Abstract
Dynactin is a multisubunit protein complex necessary for dynein function. Here, we investigated the function of dynactin in budding yeast. Loss of dynactin impaired movement and positioning of the mitotic spindle, similar to loss of dynein. Dynactin subunits required for function included p150Glued, dynamitin, actin-related protein (Arp) 1 and p24. Arp10 and capping protein were dispensable, even in combination. All dynactin subunits tested localized to dynamic plus ends of cytoplasmic microtubules, to stationary foci on the cell cortex and to spindle pole bodies. The number of molecules of dynactin in those locations was small, less than five. In the absence of dynactin, dynein accumulated at plus ends and did not appear at the cell cortex, consistent with a role for dynactin in offloading dynein from the plus end to the cortex. Dynein at the plus end was necessary for dynactin plus-end targeting. p150Glued was the only dynactin subunit sufficient for plus-end targeting. Interactions among the subunits support a molecular model that resembles the current model for brain dynactin in many respects; however, three subunits at the pointed end of brain dynactin appear to be absent from yeast.
Keywords: cell division, dynactin, dynein, mitosis, yeast
Dynactin is a widely conserved multisubunit complex necessary for the function of dynein in the cytoplasm, participating in a variety of microtubule-based transport and targeting phenomena (reviewed in 1). Biochemically, dynactin has been found to interact with microtubules, dynein and various types of cargo in different settings. In the budding yeast Saccharomyces cerevisiae, dynein’s sole known function is to position the mitotic spindle in the neck between mother and bud in preparation for cell division, and dynactin is necessary for this function (2-4). Dynein and dynactin mediate the sliding of cytoplasmic (astral) microtubules along the cortex of the cell, which pulls the mitotic spindle into the mother/bud neck and helps to keep it there (5,6).
In the current working model, dynein is targeted to the plus ends of dynamic cytoplasmic microtubules by the combined action of NudEL, LIS1, CLIP-170 and a kinesin Kip2 (7,8). When the plus end encounters a cortical target, probably marked by the protein Num1 (9,10), some dynein is ‘offloaded’ from the plus end, anchored to the cortex and then activated. Active dynein attempts to walk to the minus end of the microtubule, causing the microtubule to slide along the cortex, thus pulling the nucleus into the neck.
The structural core of dynactin is a short actin-like filament composed of Arp1 (actin-related protein) (1). The filament’s pointed end appears to interact with another Arp, Arp11, and the barbed end contains the heterodimeric capping protein for conventional actin, also known as CapZ (11,12). The subunits p62, p27 and p25 are also located at the pointed end, forming a biochemical subcomplex with Arp11. The filament has a shoulder near its barbed end (12) composed of the p150Glued, dynamitin and p24 subunit. p150Glued protrudes from the shoulder as a flexible extension.
Saccharomyces cerevisiae homologues have been described or suggested for all the dynactin subunits except for the pointed-end components p62, p27 and p25. The yeast protein Arp10 can be considered to be a member of the Arp11 family or the sole representative of its own family (13-15). Arp10 interacts with the pointed end of the Arp1 filament, based on a compelling set of genetic and biochemical evidence (16). Neither Arp10 nor the barbed-end capping protein Cap1/Cap2 has been described as important for dynein function in yeast (2,11,17). Moreover, no functional link of capping protein to dynactin has been described despite the clear biochemical association in vertebrates. The barbed end of the yeast Arp1 filament appears to interact with dynamitin/Jnm1 and p150Glued/Nip100 (17), consistent with structural and biochemical studies of the vertebrate dynactin shoulder.
Dynactin is necessary for dynein to function, but how and where dynactin contributes is not well understood. Based on genetic and cell biology results, a number of dynactin subunit genes lie in the dynein pathway, indistinguishable from genes encoding dynein, Num1 and other essential elements. For example, the dynamitin/jnm1 null mutant phenotype resembles that of the dynein/dyn1 null mutant (18), as do the phenotypes of the Arp1/arp1 null mutant (2) and the p150Glued/nip100 null mutant (19). In localization studies, dynamitin/Jnm1 was observed on the daughter-bound spindle pole body (SPB) (18) and p150Glued/Nip100 was seen on both SPBs (19). Biochemically, interactions among the Nip100, Jnm1 and Arp1 proteins have been observed (19).
Here, we investigated the function of dynactin complex with a combination of approaches. We studied a number of dynactin genes and subunits to understand their relative contributions to the process, and we investigated interactions between dynein and dynactin. Dynactin was present at the plus end of cytoplasmic microtubules along with dynein, and the presence of dynein was necessary to target dynactin to the plus end. Dynactin localized with dynein at the cortex as well, and loss of dynactin caused dynein to accumulate at the plus end and fail to appear at the cortex. Our results suggest that dynactin’s role is to offload dynein from the microtubule plus end to the cortex.
Results
Yll049w as a dynactin p24 homologue
To identify novel components and regulators of the dynein pathway, we screened the Research Genetics collection of viable null haploid mutants. Strains were grown in the cold and scored for appearance of two or more DAPI-stained bodies in single unbudded cells or mothers of small-budded cells, which reflects delayed or failed movement of the spindle into the mother-bud neck. The yll049wΔ mutant had a strong phenotype, consistent with complete loss of dynein function. The YLL049w is predicted to encode a polypeptide of 179 aa. BLAST and Pfam searching did not reveal any regions of significant similarity with known protein motifs, domains or polypeptides. YLL049w was named LDB18 based on a ‘low-dye-binding’ phenotype in a screen for genes involved in phosphorylation of oligosaccharides (20).
We performed a genetic analysis to test whether YLL049W is necessary for function of the dynein pathway. The yll049w null mutant had a strong cold phenotype, noted above, characteristic of dynein pathway mutations. Budding yeast employ two major pathways to achieve nuclear segregation - the Kar9 pathway and the dynein pathway. Null mutations in one pathway have little effect on growth, but cells lacking genes in both pathways grow very poorly. By tetrad analysis, the yll049wΔ mutation showed a synthetic growth defect with kar9Δ (11 of 11 yll049wΔ kar9Δ double mutants grew as microcolonies) but not with dyn1Δ (12 of 13 yll049wΔ dyn1Δ double mutants grew normally, and one grew as a microcolony). Thus, the genetic properties of YLL049W place it in the dynein pathway.
Dynactin is necessary for dynein function, and our screen identified several genes encoding dynactin subunits, including Arp1, p150Glued/Nip100 and dynamitin/Jnm1 (Lee, Li and Cooper, unpublished data). YLL049W was identified as an element of the dynein/dynactin pathway in a congruent gene analysis of synthetic lethal interactions in genome-wide screens (21). Yll049w was found to interact with dynamitin/Jnm1 in a genome-wide two-hybrid screen (22). Therefore, we hypothesized that Yll049w was a subunit of dynactin complex.
We asked whether Yll049w was biochemically associated with known dynactin subunits at endogenous expression levels. p150Glued/Nip100 was pulled down from a wholecell extract using a tandem-affinity purification (TAP) tag integrated at the NIP100 locus. We probed for the presence of Yll049w and known dynactin subunits. Yll049w, Arp1, Arp10/Arp11 and dynamitin/Jnm1 were detected in precipitates from the TAP-tagged p150Glued/Nip100 strain but not in precipitates from a negative control strain lacking the TAP tag (Figure S1A). As another negative control, the septin Cdc3 was not found in the precipitates.
Based on molecular mass, Yll049w was potentially homologous to any of the three small dynactin subunits - p24, p25 or p27, none of which had been identified in budding yeast. In a phylogenetic analysis, we aligned Yll049w with p24, p25 and p27 subunits from a wide range of species. Yll049w clustered with the p24 homologues in a phylogenetic tree, separated from p25 and p27 (Figure S1B). The bootstrap value for the connection of the Yll049w branch to the tree was 100%. The secondary structure of Yll049w was predicted to be mainly α-helical by several approaches (Figure S1C). p24 homologues are mainly α-helical (1), while p25 and p27 subunit homologues include regions predicted to adopt an unusual left-handed parallel β-helix fold (23). We did not find any such β-helix region in Yll049w based on the prediction program BETAWRAP and the protein-fold recognition program PHYRE.
Based on these genetic, functional, biochemical and structural properties, we conclude that Yll049w is the yeast homologue of the dynactin p24 subunit. Localization of the protein in cells, discussed next, also supports this conclusion.
Localization of dynactin
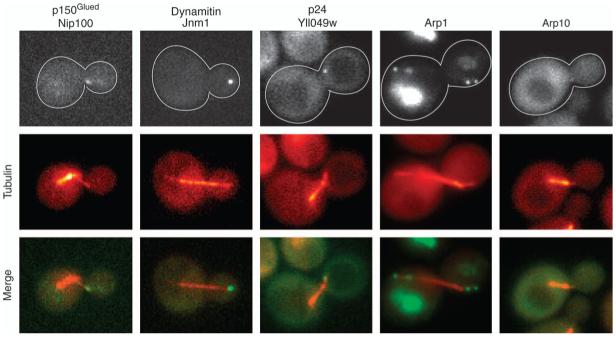
To investigate dynactin function, we localized several subunits in living cells. For high sensitivity, three tandem copies of green fluorescent protein (GFP) were fused to the C-terminus of p150Glued/Nip100, dynamitin/Jnm1, Arp10 and p24/Yll049w by integration at the endogenous locus. Haploid strains expressing the 3GFP fusions had low numbers of cells with abnormal spindle position or nuclear segregation in the cold, indicating that the dynein pathway and therefore the tagged proteins were functional. All four proteins were found at microtubule plus ends and at the SPB, which were identified by double labeling with CFP-tubulin (Figure 1).
Figure 1. Localization of dynactin in cells.
Fluorescent-tagged forms of dynactin subunits are observed at plus ends of microtubules and at SPBs. The α-tubulin subunit Tub1 was tagged with CFP for colocalization in each strain. The width of each image is 5 μm. Strain numbers: yJC3891, Yll049w-3GFP; yJC4147, Nip100-3GFP; yJC5261, Jnm1-3GFP; yJC5389, Arp10-3GFP and yJC5400, Arp1-tdimer2.
Actin fusions are rarely functional, so we tagged one copy of ARP1 in a diploid strain with tdimer2 following the approach used to visualize conventional actin/Act1 with a non-rescuing GFP fusion (24). Most of the fluorescence was seen in the vacuole, consistent with aggregation of non-functional protein. However, cytoplasmic puncta were also seen at the plus ends of microtubules and at the SPB (Figure 1). Haploid strains derived from this diploid expressing Arp1-tdimer2 as the sole form of Arp1 were viable; however, essentially, all the fluorescence in those strains appeared as aggregates in the vacuole (data not shown). Thus, all the known dynactin subunits localized to microtubule plus ends and the SPB, which are locations of dynein (8,25,26).
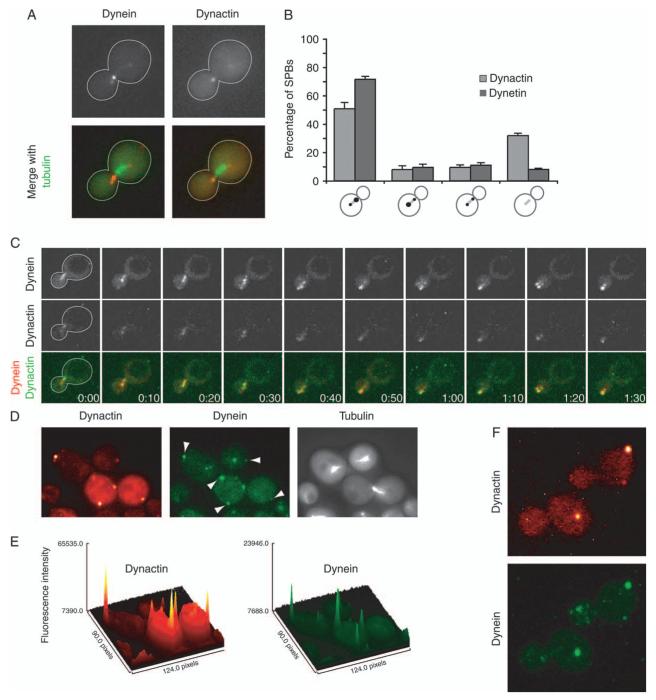
To examine the dynamics of dynactin localization and to compare it with that of dynein, we performed dual imaging of dynamitin and dynein again in living cells. Jnm1-tdimer2 was the brightest dynactin subunit fusion we tested, and brain dynactin contains four copies of dynamitin, more than any subunit other than Arp1 (1). Dyn1-3GFP and CFP-Tub1 were also present to reveal dynein and microtubules. First, dynactin and dynein colocalized at SPBs and plus ends (Figure 2A) as noted above. In Movie S1, the movement of SPBs and plus ends are quite different, allowing for their unequivocal identification. Second, we looked for polarization of localization. During early anaphase, dynactin was localized to the SPB near the bud more often than the other SPB (Figure 2B), which has been observed for dynein (27). The level of bias for dynactin was less than that for dynein, suggesting that dynactin may be following dynein, which will be discussed below. Dynein has also been seen to decorate and move along the sides of microtubules (8,25), and dynactin showed a similar behavior (Figure 2C).
Figure 2. Dynamics of dynactin and dynein localization.
A) Dynamitin/Jnm1 tagged with tdimer2 and DHC/Dyn1 tagged with 3GFP localized with CFP-labeled microtubules. Dynactin and dynein exhibit a selective association with the daughter-bound set of microtubules. Images were captured on a wide-field microscope. B) Distribution of dynamitin/Jnm1 and DHC on short bipolar spindles in early anaphase cells. From left to right, cells were scored as having fluorescence concentrated on the daughter-bound SPB, concentrated on the mother-bound SPB, on both equally or on neither. The values are the mean of four sets of 50 cells each, and error bars denote standard error of those means. C) Dynein and dynactin colocalize on the sides and ends of cytoplasmic microtubules. Jnm1-tdimer2 and Dyn1-3GFP expressed in num1Δ cells. Three confocal sections at 0.1-μm increments were captured every 10 seconds and collapsed into Z projections using ImageJ. D) Dynactin and dynein colocalize at foci on the cell cortex. Jnm1-tdimer2 and Dyn1-3GFP expressed in wild-type cells expressing CFP-labeled microtubules. Arrowheads denote cortical foci containing both Jnm1 and Dyn1. Images were captured on a wide-field microscope. E) Surface plots of Jnm1-tdimer2 and Dyn1-3GFP images shown in (D) were generated using ImageJ. F) Z projections of 16 confocal sections at 0.2-μm increments in wild-type cells expressing Jnm1-tdimer2 and Dyn1-3GFP. Strains: yJC5652 in panels A,B,D,E and F and yJC5666 in C.
Dynein is found in stationary foci at the cell cortex (8,25), and we observed dynactin at cortical foci in two-dimensional wide-field images (Figure 2D). These cortical foci were not associated with microtubules based on imaging live cells expressing fluorescent-labeled Jnm1 and tubulin. Movies of Jnm1-3GFP cells confirmed that the foci were stationary, distinguishing them from fast-moving plus ends that might have been near the cortex (Movie S1). We saw some relative differences comparing the fluorescence intensities of the dynactin and dynein cortical foci in wide-field images collected with a non-intensified CCD camera, which should provide the best representation of molecular content (Figure 2E). Cortical stationary foci of dynein are lost when dynein fails to function, such as in mutants lacking the cortical anchor Num1 or essential subunits of dynactin (8,25). Here, we observed that cortical foci of dynactin, as Jnm1-tdimer2, were also lost in null mutants lacking Num1, dynein/Dyn1 or p150Glued/Nip100 (Table S2). These observations are consistent with a working model in which dynactin accompanies dynein during transfer from the microtubule end to the cortex, and dynactin remains anchored at the cortex with dynein while the motor functions. Dynactin may function to anchor or activate dynein at the cortex in concert with Num1.
Quantification of dynactin content at plus ends and SPBs
We quantified the fluorescence intensities of individual dynactin subunits at microtubule plus ends and SPBs. These values were normalized to ones for fluorescent Cse4, which is a stable centromere protein present at two molecules per chromosome (28). After movement of DNA to the poles in anaphase, one pole contains 32 molecules of Cse4. A non-intensified CCD camera and a wide-field microscope were used to maximize linearity. For Cse4-mCitrine, the histogram of intensity values for individual foci was consistent with a single distribution (Figure 3B). The mean intensity, in arbitrary units, was 9441, which divided by 32 gave a value of 295 au/molecule of mCitrine.
Figure 3. Quantification of dynamitin fluorescence intensity.
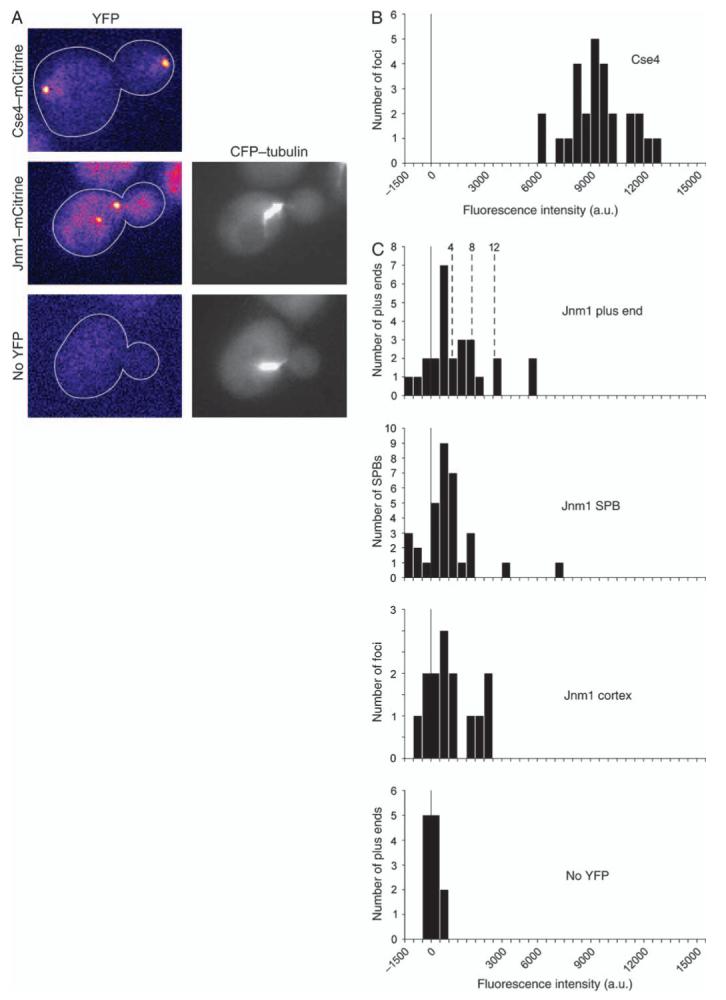
A) Representative images of cells expressing Cse4-mCitrine, Jnm1m-Citrine or CFP-Tub1. Yellow fluorescent protein images were captured using identical settings and pseudocolored with the Fire lookup table in ImageJ. B) Histogram of fluorescence intensity per focus of Cse4-mCitrine in anaphase cells. Thirty-two molecules of Cse4 are present in each spot in late anaphase (28), so the fluorescence intensity per molecule of mCitrine here is 295 arbitrary units (au). C) Histogram of fluorescence intensity of Jnm1-mCitrine in budded cells. Some values are less than zero because of background subtraction. Bud-proximal microtubule plus ends (n = 26) and SPBs (n = 33) were identified using CFP-Tub1. Dashed lines indicate the predicted fluorescence intensities for 4, 8 and 12 molecules of Jnm1-mCitrine. Cortical foci of Jnm1-mCitrine were identified by eye (n = 14). The bottom panel is a control with budded cells expressing CFP-Tub1 and no mCitrine. Here, microtubule plus ends were identified in the CFP channel, and intensity was recorded in the mCitrine channel, n = 12. Strain numbers: Cse4-mCitrine, yJC5463; Jnm1-mCitrine CFP-Tub1, yJC5349 and CFP-Tub1, yJC3883.
To quantify dynactin fluorescence, we examined strains expressing Jnm1-mCitrine. To avoid biasing the collection for bright foci, we visualized microtubules with CFP-tubulin in order to identify plus ends and SPBs. We then quantified the fluorescence at those locations in the corresponding mCitrine image and corrected for background fluorescence locally (29). We included all the corrected values whether or not a spot of fluorescence was apparent in the image. At plus ends, the distribution of Jnm1 fluorescence intensity values was broad and relatively flat compared with the Cse4 distribution (Figure 3C). Based on a value of 295 au/mCitrine, the values for 4, 8 and 12 molecules of mCitrine, which should correspond to one, two and three molecules, respectively, of dynactin complex were calculated and are plotted on Figure 3C. The observed values are consistent with a small number of dynactin molecules, which we would estimate at less than 5 to be conservative. At the SPB, the values were lower, suggesting the presence of even fewer molecules of dynactin (Figure 3C). For cortical foci of dynactin, we measured the fluorescence intensity of Jnm1-mCitrine foci of that could be identified by eye. These values were also in the range of several molecules of dynactin complex (Figure 3C).
An important parameter for this analysis is the range of values that constitute the background level of noise in the measurements. Each value was corrected for background on an individual basis. Pixel intensity values in a boxed region of interest were corrected based on the intensity values of pixels immediately outside of and adjacent to the box (29). In a control strain not expressing mCitrine, but still expressing CFP-tubulin, the corrected values were centered around zero as expected, and their distribution extended to ± approximately 800 au (Figure 3C), corresponding to less than three molecules of mCitrine. Many of the values for Jnm1 at the SPB were above the noise level, in a range consistent with one to five molecules of dynactin, assuming four subunits of Jnm1 per dynactin. Jnm1-mCitrine fluorescence included a diffuse distribution throughout the cell, consistent with a substantial cytosolic pool of protein (Figure 3A). The diffuse pool was seen when comparing Jnm1-mCitrine/Tub1-CFP cells to cells expressing only Tub1-CFP.
Next, we quantified the fluorescence intensity values for strains expressing 3GFP fusions of Jnm1, Yll049w, Nip100 and Arp10. These fusions were functional, rescuing null mutant phenotypes in haploid strains. Arp1 was not analyzed because fusions function poorly in haploids, and the fluorescence distribution included a number of extraneous foci, probably aggregates undergoing degradation, even in heterozygous diploids. At the microtubule plus end, the fluorescence distributions were broad and extended to zero for all the four subunits examined (Figure S3A). The range of values for the different subunits was similar, including their upper bounds, except for a few extremely high values. The variance of the values precludes conclusions about the stoichiometry of the subunits within the complex, although the range of the values is consistent with the estimate above of several, less than five, molecules of dynactin complex per plus end. At the SPB, 3GFP fusions of Jnm1 and Yll049w gave similar results, with broad distributions extending to zero and consistent with several molecules of dynactin complex per SPB (Figure S3B).
Roles of individual subunits in localization and composition of dynactin
To investigate the roles of the individual subunits in dynactin, we examined the localization and stability of the complex in strains lacking one subunit using fluorescence localization in cells and protein pulldowns from cell lysates. The molecular architecture of brain dynactin is shown in Figure 4 for reference (1), and the results are shown in Figures 4 and 5.
Figure 4. Localization of dynactin subunits in null mutants.
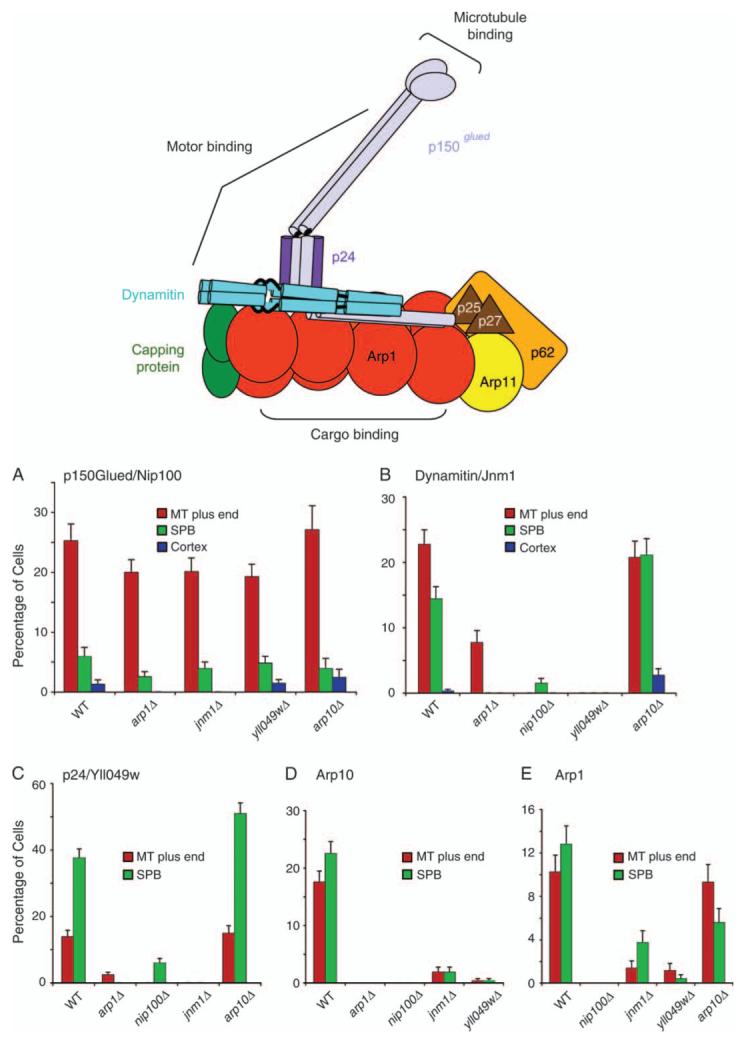
A diagram illustrates the molecular architecture of vertebrate dynactin, modified with permission from one by Trina Schroer (1). On the bar graphs, the percentage of cells that display localization of a fluorescent-tagged subunit to a given location is plotted for different dynactin null mutants. The fluorescent-tagged subunits are A) p150Glued/Nip100-3GFP, B) dynamitin/Jnm1-mCitrine, C) Yll049w-3GFP, D) Arp10-3GFP and E) Arp1-tdimer2. Strain numbers were as follows: A) Wild-type yJC4147; arp1Δ, yJC4158; jnm1Δ, yJC4156; yll049wΔ, yJC4162 and arp10Δ, yJC5377. B) Wild-type yJC5349; arp1Δ, yJC5381; nip100Δ, yJC5385; yll049wΔ, yJC5387 and arp10Δ, yJC5383. C) Wild-type yJC3891; nip100Δ, yJC4166; arp1Δ, yJC4152; jnm1Δ, yJC4154 and arp10Δ, yJC5379. D) Wild-type yJC5389; arp1Δ, yJC5416; jnm1Δ, yJC5418; nip100Δ, yJC5420 and yll049wΔ,yJC5422.E)Wild-typeyJC5400; nip100Δ/nip100Δ, yJC5483; jnm1Δ/jnm1Δ, yJC5482; yll049wΔ/yll049wΔ, yJC5484 and arp10Δ/arp10Δ, yJC5480. MT, microtubule.
Figure 5. Roles of dynactin subunits in composition of the complex.
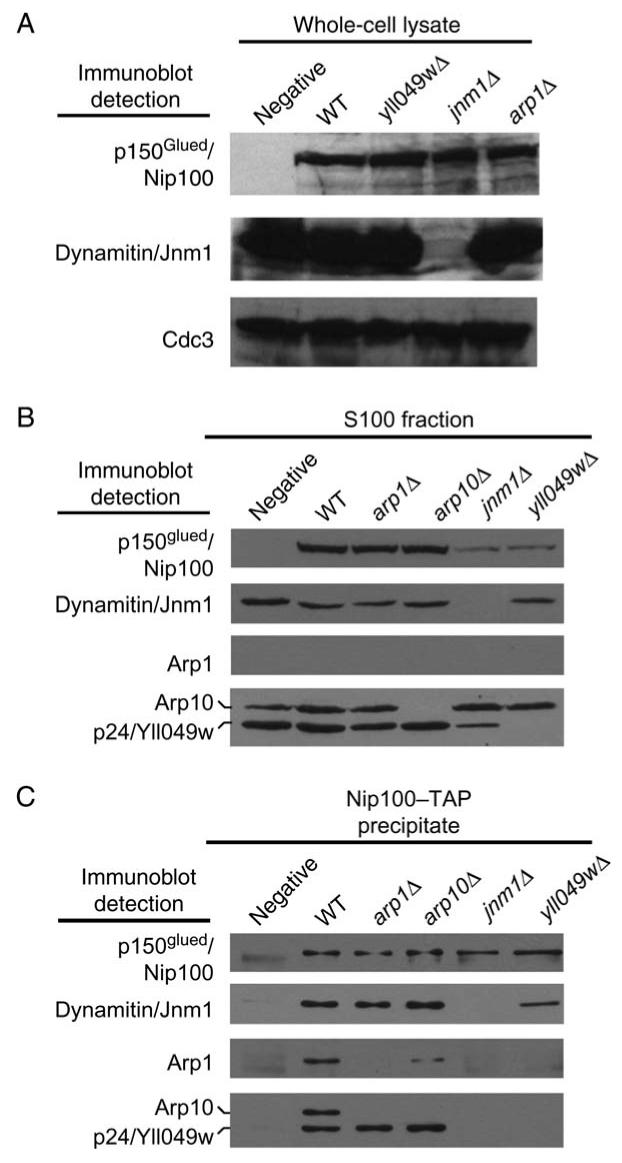
p150Glued/Nip100-TAP was tagged at the endogenous locus in haploid wild-type and various mutant strains. A) Immunoblots of whole-cell lysates. The level of TAP-tagged p150Glued/Nip100 is similar in wild-type and mutant cells. Dynamitin/Jnm1 is absent from the jnm1 mutant as expected. Cdc3 is a loading control. B) Immunoblots of high-speed supernatants, the S100 fraction, of the whole-cell lysates. The amount of p150Glued/Nip100 is decreased in the jnm1 and yll049w mutants. C) Immunoblots of Nip100-TAP precipitates loaded so that each lane received similar amounts of Nip100. Dynamitin/Jnm1 and p24/Yll049w are important for the association of Arp1 and Arp10 with Nip100. Intensities of Cap2 and Act1 bands were not reproducibly above control (not shown). Arp10 and p24/Yll049w were tagged with 13-myc. Strain numbers: untagged, yJC5813; wild-type, yJC5814; yll04wΔ, yJC5475; jnm1Δ, yJC5817; arp1Δ, yJC5815 and arp10Δ, yJC5816.
First, we counted the number of cells that displayed localization of a given GFP-tagged subunit to microtubule plus ends, to SPBs and to the cortex (Figure 4). Based on the quantification of fluorescence intensity relative to Cse4-mCitrine above, dynactin subunits tagged with three copies of GFP should permit the detection of less than five molecules of dynactin complex. The frequency of observing p150Glued/Nip100 at the plus end and the SPB in the absence of any one of the four other subunits was similar to that of wild-type (WT) cells (Figure 4A). Thus, Nip100 was sufficient for targeting to the plus end and the SPB. Conversely, in nip100 null mutant cells, none of the other subunits, including Jnm1, Yll049w, Arp1 and Arp11, was found at the plus end (Figure 4B-E). Similar results were seen for the SPB, with the possible exception of a small degree of localization of dynamitin/Jnm1 and p24/Yll049w in the nip100Δ mutant, 2% and 7% of cells, respectively (Figure 4B,C). These small values were significant and reproducibly above background, suggesting the existence of subcomplexes. Thus, p150Glued/Nip100 appears to be absolutely necessary or very important for the localization of every other subunit.
For brain dynactin, the p150Glued, dynamitin and p24 subunits interact in a subcomplex (1,11,30,31). We found that for dynamitin/Jnm1 and p24/Yll049w, the localization of each protein to the plus end and to the SPB depended on the presence of the other protein as well as on p150Glued/Nip100 (Figure 4A-C). Jnm1 formed aggregates in the absence of Yll049w or Nip100, while Yll049w showed diffuse cytoplasmic staining in the absence of Jnm1 or Nip100 (data not shown).
The actin-like filament of Arp1 is a structural core for the vertebrate dynactin complex (1). In addition to p150Glued/Nip100, Dynamitin/Jnm1 and p24/Yll049w were important but not absolutely required for the localization of Arp1, based on a substantial decrease in the percentage of cells showing targeting in the null mutants, at both the plus end and SPB (Figure 4E). In an arp1Δ mutant, Jnm1 showed partial localization to plus ends and no localization to SPBs, while Yll049w was lost from both plus ends and SPBs (Figure 4B,C), and Nip100 showed nearly normal localization (Figure 4A) as noted above. Biochemical sedimentation analysis confirmed the importance of Arp1 for complex formation (Figure S4).
The localization of Arp10 depended completely on p150Glued/Nip100 and Arp1 and partially on dynamitin/Jnm1 and p24/Yll049w (Figure 4D). Arp10 was not required for the proper localization of any other dynactin subunit to the plus end or the SPB (Figure 4A-C), with the possible exception of Arp1 at the SPB (Figure 4E).
The subunits were largely associated with each other. However, for wild-type cells in this assay, the ratio of the plus-end value to the SPB value was greater than one for p150Glued/Nip100 (Figure 4A) and less than one for p24/Yll049w (Figure 4C) on a consistent basis. The ratios for Jnm1, Arp10 and Arp1 were near unity (Figure 4B,D,E). Biochemical sedimentation analysis of wild-type cell extracts revealed significant amounts of Jnm1, p24 and Arp10 outside of the intact complex (Figure S4). Thus, subunits may exist independent of other subunits.
As a complementary approach, we performed protein precipitation assays with cell lysates of the various mutants to test for the association of other subunits with p150Glued/Nip100. Endogenous p150Glued/Nip100 was tagged with TAP at its C-terminus and precipitated with immunoglobulin beads added to a high-speed supernatant, an S100 fraction, of cell lysates. First, we observed that the level of p150Glued/Nip100-TAP in the S100 input was much less when dynamitin/Jnm1 or p24/Yll049w was absent (Figure 5B), even though the level of p150Glued/Nip100-TAP in whole-cell extracts was not affected (Figure 5A). Most likely, a fraction of Nip100 is unstable and aggregates in the absence of Jnm1 or Yll049w. In Neurospora, a p24 mutant, ro-10, showed loss of p150Glued/RO3 based on immunoblots of cell extracts (32).
To compare the subunit composition of dynactin complex in the mutants, we adjusted the volume of each sample loaded on the gel to achieve equal amounts of precipitated p150Glued/Nip100-TAP in each lane. Dynamitin/Jnm1, Arp1, Arp11/Arp10 and p24/Yll049w were present in the precipitates from lysates of TAP-tagged strains but not strains without a TAP tag (Figure 5C). Capping protein and conventional actin are found in biochemical preparations of vertebrate dynactin (1). We looked for their presence in these samples with polyclonal antibodies to the yeast capping protein β subunit, Cap2, and to yeast conventional actin, Act1. Neither protein was detected (not shown). Furthermore, capping protein was not detected in association with dynactin in a sedimentation analysis (Figure S4).
Dynamitin and p24 form a ‘shoulder’ complex with p150Glued in vertebrate dynactin (12,30,31). Here, loss of dynamitin/Jnm1 led to the complete failure of p24/Yll049w to precipitate with p150Glued/Nip100. Conversely, loss of p24/Yll049w led to a large but not complete loss of dynamitin/Jnm1 from the precipitate (Figure 5C). Thus, the shoulder complex requires all three of its components for optimal stability, and dynamitin is more proximal to p150Glued than is p24. The shoulder complex was important for the association of the Arp1 filament with p150Glued; little or no Arp1 or Arp10 was found in the p150Glued precipitate in the absence of dynamitin or p24. In the cell assay where foci of accumulation were observed and counted in cells, the values for Arp1 and Arp10 were greatly decreased, but not to zero, especially for Arp1 (Figure 4). One might expect the precipitation assay to be a more rigorous test of the stability of the association because proteins that dissociate from the pellet are washed away with buffer, while in the cell, the unbound proteins remain present in solution in the cytoplasm. Thus, Arp1 appears to be proximal to Arp10 with respect to the shoulder complex. These results are consistent with the model proposed for vertebrate dynactin (Figure 4A) (1).
Plus-end targeting of dynactin depends on dynein
We investigated how dynactin is targeted to the microtubule plus end and SPB by examining Nip100-3GFP localization in various mutants. We analyzed the fluorescence images in two ways - by counting cells in which foci of Nip100-3GFP were observed by eye (Figure 6A-D) and by quantifying the fluorescence intensity from digital video images (Figure 6E,F and Table S2).
Figure 6. Dynein is important for targeting of p150Glued/Nip100 to the microtubule plus end.
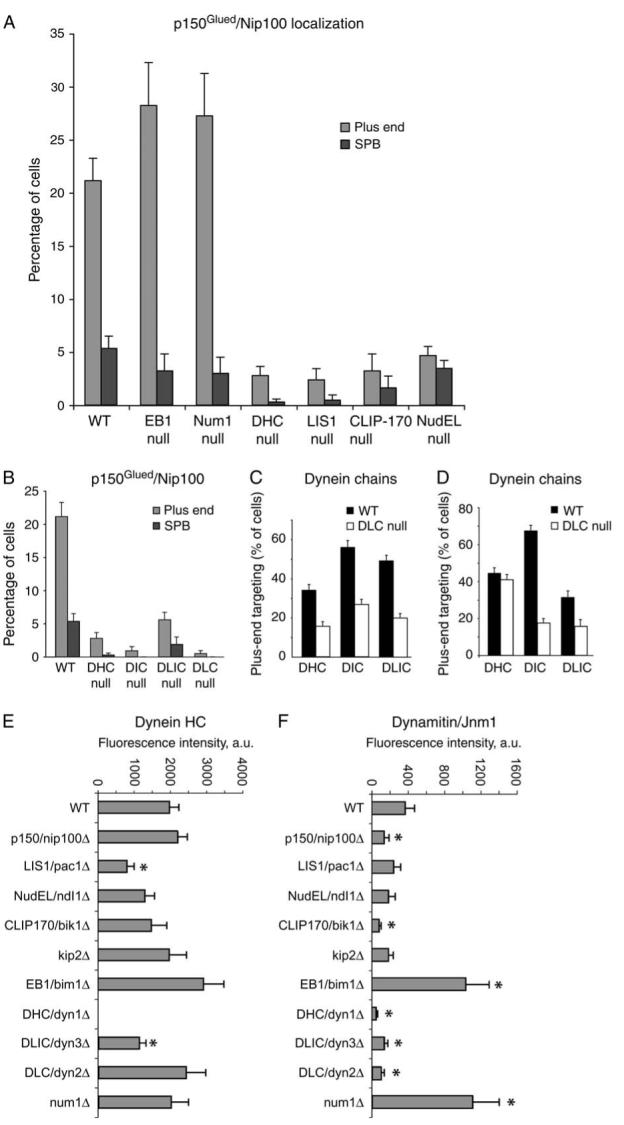
A and B) Percentage of cells with Nip100-GFP at a plus end or an SPB in various mutants. DHC, Dyn1. DIC, Pac11. DLIC, Dyn3. DLC, Dyn2. C) Plus-end targeting of GFP-tagged dynein chains in a DLC null mutant, dyn2Δ. D)Plus-end targeting of dynein chains in a DLIC null mutant, dyn3Δ. Dynein heavy chain and DIC require each other for stability and thus for targeting, and no dynein chains target to plus ends in the absence of DIC or intermediate chain. E and F) Fluorescence intensity of Dyn1-3GFP and Jnm1-tdimer2 at the plus end. Values are the means of the fluorescence intensity of the most bud-proximal cytoplasmic microtubule in G2/M cells expressing CFP-Tub1. Error bars denote standard error of the mean. Z-series images were collected for G2/M cells, identified by spindle length and grown in log-phase cultures. Microtubule ends were identified in the CFP-Tub1 image, and intensity measurements were taken from the corresponding plane of the GFP or tdimer2 stack. Asterisks mark results significantly different from wildtype (p < 0.05). Strain numbers and numbers of cells were as follows: A) WT, yJC4147, 369; EB1 null, yJC4177, 124; Num1 null, yJC4316, 301; DHC null, yJC4180, 361; LIS1 null, yJC4178, 210; CLIP-170 null, yJC4182, 124 and NudEL null, yJC4253, 581. B) WT, yJC4147, 369; DHC null, yJC4180, 361; DIC null, yJC5264, 222; DLIC null, yJC4314, 323 and DLC null, yJC43112, 211. C) DHC/WT, yJC2914, 261; DHC/DLC null, yJC4310, 195; DIC/WT, yJC3499, 242; DIC/DLC null, yJC4369, 280; DLIC/WT, yJC3369, 294 and DLIC/DLC null, yJC4367, 270. D) DHC/WT, yJC2914, 261; DHC/DLIC null, yJC4458, 291; DIC/WT, yJC3499, 242; DIC/DLIC null, yJC5266, 235; DLC WT, yJC4555, 166 and DLC/DLIC null, yJC4951, 96. E and F) WT, yJC5652, 23; nip100Δ, yJC5661, 32; pac1Δ, yJC5664, 22; ndl1Δ, yJC5665, 20; bik1Δ, yJC5662, 23; kip2Δ, yJC5663, 13; bim1Δ, yJC5678, 15; dyn1Δ, yJC5669, 16; dyn3Δ, yJC5667, 21; dyn2Δ, yJC5668, 12 and num1Δ, yJC5666, 17.
Loss of dynein heavy chain (DHC) caused a severe loss of plus-end targeting of dynactin (Figure 6A,F). Dynein plus-end targeting depends on the combined action of two pathways - NudEL/Ndl1 working with LIS1/NudF/Pac1 and CLIP-170/Bik1 working with the kinesin Kip2 (7,33). Here, we found that the loss of Ndl1, Pac1, Bik1 or Kip2 had a strong inhibitory effect on the plus-end targeting of dynactin in both assays (Figure 6A,F), consistent with their roles in dynein targeting. Conversely, the amount of dynein at the plus end is increased by loss of the cortical protein Num1 (25,26). Here, loss of Num1 also increased the level of plus-end targeting of dynactin in both assays (Figure 6A,F). All the results are therefore consistent with the dynein content of the plus end being a critical variable for dynactin targeting.
We considered the possibility that the loss of dynein may disrupt the localization of dynactin by destabilizing the complex. However, we found that mutants lacking dynein did not exhibit a noticeable reduction in the levels of dynactin subunits in high-speed supernatants or in the stability of the dynactin complex determined by precipitation of Nip100-TAP (Figure S5).
EB1/Bim1 is a microtubule-interacting protein with moderate targeting to the plus end (34-36). The loss of EB1 caused increased targeting of dynactin to the plus end in the foci-counting assay (Figure 6A) and the fluorescence intensity assay (Figure 6F). The level of dynein at the plus end was also increased in the bim1Δ mutant by a relatively smaller amount, which was statistically significant in the foci-counting assay (Figure 6A) but not the fluorescence intensity assay (Figure 6E). In previous studies, dynein targeting to the plus end was not observed to change in a bim1Δ mutant with less quantitative methods (25,26).
Dynein has four subunits - the heavy, intermediate, light intermediate and light chains. To investigate which subunits are involved in the targeting of dynactin, we examined the localization of p150Glued/Nip100-GFP in null mutant strains (Figure 6). Loss of dynein light chain (DLIC/Dyn2) or light intermediate chain (DLIC/Dyn3) caused a large decrease in plus-end targeting of dynactin assayed by counting foci (Figure 6B) and measuring fluorescence intensity (Figure 6F), with substantially less to no effect on the targeting of the other dynein subunits assayed by foci counting (Figure 6C,D) or fluorescence intensity (Figure 6E). Thus, the DLC and the DLIC have effects on dynactin targeting that are greater than would be expected from their effects on targeting of dynein as a whole, suggesting that they play a primary role.
Loss of DHC or dynein intermediate chain (DIC) caused a large loss of dynactin targeting; however, all dynein chains fail to localize to plus ends in those mutants [Figure 6 and (8)], so the primacy of the DLC and DIC cannot be determined.
Figure 8. Model for the function and localization of dyneindynactin with respect to cytoplasmic microtubules and the cell cortex.
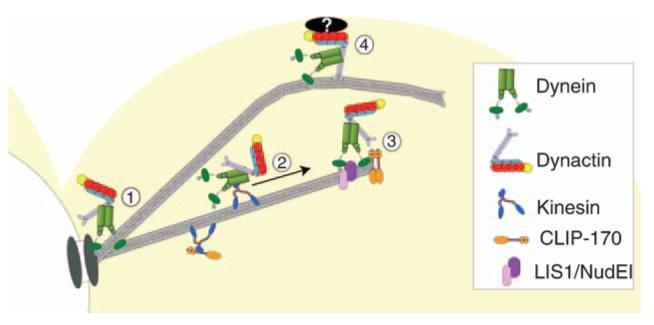
1) Dynactin localizes to cytoplasmic microtubules through its interaction with dynein. 2) Recruitment of the dynein-dynactin complex to microtubule plus ends. 3) Retention of dynein-dynactin complex at plus ends through interactions with CLIP-170/Bik1, LIS1/Pac1 and NudEL/Ndl1. 4) Dynein and dynactin are offloaded to a cortical-binding site, and dynein is activated to power microtubule sliding.
Dynein localization in the absence of dynactin
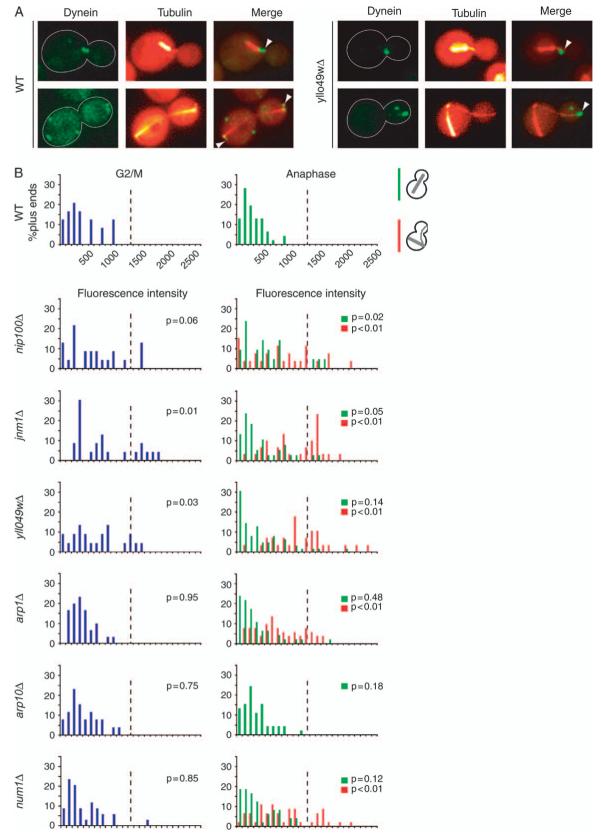
We investigated the contribution of dynactin complex to the function of dynein by examining the localization of dynein in the absence of dynactin. In wild-type cells, dynein is observed at the highly mobile distal plus ends of cytoplasmic microtubules and at stationary foci at the cell cortex. Our working model predicts that dynein is targeted to the plus end, transfers to the cortex on contact, becomes anchored and activated, and then pulls on cytoplasmic microtubules. In previous studies of null mutants lacking the cortical anchor Num1 or certain dynactin subunits (including p150Glued/Nip100, dynamitin/Jnm1 and Arp1), dynein intensity was found to be increased at plus ends (25,26). Here, we found that in several dynactin mutants, including yll049wΔ, dynein was increased in intensity at microtubule plus ends (Figure 7) as expected. Furthermore, dynein was not seen as stationary puncta at the cell cortex, consistent with dynactin being necessary for dynein to transfer from plus ends or to anchor to the cortex.
Figure 7. Dynein accumulation at microtubule plus ends in the absence of dynactin.
A) Images of DHC/Dyn1 tagged with 3GFP in wild-type and yll049wΔ cells expressing CFP-labeled microtubules. Arrowheads mark Dyn1-3GFP located at microtubule plus ends. B) Histograms of Dyn1-3GFP fluorescence intensity at microtubule plus ends in wild-type and mutant cells. The fluorescence intensity of Dyn1-3GFP per cytoplasmic microtubule plus end was determined in G2/M and anaphase cells expressing CFP-Tub1-labeled microtubules. Cell cycle stage was assessed from spindle length. Spindles ∼1-1.25 μm in length were scored as G2/M, and longer spindles were scored as anaphase. Blue bars represent microtubule plus ends in G2/M cells. Green bars represent plus ends of cells with properly aligned anaphase spindles that traverse the bud neck. Red bars represent plus ends of cells with anaphase spindles that are contained entirely within the mother cell and misaligned with respect to the axis of division. The wild-type and arp10Δ strains displayed only correctly positioned anaphase spindles. Z-series images were captured from asynchronous cultures of wild-type (yJC4149), nip100Δ (yJC4145), jnm1Δ (yJC4150), yll049wΔ (yJC4160), arp1Δ (yJC4143), arp10Δ (yJC4547) and num1Δ (yJC4164) strains. Microtubule ends were identified using CFP-Tub1, and intensity measurements were taken on the corresponding plane of the GFP stack using ImageJ (Materials and Methods). For G2/M cells, 22-34 microtubule ends were scored per sample, and for anaphase, 21-62 were scored. The p values were generated by t-tests comparing the result for a given mutant with the wild-type result in either G2/M or anaphase. The dashed line on each chart is drawn at the same position for reference.
If dynein is transferred from the plus end to the cortex and its accumulation is because of a failure of transfer, then one would expect the intensity of plus-end dynein to be greater at later stages of the cell cycle. To test this prediction, we quantified the fluorescence intensity of dynein at every plus end comparing G2/M and anaphase cells. The majority of wild-type and mutant cells exhibited a similar level of Dyn1-3GFP fluorescence intensity during G2/M, however we did detect increased signal in a subset of nip100Δ, jnm1Δ and yll049wΔ cells. During anaphase, the intensity observed in each mutant, with the exception of arp10Δ, was slightly greater than it was in G2/M cells(Figure 7B). In addition, when cells in anaphase were divided into groups where the mitotic spindle was mispositioned in the mother, as opposed to occupying a normal position in the neck, the dynein fluorescence intensity was significantly greater in the misoriented spindle group (Figure 7B, red versus green). This result is consistent with dynein accumulating at the plus end when cytoplasmic microtubules fail to make productive interactions with the cortex.
Cappers of the Arp1 filament
We asked if the caps on the ends of the actin-like Arp1 filament were important for dynactin function in cells. The length of the actin-like filament of purified brain dynactin is characteristically short and constant, with an apparently invariant number of Arp1 subunits per complex (1,12). Purified brain Arp1 can form actin-like filaments that have a similar short length, and the length variation is small but not to the high degree observed for purified dynactin (37). Capping protein is known to be a biochemical component of dynactin complex in vertebrate systems (1); however, in yeast, no evidence links capping protein with dynactin. We tested null mutants lacking Cap1 or Cap2, the α and β subunits of the obligate heterodimer of capping protein. The mutants showed only a small increase in the number of cells with aberrant spindle position, well below the level seen for loss of function of the dynein pathway (Figure S2). The Kar9 pathway also positions the spindle by a mechanism complementary with that of dynein. Kar9 moves microtubule ends along polarized actin cables (38), which are somewhat impaired in capping protein mutants (39), and this may account for the mild phenotype of the capping protein mutants here. Cells lacking dynein and Kar9 function grow extremely poorly with a greatly enhanced defect in spindle position (40). Here, a cap2Δ kar9Δ double mutant resembled a kar9Δ single mutant in assays of growth and spindle position (Figure S2). Thus, capping protein is not important for dynein function.
In Neurospora, the Arp11 homologue ro-7 is important for dynein function (41). In yeast, Arp10 interacts with the pointed end of the Arp1 filament (16). We found that an arp10 null mutant had only a small defect in spindle position, less than that of dynein or kar9Δ mutants (Figure S2), consistent with previous results (11). In addition, an arp10Δ cap2Δ double mutant showed no enhancement of the phenotype. Thus, the presumed cappers of the Arp1 filament are not crucial for dynactin function either alone or together. Arp1 is essential for dynactin function, so the intrinsic ability of Arp1 to regulate its length may be sufficient for function in the context of these assays.
Discussion
In nearly every eukaryotic cell, the dynactin complex and the cytoplasmic dynein motor work to organize the cytosol by manipulating the position of a variety of cargoes. Budding yeast employ dynein and dynactin for a single task - drawing the mitotic spindle across the plane where cytokinesis will occur. Here, we examined the role of dynactin in this process in order to gain insight into the contributions of individual subunits to the structure and function of the complex and to determine how dynactin contributes to the function of dynein.
We performed a systematic characterization of the subunits of the complex, including the newly discovered p24 subunit homologue. Most subunits, but not all, were necessary for dynactin to function with dynein. The p150Glued subunit was necessary and sufficient for targeting dynactin to the plus end. The amount of dynactin complex at the plus end and the SPB was remarkably small, less than five molecules at each location. We found that dynactin was not required to recruit dynein to the microtubule plus end but that dynein was needed for dynactin recruitment. In the absence of dynactin, the amount of dynein at the plus end increased. Dynactin and dynein were both present as stationary foci at the cell cortex, but neither protein appeared at the cortex when the other did not function. Together, the results are consistent with dynactin’s role being to mediate offloading of dynein from the plus end to the cortex or to assist with anchoring or activation of dynein at the cortex.
The function of dynactin
Dynactin appears to follow dynein in the cell, being targeted to the dynamic plus ends of microtubules and to foci anchored on the cell cortex. In the absence of dynactin, dynein is still targeted to the plus end, and its level there increases when the mitotic spindle fails to move. These observations are consistent with a role for dynactin in the transfer of dynein from the plus end to the cortex by direct contact, but the possibility of transfer through the cytoplasm cannot be excluded.
Another potential role for dynactin is to help anchor dynein to the cortex, which would be analogous to an accepted role for dynactin in vertebrates, if the cell cortex is considered as the cargo of the dynein motor. However, in the absence of dynein, dynactin was not seen at the cortex, contrary to the prediction from this hypothesis. Another potential role for dynactin is to promote the activity of dynein at the cortex. In biochemical studies in other systems, dynactin increases the processivity of dynein (42), and the yeast microtubule generally slides along the cortex in one smooth motion (5). If this was the primary role of dynactin, then dynein might be present at the cortex, but inactive, in the absence of dynactin. That is not the case. Thus, our results support a role for dynactin in offloading, and the alternative functions of anchoring and activation are not supported but not excluded.
The subunits of the dynactin complex
We found that the p150Glued, dynamitin, Arp1 and p24 subunits are all essential for the ability of dynactin to function in the dynein pathway, which supports spindle position and thus chromosome segregation. All the subunits localize in similar patterns. The Arp10 subunit was not necessary for function, but it localized and associated with the other subunits.
Our studies of the localization, coimmunoprecipitation and stoichiometry of the subunits are largely consistent with yeast dynactin having a composition and architecture similar to that of vertebrate dynactin (1) and with studies of Arp1 and Arp10 in yeast (16,17). We found that the shoulder complex on the side of the Arp1 filament, near its barbed end, is formed by the trimolecular association of p24, dynamitin and p150. p24 needs dynamitin in order to associate with p150Glued, and p150Glued needs dynamitin and p24 for optimal protein stability in solution.
Furthermore, both dynamitin and p24 were necessary for the Arp1 filament, including Arp10, to interact biochemically with p150Glued. In cells, the absence of any one of the three shoulder proteins caused substantial decreases in the association of Arp1 and Arp10 with plus ends and SPBs. Arp10 depended on Arp1 for association with the complex as expected (16,17). Conversely, the level of Arp1 associated with p150Glued was decreased in the absence of Arp10 by a large amount in the protein precipitation assay and by a small amount in the cell localization assay. This discrepancy between the assays is not unreasonable; the protein precipitation assay should be a more stringent test of subunit affinity for the complex than the cell localization assay because the precipitate is washed with buffer. The cell localization assay results are also consistent with the genetic results that Arp1 is necessary for dynactin function, whereas Arp10 is not. If Arp1 was lost from the complex when Arp10 was removed, then one would expect the arp10 mutant to show loss of dynactin function.
The three subunits that make up the pointed-end subcomplex of brain dynactin - p62, p27 and p25 - have not been identified in budding yeast. A number of genome-wide searches, based on sequence, function and biochemistry, have failed to produce any positive results. In addition, the sedimentation coefficient of yeast dynactin is 15S, while that of brain dynactin is 20S [Figure S4; (1,19)]. Most likely, these subunits simply do not exist in yeast; however, the negative evidence against their existence does have certain limitations. Most important, purification of yeast dynactin complex to near homogeneity has not been achieved, which was critical for defining the composition of brain dynactin (1). In addition, searches for sequence similarity fail to identify several of the yeast dynactin subunits that have been recognized as homologues by other criteria. Finally, if pointed-end capping is dispensable for function, as indicated by the normalcy of the arp10Δ mutant, then genes encoding p62, p27 and p25 would have been missed in functional screens.
In Neurospora, pointed-end complex proteins are present, and they are important for function (41). Strains lacking those proteins have slightly different phenotypes than null mutants deficient in other dynactin components, suggesting that the pointed-end complex may provide for the interaction of dynactin and dynein with specific cargoes, such as membranous organelles. In budding yeast, dynein appears to have no role other than to position the mitotic spindle, so its ability to interact with cargoes other than the cell cortex may have been lost along with pointed-end complex genes.
Capping protein and conventional actin have also been defined as biochemical components of dynactin purified from brain (1). Neither protein has been implicated in the function of dynactin or dynein in yeast or any other system to our knowledge (43). Here, we found no evidence for either protein of biochemical association with dynactin or of function in the dynein pathway. However, capping protein and actin are present in relatively high concentrations in cells, so their biochemical presence in dynactin complex may have been difficult to detect. Likewise, dynein-related phenotypes may have been masked by the roles of the proteins in the function of the conventional actin cytoskeleton.
To estimate the number of molecules of dynactin that are targeted to the plus end, we quantified the fluorescence of tagged subunits using Cse4 as an internal standard, a novel approach developed for the study of the kineto-chore (28). The fluorescence intensity values for Cse4 were well above the level of noise in the system, and the distribution of those values was unimodal as expected. The values for the dynactin subunits were much broader and extended to zero, suggesting a small and perhaps variable number of molecules. Our data are consistent with plus ends containing at most several, generally less than five, molecules of dynactin if one assumes that yeast dynactin contains four copies of the dynamitin subunit. The analysis identified plus ends and SPBs from images of microtubules, and all the plus ends and SPBs were included. Based on the level of noise, some plus ends and SPBs appear to contain zero molecules of dynactin. The analysis offers little information about the relative stoichiometry of subunits within the complex because of the wide range of the values. Additional biochemical and cell biological studies will be necessary for definitive conclusions about stoichiometry.
Subcomplexes
One interesting question for the biology of dynactin is whether subunits form subcomplexes with discrete functions in cells. For vertebrate dynactin complex, dynamitin and p24 form a stable complex in vitro, with stoichiometry identical to that of the native shoulder-sidearm complex (30,31). Here, sedimentation of cell extracts from wild-type cells revealed pools of dynamitin, p24 and Arp10 that did not sediment with dynactin, while all of p150Glued and some of each of those subunits sedimented as one large complex. In mutants, p150Glued was able to localize to microtubules on its own, and dynamitin remained at microtubule ends in the absence of Arp1. Biochemically, the interaction of dynamitin and p24 with p150Glued was not disturbed by the loss of Arp1.
Dependence between dynactin and dynein
In many systems, including yeast, dynein and dynactin localize to microtubule plus ends and the SPB/centrosome. In yeast, plus-end tracking of dynein depends on NudE, LIS1 and CLIP-170 (25,26,33).
Here, we found that the plus-end tracking of dynactin depended on dynein and not vice versa. Dependency between dynein and dynactin varies quite substantially in different systems. In Aspergillus, dynein and dynactin depend on each other for plus-end targeting, based on localization in cells (44), and in Neurospora, they depend on each other for interaction with membranes, based on biochemical fractionation experiments (45). In Ustilago maydis, a filamentous fungus where offloading of dynein from the plus end to the cortex has been visualized directly (46), dynactin and dynein are targeted to plus ends, and loss of dynactin leads to a loss of dynein (47).
In vertebrate systems, dynactin has been found to be responsible for targeting of dynein to multiple locations, including kinetochores, membranous organelles and microtubules (48). In COS-7 cells, dynactin is present on the distal segments of interphase microtubules, but dynein was not (49), suggesting that dynactin targeting did not depend on dynein. In this study, dynein was observed to associate with the distal segments at low temperatures, consistent with dynactin targeting dynein. Also in mammalian cells, dynactin was found to associate with pronuclei independent of dynein during fertilization (50).
In Drosophila oogenesis, dynactin depends on dynein (51), as seen here for yeast. Dynein heavy chain mutations that impaired dynein function also disrupted dynactin localization, as did mutations in other genes that disrupted dynein function. Dynein localization in the absence of dynactin was not examined in that study; however, a biochemical analysis of dynein association with membranes from Drosophila larval brains found that dynactin was not required (52). These results are consistent with our conclusions regarding plus-end targeting in yeast that dynactin depends on dynein but dynein does not depend on dynactin.
We investigated which subunits were important for the dynein-dependent targeting of dynactin, and we found that p150Glued was the only subunit of dynactin that was both necessary and sufficient for targeting. In COS7 cells, p150Glued is able to localize normally to distal segments of microtubules in the absence of Arp1, as induced by dynamitin overexpression (49); however, as noted above, dynein was not accumulated at these locations. In terms of dynein, we found that the DLC and DLIC were necessary for plus-end targeting of dynactin. Null mutants lacking these chains still showed targeting of DHC and DIC. However, mutants lacking DHC or DIC showed a failure of all dynein subunits to target as well as dynactin, so the individual roles of the DHC and DIC could not be assessed. In mammals and Drosophila, p150Glued interacts biochemically with DIC (53,54), and the interaction is known to be functionally relevant in Drosophila (55).
Interactions among p150Glued, EB1 and CLIP-170
In vertebrates, EB1 and CLIP-170 have been shown to be important for the recruitment of dynactin to microtubule plus ends. In HeLa cells, siRNA knockdown of EB1 or CLIP-170 caused a loss of p150Glued from microtubule plus ends (56). Biochemically, EB1 was found to associate with p150Glued in extracts of cultured human cells (57), and EB1 interacted with p150Glued in experiments with purified recombinant proteins (58). In the latter study, EB1 also interacted directly with CLIP-170, and their interaction excluded the interaction of EB1 with p150Glued.
Here, in budding yeast, EB1 was not necessary for dynactin recruitment to plus ends. Moreover, loss of EB1 led to an increase in the amount of dynactin at the plus end, suggesting that EB1’s presence inhibits dynactin recruitment. The CLIP-170 homologue Bik1 was important for dynactin targeting to plus ends, but that effect could be explained by the decrease in dynein targeting caused by the loss of Bik1.
The difference between vertebrate and yeast systems in this regard may reflect differences in the CAP-Gly domain of p150Glued. The interaction between p150Glued and EB1 in vertebrates involves amino acid residues Ala49, Leu51, Thr54 and Lys56 in the β2-β3 loop of the CAP-Gly domain (59,60). An alignment of yeast and metazoan p150Glued sequences showed that these residues are conserved across vertebrates, but not in yeast, where the residues are Glu, Gln, Lys and Ile, respectively (not shown).
Model for the dynamics of dynactin-dynein
Our working model for the dynamic localization of dynactin and dynein is depicted in Figure 8. Dynein and dynactin localize to microtubules, concentrating at the plus end and the SPB. Dynactin depends on dynein for plus-end targeting, and dynein depends on CLIP-170, LIS1 and NudEL. Whether the initial recruitment of dynein to the microtubule occurs at the plus or minus ends or along the length of the filament is yet unresolved. We know that the localization of dynein to microtubules is lost when the microtubule-binding domain within DHC is deleted (Moore unpublished data).
Little is known about how dynein is targeted to these locations. Dynein is primarily a minus-end-directed motor in vitro, so motor activity may promote accumulation to minus ends near the SPB. However, the plus-end targeting of dynein may be explained by the motor walking in the opposite direction, which has been seen for brain dynein (61,62). Alternatively, other plus-end proteins may hold dynein, or the plus-end kinesin Kip2 may carry dynein (7). Obviously, much remains to be learned about dynein targeting.
In the model, dynein at the plus end moves to the cortex when a dynamic plus end encounters a focus of the cortical anchor Num1 (9,10,25,26). Here, dynactin was also observed at cortical dynein foci, raising the possibility that dynactin may be important for controlling the motor activity of dynein during the microtubule-sliding process.
Materials and Methods
Chemicals and reagents were from Fisher Scientific and Sigma-Aldrich unless stated otherwise.
Yeast strains and manipulation
General yeast manipulation, media and transformation were performed by standard methods (63). Strains are listed in Table S1. Gene-deletion and epitope-tag strains were constructed by polymerase chain reaction product-mediated transformation. Tags were placed at the C-terminus of the protein with a nine-residue alanine linker, and fusion proteins were functional in assays for dynein function in haploids that expressed only the tagged version of the protein. For ARP1, fusions did not rescue haploid null mutations, so we tagged one endogenous copy of the gene in a diploid strain following the approach used to visualize conventional actin/Act1 with a non-rescuing GFP fusion (24). We integrated tdimer2 (12), which consists of two tandem repeats of DsRed separated by a 12-aa linker (64). The tdimer2 does not aggregate, and it matures with a half-time of less than 1 h.
Sequence analysis
Homologues of dynactin p24, p25 and p27 subunits, also known as dynactin3, dynactin5 and dynactin6 (1), were identified in multiple organisms by BLAST and text searching through NCBI Entrez. Their amino acid sequences and that of Yll049w were aligned with Clustalx/CLUSTALW 1.83.1 (European Bioinformatics Institute, Cambridge, UK, ftp.ebi.ac.uk) and bootstrap analysis. Unrooted phylogenetic trees were produced with Drawtree in Phylip3.66 (Felsenstein J, 2005, University of Washington, Seattle, evolution.gs.washington.edu/phylip.html). Secondary structure prediction for multiple sequences was performed with a neural network training algorithm in the program Jnet, online at www.compbio.dundee.ac. uk/∼www-jpred (65). To look for the β-helix fold, we used the program Betawrap, online at betawrap.lcs.mit.edu (66), and the program phyre, online at http://www.sbg.bio.ic.ac.uk/phyre/ (67).
Fluorescence microscopy
Images were collected on an Olympus IX70 inverted fluorescence microscope with a ×100 N.A. 1.35 oil objective lens and a CoolSNAP HQ camera (Roper Scientific) using qed software (Media Cybernetics). Living cells from an asynchronous culture at early log phase were suspended in non-fluorescent medium and placed on an agarose pad as described (68). Microtubules were visualized with CFP-Tub1 expressed from the tub1 promoter integrated at the ura3 locus as described (25) using plasmid pAFS125C, a gift from D. Beach and K. Bloom (University of North Carolina, Chapel Hill). Dual fluorescence images were collected with an 86002bs v1 beam splitter cube (Chroma) to capture fluorescence from GFP and CFP sequentially. To image Arp1-tdimer2 CFP-Tub1 cells, we used a rhodamine filter cube and a CFP cube in sequence. Fluorescence intensities of microtubule plus ends and SPBs were measured and corrected for background fluorescence with ImageJ [(rsb.info.nih.gov/ij/, Wayne Rasband, National Institutes of Health (NIH)]. Pixels immediately adjacent to the region of interest were used for background subtraction as described (29), with a macro written by Dennis Chuang and Brian Galletta in our laboratory.
Immunoprecipitation
Cells from a 20-mL overnight culture were suspended in 1 mL of cold lysis buffer (50 mm Tris-HCl pH 8.0, 150 mm NaCl, 2 mm MgCl2, 0.2% Tween-20 and 0.5 m m DTT) with 20 μL yeast protease inhibitor cocktail, 10 mLof 100 mm phenylmethylsulphonyl fluoride (Sigma) and 0.5 mL acid-washed glass beads. Cells were lysed in a chilled minibeadbeater™ (Cole-Parmer) for 8 × 1 min, resulting in >95% lysis by phase contrast microscopy. Lysates were spun in a tabletop microcentrifuge at 13 200 r.p.m. for 15 min at 4°C, and the supernatant was spun at 50 000 r.p.m. in a Beckman TLA100.3 rotor (103 320 × g) for 1 h at 4°C. The protein content of the resulting high-speed supernatant was measured by Bradford assay (BioRad), and 2 mg of protein was mixed with 30 μL immunoglobulin G (IgG) Sepharose 6 fast-flow beads (Amersham Biosciences) for 2 h at 4°C. Beads were washed three times with 1 mL washing buffer (50 mm Tris-HCl pH 8.0, 250 mm NaCl, 2 mm MgCl2 and 0.4% Tween-20) and three times with 1 mL lysis buffer without DTT. Bound proteins were eluted from the beads by adding 50 μL sodium dodecyl sulfate (SDS) loading buffer without reducing agent (to minimize dissociation of IgG chains) and boiling for 5 min. The supernatant was collected, and 2.5 μL β-mercaptoethanol was added.
For immunoblots, a pilot assay was performed to determine loading amounts. Ten microliters of each sample was applied to a 10% SDS-polyacrylamide gel, electrophoresed and transferred to a nitrocellulose membrane. The membrane was blocked with TTBS (50 mm Tris-HCl pH 8.0, 150 mm NaCl and 0.05% Tween-20) with 5% non-fat milk. The membrane was first probed with rabbit peroxidase-anti-peroxidase (Sigma) to visualize Nip100-TAP (the protein A region of the TAP tag binds rabbit IgG). The amount of each sample loaded on the gel was then adjusted so that every lane contained a similar amount of Nip100.
Antibodies and dilutions for immunoblots were as follows: rabbit anti-hemagglutinin (Invitrogen), 1:10 000, or rabbit peroxidase-anti-peroxidase, 1:200, for TAP-tagged Nip100; mouse monoclonal antibody 9E10 (Covance), 1:2000, for Arp10-myc and Yll049w-myc; rabbit anti-Cdc3 (a gift from Mark Longtine, Washington University, St Louis, MO, USA), 1:5000; rabbit anti-Jnm1, Unc52, (a gift from Kelly Tatchell, LSU Medical Center, Shreveport, LA, USA); affinity-purified rabbit anti-Cap2, R12 from our laboratory (69), 1:5000; rabbit anti-Arp1, R31 from our laboratory (2), 1:2000; goat anti-yeast actin/Act1, G2 from our laboratory (70), 1:5000, and affinity-purified rabbit anti-Yll049w, R1783 produced for this study, 1:1000. Secondary antibodies were horseradish peroxidase-conjugated goat anti-mouse (Jackson ImmunoResearch), goat anti-rabbit (Biosource) and donkey anti-goat (Santa Cruz Biotechnology) at 1:10 000. Signals were detected by enhanced chemiluminescence.
To produce antibodies against Yll049w, rabbits were immunized with purified His-tagged Yll049w protein and expressed in bacteria with pHis-Parallel2 (71). Antibodies were affinity purified from antisera with immunoblots (72). Affinity-purified antibody did not detect Yll049w at its endogenous level, so we constructed a yeast strain that overexpressed Yll049w from a GAL1/10 promoter to test the specificity of the antibody. In whole-cell extracts from this strain, induced with galactose, only Yll049w was detected by immunoblot.
Supplementary Material
Movie S1: Dynactin as a stationary focus at the cortex. A cell expressing Jnm1-3GFP and CFP-Tub1 shows a stationary cortical focus in the lower left. In order to visualize the spindle and cytoplasmic microtubules, a GFP filter set that allowed CFP-Tub1 crossover at a low level was employed. Images were captured on a wide-field fluorescence microscope at 5-second intervals for 4 min, and the movie plays at six frames per second, thus 30-fold faster than real time. The strain is yJC5260.
A) Coprecipitation of dynactin subunits. Yll049w precipitated with TAP-tagged p150Glued/Nip100, as did the dynactin subunits dynamitin/Jnm1, Arp1 and Arp10. The septin Cdc3 served as a negative control. For Western blot detection, antibodies against endogenous proteins were used for Yll049w, Jnm1, Arp1 and Cdc3. Arp10 was tagged with Myc and detected with anti-Myc. The specific bands for Nip100, Arp1 and Yll049w are only seen in the final lane on the right, while Jnm1 and Arp10 are also observed in the total and supernatant lanes, owing to different sensitivities of the antibodies. B) Phylogenetic tree of Yll049w with p24, p25 and p27 subunits from various species. The bootstrap value for the Yll049w branch was 100%. Abbreviations: Mm, mouse; Hs, human; Gg, chicken; Xl, Xenopus; Dr, zebra fish; Sp, sea urchin; Sj, Schistosoma japonicum; Dm, Drosophila melanogaster; Dd, Dictyostelium discoideum; Nc, Neurospora crassa. C) Sequence alignment of Yll049w with two p24 homologues from human, Hs1, and D. melanogaster, Dm. Identity with the majority sequence is indicated by a period. Secondary structure features common to the three sequences are indicated in the bottom row, where H is helical and E is extended.
A) GFP-labeled microtubules in yll049wΔ mutant cells after a 20-h incubation at12°C. Some mother cells contained long anaphase spindles (solid outlines) or two or more distinct microtubule organizing centers (MTOCs) (dashed outline). B) The percentage of cells with either multiple MTOCs or long anaphase spindles located within the mother cell after incubation at 12°C. Strains: wild-type yJC3638; dyn1Δ, yJC4401; nip100Δ, yJC5740; jnmΔ, yJC5741; yll049wΔ, yJC5659; arp1Δ, yJC5742; arp10Δ, yJC5743; cap1Δ, yJC5745; cap2Δ, yJC5739; cap2Δ arp10Δ, yJC5744; kar9Δ, yJC5677 and cap2Δ kar9Δ, yJC5738. For each strain, n > 500 cells. Values are the means of five sets of 50 cells, and error bars are standard error of those means.
A) Plus ends. B) SPBs. Histograms of fluorescence intensity per focus for dynactin components at one plus end or SPB are shown. Plus ends and SPBs were identified in budded cells using CFP-Tub1, and fluorescence intensity values were measured on the corresponding GFP images. As a negative control, GFP fluorescence intensity at SPBs was measured using the identical procedure in cells expressing CFP-Tub1 but not GFP-tagged proteins. Strain numbers: Jnm1-3GFP, yJC4198; Yll049w-3GFP, yJC3891; Nip100-3GFP, yJC4147; Arp10-3GFP, yJC5389 and CFP-Tub1 alone, yJC3883. Yll049w-3GFP values in (B) were collected from the strains yJC3891, yJC3892, yJC4805, yJC5298 and yJC5299.
A) Rate-zonal sedimentation of dynactin subunits. High-speed supernatant prepared from wild-type cells expressing Nip100-TAP, Arp10-myc and Yll049w-myc was centrifuged across a 5-20% sucrose gradient. Twelve equal fractions were collected and analyzed by immunoblotting. Fraction 1 represents the top of the gradient. B) Sedimentation of dynactin subunits from arp1Δ mutant cells. C) Sedimentation of thyroglobulin, catalase and yeast alcohol dehydrogenase standards. D) The S value of standards plotted against sedimentation. Strains: wild-type, yJC5814 and arp1Δ, yJC5815.
p150glued/Nip100-TAP was immunoprecipitated from wild-type cells or dyn1Δ mutants. Immunoblots of the S100 fraction and precipitates are shown. Cdc3 was used as a loading control. Strains: wild-type, yJC5814 and dyn1Δ, yJC6054.
Acknowledgments
We thank Kelly Tatchell, Mark Longtine and Kerry Bloom for strains, plasmids and antibodies. This work was supported by NIH grants GM47337 and GM69895 to J. A. C. J. L. was supported by a postdoctoral fellowship AHA40390 from the American Heart Association. J. K. M. was supported by a postdoctoral fellowship from the Molecular Oncology program of the Siteman Cancer Center at Washington University and funded by NIH T-32-CA113275. We thank Scott Nelson and Mark Longtine for advice and suggestions.
References
- 1.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 2.Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- 3.Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, vanVliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci U S A. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci U S A. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho P, Gupta ML, Jr, Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Lee WL, Kaiser MA, Cooper JA. The offloading model for dynein function: differential function of motor subunits. J Cell Biol. 2005;168:201–207. doi: 10.1083/jcb.200407036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farkasovsky M, Kuntzel H. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J Cell Biol. 2001;152:251–262. doi: 10.1083/jcb.152.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heil-Chapdelaine RA, Oberle JR, Cooper JA. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol. 2000;151:1337–1344. doi: 10.1083/jcb.151.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the Arp1 minifilament pointed end. J Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai H, Narita A, Schroer TA, Maeda Y. Two-dimensional averaged images of the dynactin complex revealed by single particle analysis. J Mol Biol. 2006;359:833–839. doi: 10.1016/j.jmb.2006.03.071. [DOI] [PubMed] [Google Scholar]
- 13.Poch O, Winsor B. Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast. 1997;13:1053–1058. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1053::AID-YEA164>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Goodson HV, Hawse WF. Molecular evolution of the actin family. J Cell Sci. 2002;115:2619–2622. doi: 10.1242/jcs.115.13.2619. [DOI] [PubMed] [Google Scholar]
- 15.Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. 2005;16:5736–5748. doi: 10.1091/mbc.E05-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark SW, Rose MD. Arp10p is a pointed-end-associated component of yeast dynactin. Mol Biol Cell. 2006;17:738–748. doi: 10.1091/mbc.E05-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark SW, Rose MD. Alanine scanning of Arp1 delineates a putative binding site for Jnm1/dynamitin and Nip100/p150glued. Mol Biol Cell. 2005;9:3999–4012. doi: 10.1091/mbc.E05-02-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMillan JN, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahana JA, Schlenstedt G, Evanchuk DM, Geiser JR, Hoyt MA, Silver PA. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol Biol Cell. 1998;9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbacho I, Olivero I, Hernandez LM. A genome-wide screen for Saccharomyces cerevisiae nonessential genes involved in mannosyl phosphate transfer to mannoprotein-linked oligosaccharides. Fungal Genet Biol. 2005;42:773–790. doi: 10.1016/j.fgb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Ye P, Peyser BD, Pan X, Boeke JD, Spencer FA, Bader JS. Gene function prediction from congruent synthetic lethal interactions in yeast. Mol Syst Biol. 2005;1:2005–0026. doi: 10.1038/msb4100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parisi G, Fornasari MS, Echave J. Dynactins p25 and p27 are predicted to adopt the LbetaH fold. FEBS Lett. 2004;562:1–4. doi: 10.1016/S0014-5793(04)00165-6. [DOI] [PubMed] [Google Scholar]
- 24.Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci U S A. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 27.Grava S, Schaerer F, Faty M, Philippsen P, Barral Y. Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell. 2006;10:425–439. doi: 10.1016/j.devcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier KC, Godfrey JE, Echeverri CJ, Cheong FK, Schroer TA. Dynamitin mutagenesis reveals protein-protein interactions important for dynactin structure. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00702.x. (Online Accepted Articles). doi:10.1111/j.1600-0854.2008.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melkonian KA, Maier KC, Godfrey JE, Rodgers M, Schroer TA. Mechanism of dynamitin-mediated disruption of dynactin. J Biol Chem. 2007;282:19355–19364. doi: 10.1074/jbc.M700003200. [DOI] [PubMed] [Google Scholar]
- 32.Minke PF, Lee IH, Tinsley JH, Bruno KS, Plamann M. Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol Microbiol. 1999;32:1065–1076. doi: 10.1046/j.1365-2958.1999.01421.x. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muhua L, Adames NR, Murphy MD, Shields CR, Cooper JA. A cytokinesis checkpoint requiring the yeast homolog of an APC-binding protein. Nature. 1998;393:487–491. doi: 10.1038/31014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz K, Richards K, Botstein D. BIM1 encodes a microtubule-binding protein in yeast. Mol Biol Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tirnauer JS, O'Toole E, Berrueta L, Bierer BE, Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bingham JB, Schroer TA. Self-regulated polymerization of the actin-related protein Arp1. Curr Biol. 1999;9:223–226. doi: 10.1016/s0960-9822(99)80095-5. [DOI] [PubMed] [Google Scholar]
- 38.Hwang E, Kusch J, Barral Y, Huffaker TC. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amatruda JF, Gattermeir DJ, Karpova TS, Cooper JA. Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution and polarized secretion in yeast. J Cell Biol. 1992;119:1151–1162. doi: 10.1083/jcb.119.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haarer BK, Helfant AH, Nelson SA, Cooper JA, Amberg DC. Stable preanaphase spindle positioning requires Bud6p and an apparent interaction between the spindle pole bodies and the neck. Eukaryot Cell. 2007;5:797–807. doi: 10.1128/EC.00332-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee IH, Kumar S, Plamann M. Null mutants of the neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol Biol Cell. 2001;12:2195–2206. doi: 10.1091/mbc.12.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 43.Heil-Chapdelaine RA, Tran NK, Cooper JA. Dynein-dependent movements of the mitotic spindle in Saccharomyces cerevisiae do not require filamentous actin. Mol Biol Cell. 2000;11:863–872. doi: 10.1091/mbc.11.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Li S, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell. 2003;14:1479–1488. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, Zhou Y, Plamann M. Dynactin-membrane interaction is regulated by the C-terminal domains of p150(Glued) EMBO Rep. 2001;2:939–944. doi: 10.1093/embo-reports/kve202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fink G, Schuchardt I, Colombelli J, Stelzer E, Steinberg G. Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J. 2006;25:4897–4908. doi: 10.1038/sj.emboj.7601354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallee RB, Vaughan KT, Echeverri CJ. Targeting of cytoplasmic dynein to membranous organelles and kinetochores via dynactin. Cold Spring Harb Symp Quant Biol. 1995;60:803–811. doi: 10.1101/sqb.1995.060.01.086. [DOI] [PubMed] [Google Scholar]
- 49.Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci. 1999;112:1437–1447. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- 50.Payne C, Rawe V, Ramalho-Santos J, Simerly C, Schatten G. Preferentially localized dynein and perinuclear dynactin associate with nuclear pore complex proteins to mediate genomic union during mammalian fertilization. J Cell Sci. 2003;116:4727–4738. doi: 10.1242/jcs.00784. [DOI] [PubMed] [Google Scholar]
- 51.McGrail M, Gepner J, Silvanovich A, Ludmann S, Serr M, Hays TS. Regulation of cytoplasmic dynein function in vivo by the Drosophila glued complex. J Cell Biol. 1995;131:411–425. doi: 10.1083/jcb.131.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haghnia M, Cavalli V, Shah SB, Schimmelpfeng K, Brusch R, Yang G, Herrera C, Pilling A, Goldstein LS. Dynactin is required for coordinated bidirectional motility, but not for dynein membrane attachment. Mol Biol Cell. 2007;18:2081–2089. doi: 10.1091/mbc.E06-08-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- 54.Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and Pl50(Glued) J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boylan K, Serr M, Hays T. A molecular genetic analysis of the interaction between the cytoplasmic dynein intermediate chain and the glued (dynactin) complex. Mol Biol Cell. 2000;11:3791–3803. doi: 10.1091/mbc.11.11.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watson P, Stephens DJ. Microtubule plus-end loading of p150(Glued) is mediated by EB1 and CLIP-170 but is not required for intracellular membrane traffic in mammalian cells. J Cell Sci. 2006;119:2758–2767. doi: 10.1242/jcs.02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berrueta L, Tirnauer JS, Schuyler SC, Pellman D, Bierer BE. The APC-associated protein EB1 associates with components of the dynactin complex and cytoplasmic dynein intermediate chain. Curr Biol. 1999;9:425–428. doi: 10.1016/s0960-9822(99)80190-0. [DOI] [PubMed] [Google Scholar]
- 58.Ligon LA, Shelly SS, Tokito MK, Holzbaur EL. Microtubule binding proteins CLIP-170, EB1, and p150Glued form distinct plus-end complexes. FEBS Lett. 2006;580:1327–1332. doi: 10.1016/j.febslet.2006.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayashi I, Wilde A, Mal TK, Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol Cell. 2005;19:449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 60.Honnappa S, Okhrimenko O, Jaussi R, Jawhari H, Jelesarov I, Winkler FK, Steinmetz MO. Key interaction modes of dynamic +TIP networks. Mol Cell. 2006;23:663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 61.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by Tau. Science. 2008 doi: 10.1126/science.1152993. Epub ahead of print. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 63.Amberg DC, Burke D, Strathern JN. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. [Google Scholar]
- 64.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 66.Bradley P, Cowen C, Menke M, King J, Berger B. Predicting the Beta-Helix Fold from Protein Sequence Data; RECOMB; the Fifth Annual International Conference on Computational Molecular Biology; ACM Press, New York, NY. 2001; 2001. [DOI] [PubMed] [Google Scholar]
- 67.Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 68.Waddle JA, Karpova TS, Waterston RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim K, Yamashita A, Wear MA, Maeda Y, Cooper JA. Capping protein binding to actin in yeast: biochemical mechanism and physiological relevance. J Cell Biol. 2004;164:567–580. doi: 10.1083/jcb.200308061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karpova TS, Lepetit MM, Cooper JA. Mutations that enhance the cap2 null mutant phenotype in Saccharomyces cerevisiae affect the actin cytoskeleton, morphogenesis and pattern of growth. Genetics. 1993;135:693–709. doi: 10.1093/genetics/135.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 72.Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- 73.Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: Dynactin as a stationary focus at the cortex. A cell expressing Jnm1-3GFP and CFP-Tub1 shows a stationary cortical focus in the lower left. In order to visualize the spindle and cytoplasmic microtubules, a GFP filter set that allowed CFP-Tub1 crossover at a low level was employed. Images were captured on a wide-field fluorescence microscope at 5-second intervals for 4 min, and the movie plays at six frames per second, thus 30-fold faster than real time. The strain is yJC5260.
A) Coprecipitation of dynactin subunits. Yll049w precipitated with TAP-tagged p150Glued/Nip100, as did the dynactin subunits dynamitin/Jnm1, Arp1 and Arp10. The septin Cdc3 served as a negative control. For Western blot detection, antibodies against endogenous proteins were used for Yll049w, Jnm1, Arp1 and Cdc3. Arp10 was tagged with Myc and detected with anti-Myc. The specific bands for Nip100, Arp1 and Yll049w are only seen in the final lane on the right, while Jnm1 and Arp10 are also observed in the total and supernatant lanes, owing to different sensitivities of the antibodies. B) Phylogenetic tree of Yll049w with p24, p25 and p27 subunits from various species. The bootstrap value for the Yll049w branch was 100%. Abbreviations: Mm, mouse; Hs, human; Gg, chicken; Xl, Xenopus; Dr, zebra fish; Sp, sea urchin; Sj, Schistosoma japonicum; Dm, Drosophila melanogaster; Dd, Dictyostelium discoideum; Nc, Neurospora crassa. C) Sequence alignment of Yll049w with two p24 homologues from human, Hs1, and D. melanogaster, Dm. Identity with the majority sequence is indicated by a period. Secondary structure features common to the three sequences are indicated in the bottom row, where H is helical and E is extended.
A) GFP-labeled microtubules in yll049wΔ mutant cells after a 20-h incubation at12°C. Some mother cells contained long anaphase spindles (solid outlines) or two or more distinct microtubule organizing centers (MTOCs) (dashed outline). B) The percentage of cells with either multiple MTOCs or long anaphase spindles located within the mother cell after incubation at 12°C. Strains: wild-type yJC3638; dyn1Δ, yJC4401; nip100Δ, yJC5740; jnmΔ, yJC5741; yll049wΔ, yJC5659; arp1Δ, yJC5742; arp10Δ, yJC5743; cap1Δ, yJC5745; cap2Δ, yJC5739; cap2Δ arp10Δ, yJC5744; kar9Δ, yJC5677 and cap2Δ kar9Δ, yJC5738. For each strain, n > 500 cells. Values are the means of five sets of 50 cells, and error bars are standard error of those means.
A) Plus ends. B) SPBs. Histograms of fluorescence intensity per focus for dynactin components at one plus end or SPB are shown. Plus ends and SPBs were identified in budded cells using CFP-Tub1, and fluorescence intensity values were measured on the corresponding GFP images. As a negative control, GFP fluorescence intensity at SPBs was measured using the identical procedure in cells expressing CFP-Tub1 but not GFP-tagged proteins. Strain numbers: Jnm1-3GFP, yJC4198; Yll049w-3GFP, yJC3891; Nip100-3GFP, yJC4147; Arp10-3GFP, yJC5389 and CFP-Tub1 alone, yJC3883. Yll049w-3GFP values in (B) were collected from the strains yJC3891, yJC3892, yJC4805, yJC5298 and yJC5299.
A) Rate-zonal sedimentation of dynactin subunits. High-speed supernatant prepared from wild-type cells expressing Nip100-TAP, Arp10-myc and Yll049w-myc was centrifuged across a 5-20% sucrose gradient. Twelve equal fractions were collected and analyzed by immunoblotting. Fraction 1 represents the top of the gradient. B) Sedimentation of dynactin subunits from arp1Δ mutant cells. C) Sedimentation of thyroglobulin, catalase and yeast alcohol dehydrogenase standards. D) The S value of standards plotted against sedimentation. Strains: wild-type, yJC5814 and arp1Δ, yJC5815.
p150glued/Nip100-TAP was immunoprecipitated from wild-type cells or dyn1Δ mutants. Immunoblots of the S100 fraction and precipitates are shown. Cdc3 was used as a loading control. Strains: wild-type, yJC5814 and dyn1Δ, yJC6054.