room temperature in a total volume of 10 μl. The reaction buffer contained 10 mm Tris-HCl (pH 8.0), 10 mm MgCl2, 50 mm KCl, 1 mm dithiothreitol, 0.025 mg/ml poly(dI-dC)·poly(dI-dC), 0.2 mg/ml bovine serum albumin, 0.04% IGEPAL CA-630, and 10% glycerol. Following incubation, the reaction mixture was resolved on a 6% polyacrylamide gel (1.5-mm thickness) in 0.5× Tris borate-EDTA buffer at 100 V for 45 min. Gels were dried onto blotting paper, and the radioactive signals were visualized by phosphorimaging analysis.
TABLE 2.
Oligonucleotides used for electrophoretic mobility shift assays
| Element | Annealed oligonucleotidesa |
|---|---|
| ZRE1 | 5′-GTAACCTCCTTCACTTTAGAaca-3′ |
| 3′-CATTGGAGGAAGTGAAATCTTGA-5′ | |
| ZRE2 | 5′-TCTTTGTTCTTTATGGTATAata-3′ |
| 3′-AGAAACAAGAAATACCATATTAT-5′ | |
| ZRE3 | 5′-GGTTAAATCTCGAAGAGACAgaa-3′ |
| 3′-CCAATTTAGAGCTTCTCTGTCTT-5′ |
Underlined sequences are putative ZRE sites. The lower case letters indicate the nucleotides filled with the Klenow fragment.
Preparation of Cell Extracts and Protein Determination—All steps were performed at 5 °C. Yeast cells were disrupted with glass beads with a Mini BeadBeater-16 (Biospec Products) in 50 mm Tris malate buffer (pH 7.0) containing 1 mm EDTA, 0.3 m sucrose, 10 mm 2-mercaptoethanol, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin (53). Glass beads and cell debris were removed by centrifugation at 1,500 × g for 10 min, and the supernatant was used as the cell extract. Protein concentration was determined by the Coomassie Blue dye binding assay of Bradford (54) using bovine serum albumin as the standard.
Enzyme Assays—Choline kinase activity was measured in a reaction mixture that contained 67 mm glycine-NaOH buffer (pH 9.5), 5 mm [methyl-14C]choline (2,000 cpm/nmol), 5 mm ATP, 10 mm MgSO4, 1.3 mm dithiothreitol, and enzyme protein in a final volume of 60 μl (55). The radiolabeled product, phosphocholine, was separated from the radiolabeled substrate by precipitation of choline as a reineckate salt (56). A unit of choline kinase activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of phosphocholine/min. β-Galactosidase activity was measured for 5 min at room temperature by following the release of o-nitrophenol from o-nitrophenyl β-d-galactopyranoside at 410 nm. The reaction mixture contained 100 mm sodium phosphate buffer (pH 7.0), 1 mm MgCl2, 100 mm 2-mercaptoethanol, 3 mm o-nitrophenyl-β-d-galactopyranoside, and enzyme in a total volume of 0.1 ml. A unit of β-galactosidase activity was defined as the amount of enzyme that catalyzed the formation of 1 μmol of o-nitrophenol/min. Specific activity was defined as units/mg of total cellular protein. All enzyme assays were conducted in triplicate, and the average standard deviation of the assays was ±5%. Enzyme reactions were linear with time and protein concentration.
Labeling and Analysis of CDP-Choline Pathway Intermediates and PC—Exponential phase cells were labeled for five to six generations with 10 μm [methyl-14C]choline (0.2 μCi/ml). The CDP-choline pathway intermediates and PC were extracted from whole cells by a chloroform:methanol:water extraction, followed by separation of the aqueous and chloroform phases (57). The aqueous and chloroform phases that contained the CDP-choline pathway intermediates and PC, respectively, were dried in vacuo and then dissolved in 100 μl of methanol:water (1:1, v/v) and 100 μl of chloroform, respectively. The CDP-choline pathway intermediates and PC were subjected to TLC on silica gel plates using the solvent systems methanol, 0.6% sodium chloride, ammonium hydroxide (10:10:1, v/v) and chloroform, pyridine, 88% formic acid, methanol, water (60:35:10: 5:2, v/v), respectively. The positions of the labeled compounds on chromatograms were determined by phosphorimaging and compared with standards. The amount of each labeled compound was determined by liquid scintillation counting and normalized based on cell number.
Data Analyses—The Student's t test (SigmaPlot software) was used to determine statistical significance, and p values <0.05 were taken as a significant difference.
RESULTS
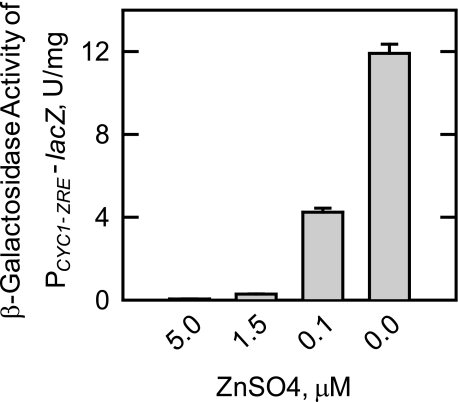
Effect of Zinc Depletion on the Expression of CKI1—In this and in subsequent experiments, cells were grown in medium that lacked inositol supplementation to obviate the regulatory effects that inositol has on phospholipid metabolism (36, 40, 58). Cells were grown in a defined zinc-free medium that contained 1.5 μm zinc sulfate. This concentration of zinc was equivalent to that found in standard synthetic growth media (46). To confirm that our growth conditions depleted the intracellular level of zinc, we made use of a PCYC1-ZRE-lacZ reporter gene assay that is sensitive to the labile pool of intracellular zinc (11, 17). In this assay, when the total intracellular concentration of zinc is limiting (∼10 pmol/106 cells),4 the transcriptional activity of Zap1p is up-regulated, and this results in elevated levels of β-galactosidase activity (11, 18). As described previously (11), the β-galactosidase activity of wild type zinc-depleted cells was greatly induced when compared with cells that were grown with zinc (Fig. 2).
FIGURE 2.
Effect of zinc depletion on the expression of β-galactosidase activity in wild type cells bearing the PCYC1-ZRE-lacZ reporter gene. Wild type cells bearing the PCYC1-ZRE-lacZ reporter plasmid pDg2 were grown to the exponential phase of growth in the presence of the indicated concentrations of ZnSO4. Cell extracts were prepared and assayed for β-galactosidase activity. Each data point represents the average of triplicate determinations from two independent experiments ± S.D.
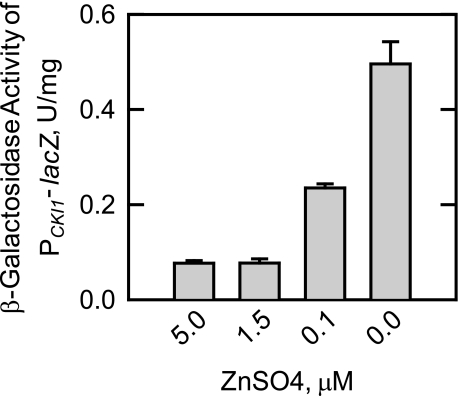
The effect of zinc depletion on the expression of the CKI1 gene was examined by use of a PCKI1-lacZ reporter gene. This reporter gene was constructed by fusing the CKI1 promoter in-frame with the coding sequence of the E. coli lacZ gene. Thus, the expression of β-galactosidase activity was dependent on transcription driven by the CKI1 promoter. Wild type cells bearing the PCKI1-lacZ reporter gene were grown to the exponential phase in the absence or presence of increasing concentrations of zinc. Following growth, cell extracts were prepared and used for the assay of β-galactosidase activity. The depletion of zinc from the growth medium resulted in a concentration-dependent increase in CKI1 expression (Fig. 3). The β-galactosidase activity in zinc-depleted cells was 7-fold greater than the activity found in cells grown with 1.5 μm zinc. CKI1 expression was not further affected by 5 μm zinc.
FIGURE 3.
Effect of zinc depletion on the expression of β-galactosidase activity in wild type cells bearing the PCKI1-lacZ reporter gene. Wild type cells bearing the PCKI1-lacZ reporter plasmid pKSK11 were grown to the exponential phase of growth in the presence of the indicated concentrations of ZnSO4. Cell extracts were prepared and assayed for β-galactosidase activity. Each data point represents the average of triplicate determinations from two independent experiments ± S.D.
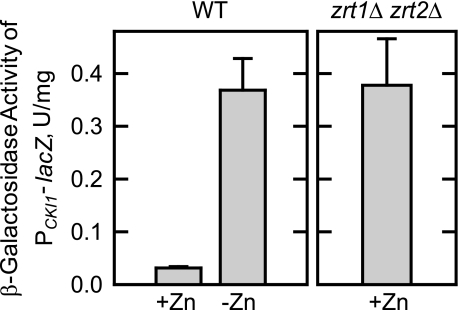
To confirm that the zinc-mediated regulation of CKI1 expression was due to the intracellular level of zinc, PCKI1-lacZ reporter gene activity was examined in a zrt1Δ zrt2Δ double mutant that was grown in the presence of zinc. This mutant lacks both the high (Zrt1p) and low (Zrt2p) affinity plasma membrane zinc transporters and contains a low cytoplasmic level of zinc even when the growth medium contains a standard amount of zinc (6, 7). The zrt1Δ zrt2Δ double mutant exhibited a high level of β-galactosidase activity that was similar to that shown for wild type cells grown without zinc (Fig. 4). Thus, the induction of CKI1 expression was mediated by a low intracellular level of zinc.
FIGURE 4.
Effect of zrt1Δ zrt2Δ mutations on the expression of the PCKI1- lacZ reporter gene. Wild type (WT) and zrt1Δ zrt2Δ mutant cells bearing the PCKI1-lacZ reporter plasmid pKSK11 were grown to the exponential phase of growth in the absence and presence of 1.5 μm ZnSO4. Cell extracts were prepared and used for the assay of β-galactosidase activity. Each data point represents the average of triplicate enzyme determinations from a minimum of two independent experiments ± S.D.
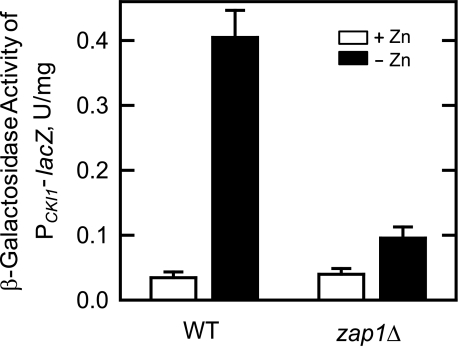
Effect of the zap1Δ Mutation on the Zinc-mediated Regulation of CKI1 Expression—The CKI1 promoter contains putative UASZRE sequences (see below) that are potential binding sites for the Zap1p transcription factor (17). Accordingly, we questioned whether the induction of CKI1 expression by zinc depletion was dependent on Zap1p function. To address this question, the PCKI1-lacZ reporter gene activity was examined in zap1Δ mutant cells that were grown in the presence and absence of zinc. The β-galactosidase activity of zap1Δ mutant cells grown with zinc was similar to the activity found in wild type cells grown with zinc (Fig. 5). However, when cells were grown without zinc, the β-galactosidase activity in the zap1Δ mutant was 4.4-fold lower than that found in the wild type control (Fig. 5). Thus, the zap1Δ mutation attenuated the induction of CKI1 expression when cells were depleted for zinc.
FIGURE 5.
Effect of the zap1Δ mutation on the expression of the PCKI1-lacZ reporter gene in response to zinc depletion. Wild type (WT) and zap1Δ mutant cells bearing the PCKI1-lacZ reporter plasmid pKSK11 were grown to the exponential phase of growth in the absence and presence of 1.5 μm ZnSO4. Cell extracts were prepared and assayed for β-galactosidase activity. Each data point represents the average of triplicate determinations from two independent experiments ± S.D.
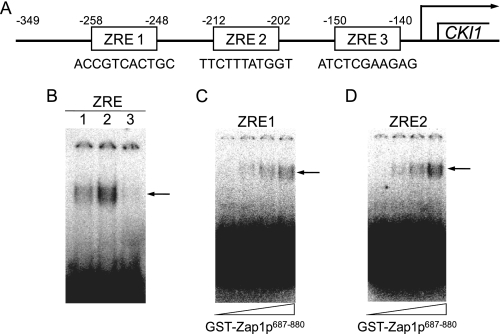
Binding of Zap1p to Putative UASZRE Sequences in the CKI1 Promoter—There are three putative UASZRE sequences in the CKI1 promoter for Zap1p binding (Fig. 6A). The percent identities of ZRE1, ZRE2, and ZRE3 to the consensus UASZRE sequence (ACCTTNAAGGT) (17) are 64, 73, and 64%, respectively. We questioned whether Zap1p would bind to labeled oligonucleotides containing these sequences using the electrophoretic mobility shift assay. Purified recombinant GST-Zap1p687-880 (amino acids 687-880 of the Zap1p binding domain (59)) interacted with ZRE1 and ZRE2, but not with ZRE3 (Fig. 6B). The interaction of GST-Zap1p687-880 with ZRE2 was 2.5-fold greater when compared with ZRE1. The specificity of GST-Zap1p687-880 binding to ZRE1 and ZRE2 was examined further. The formation of the GST-Zap1p687-880-ZRE1 and GST-Zap1p687-880-ZRE2 complexes was dependent on the concentration of GST-Zap1p687-880 (Fig. 6, C and D, respectively). Moreover, unlabeled ZRE1 and ZRE2 probes competed with their labeled probe counterparts for GST-Zap1p687-880 interactions, and mutations in ZRE1 and ZRE2 to a nonconsensus UASZRE sequence abolished interactions with GST-Zap1p687-880 (data not shown).
FIGURE 6.
Interactions of GST-Zap1p687-880 with putative ZRE sequences in the CKI1 promoter. A, the locations and sequences of the putative ZRE sites in the CKI1 promoter are shown in the figure. B, samples (1 pmol) of radiolabeled double-stranded synthetic oligonucleotides (2.0 × 105 cpm/pmol) with sequences for ZRE1 (lane 1), ZRE2 (lane 2), and ZRE3 (lane 3) in the CKI1 promoter were incubated with 0.5 μg of purified recombinant GST-Zap1p687-880. The ZRE1 (C) and ZRE2 (D) radiolabeled probes were incubated with 0, 0.15, 0.3, and 0.5 μg of recombinant GST-Zap1p687-880. Interaction of GST-Zap1p687-880 with the labeled oligonucleotides was determined by electrophoretic mobility shift assay using a 6% polyacrylamide gel. The data shown are representative of two independent experiments. The arrow indicates the position of the GST-Zap1p687-880-ZRE complex.
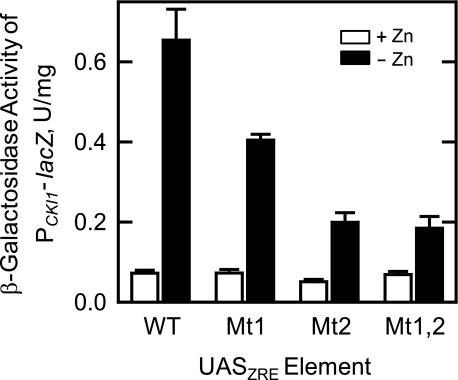
Effects of ZRE1 and ZRE2 Mutations on the Zinc-mediated Regulation of CKI1 Expression—We sought evidence that the induction of CKI1 in response to zinc depletion was dependent on the UASZRE sequences in the CKI1 promoter. The ZRE1 and ZRE2 sequences within the PCKI1-lacZ reporter gene were mutated to the nonconsensus UASZRE sequence 5′-AAAAAAAAAAA-3′. Cells bearing the wild type or mutant PCKI1-lacZ reporter genes were grown in the presence and absence of zinc; cell extracts were prepared and assayed for β-galactosidase activity. The ZRE1 and ZRE2 mutations caused the attenuation of CKI1 induction in response to zinc depletion (Fig. 7). The β-galactosidase activity of zinc-depleted cells bearing the PCKI1-lacZ reporter gene with the ZRE1 and ZRE2 mutations was 1.6- and 3.2-fold lower, respectively, when compared with cells bearing the control reporter gene. These results supported the conclusion that the zinc-mediated regulation of CKI1 expression was dependent on ZRE1 and ZRE2. These results were consistent with the electrophoretic mobility shift assays that indicated that the regulation was primarily governed by ZRE2. We considered the possibility that the full extent of induction was dependent on interaction of Zap1p with ZRE1 and ZRE2 together. However, the effects of the ZRE1 and ZRE2 mutations on the zinc-mediated regulation of CKI1 expression were not additive or synergistic. The β-galactosidase activity in zinc-depleted cells bearing the reporter gene with the ZRE1 and ZRE2 mutations made in combination was the same as that found in cells bearing the reporter gene with the ZRE2 mutation alone (Fig. 7).
FIGURE 7.
Effects of mutations in ZRE1 and ZRE2 in the CKI1 promoter on the expression of the PCKI1-lacZ reporter gene in response to zinc depletion. Wild type cells bearing the PCKI1-lacZ reporter plasmids pKSK11 (WT), pCK-ZRE1 (Mt1, mutation of ZRE1), pCK-ZRE2 (Mt2, mutation of ZRE2), and pCK-ZRE1,2 (Mt1,2, mutations of ZRE1 and ZRE2) were grown to the exponential phase of growth in the absence and presence of 1.5 μm ZnSO4. Cell extracts were prepared and assayed for β-galactosidase activity. Each data point represents the average of triplicate determinations from two independent experiments ± S.D.
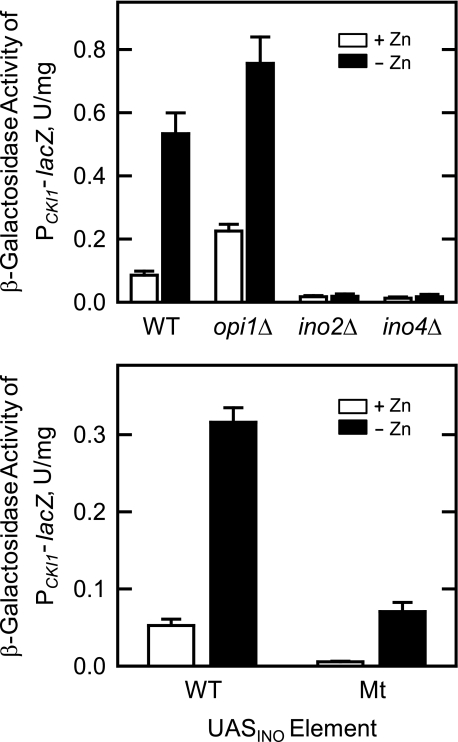
Effects of opi1Δ, ino2Δ, and ino4Δ Mutations, and a UASINO Element Mutation in the CKI1 Promoter on the Zinc-mediated Regulation of CKI1 Expression—The CKI1 promoter contains a UASINO element (-189 to -197) that is four bases away from ZRE2. The UASINO element is a binding site for the transcriptional activator Ino2p-Ino4p complex (60) that is responsible for maximum expression of phospholipid synthesis UASINO-containing genes (36, 40, 58, 61). Expression of the UASINO-containing genes is balanced by the Opi1p repressor (62), which attenuates transcription through its interaction with Ino2p (36, 40, 58, 61, 63). Given the close proximity of ZRE2 with the UASINO element in the CKI1 promoter, we questioned whether the functions of Opi1p, Ino2p, or Ino4p affected the zinc-mediated regulation of CKI1 expression. As expected, the expression of the PCKI1-lacZ reporter gene was elevated (2.6-fold) in the opi1Δ mutant and reduced (5-fold) in the ino2Δ and ino4Δ mutants (Fig. 8A). Thus, Ino2p and Ino4p played a positive role in CKI1 expression and Opi1p played a negative role in expression. The opi1Δ mutation did not have a major effect on the induced expression of the PCKI1-lacZ reporter gene in response to zinc depletion (Fig. 8A), indicating that Opi1p did not play a role in the zinc-mediated regulation of CKI1. However, the depletion of zinc from the growth medium of the ino2Δ and ino4Δ mutants did not result in the induction of the PCKI1-lacZ reporter gene (Fig. 8A). This indicated that Ino2p and Ino4p might play a role in the zinc-mediated regulation of CKI1.
FIGURE 8.
Effects of the opi1Δ, ino2Δ, and ino4Δ mutations, and a mutation in the UASINO element in the CKI1 promoter on the expression of the PCKI1-lacZ reporter gene in response to zinc depletion. Wild type (WT), opi1Δ, ino2Δ, and ino4Δ cells bearing the PCKI1-lacZ reporter plasmid pKSK11 (A), and wild type cells bearing the PCKI1-lacZ reporter plasmids pKSK11 (WT) and pCK-UASino (Mt, mutation of UASINO) (B) were grown to exponential phase of growth in the absence and presence of 1.5 μm ZnSO4. The growth medium for the ino2Δ and ino4Δ mutants was supplemented with 75 μm inositol. Cell extracts were prepared and used for the assay of β-galactosidase activity. Each data point represents the average of triplicate enzyme determinations from a minimum of two independent experiments ± S.D.
The inositol auxotrophic ino2Δ and ino4Δ mutants grew very slow in synthetic medium despite the supplementation of the medium with 75 μm inositol. The depletion of zinc from this growth medium further reduced the growth of the mutants. We were concerned that the repressive effects of inositol supplementation on CKI1 expression (41) and the added stress of zinc depletion on the growth of the mutants may have affected the zinc-mediated regulation of CKI1 expression. Accordingly, we utilized another approach to examine whether the UASINO element played a role in the zinc-mediated regulation of CKI1. The UASINO element within the PCKI1-lacZ reporter gene was mutated to a nonconsensus sequence (5′-TATTTTTTTT-3′), and the plasmid was expressed in wild type cells that were grown in the presence and absence of zinc. The β-galactosidase activity of cells bearing the mutant reporter gene was reduced (9.5-fold with zinc and 5-fold without zinc) when compared with the activity of cells bearing the wild type reporter gene (Fig. 8B). This result was consistent with the notion that the UASINO element was required for maximum expression of CKI1. The depletion of zinc from the growth medium resulted in a 10-fold induction of β-galactosidase activity driven by the reporter gene with the UASINO mutation (Fig. 8B). These results supported the conclusion that the UASINO element was not involved with the zinc-mediated regulation of CKI1.
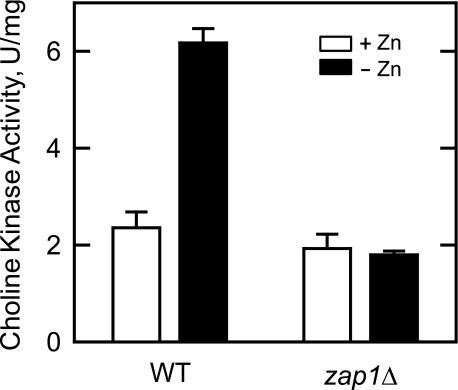
Effect of Zinc Depletion on Choline Kinase Activity, and on the Incorporation of Choline into PC via the Kennedy Pathway—We sought evidence that the transcriptional regulation of CKI1 in response to zinc depletion translated into changes in choline kinase activity. Wild type cells were grown in the presence and absence of zinc, cell extracts were prepared, and choline kinase activity was measured by following the incorporation of [methyl-14C]choline into phosphocholine. The specific activity of choline kinase in zinc-depleted cells was 2.7-fold greater when compared with the activity from cells grown with zinc (Fig. 9). This level of induced choline kinase activity is not as great as that observed for the induction of PCKI1-lacZ reporter gene activity (Fig. 5). This difference may be attributed to the stable nature of lacZ fusion proteins that are not subject to proteolytic turnover when expressed in yeast (64). We also examined the expression of choline kinase activity in zap1Δ mutant cells. This analysis showed that the induction of choline kinase activity that occurred in wild type cells in response to zinc depletion was precluded by the zap1Δ mutation (Fig. 9). These results provided further evidence for the role of Zap1p in the regulation of CKI1 expression in response to zinc.
FIGURE 9.
Effect of the zap1Δ mutation on choline kinase activity in response to zinc depletion. Wild type (WT) and zap1Δ mutant cells were grown to the exponential phase of growth in the absence and presence of 1.5 μm ZnSO4. Cell extracts were prepared and assayed for choline kinase activity. Each data point represents the average of triplicate determinations from two independent experiments ± S.D.
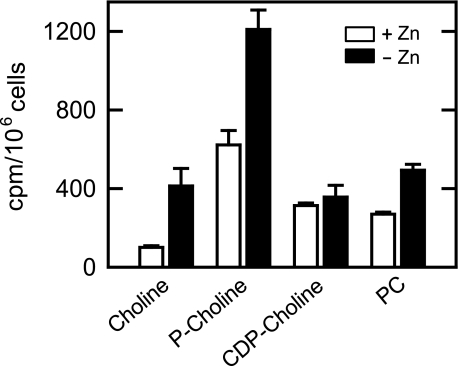
We questioned whether the induction of choline kinase activity by zinc depletion was reflected in vivo. To address this question, zinc-depleted cells were labeled to steadystate with [methyl-14C]choline followed by the extraction and analysis of phosphocholine by TLC. The amount of label incorporated into phosphocholine in zinc-depleted cells was 2-fold greater when compared with cells grown in the presence of zinc (Fig. 10). The choline label was also incorporated into CDP-choline and the end product of the Kennedy pathway PC. Whereas zinc depletion did not affect the steady-state level of CDP-choline, it caused a 1.8-fold increase in the level of PC that was derived from CDP-choline (Fig. 10). That CDP-choline was not affected might be a reflection that it is a rate-limiting intermediate in the Kennedy pathway. Zinc depletion also caused a 4-fold increase in the cellular content of free choline (Fig. 10).
FIGURE 10.
Effect of zinc depletion on the composition of the CDP-choline pathway intermediates and on PC. Wild type cells were grown to the exponential phase of growth in the absence or presence of 1.5 μm ZnSO4. The cells were labeled for five to six generations with [methyl-14C]choline (0.2 μCi/ml). The CDP-choline pathway intermediates and PC were extracted and analyzed by thin-layer chromatography. The values reported were the average of four separate experiments ± S.D.
DISCUSSION
The synthesis of phospholipids is coordinately regulated with the expression of zinc transporters that control zinc homeostasis in S. cerevisiae (1). The depletion of zinc from the growth medium results in the induced expression of zinc transporters (e.g. Zrt1p, Zrt2p, Fet4p, and Zrt3p), and their transport functions increase the cytoplasmic levels of zinc (5, 15). At the same time, zinc depletion causes alterations in membrane phospholipid composition (e.g. an increase in PI and decreases in PE, phosphatidate, and diacylglycerol pyrophosphate) that are brought by changes in the expression of phospholipid synthesis enzyme activities (1, 37, 38, 65, 66). The PC content of cellular membranes is not altered by zinc depletion even though the enzyme activities responsible for its synthesis via the CDP-DAG pathway are repressed (37). In this study, we provided a mechanistic explanation for this observation. The decrease in PC synthesis via the CDP-DAG pathway was compensated by an increase in PC synthesis via the CDP-choline branch of the Kennedy pathway. Data indicated that the Zap1p-mediated induction of the CKI1-encoded choline kinase played an important role in this regulation. This compensatory mechanism complements the Zap1p-mediated induction of the EKI1-encoded ethanolamine kinase (Fig. 1) for increased PC synthesis in response to zinc depletion (65).
Analysis of PCKI1-lacZ reporter gene activity indicated that the expression of CKI1 was induced in response to zinc depletion, a growth condition that resulted in a limiting intracellular concentration of zinc (11, 18). That a limiting cellular zinc concentration was responsible for this regulation was confirmed by the induced expression of PCKI1-lacZ reporter gene activity in zinc-supplemented zrt1Δ zrt2Δ mutant cells that lack the major plasma membrane zinc transporters Zrt1p and Zrt2p (6, 7). The induction of PCKI1-lacZ reporter gene activity was precluded by the zap1Δ mutation. This indicated that CKI1 expression was mediated by Zap1p, a positive transcription factor that is induced in zinc-depleted cells and repressed in zinc-replete cells (18).
Zap1p interacts with a UASZRE in the promoters of several genes to activate transcription when wild type cells are depleted for zinc. These include zinc transporter genes (e.g. ZRT1, ZRT2, ZRT3, and FET4) that control intracellular stores of zinc (5, 8, 12, 67, 68) and phospholipid synthesis genes (e.g. DPP1, PIS1, and EKI1) that control membrane phospholipid composition (38, 47, 65). Electrophoretic mobility shift assays indicated that purified GST-Zap1p687-880 interacted with two of the three putative UASZRE sequences (i.e. ZRE1 and ZRE2) within the CKI1 promoter. The in vitro interactions of GST-Zap1p687-880 with ZRE1 and ZRE2 were specific and could be abolished by mutations to a nonconsensus UASZRE sequence. Moreover, mutations of ZRE1 and ZRE2 to a nonconsensus UASZRE sequence attenuated the induced expression of the PCKI1-lacZ reporter gene activity when cells were depleted for zinc.
The Zap1p binding sites (i.e. ZRE1 and ZRE2) in the CKI1 promoter were not identical (64 and 73% identity, respectively) to the core consensus UASZRE that is based on the promoters of ZRT1, ZRT2, and ZAP1 (17). It is known that deviations from the core sequence reduce the interaction of Zap1p to the UASZRE sequence (38, 65). This provides an explanation as to why the Zap1p-mediated induction of CKI1 was not as great as that observed for Zap1p-mediated induction of zinc transporter genes (e.g. ZRT1, ZRT2, and ZRT3) or for the phospholipid metabolism gene DPP1 that contain UASZRE sequences with high identity to the core sequence (6, 7, 11, 47). We speculate that differences in Zap1p binding efficiency based on the UASZRE sequence provide a mechanism to control the relative induction of various genes in response to zinc depletion.
The UASINO element is found in several phospholipid synthesis genes in S. cerevisiae (35, 39, 40, 58, 61). It is the binding site for the transcriptional activator complex Ino2p-Ino4p that stimulates transcription of most UASINO-containing genes (36, 40, 58, 61).5 When exponential phase wild type cells are supplemented with inositol, or depleted for zinc, the repressor Opi1p interacts with Ino2p to attenuate transcription of UASINO-containing genes such as CHO1 and INO1 (1, 36). CKI1 contains a UASINO, and the work presented here using the PCKI1-lacZ reporter gene showed that this element was responsible for stimulating CKI1 expression. However, the UASINO element was not involved with the regulation of CKI1 expression in response to zinc depletion.
Choline kinase activity is the functional product of the CKI1 gene (55, 69). The Zap1p-mediated induction of CKI1 expression in response to zinc depletion translated into a 2.7-fold increase in choline kinase activity. This conclusion was confirmed by the loss of induced choline kinase activity in zap1Δ mutant cells. Moreover, the in vitro data showing the zinc-regulated induction of choline kinase activity was mirrored in vivo. The incorporation of labeled choline into phosphocholine was elevated by 2-fold in response to zinc depletion. In addition, the increase in CKI1 expression and choline kinase activity correlated with a 1.8-fold increase in the amount of PC that was synthesized from choline. Thus, the synthesis of PC via the Kennedy pathway was elevated in zinc-depleted cells, and choline kinase played an important role in this regulation. We also showed that zinc depletion caused an increase in the cellular choline content. This result might reflect an increase in choline transporter activity and/or an increase in the turnover of PC that was synthesized via the Kennedy pathway. Additional work will be required to address these hypotheses.
This work advances our understanding of the regulation of phospholipid synthesis in S. cerevisiae by zinc availability, and in particular, the transcriptional regulation of the CKI1-encoded choline kinase. The importance of understanding choline kinase regulation is highlighted by the fact that the enzyme in mice is essential to embryonic development (α form (70)) and to normal muscular development (β form (71)). Moreover, unregulated levels of choline kinase play a role in the generation of human tumors by ras oncogenes (72-75). By catalyzing the committed step in the Kennedy pathway, choline kinase regulation governs PC content. Whether PC content per se or whether another membrane phospholipid(s) (e.g. PE and PI) regulates zinc transporter function to control zinc homeostasis are important questions that warrant further investigation.
Acknowledgments
We thank Michael C. Kersting and Gil-Soo Han for support and advice during the course of this work. Keunsung Kim is acknowledged for construction of the PCKI1-lacZ reporter gene. We also thank William Dowhan, David J. Eide, and Susan A. Henry for plasmids and mutants used in this study.
This work was supported in part by United States Public Health Service, National Institutes of Health Grants GM-28140 (to G. M. C.) and GM-75378 (to A. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PC, phosphatidylcholine; CDP-DAG, CDP-diacylglycerol; PS, phosphatidylserine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; UASINO, upstream activating sequence inositol-responsive element; UASZRE, upstream activating sequence zinc-responsive element; GST, glutathione S-transferase.
The decrease in PE content is attributed to reductions in both PS synthase and PS decarboxylase activities (37).
It is technically difficult to quantify the labile pool of zinc (11).
One exception is the PIS1 gene, which contains a UASINO element but the element does not control PIS1 expression (76).
References
- 1.Carman, G. M., and Han, G. S. (2007) Biochim. Biophys. Acta 1771 322-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vallee, B. L., and Falchuk, K. H. (1993) Physiol. Rev. 73 79-118 [DOI] [PubMed] [Google Scholar]
- 3.Schwabe, J. W., and Klug, A. (1994) Nat. Struct. Biol. 1 345-349 [DOI] [PubMed] [Google Scholar]
- 4.Ellis, C. D., Wang, F., MacDiarmid, C. W., Clark, S., Lyons, T., and Eide, D. J. (2004) J. Cell Biol. 166 325-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eide, D. J. (2003) J. Nutr. 133 1532S-1535S [DOI] [PubMed] [Google Scholar]
- 6.Zhao, H., and Eide, D. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 2454-2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao, H., and Eide, D. (1996) J. Biol. Chem. 271 23203-23210 [DOI] [PubMed] [Google Scholar]
- 8.Waters, B. M., and Eide, D. J. (2002) J. Biol. Chem. 277 33749-33757 [DOI] [PubMed] [Google Scholar]
- 9.Ellis, C. D., MacDiarmid, C. W., and Eide, D. J. (2005) J. Biol. Chem. 280 28811-28818 [DOI] [PubMed] [Google Scholar]
- 10.Muhlenhoff, U., Stadler, J. A., Richhardt, N., Seubert, A., Eickhorst, T., Schweyen, R. J., Lill, R., and Wiesenberger, G. (2003) J. Biol. Chem. 278 40612-40620 [DOI] [PubMed] [Google Scholar]
- 11.MacDiarmid, C. W., Gaither, L. A., and Eide, D. (2000) EMBO J. 19 2845-2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacDiarmid, C. W., Milanick, M. A., and Eide, D. J. (2003) J. Biol. Chem. 278 15065-15072 [DOI] [PubMed] [Google Scholar]
- 13.Miyabe, S., Izawa, S., and Inoue, Y. (2001) Biochem. Biophys. Res. Commun. 282 79-83 [DOI] [PubMed] [Google Scholar]
- 14.Devirgiliis, C., Murgia, C., Danscher, G., and Perozzi, G. (2004) Biochem. Biophys. Res. Commun. 323 58-64 [DOI] [PubMed] [Google Scholar]
- 15.Guerinot, M. L., and Eide, D. (1999) Curr. Opin. Plant Biol. 2 244-249 [DOI] [PubMed] [Google Scholar]
- 16.Lyons, T. J., Gasch, A. P., Gaither, L. A., Botstein, D., Brown, P. O., and Eide, D. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7957-7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao, H., Butler, E., Rodgers, J., Spizzo, T., Duesterhoeft, S., and Eide, D. (1998) J. Biol. Chem. 273 28713-28720 [DOI] [PubMed] [Google Scholar]
- 18.Zhao, H., and Eide, D. J. (1997) Mol. Cell. Biol. 17 5044-5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gitan, R. S., Luo, H., Rodgers, J., Broderius, M., and Eide, D. (1998) J. Biol. Chem. 273 28617-28624 [DOI] [PubMed] [Google Scholar]
- 20.Stryer, L. (1995) Biochemistry, Fourth Ed., W. H. Freeman and Co., New York
- 21.Becker, G. W., and Lester, R. L. (1980) J. Bacteriol. 142 747-754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menon, A. K., and Stevens, V. L. (1992) J. Biol. Chem. 267 15277-15280 [PubMed] [Google Scholar]
- 23.Lester, R. L., and Dickson, R. C. (1993) Adv. Lipid Res. 26 253-274 [PubMed] [Google Scholar]
- 24.Fankhauser, C., Homans, S. W., Thomas-Oates, J. E., McConville, M. J., Desponds, C., Conzelmann, A., and Ferguson, M. A. (1993) J. Biol. Chem. 268 26365-26374 [PubMed] [Google Scholar]
- 25.Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M., Noda, T., and Ohsumi, Y. (2000) Nature 408 488-492 [DOI] [PubMed] [Google Scholar]
- 26.Exton, J. H. (1994) Biochim. Biophys. Acta 1212 26-42 [DOI] [PubMed] [Google Scholar]
- 27.Bogdanov, M., and Dowhan, W. (1999) J. Biol. Chem. 274 36827-36830 [DOI] [PubMed] [Google Scholar]
- 28.Bogdanov, M., Sun, J., Kaback, H. R., and Dowhan, W. (1996) J. Biol. Chem. 271 11615-11618 [DOI] [PubMed] [Google Scholar]
- 29.Dowhan, W. (1997) Annu. Rev. Biochem. 66 199-232 [DOI] [PubMed] [Google Scholar]
- 30.Chen, C. C., and Wilson, T. H. (1984) J. Biol. Chem. 259 10150-10158 [PubMed] [Google Scholar]
- 31.Wang, X., Bogdanov, M., and Dowhan, W. (2002) EMBO J. 21 5673-5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogdanov, M., Heacock, P. N., and Dowhan, W. (2002) EMBO J. 21 2107-2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, W., Bogdanov, M., Pi, J., Pittard, A. J., and Dowhan, W. (2003) J. Biol. Chem. 278 50128-50135 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, W., Campbell, H. A., King, S. C., and Dowhan, W. (2005) J. Biol. Chem. 280 26032-26038 [DOI] [PubMed] [Google Scholar]
- 35.Paltauf, F., Kohlwein, S. D., and Henry, S. A. (1992) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression (Jones, E. W., Pringle, J. R., and Broach, J. R., eds) pp. 415-500, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 36.Carman, G. M., and Henry, S. A. (2007) J. Biol. Chem. 282 37293-37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwanyshyn, W. M., Han, G. S., and Carman, G. M. (2004) J. Biol. Chem. 279 21976-21983 [DOI] [PubMed] [Google Scholar]
- 38.Han, S.-H., Han, G.-S., Iwanyshyn, W. M., and Carman, G. M. (2005) J. Biol. Chem. 280 29017-29024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carman, G. M., and Henry, S. A. (1989) Annu. Rev. Biochem. 58 635-669 [DOI] [PubMed] [Google Scholar]
- 40.Carman, G. M., and Henry, S. A. (1999) Prog. Lipid Res. 38 361-399 [DOI] [PubMed] [Google Scholar]
- 41.Hosaka, K., Murakami, T., Kodaki, T., Nikawa, J., and Yamashita, S. (1990) J. Bacteriol. 172 2005-2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jesch, S. A., Zhao, X., Wells, M. T., and Henry, S. A. (2005) J. Biol. Chem. 280 9106-9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jesch, S. A., Liu, P., Zhao, X., Wells, M. T., and Henry, S. A. (2006) J. Biol. Chem. 281 24070-24083 [DOI] [PubMed] [Google Scholar]
- 44.Yu, Y., Sreenivas, A., Ostrander, D. B., and Carman, G. M. (2002) J. Biol. Chem. 277 34978-34986 [DOI] [PubMed] [Google Scholar]
- 45.Choi, M.-G., Kurnov, V., Kersting, M. C., Sreenivas, A., and Carman, G. M. (2005) J. Biol. Chem. 280 26105-26112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose, M. D., Winston, F., and Heiter, P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 47.Han, G.-S., Johnston, C. N., Chen, X., Athenstaedt, K., Daum, G., and Carman, G. M. (2001) J. Biol. Chem. 276 10126-10133 [DOI] [PubMed] [Google Scholar]
- 48.Culbertson, M. R., and Henry, S. A. (1975) Genetics 80 23-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning, A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 50.Innis, M. A., and Gelfand, D. H. (1990) in PCR Protocols, A Guide to Methods and Applications (Innis, M. A., Gelfand, D. H., Sninsky, J. J., and White, T. J., eds) pp. 3-12, Academic Press, Inc., San Diego
- 51.Ito, H., Yasuki, F., Murata, K., and Kimura, A. (1983) J. Bacteriol. 153 163-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schiestl, R. H., and Gietz, R. D. (1989) Curr. Genet. 16 339-346 [DOI] [PubMed] [Google Scholar]
- 53.Klig, L. S., Homann, M. J., Carman, G. M., and Henry, S. A. (1985) J. Bacteriol. 162 1135-1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 55.Kim, K.-H., Voelker, D. R., Flocco, M. T., and Carman, G. M. (1998) J. Biol. Chem. 273 6844-6852 [DOI] [PubMed] [Google Scholar]
- 56.Porter, T. J., and Kent, C. (1992) Methods Enzymol. 209 134-146 [DOI] [PubMed] [Google Scholar]
- 57.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37 911-917 [DOI] [PubMed] [Google Scholar]
- 58.Greenberg, M. L., and Lopes, J. M. (1996) Microbiol. Rev. 60 1-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bird, A., Evans-Galea, M. V., Blankman, E., Zhao, H., Luo, H., Winge, D. R., and Eide, D. J. (2000) J. Biol. Chem. 275 16160-16166 [DOI] [PubMed] [Google Scholar]
- 60.Ambroziak, J., and Henry, S. A. (1994) J. Biol. Chem. 269 15344-15349 [PubMed] [Google Scholar]
- 61.Henry, S. A., and Patton-Vogt, J. L. (1998) Prog. Nucleic Acids Res. 61 133-179 [DOI] [PubMed] [Google Scholar]
- 62.White, M. J., Hirsch, J. P., and Henry, S. A. (1991) J. Biol. Chem. 266 863-872 [PubMed] [Google Scholar]
- 63.Wagner, C., Dietz, M., Wittmann, J., Albrecht, A., and Schuller, H. J. (2001) Mol. Microbiol. 41 155-166 [DOI] [PubMed] [Google Scholar]
- 64.Guarente, L. (1983) Methods Enzymol. 101 181-191 [DOI] [PubMed] [Google Scholar]
- 65.Kersting, M. C., and Carman, G. M. (2006) J. Biol. Chem. 281 13110-13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han, G.-S., Johnston, C. N., and Carman, G. M. (2004) J. Biol. Chem. 279 5338-5345 [DOI] [PubMed] [Google Scholar]
- 67.Manolson, M. F., Proteau, D., Preston, R. A., Stenbit, A., Roberts, B. T., Hoyt, M. A., Preuss, D., Mulholland, J., Botstein, D., and Jones, E. W. (1992) J. Biol. Chem. 267 14294-14303 [PubMed] [Google Scholar]
- 68.Miyabe, S., Izawa, S., and Inoue, Y. (2000) Biochem. Biophys. Res. Commun. 276 879-884 [DOI] [PubMed] [Google Scholar]
- 69.Hosaka, K., Kodaki, T., and Yamashita, S. (1989) J. Biol. Chem. 264 2053-2059 [PubMed] [Google Scholar]
- 70.Wu, G., Aoyama, C., Young, S. G., and Vance, D. E. (2007) J. Biol. Chem. 283 1456-1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sher, R. B., Aoyama, C., Huebsch, K. A., Ji, S., Kerner, J., Yang, Y., Frankel, W. N., Hoppel, C. L., Wood, P. A., Vance, D. E., and Cox, G. A. (2006) J. Biol. Chem. 281 4938-4948 [DOI] [PubMed] [Google Scholar]
- 72.Hernandez-Alcoceba, R., Saniger, L., Campos, J., Nunez, M. C., Khaless, F., Gallo, M. A., Espinosa, A., and Lacal, J. C. (1997) Oncogene 15 2289-2301 [DOI] [PubMed] [Google Scholar]
- 73.Hernandez-Alcoceba, R., Fernandez, F., and Lacal, J. C. (1999) Cancer Res. 59 3112-3118 [PubMed] [Google Scholar]
- 74.Nakagami, K., Uchida, T., Ohwada, S., Koibuchi, Y., and Morishita, Y. (1999) Jpn. J. Cancer Res. 90 1212-1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakagami, K., Uchida, T., Ohwada, S., Koibuchi, Y., Suda, Y., Sekine, T., and Morishita, Y. (1999) Jpn. J. Cancer Res. 90 419-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderson, M. S., and Lopes, J. M. (1996) J. Biol. Chem. 271 26596-26601 [DOI] [PubMed] [Google Scholar]
- 77.Thomas, B., and Rothstein, R. (1989) Cell 56 619-630 [DOI] [PubMed] [Google Scholar]
- 78.Lamping, E., Luckl, J., Paltauf, F., Henry, S. A., and Kohlwein, S. D. (1995) Genetics 137 55-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nikawa, J., Hosaka, K., and Yamashita, S. (1993) Mol. Microbiol. 10 955-961 [DOI] [PubMed] [Google Scholar]