Abstract
To identify key regulators of subminimum inhibitory concentration (sub-MIC) antibiotic response in the Pasteurella multocida proteome, we applied systems approaches. Using 2D-LC-ESI-MS2, we achieved 53% proteome coverage. To study the differential protein expression in response to sub-MIC antibiotics in the context of protein interaction networks, we inferred P. multocida Pm70 protein interaction network from orthologous proteins. We then overlaid the differential protein expression data onto the P. multocida protein interaction network to study the bacterial response. We identified proteins that could enhance antimicrobial activity. Overall compensatory response to antibiotics was characterized by altered expression of proteins involved in purine metabolism, stress response, and cell envelope permeability.
1. Introduction
The use of antibiotics is continually being challenged by the emergence of resistant strains of bacteria [1]. Resistance has resulted both from mutations associated with bacterial DNA replication and from horizontal gene transfer [2, 3]. However, while antibiotic resistance is increasing, progress in the discovery of new antimicrobial compounds is not [4]. The current trends in drug resistance and drug discovery could result in future crises in treating microbial infections.
Currently, antibiotic therapy is based on achieving and exceeding a minimum inhibitory concentration (MIC) for a sufficient amount of time in infected tissues [5]. The ability to surpass and exceed greater than MIC in target tissues is influenced by the susceptibility of the pathogen to a given antibiotic and the ability of the antibiotic to partition into the target tissue. However, when faced with an infection caused by an antibiotic-resistant bacterial strain, it may not be possible to surpass the MIC in the target tissue. In this situation, one therapeutic option is treatment with a sub-MIC concentration of the antibiotic. Sub-MIC antibiotic therapies can, however, lead to treatment failure and antibiotic resistance [2]. Therefore, careful evaluation of sub-MIC effects on bacterial physiology is needed prior to therapeutic use of sub-MICs.
Pasteurella multocida is an important Gram-negative zoonotic respiratory bacterial pathogen with a broad host range [6–10], and it is a particularly good model organism to study antibiotic effects because it has a Gram-negative envelope that is permeable to hydrophobic molecules (including antibiotics) [11, 12]. Therefore, investigating antibiotic effects is possible with fewer confounding effects caused by permeability differences between antibiotics.
Sub-MICs of antibiotics model the conditions that bacteria face in wild environments and have evolved to survive [13]. Sub-MICs of antibiotics cause defined effects on, and responses in, bacterial physiology. For example, although sub-MICs of chlortetracycline reduced virulence factor expression in the bovine respiratory pathogen Mannheimia haemolytica [14] they caused secondary or “nontarget” effects, in addition to their primary target effects, in Pasteurella multocida [15]. Some of these secondary effects may enhance the primary activity of the antibiotic, while others, such as an increased expression of recombinase A (RecA) in response to enrofloxacin, are apparent compensatory mechanisms.
Molecular systems level analysis including interaction networks can identify not only direct and indirect global responses of bacterial genes to sub-MICs, but may also identify key elements in genetic networks (regulatory hubs) that, when altered, change the fundamental properties of these networks [16]. Identification of key targets in the context of sub-MIC antibiotic effects on virulence factors necessary for pathogenesis, and leading to antimicrobial resistance, can help assess the benefit versus risk of sub-MIC therapeutic usage. Systems approaches can also identify key molecules as potential targets for novel therapeutic combinations.
Here, we report a systems analysis of the Pasteurella multocida Pm70 response to sub-MICs of three different antibiotics that differ in their mode of action: amoxicillin (AMX), which is a cell wall biosynthesis inhibitor; chlortetracycline (CTC), which inhibits protein synthesis; enrofloxacin (ENR), which is a quinolone that inhibits DNA gyrase and DNA topoisomerase IV [17]. We used three different classes of antibiotics to find common themes to antibiotic effects and the bacterial adaptive response. Of these three antibiotics, resistant strains of P. multocida are described for penicillins and tetracyclines [18]. Currently, no fluoroquinolone resistant P. multocida are described; however, resistance to this antibiotic is documented for other Gram-negative pathogens [19]. Using a liquid chromatography tandem mass spectrometric proteomics approach, we identified 53% of the predicted P. multocida proteome expressed under our experimental conditions. The protein expression data was analyzed in the context of interaction networks using Pathway Studio (Ariadne, Rockville, Md, USA), which has canonical bacterial interaction networks. Overall compensatory response of P. multocida to sub-MICs was characterized by altered expression of virulence factors; proteins involved in purine metabolism, stress response, and cell envelope permeability.
2. Materials and Methods
2.1. Pasteurella Multocida Culture
Pasteurella multocida strain Pm70, a serotype A : 1 poultry isolate that has a fully sequenced genome [20], was used in this study. Pm70 was cultivated in brain heart infusion (BHI) broth at 37°C with rotary aeration. Minimum inhibitory concentrations (MICs) of AMX, CTC, and ENR for Pm70 are 0.5 μg/mL, 4 μg/mL, and 0.031 μg/mL, respectively [15]. Growth kinetics of Pm70 in the presence of 1/4 MIC of the three antibiotics were previously described [15]. Stationary phase cultures of Pm70 were used to inoculate 50 mL BHI to an initial A600 of 0.05; antibiotic treated cultures contained 1/4 MIC of AMX, CTC, or ENR, and control cultures were grown without antibiotics. All cultures were grown in triplicate to mid-log phase (A600 of 0.8) and harvested by centrifugation (10 000 × g, 10 minutes, 4°C). Pellets were stored at −80°C.
2.2. Proteomics
Protein extraction, quantification, and trypsin digestion were done exactly as described [14] from three biological replicates. Briefly, protein solutions (100 μg; <1 M urea; 50 mM Tris-cl pH 8.0) from untreated control and antibiotic-treated bacteria were reduced (5 mM dithiothreitol, 65°C, 5 minutes), alkylated (10 mM iodoacetamide, 30°C, 30 minutes), and then trypsin-digested until there was no visible pellet (1 : 50 w/w 37°C, 16 hours). Peptides were desalted using a peptide macrotrap (Michrom BioResources, Inc., Auburn, Calif, USA) and eluted using a 0.1% triflouroacetic acid, 95% ACN solution. Desalted peptides were vacuum dried and resuspended in 20 μl of 0.1% formic acid.
Two-dimensional liquid chromatography (LC) analysis was done by strong cation exchange (SCX) followed by reverse phase (RP) coupled directly in line with an electrospray ionization (ESI) ion trap tandem mass spectrometer (LCQ; ThermoElectron Corp., San Jose, Calif, USA) essentially as described in [15]. The salt gradient applied in this study was different from the published method and was applied in steps of 0, 10, 15, 20, 25, 30, 35, 40, 45, 50, 57, 64, 90, and 700 mM ammonium acetate in 5% acetonitrile (ACN) and 0.1% formic acid. The reverse phase gradient used 0.1% formic acid in ACN and increased the ACN concentration in a linear gradient from 5% to 30% in 20 minutes and then 30% to 95% in 7 minutes, followed by 5% for 10 minutes for 0, 10, 15, 25, 30, 45, 64, 90, and 700 mM salt gradient steps. For 20, 35, 40, 50, and 57 mM salt gradient steps, ACN concentration was increased in a linear gradient from 5% to 40% in 65 minutes, 95% for 15 minutes, and 5% for 20 minutes.
All database searches of tandem mass spectra were done using TurboSEQUEST (Bioworks Browser 3.2; ThermoElectron) [21]. Mass spectra and tandem mass spectra were searched against an in silico trypsin-digested protein database of P. multocida Pm70 downloaded from National Center for Biotechnology Institute (NCBI). Cysteine carbamidomethylation and methionine oxidation (single and double) were included in the search criteria. We used the reverse database functionality in Bioworks 3.2 and searched tandem MS (MS2) data against a reversed Pm70 database using the same search criteria as described above. Peptide identifications from Pm70 protein database that were >5 amino acids long with Xcorr ≥ 1.5, 2.0, and 2.5 for +1, +2, and +3 charged ions, respectively, and with delta Cn values of ≥0.1 were used for protein identifications [15]. None of the proteins identified at the applied Xcorr and delta Cn filters had peptides identified from the reversed Pm70 database. Protein identifications have been submitted to PRoteomics IDEntifications (PRIDE) database [22] and the accession numbers are 1751, 1752, 1753, and 1754. PRIDE submission requirements are based on the proposed guidelines by proteomics standards initiative [23] and include all the peptides identified for each protein with their sequence, charge state, Xcorr, and delta cn.
We used an isotope-free quantification method [14] and a custom program ProtQuant [24] to identify differences in protein expression between control and sub-MIC antibiotic treated P. multocida. ProtQuant is a java-based tool for label-free quantification that uses a spectral counting method [25] with increased specificity. ProtQuant includes the quantitative aspects of the Sequest cross correlation (XCorr) into the spectral counting method and computes the statistical significance of differential protein expression using one-way ANOVA (α ≤ 0.05). This method requires at least 3 peptides identified from either the control or treatment datasets.
2.3. Systems Modeling: Pasteurella Multocida Pm70 Molecular Interaction Database
To study the effects of antibiotics on Pm70 using systems biology approaches, we built an information rich predicted protein interaction network using Pathway Studio (Ariadne, Rockville, Md, USA) using a bacterial molecular interaction database. Multiple aspects of protein function, including protein modifications, cellular location, protein-protein interactions, gene expression regulation, molecular transport and synthesis, and regulation of various cellular processes are included [26].
Although the bacterial molecular interaction database contains data for Gram-negative and Gram-positive bacteria, it does not include P. multocida. Therefore, to append the bacteria database with P. multocida, all 2015 Pm70 proteins were mapped to their corresponding orthologs in the database by identifying reciprocal-best-BLAST hits with greater than 30% similarity. Gram-positive orthologs were removed to ensure that protein interaction networks were derived only from Gram-negative species. The resulting ortholog map file was imported into Pathway Studio to allow prediction of interactions between P. multocida proteins. The Gram-negative ortholog-only network was used to analyze sub-MIC antibiotic effects on the P. multocida proteome. Our protein quantification method did not measure absolute fold changes in protein expression, but instead indicated whether the increase or decease in expression was significant. Therefore, for each antibiotic treatment, we imported all identified proteins into Pathway Studio along with the expression values indicating the significance of expression change; we represented a significant increase in protein expression compared to the untreated control as +1, while a significant decrease in expression was represented as −1.
As an initial screening method, we used the Pathway Studio “Find groups” tool to identify Gene Ontology (GO) groups that had a significant number of identified proteins within each paired antibiotic treatment/control protein set. By doing so we identified GO groups that had good proteome coverage for analysis of antibiotic effects. Pathway Studio calculates the statistical significance of the overlap between the protein list and a GO group using the Fisher exact test. Thus, the calculated p-value depends on the extent of overlap between the protein list and a group as well as the sizes of the list and a group. We used P ≤ .05 to select GO groups with significant protein coverage.
We built interaction networks in Pathway Studio with proteins of interest including the upstream regulators and downstream targets. In the interaction networks, different colors were used for the nodes to indicate whether a protein was present in the dataset (pink) and whether there was a significant increase (red), or decrease (green) in protein expression in response to sub-MIC of antibiotic. Entities in the interaction map that were not from Pm70 were shown in gray color. For each of the three antibiotics, protein expression was compared between the control (no antibiotic) and the antibiotic treatment and overlaid onto the Pm70 interaction network. The individual responses to each antibiotic were compared to identify overall trends in Pm70 response to the different antibiotics.
3. Results and Discussion
3.1. Proteome Coverage
We identified 1064 (53%) of the 2015 predicted Pm70 proteins [20] from our control (no antibiotic) and antibiotic treated datasets. The number of identifications with two or more peptides in at least one dataset was 572 (56%) (PRIDE datasets (supplementary Table 1)) and this compares favorably with reported MudPIT results which have as few as 20% of proteins identified by two or more peptides [27]. Furthermore, compared to the 20% coverage of the accessible proteome that we reported with ICAT [15], we achieved 2.5 fold higher proteome coverage in this study. This level of proteome coverage gave us confidence that we could conduct a systems-wide analysis to formulate new hypothesis for sub-MIC antibiotic effects on Pm70 physiology.
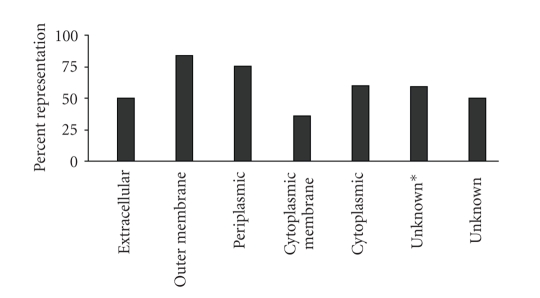
Furthermore, because protein function depends on cellular location, a systems-wide analysis requires that we identify proteins from all cell locations. We used the PSORTb version 2.0 bacterial protein subcellular localization prediction tool to classify all proteins in our dataset [28]. PSORTb has an overall precision of 96% and places all Pm70 proteins into seven categories: extracellular (4), outer membrane (49), periplasmic (61), cytoplasmic membrane (411), cytoplasmic (774), unknown (with multiple localization sites) (37), and unknown (679). To determine the percent coverage in each of these categories, the number of proteins identified in each category was compared to the total number of proteins predicted to be in that category from the genome (Figure 1). The average coverage for each subcellular compartment in our data was 59%, and although proteins from all subcellular compartments were represented, not all subcellular compartments were equally represented. We identified more outer membrane and periplasmic proteins (84% and 75%, resp.), than cytoplasmic membrane proteins (36%).
Figure 1.
Representation of P. multocida proteins in PSORTb subcellular locations. Percent coverage for each PSORTb predicted subcellular localization for P.multocida proteins identified in this study is shown. Percent representation was calculated by comparing the number of proteins identified in each category to the total number of proteins predicted to be in that category from P. multocida genome. PSORTb prediction “unknown*” indicates that the proteins may have multiple subcellular localizations while unknown refers to location unknown.
To investigate the lower coverage of cytoplasmic membranes, we estimated the total abundance of cytoplasmic membrane and outer membrane proteins based on ΣXcorr [14]. The outer membrane proteins had a ΣXcorr of 1599, while cytoplasmic membrane proteins had a ΣXcorr of 1424. Therefore, although a lower percentage of cytoplasmic membrane proteins were identified, there was a very similar amount of protein isolated from the cytoplasmic and outer membrane fractions. This suggests that although the Pm70 cytoplasmic membrane contains a greater variety of proteins than the outer membrane, cytoplasmic membrane proteins are present in relatively low abundance compared to the outer membrane. Because we used the mass spectrometer in a data-dependent way to maximize proteome coverage, it is logical that we achieved greater coverage of bacterial compartments that have proteins with higher abundance. These results also show that our methods for protein isolation solubilized relatively hydrophobic membrane proteins. Our previously published ICAT methodology [15] and the current label-free 2D-LC ESI MS2 approach utilized the same protein isolation method. Nonisotopic relative quantitative proteomics methods are as good (or better) than isotopic methods [14, 29] but have the advantage of far greater proteome coverage. We confirm this observation in the current work; compared to our previous ICAT study we identified tenfold more membrane proteins, which is 38.6% of the predicted membrane proteome of P. multocida compared with 3.7% identified using the ICAT method.
3.2. Pasteurella Multocida Pm70 Molecular Interaction Database
Predicted protein interaction networks for P. multocida (based on computational comparative genomics) are available [30]. However, these interactions have edges that have no directionality or any biological information. System analysis can be a powerful tool to identify global trends so long as the protein interaction network under investigation is information rich. Therefore, to enable system analysis of our protein expression data, we chose to build a protein interaction network for P. multocida where all nodes had all possible biological information associated with them. Reciprocal-BLASTp searches of Pm70 proteins against both Gram-negative and Gram-positive bacterial proteins identified unique orthologs for only 1595 proteins. Of these, 1525 were from Gram-negative bacteria (Escherichia coli CFT 073, E. coli K12, E. coli O157:H7, Synechocystis sp. PCC6803 (mean e-value and percent identity were 2.03e-06 and 65.2%) of which 848 orthologs were identified in our datasets. We constructed a network with 848 proteins in which the nodes (proteins) had predicted functional annotation and the edges (links) between proteins described regulation, expression, binding, and chemical reactions.
3.3. Systems Modeling: Data Analysis
To determine the effects of 1/4 MIC of each of the three antibiotics, AMX, CTC, and ENR, on the Pm70 proteome, protein expression from each of the three antibiotic treatments was compared to protein expression from the nonantibiotic control. We did not compare protein expression changes between antibiotic treatments. The paired control antibiotic-treated samples contained 913, 781, and 762 proteins, representing 143, 133, and 137 GO groups, from AMX-, CTC-, and ENR-treated cultures, respectively. Each GO group is a unique GO term that corresponds to a biological process, molecular function, or a cellular component annotation.
At P ≤ .05, 69, 61, and 58 GO groups for AMX, CTC, and ENR, respectively had significant proteome coverage. Of these, 46 GO groups were common to all three antibiotics (Table 1). To identify the underlying higher level themes involved in the response to antibiotics, we used the GoSlimViewer at AgBase [31] and found that the 46 common GO groups had 16 higher level GO terms (Table 2).
Table 1.
Significantly overrepresented GO groups in response to sub-MIC AMX, CTC and ENR.
| GO ID | Name |
|---|---|
| GO:0009257 | 10-formyltetrahydrofolate biosynthesis |
| GO:0045733 | Acetate catabolism |
| GO:0006086 | Acetyl-CoA biosynthesis from pyruvate |
| GO:0006418 | Amino acid activation |
| GO:0009063 | Amino acid catabolism |
| GO:0046349 | Amino sugar biosynthesis |
| GO:0009061 | Anaerobic respiration |
| GO:0015986 | ATP synthesis coupled proton transport |
| GO:0030113 | Capsule (sensu Bacteria) |
| GO:0016052 | Carbohydrate catabolism |
| GO:0042280 | Cell surface antigen activity, host-interacting |
| GO:0017004 | Cytochrome biogenesis |
| GO:0005737 | Cytoplasm |
| GO:0009281 | Cytosolic ribosome (sensu Bacteria) |
| GO:0006308 | DNA catabolism |
| GO:0006261 | DNA dependent DNA replication |
| GO:0006310 | DNA recombination |
| GO:0006113 | Fermentation |
| GO:0006012 | Galactose metabolism |
| GO:0006094 | Gluconeogenesis |
| GO:0006096 | Glycolysis |
| GO:0009436 | Glyoxylate catabolism |
| GO:0009089 | Lysine biosynthesis via diaminopimelate |
| GO:0009086 | Methionine biosynthesis |
| GO:0006777 | Mo-molybdopterin cofactor biosynthesis |
| GO:0015949 | Nucleobase, nucleoside and nucleotide interconversion |
| GO:0009052 | Pentose-phosphate shunt, non-oxidative branch |
| GO:0009051 | Pentose-phosphate shunt, oxidative branch |
| GO:0008233 | Peptidase activity |
| GO:0009252 | Peptidoglycan biosynthesis |
| GO:0000270 | Peptidoglycan metabolism |
| GO:0042597 | Periplasmic space |
| GO:0006412 | Protein biosynthesis |
| GO:0006457 | Protein folding |
| GO:0006508 | Proteolysis and peptidolysis |
| GO:0006164 | Purine nucleotide biosynthesis |
| GO:0009152 | Purine ribonucleotide biosynthesis |
| GO:0042867 | Pyruvate catabolism |
| GO:0009269 | Response to dessication |
| GO:0006401 | RNA catabolism |
| GO:0009451 | RNA modification |
| GO:0003735 | Structural constituent of ribosome |
| GO:0009088 | Threonine biosynthesis |
| GO:0006350 | Transcription |
| GO:0009386 | Translational attenuation |
| GO:0006099 | Tricarboxylic acid cycle |
AMX: amoxicillin; CTC: chlortetracycline; ENR: enrofloxacin.
Table 2.
GO slims common to sub-MIC AMX, CTC, and ENR response of P. multocida.
| GO ID | GO TERM |
|---|---|
| Cellular Component | |
| GO:0005737 | Cytoplasm |
| GO:0005829 | Cytosol |
| GO:0030312 | External encapsulating structure |
|
| |
| Biological Process | |
| GO:0006519 | Amino acid and derivative metabolism |
| GO:0005975 | Carbohydrate metabolism |
| GO:0009056 | Catabolism |
| GO:0006259 | DNA metabolism |
| GO:0006091 | Generation of precursor metabolites and energy |
| GO:0008152 | Metabolism |
| GO:0006139 | Nucleobase, nucleoside, nucleotide and nucleic acid metabolism |
| GO:0006412 | Protein biosynthesis |
| GO:0019538 | Protein metabolism |
| GO:0006950 | Response to stress |
| GO:0006350 | Transcription |
|
| |
| Molecular Function | |
| GO:0008233 | Peptidase activity |
| GO:0005198 | Structural molecule activity |
AMX: amoxicillin; CTC: chlortetracycline; ENR: enrofloxacin.
We identified 147, 126, and 134 significant changes in protein expression with sub-MIC AMX, CTC, and ENR, respectively (Table 3, and supplementary Table 2, all proteins were identified with at least three peptides in one dataset). A number of proteins differentially expressed in response to sub-MIC antibiotics in our previous ICAT study were also identified in this study. Although the general trend of expression was similar (i.e., increase or decrease) compared to ICAT, many of the differences were not statistically significant in this study. This could be due to the data dependent acquisition inherent to ESI MS2 methodology combined with the differences in both the amount of the proteome accessible by ICAT and label-free proteomics and the difference in total coverage (numbers of peptides/protein). We suggest that this nonisotopic proteomics workflow and analysis offer more comprehensive coverage and a greater probability of accurate representation of the Pm70 proteome than did our ICAT study.
Table 3.
Differential P. multocida protein expression in response to a quarter-MIC AMX, CTC, and ENR in different GO categories.
| Accession | GO Slims | ∑Xcorr | |||
|---|---|---|---|---|---|
| Pm70 | AMX | CTC | ENR | ||
| Glycolysis/Gluconeogenesis | |||||
| 15603407 | Phosphoenolpyruvate carboxykinase, PckA | 264.2 | ns | 120 | 112.9 |
| 15603725 | Phosphoglycerate kinase, Pgk | 218.1 | ns | 170.2 | 161 |
| 15603726 | Fructose-bisphosphate aldolase, FbaA | 504.9 | 566.6 | 450.1 | 436.8 |
| 15602789 | Glyceraldehyde-3-phosphate dehydrogenase, GapA | 253.6 | ns | 125.5 | 70.1 |
| 15603371 | Phosphoglyceromutase, GpmA | 98.3 | 124.9 | 130.3 | ns |
| 15602518 | Pyruvate kinase, PykA | 28.6 | 51.4 | 54.9 | ns |
| 15602688 | Fumarate hydratase, FumC | 42.2 | 80.5 | 19.1 | ns |
| 15602146 | SucD | bdt | 7.8 | ns | ns |
|
| |||||
| Response to stress | |||||
| 15602972 | Chaperonin GroEL | 148.6 | 246.8 | ns | 177.1 |
| 15602971 | Co-chaperonin GroES | 23 | 42.7 | 56.3 | 56.6 |
| 15602601 | DnaK | 97.6 | 132.7 | ns | 72.6 |
| 15602303 | FtsH | 21.9 | 7.9 | 4.7 | 3.9 |
|
| |||||
| Peptidoglycon biosynthesis and cell envelope permeability | |||||
| 15603596 | D-fructose-6-phosphate amidotransferase, GlmS | bdt | 31.3 | 12.4 | 6.8 |
| 15603671 | GlmU | 8.5 | 20.2 | ns | ns |
| 15602548 | NagC | 14.4 | ns | 5.3 | 5 |
| 15602651 | Hypothetical protein PM0786, OmpA | 262.8 | ns | 172.5 | 76 |
| 15602419 | Lpp | 7.2 | ns | 18.7 | ns |
|
| |||||
| Purine metabolism | |||||
| 15602149 | Adenylate kinase, Adk | 2.8 | 22.3 | 42.2 | 11.3 |
| 15602803 | Adenylosuccinate synthetase, PurA | 2.4 | ns | 11.2 | 19.2 |
| 15602109 | Ribose-phosphate pyrophosphokinase, PrsA | bdt | 9.2 | 8.4 | ns |
|
| |||||
| RNA metabolism | |||||
| 15603255 | DNA-directed RNA polymerase alpha subunit, RpoA | 32.8 | 76.1 | 51.9 | ns |
| 15603602 | DNA-directed RNA polymerase beta subunit, RpoB | 17 | 34.5 | ns | ns |
| 15603601 | DNA-directed RNA polymerase beta~ subunit, RpoC | 56.2 | 85.2 | ns | ns |
| 15602786 | DNA-directed RNA polymerase omega subunit, RpoZ | bdt | 11 | ns | 9.6 |
| 15602119 | Lrp | 8.7 | ns | 25.1 | ns |
| 15603209 | DeoR | bdt | 10.6 | ns | ns |
| 15603785 | Transcription termination factor Rho | 42.4 | 15.9 | 20.5 | 21.3 |
|
| |||||
| Xenobiotic metabolism | |||||
| 15601866 | SodA | 153.5 | ns | 66 | 85.4 |
| 15601897 | HktE/KatE | 26.1 | ns | 3 | 6.1 |
|
| |||||
| Lipopolysaccharide biosynthesis | |||||
| 15603862 | Lipid-A-disaccharide synthase, LpxB | bdt | 12.1 | ns | ns |
| 15602423 | 2-dehydro-3-deoxyphosphooctonate aldolase, KdsA | 9.2 | 23.2 | ns | ns |
| 15602749 | RfaE | bdt | 12.9 | ns | ns |
| 15603154 | GalU | 3.7 | 18.7 | ns | ns |
|
| |||||
| Cell division | |||||
| 15603384 | FtsY | bdt | 8.2 | ns | ns |
| 15602012 | Cell division protein FtsZ | 11.5 | 22.1 | ns | ns |
|
| |||||
| Protein translation | |||||
| 15603222 | Elongation factor Tu | 875.7 | ns | 936 | ns |
| 15602977 | DeaD | 4.7 | ns | 13.3 | ns |
| 15602168 | Methionyl-tRNA synthetase | 2.4 | ns | 11.5 | ns |
|
| |||||
| DNA repair | |||||
| 15603682 | Recombinase A, RecA | bdt | ns | ns | 19.5 |
AMX: amoxicillin; CTC: chlortetracycline; ENR: enrofloxacin.
Pm70: P. multocida cultured without sub-MICs.
bdt: below detectable threshold.
ns: differential expression not significant compared to Pm70.
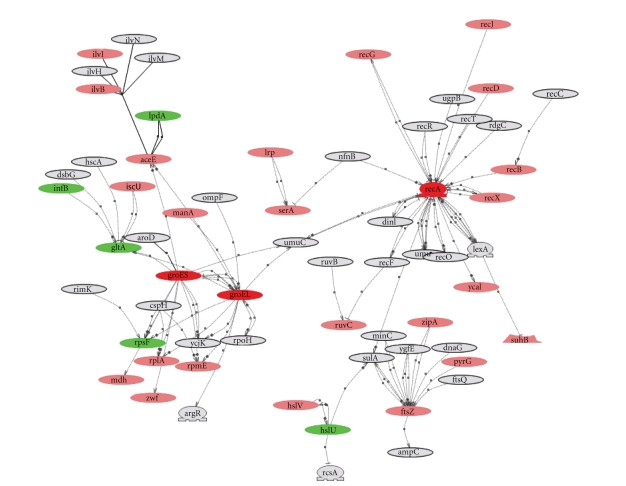
To identify common themes in the response to sub-MICs of antibiotics, we superimposed significant changes in protein expression for each antibiotic onto the Pm70 protein interaction network (supplementary Figure ). For more detailed analysis of selected proteins, we used the Pm70 protein interaction network to iteratively build and visualize networks around the proteins of interest. The network built around RecA in response to ENR is shown in Figure 3 as an example of the type of analysis that was used to identify the trends described in the following paragraphs.
Figure 3.
P. multocida RecA protein interaction network. P. multocida sub-MIC ENR response was marked by significant change in RecA expression. We built interaction network iteratively with RecA as primer and identified RecA, GroEL, and GroES subnetwork. Red nodes are proteins with increased expression and green nodes are proteins with decreased expression in response to ENR. Proteins with no significant changes in expression are shown in pink, and gray nodes are proteins from P. multocida interaction network that were not identified in our dataset.
3.4. Common Adaptive Responses to AMX, CTC, and ENR
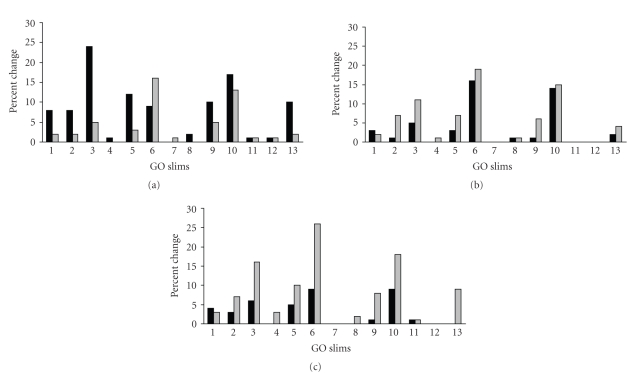
We examined the overall trends of antibiotic effects on the 16 GO groups that were identified by GO SlimViewer (Table 2). After excluding general GO terms like cytoplasm, metabolism and so forth, 13 GO groups were evaluated. For each GO group, we calculated the percentage of proteins whose expression either increased or decreased, compared to the total entities in that GO group, for each antibiotic treatment. The trend of AMX was an overall increase in protein expression, which could indicate the induction of an adaptive response (Figure 2). Conversely, CTC and ENR had an overall suppressive effect on protein expression, potentially indicating that CTC and ENR detrimentally affect P. multocida fitness at doses below MIC. Alternatively, this could be a compensatory response by slowing metabolism.
Figure 2.
Overall trends in protein expression in GO Slims common to sub-MICs. For each GO Slim, the percentage of proteins whose expression was either up (black bars) or down regulated (gray bars) is compared to the total entities in that GO Slim are shown for amoxicillin (a), chlortetracycline (b), and enrofloxacin (c). GO Slim categories are as follows: 1, external encapsulating structure; 2, DNA metabolism; 3, nucleobase, nucleoside, nucleotide, and nucleic acid metabolism; 4, protein metabolism; 5, transcription; 6, protein biosynthesis; 7, response to stress; 8, carbohydrate metabolism; 9, catabolism; 10, structure molecule activity; 11, peptidase activity; 12, amino acid derivative metabolism; 13, generation of precursor metabolites and energy.
The overall suppressive effects of CTC and ENR on P. multocida metabolism are further substantiated by their effects on individual protein expression in central metabolic pathways. In glycolysis/gluconeogenesis, CTC and ENR significantly decreased the expression of phosphoenolpyruvate carboxykinase, phosphoglycerate kinase, fructose bisphosphate aldolase, and glyceraldehyde-3-phosphate dehydrogenase. By contrast, AMX significantly increased the expression of fructose bisphosphate aldolase, pyruvate kinase, and phosphoglyceromutase, and it significantly increased expression of fumarate hydratase and succinyl-CoA synthetase from the TCA cycle (Table 3).
The differential protein expression profiles of P. multocida in response to antibiotic sub-MICs were analyzed to delineate the underlying physiological response. A predictable stress response was indicated by increased GroES expression after treatment with all three antibiotics and increased GroEL and DnaK expression after AMX treatment. The GroEL/GroES chaperone system is induced by different forms of environmental stress in various bacterial species and functions to maintain appropriate protein folding [32]. All antibiotics reduced the expression of FtsH, a zinc metalloprotease that degrades cellular proteins, including heat shock promoter protein sigma 32 (RpoH) [33]. Therefore, sub-MICs of antibiotics may reduce expression of P. multocida genes under the control of heat shock promoters.
Sub-MICs of antibiotics induce a response that appears to decrease cell envelope permeability by increasing the availability of N-acetyl glucosamine, a necessary precursor for peptidoglycan and lipid A biosynthesis. E. coli mutants that are defective in lipid A biosynthesis are permeable to hydrophobic molecules and highly susceptible to antibiotics [34, 35]; therefore, increasing the amount of lipid A would be a logical response to decrease permeability. All three antibiotics increased GlmS expression, which catalyzes the first reaction in the pathway for N-acetylglucosamine biosynthesis, and AMX increased GlmU expression, which catalyzes the last two reactions in the pathway. NagC, is a transcriptional regulator controlling glmUS operon expression [36], its expression decreased after CTC and ENR treatment. In addition to increasing N-acetylglucosamine availability, CTC and ENR decreased expression of OmpA, a multifunctional outer membrane porin that allows diffusion of small solutes across the outer membrane [37, 38] (Table 3).
Purine metabolism was another common cellular function affected by all three antibiotics. AMX, CTC, and ENR increased adenylate kinase expression. CTC and ENR increased adenylosuccinate synthetase expression. Adenylate kinase and adenylosuccinate synthetase catalyze the first and last steps in the conversion of inosine monophosphate to adenosine diphosphate [39], respectively. AMX and CTC also significantly increased expression of PrsA, which catalyzes formation of phosphoribosyl pyrophosphate, a necessary metabolite for synthesis of purines, pyrimidines, and histidine.
RNA metabolism was affected by antibiotics; all increased RNA polymerase expression; AMX increased RpoA, RpoB, RpoC, and RpoZ expression; CTC increased RpoA expression; and ENR increased RpoZ expression. In addition, all three antibiotics decreased expression of transcriptional terminator Rho, which may cause polarity suppression. Decreased expression of Rho may also decrease the half life of mRNA in the bacterial cell [40].
It is possible that these protein expression trends in P. multocida that were common for all three antibiotics could reflect a general response to stress. These changes could be induced by other nonantibiotic stressors; further experiments with nonantibiotic stress conditions would be required to confirm that these trends are in fact a common response to antibiotic-induced stress.
3.5. Virulence Factor Expression
Sub-MICs of antibiotics can potentially impact the outcome of infection by altering bacterial virulence factor expression [14, 41–43]. In the current study, antibiotics altered expression of known and putative virulence proteins in Pasteurella multocida. For example, CTC and ENR reduced the expression of OmpA, which in addition to allowing small molecule diffusion across the outer membrane, is also a known virulence factor of P. multocida and is involved in binding to host cells [44]. In addition, sub-MICs of CTC and ENR decreased expression of detoxifying enzymes superoxide dismutase and catalase, which constitute a major defense mechanism of bacteria against reactive oxygen species and play a role in pathogenesis in certain bacteria [45, 46]. Other putative P. multocida virulence factors whose expression was decreased by CTC and ENR included glyceraldehyde-3-phosphate dehydrogenase, which is a virulence factor in Gram-positive bacteria and has been recently implicated in pathogenesis of Gram-negative bacteria [47], and phosphoenolpyruvate carboxykinase, which is required for M. bovis virulence in mice [48]. Stress response chaperone protein Hsp90 was decreased by CTC and ENR, and DnaK was decreased by ENR. These proteins are required for virulence in a number of bacterial pathogens including S. enterica and L. pneumophila [49, 50].
3.6. Amoxicillin Specific Effects
The AMX mode of action involves binding and inactivation of peptidoglycan cross-linking transpeptidases with subsequent inhibition of cell wall biosynthesis. The transpeptidases, also known as penicillin binding proteins (Pbps), are known targets of AMX [51] and the P. multocida genome has eight Pbps [20]. Logically, the bacterial compensatory response to AMX could involve increasing Pbp expression. Though we identified 5 Pbps in our dataset; there were no significant changes in expression of any of these. Therefore, it appears that the adaptive response to AMX treatment does not entail over expression of Pbps. Alternatively, more than 1/4 MIC of AMX may be required to elicit over expression of Pbps in P. multocida.
There was good evidence that the P. multocida compensatory response to AMX involves decreasing cell envelope permeability. In addition to the previously described effects on GlmU and GlmS, expression of several enzymes responsible for synthesis of lipopolysaccharide was significantly increased. LpxB, an enzyme in the lipid A biosynthetic pathway, had significantly increased expression. KdsA and RfaE, both of which are involved in synthesis of core oligosaccharide [52], were significantly increased. Expression of GalU, which is responsible for the synthesis of cell envelope precursor UDP-galactose, was also increased.
The compensatory response to AMX also appeared to involve upregulation of cell division proteins. Both FtsZ (essential for cell division) and FtsY had increased expression. AMX also caused increased expression of DeoR: a DNA-binding transcriptional regulator involved in the negative regulation of genes encoding nucleotide catabolism enzymes.
3.7. Chlortetracycline Specific Effects
As could be predicted, the response to CTC involved the compensation against its inhibition of protein translation. Elongation factor Tu was increased, as was methionyl-tRNA synthetase (MetG), which initiates translation through tRNA(fMet) aminoacylation. In addition, DeaD (CsdA), which participates in the assembly of the large subunit of the ribosome [53], also had increased expression.
CTC also appeared to induce a protective response for the cell envelope. In addition to its effects on GlmS, NagC, and OmpA, it caused an increase in expression of murein lipoprotein Lpp, which is required for stabilization and integrity of the cell envelope [54]. Lpp mutants are hypersensitive to toxic compounds [55]. Finally, CTC caused increased expression of Lrp, a transcriptional regulator that is a global mediator of the leucine response, specifically branched amino acid transport [56].
3.8. Enrofloxacin Specific Effects
Results from the current study agree with our earlier findings [15] that the compensatory response to ENR entails recruitment of double stranded DNA damage repair machinery to overcome the quinolone-mediated block on progression of the replication fork. This response is logical because blockage of the replication fork by quinolones is caused by accumulation of inactive quinolone-bound intermediate with double stranded DNA breaks. We previously reported an increase in expression of RecA, which catalyzes DNA strand exchange during homologous recombination and double-stranded DNA break repair [15], in response to ENR. Here we found that expression of RecA and RecN, another enzyme required for repair of double stranded breaks in the chromosome, increased.
4. Conclusions
We achieved greatly improved coverage of the P. multocida proteome by detecting unlabeled peptides using 2D LC with ESI MS2 compared to a similar study we conducted using isotope coded affinity tag labeled (ICAT) peptides [15]. The advantage of ICAT labeling is that we were able to report fold changes in protein expression; however, the increased proteome coverage in the current study enabled us to conduct systems analysis, which was not possible in the ICAT study.
Systems analysis improved our study of P. multocida protein expression and enabled the identification of antibiotic-mediated mechanisms and pathways. In particular, visualizing proteins as interacting networks enabled us to identify proteins with high connectivity that could be important targets for modulation. However, although proteome coverage was not a major limitation in conducting systems analysis with our P. multocida protein expression data, the paucity of experimental evidence from this nonmodel species did limit our analysis. The strategy we used to identify orthologs and import experimental evidence based on E. coli and other model species greatly enhanced our analysis, but extrapolation of protein functions across species must be interpreted in the light of this electronic inference.
Despite this limitation, our systems analysis identified several proteins to target in future studies. In particular, we speculate that inactivating P. multocida proteins in the heat shock response, cell envelope biosynthesis/integrity, purine metabolism, or RNA metabolism may be viable strategies to enhance antimicrobial activity. In addition, inactivation of specific cell division proteins, translation proteins, and DNA repair enzymes may impair bacterial adaptation to antibiotics in the penicillin, tetracycline, and quinolone classes, respectively. Inactivation of specific transcriptional regulators, such as DeoR or Lrp, may also be an effective method to impair the P. multocida adaptive response against AMX and CTC. These targets would not have been predicted a priori based on the known mechanisms of action of these antibiotics; their identification was facilitated by our global systems analysis.
In summary, our findings could lead to the development of new antimicrobial potentiating drugs aimed at inactivating antibiotic compensatory mechanisms to prevent occurrence and emergence of antimicrobial resistance. In addition, our results indicate that systems analysis in nonmodel bacterial species can be used with high-throughput proteomics data to identify protein targets for further functional investigations.
Supplementary Material
All proteins identified in this study are listed in Supplementary Table 1. Proteins with significant quantitative changes in expression in response to sub-MICs of AMX, CTC, or ENR are listed in Supplementary Table 2. Protein interaction networks built with significant changes in expression with sub-MICs shown in Table 3 are included in the supplementary figure. This figure shows changes in protein expression in response to sub-MICs of AMX, CTC, and ERN. Red and green indicate significant increase and decrease in expression, respectively. Proteins whose expression did not change (pink) and proteins in the network not identified in our datasets (gray) are also included.
Acknowledgments
This project was partially supported by a grant from the National Science Foundation (EPS-0556308-06040293) and a competitive grant from Mississippi State University Life Sciences and Biotechnology Institute (LSBI). We acknowledge Dr. Vivek Kapur for providing P. multocida pm70 strain, Tibor Pechan, and Anton Yuryev for technical help. Mass spectrometry analysis was carried out at LSBI, Mississippi State University. This is a Mississippi Agricultural and Forestry Experiment Station publication (J11226), part of the publication costs were provided by the Department of Basic Sciences (CVM/MSU).
Abbreviations
- MIC:
Minimum inhibitory concentration
- CTC:
Chlortetracycline
- AMX:
Amoxicillin
- ENR:
Enrofloxacin
- BHI:
Brain heart infusion
- GO:
Gene ontology
- NCBI:
National Center for Biotechnology Institute
- BRD:
Bovine respiratory disease
- BLAST:
Basic local alignment search tool
- ACN:
Acetonitrile
- 2D:
Two dimensional
- LC:
Liquid chromatography
- ESI:
Electrospray ionization
- MS:
Mass spectrometry
- MS2:
Tandem MS
- RP:
Reverse phase
- RecA:
Recombinase A
References
- 1.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nature Reviews Microbiology. 2007;5(3):175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 2.Roe MT, Pillai SD. Monitoring and identifying antibiotic resistance mechanisms in bacteria. Poultry Science. 2003;82(4):622–626. doi: 10.1093/ps/82.4.622. [DOI] [PubMed] [Google Scholar]
- 3.Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clinical Microbiology and Infection. 2007;13(1):5–18. doi: 10.1111/j.1469-0691.2006.01492.x. [DOI] [PubMed] [Google Scholar]
- 4.Weber JT, Courvalin P. An emptying quiver: antimicrobial drugs and resistance. Emerging Infectious Diseases. 2005;11(6):791–793. doi: 10.3201/eid1106.050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rapp RP. Antimicrobial resistance in Gram-positive bacteria: the myth of the MIC. Minimum inhibitory concentration. Pharmacotherapy. 1999;19(8, part 2):112S–119S. doi: 10.1592/phco.19.12.112s.31703. discussion 133S–137S. [DOI] [PubMed] [Google Scholar]
- 6.Christensen JP, Bisgaard M. Fowl cholera. Revue Scientifique et Technique. 2000;19(2):626–637. doi: 10.20506/rst.19.2.1236. [DOI] [PubMed] [Google Scholar]
- 7.Hunt ML, Adler B, Townsend KM. The molecular biology of Pasteurella multocida . Veterinary Microbiology. 2000;72(1-2):3–25. doi: 10.1016/s0378-1135(99)00183-2. [DOI] [PubMed] [Google Scholar]
- 8.Klein NC, Cunha BA. Pasteurella multocida pneumonia. Seminars in Respiratory Infections. 1997;12(1):54–56. [PubMed] [Google Scholar]
- 9.Weiser GC, DeLong WJ, Paz JL, Shafii B, Price WJ, Ward ACS. Characterization of pasteurella multocida associated with pneumonia in bighorn sheep. Journal of Wildlife Diseases. 2003;39(3):536–544. doi: 10.7589/0090-3558-39.3.536. [DOI] [PubMed] [Google Scholar]
- 10.Welsh RD, Dye LB, Payton ME, Confer AW. Isolation and antimicrobial susceptibilities of bacterial pathogens from bovine pneumonia: 1994–2002. Journal of Veterinary Diagnostic Investigation. 2004;16(5):426–431. doi: 10.1177/104063870401600510. [DOI] [PubMed] [Google Scholar]
- 11.Darnell KR, Hart ME, Champlin FR. Variability of cell surface hydrophobicity among Pasteurella multocida somatic serotype and Actinobacillus lignieresii strains. Journal of Clinical Microbiology. 1987;25(1):67–71. doi: 10.1128/jcm.25.1.67-71.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart ME, Champlin FR. Susceptibility to hydrophobic molecules and phospholipid composition in Pasteurella multocida and Actinobacillus lignieresii . Antimicrobial Agents and Chemotherapy. 1988;32(9):1354–1359. doi: 10.1128/aac.32.9.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends in Microbiology. 2000;8(3):128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 14.Nanduri B, Lawrence ML, Vanguri S, Burgess SC. Proteomic analysis using an unfinished bacterial genome: the effects of subminimum inhibitory concentrations of antibiotics on Mannheimia haemolytica virulence factor expression. Proteomics. 2005;5(18):4852–4863. doi: 10.1002/pmic.200500112. [DOI] [PubMed] [Google Scholar]
- 15.Nanduri B, Lawrence ML, Boyle CR, Ramkumar M, Burgess SC. Effects of subminimum inhibitory concentrations of antibiotics on the Pasteurella multocida proteome. Journal of Proteome Research. 2006;5(3):572–580. doi: 10.1021/pr050360r. [DOI] [PubMed] [Google Scholar]
- 16.He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genetics. 2006;2(6):e88. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh C. Antibiotics: Actions, Origins, Resistance. Washington, DC, USA: ASM Press; 2003. [Google Scholar]
- 18.Singer RS, Case JT, Carpenter TE, Walker RL, Hirsh DC. Assessment of spatial and temporal clustering of ampicillin- and tetracycline-resistant strains of Pasteurella multocida and P haemolytica isolated from cattle in California. Journal of the American Veterinary Medical Association. 1998;212(7):1001–1005. [PubMed] [Google Scholar]
- 19.Cooke CL, Singer RS, Jang SS, Hirsh DC. Enrofloxacin resistance in Escherichia coli isolated from dogs with urinary tract infections. Journal of the American Veterinary Medical Association. 2002;220(2):190–192. doi: 10.2460/javma.2002.220.190. [DOI] [PubMed] [Google Scholar]
- 20.May BJ, Zhang Q, Li LL, Paustian ML, Whittam TS, Kapur V. Complete genomic sequence of Pasteurella multocida, Pm70. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(6):3460–3465. doi: 10.1073/pnas.051634598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 22.Jones P, Côté RG, Martens L, et al. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Research. 2006;34, database issue:D659–D663. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orchard S, Jones AR, Stephan C, Binz P-A. The HUPO Pre-Congress Proteomics Standards Initiative Workshop HUPO 5th Annual World Congress Long Beach, CA, USA 28 October-1 November 2006. Proteomics. 2007;7(7):1006–1008. doi: 10.1002/pmic.200700014. [DOI] [PubMed] [Google Scholar]
- 24.Bridges SM, Magee GB, Wang N, Williams WP, Burgess SC, Nanduri B. ProtQuant: a tool for the label-free quantification of MudPIT proteomics data. BMC Bioinformatics. 2007;8, supplement 7:S24. doi: 10.1186/1471-2105-8-S7-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical Chemistry. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 26.Novichkova S, Egorov S, Daraselia N. MedScan, a natural language processing engine for MEDLINE abstracts. Bioinformatics. 2003;19(13):1699–1706. doi: 10.1093/bioinformatics/btg207. [DOI] [PubMed] [Google Scholar]
- 27.Kim JY, Lee JH, Park GW, et al. Utility of electrophoretically derived protein mass estimates as additional constraints in proteome analysis of human serum based on MS/MS analysis. Proteomics. 2005;5(13):3376–3385. doi: 10.1002/pmic.200401220. [DOI] [PubMed] [Google Scholar]
- 28.Gardy JL, Spencer C, Wang K, et al. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Research. 2003;31(13):3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turck CW, Falick AM, Kowalak JA, et al. The association of biomolecular resource facilities proteomics research group 2006 study: relative protein quantitation. Molecular and Cellular Proteomics. 2007;6(8):1291–1298. doi: 10.1074/mcp.M700165-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Mellor JC, Yanai I, Clodfelter KH, Mintseris J, DeLisi C. Predictome: a database of putative functional links between proteins. Nucleic Acids Research. 2002;30(1):306–309. doi: 10.1093/nar/30.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy FM, Bridges SM, Wang N, et al. AgBase: a unified resource for functional analysis in agriculture. Nucleic Acids Research. 2007;35, database issue:D599–D603. doi: 10.1093/nar/gkl936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis RJ. Protein folding: inside the cage. Nature. 2006;442(7101):360–362. doi: 10.1038/442360a. [DOI] [PubMed] [Google Scholar]
- 33.Herman C, Thevenet D, D'Ari R, Bouloc P. Degradation of σ 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(8):3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaara M, Nurminen M. Outer membrane permeability barrier in Escherichia coli mutants that are defective in the late acyltransferases of lipid A biosynthesis. Antimicrobial Agents and Chemotherapy. 1999;43(6):1459–1462. doi: 10.1128/aac.43.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vuorio R, Vaara M. The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrobial Agents and Chemotherapy. 1992;36(4):826–829. doi: 10.1128/aac.36.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plumbridge JA. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Molecular Microbiology. 1991;5(8):2053–2062. doi: 10.1111/j.1365-2958.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- 37.Datta DB, Arden B, Henning U. Major proteins of the Escherichia coli outer cell envelope membrane as bacteriophage receptors. Journal of Bacteriology. 1977;131(3):821–829. doi: 10.1128/jb.131.3.821-829.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugawara E, Nikaido H. Pore-forming activity of OmpA protein of Escherichia coli . Journal of Biological Chemistry. 1992;267(4):2507–2511. [PubMed] [Google Scholar]
- 39.He B, Zalkin H. Regulation of Escherichia coli purA by purine repressor, one component of a dual control mechanism. Journal of Bacteriology. 1994;176(4):1009–1013. doi: 10.1128/jb.176.4.1009-1013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sozhamannan S, Stitt BL. Effects on mRNA degradation by Escherichia coli transcription termination factor Rho and pBR322 copy number control protein Rop. Journal of Molecular Biology. 1997;268(4):689–703. doi: 10.1006/jmbi.1997.1004. [DOI] [PubMed] [Google Scholar]
- 41.Molinari G, Guzman CA, Pesce A, Schito GC. Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. Journal of Antimicrobial Chemotherapy. 1993;31(5):681–688. doi: 10.1093/jac/31.5.681. [DOI] [PubMed] [Google Scholar]
- 42.Nagino K, Kobayashi H. Influence of macrolides on mucoid alginate biosynthetic enzyme from Pseudomonas aeruginosa . Clinical Microbiology and Infection. 1997;3(4):432–439. doi: 10.1111/j.1469-0691.1997.tb00279.x. [DOI] [PubMed] [Google Scholar]
- 43.Wagner T, Soong G, Sokol S, Saiman L, Prince A. Effects of azithromycin on clinical isolates of Pseudomonas aeruginosa from cystic fibrosis patients. Chest. 2005;128(2):912–919. doi: 10.1378/chest.128.2.912. [DOI] [PubMed] [Google Scholar]
- 44.Dabo SM, Confer AW, Quijano-Blas RA. Molecular and immunological characterization of Pasteurella multocida serotype A:3 OmpA: evidence of its role in P. multocida interaction with extracellular matrix molecules. Microbial Pathogenesis. 2003;35(4):147–157. doi: 10.1016/s0882-4010(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 45.Lee J-S, Heo Y-J, Lee JK, Cho Y-H. KatA, the major catalase, is critical for osmoprotection and virulence in Pseudomonas aeruginosa PA14. Infection and Immunity. 2005;73(7):4399–4403. doi: 10.1128/IAI.73.7.4399-4403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poyart C, Pellegrini E, Gaillot O, Boumaila C, Baptista M, Trieu-Cuot P. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae . Infection and Immunity. 2001;69(8):5098–5106. doi: 10.1128/IAI.69.8.5098-5106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egea L, Aguilera L, Giménez R, et al. Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: interaction of the extracellular enzyme with human plasminogen and fibrinogen. The International Journal of Biochemistry & Cell Biology. 2007;39(6):1190–1203. doi: 10.1016/j.biocel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 48.Liu K, Yu J, Russell DG. pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiology. 2003;149(7):1829–1835. doi: 10.1099/mic.0.26234-0. [DOI] [PubMed] [Google Scholar]
- 49.Garduño RA, Garduño E, Hoffman PS. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infection and Immunity. 1998;66(10):4602–4610. doi: 10.1128/iai.66.10.4602-4610.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takaya A, Tomoyasu T, Matsui H, Yamamoto T. The DnaK/DnaJ chaperone machinery of Salmonella enterica serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages, leading to systemic infection. Infection and Immunity. 2004;72(3):1364–1373. doi: 10.1128/IAI.72.3.1364-1373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poole K. Resistance to β-lactam antibiotics. Cellular and Molecular Life Sciences. 2004;61(17):2200–2223. doi: 10.1007/s00018-004-4060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yethon JA, Whitfield C. Lipopolysaccharide as a target for the development of novel therapeutics in Gram-negative bacteria. Current Drug Targets. Infectious Disorders. 2001;1(2):91–106. doi: 10.2174/1568005014606143. [DOI] [PubMed] [Google Scholar]
- 53.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Research. 2004;32(9):2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cascales E, Bernadac A, Gavioli M, Lazzaroni J-C, Lloubes R. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. Journal of Bacteriology. 2002;184(3):754–759. doi: 10.1128/JB.184.3.754-759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki H, Nishimura Y, Yasuda S, Nishimura A, Yamada M, Hirota Y. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Molecular and General Genetics. 1978;167(1):1–9. doi: 10.1007/BF00270315. [DOI] [PubMed] [Google Scholar]
- 56.Haney SA, Platko JV, Oxender DL, Calvo JM. Lrp, a leucine-responsive protein, regulates branched-chain amino acid transport genes in Escherichia coli . Journal of Bacteriology. 1992;174(1):108–115. doi: 10.1128/jb.174.1.108-115.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All proteins identified in this study are listed in Supplementary Table 1. Proteins with significant quantitative changes in expression in response to sub-MICs of AMX, CTC, or ENR are listed in Supplementary Table 2. Protein interaction networks built with significant changes in expression with sub-MICs shown in Table 3 are included in the supplementary figure. This figure shows changes in protein expression in response to sub-MICs of AMX, CTC, and ERN. Red and green indicate significant increase and decrease in expression, respectively. Proteins whose expression did not change (pink) and proteins in the network not identified in our datasets (gray) are also included.