Abstract
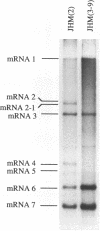
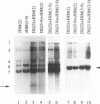
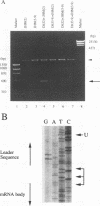
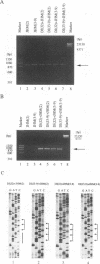
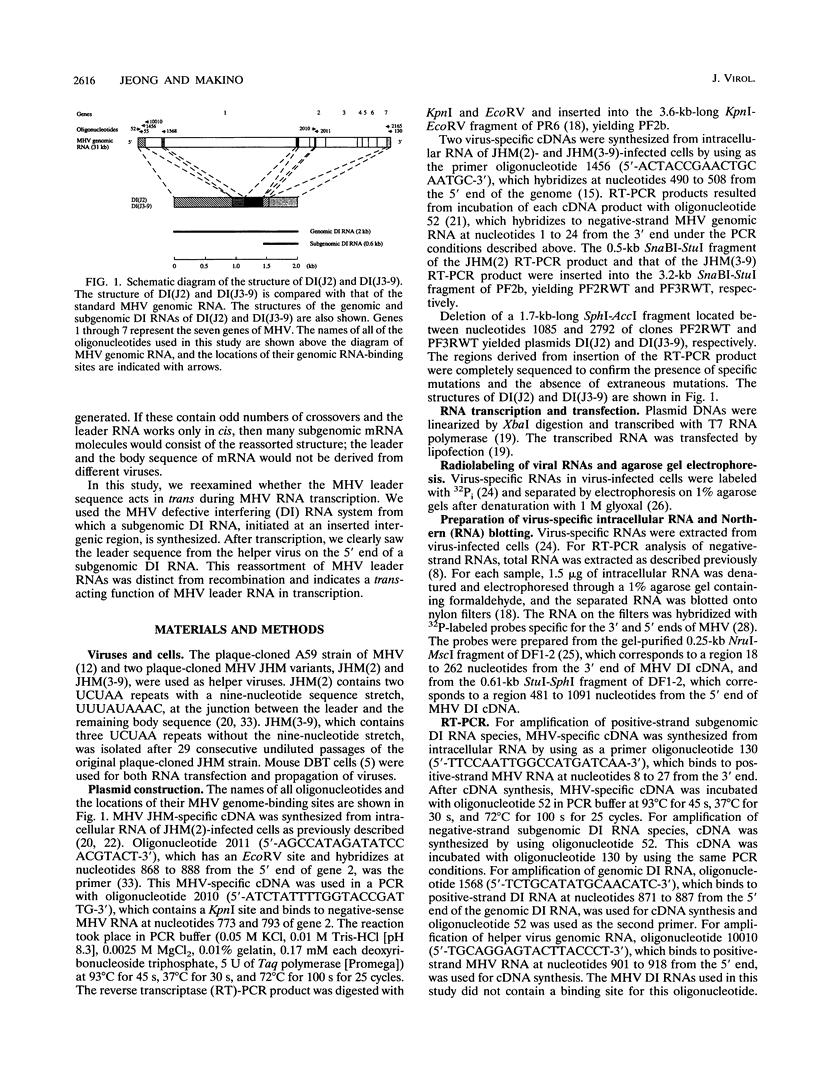
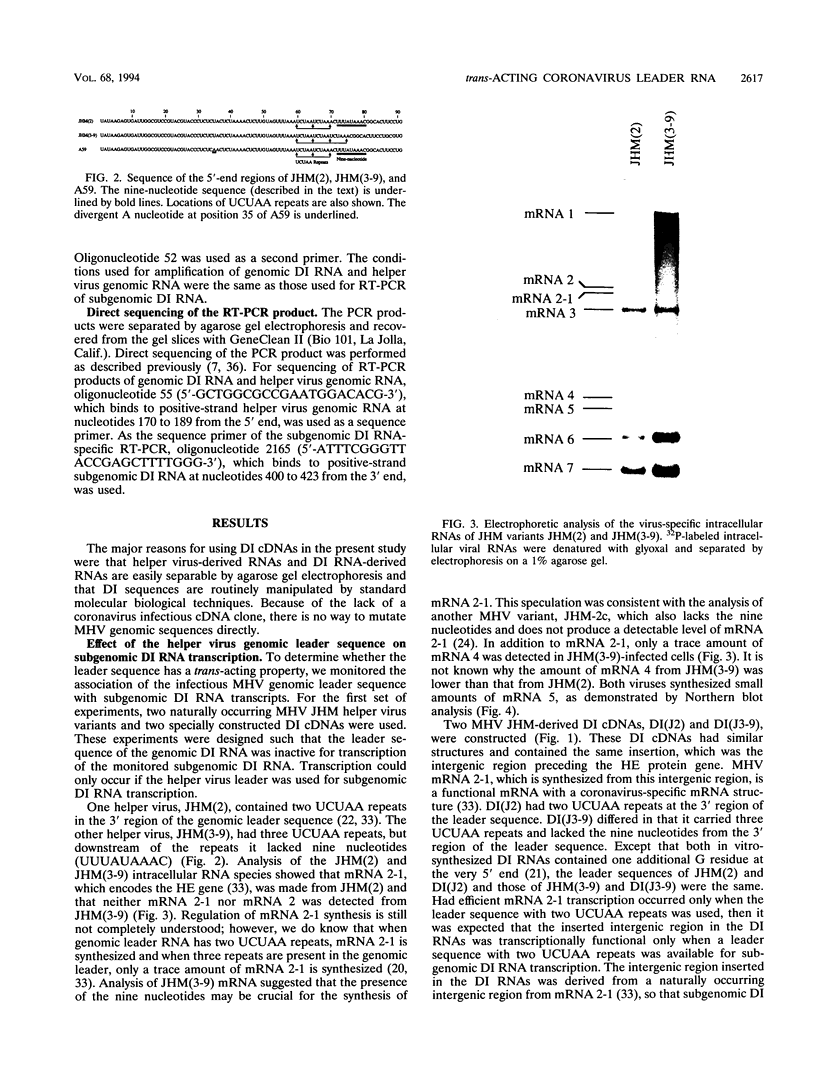
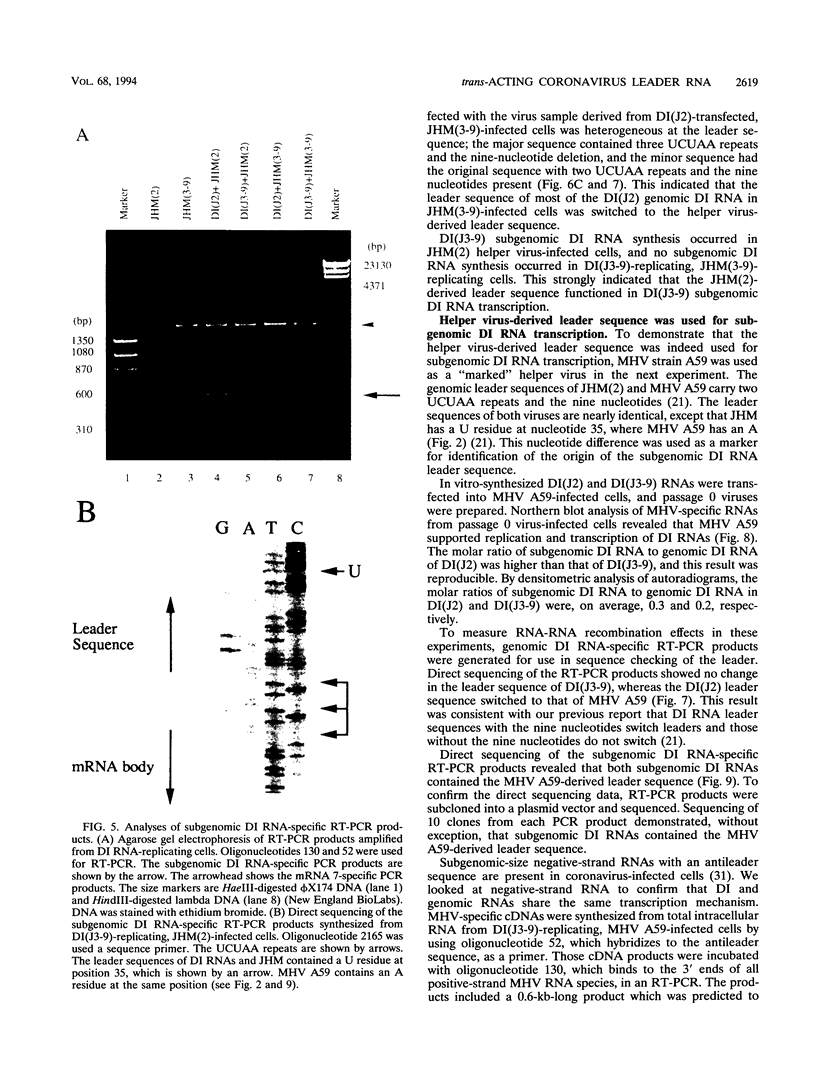
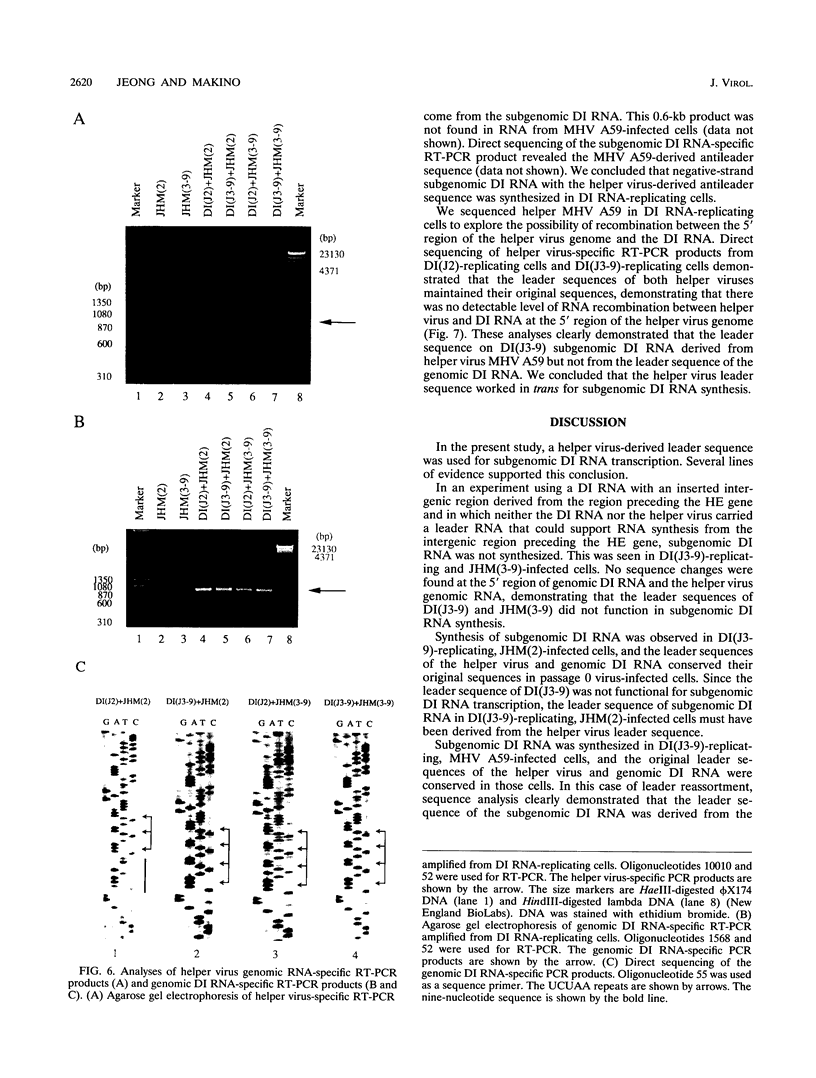
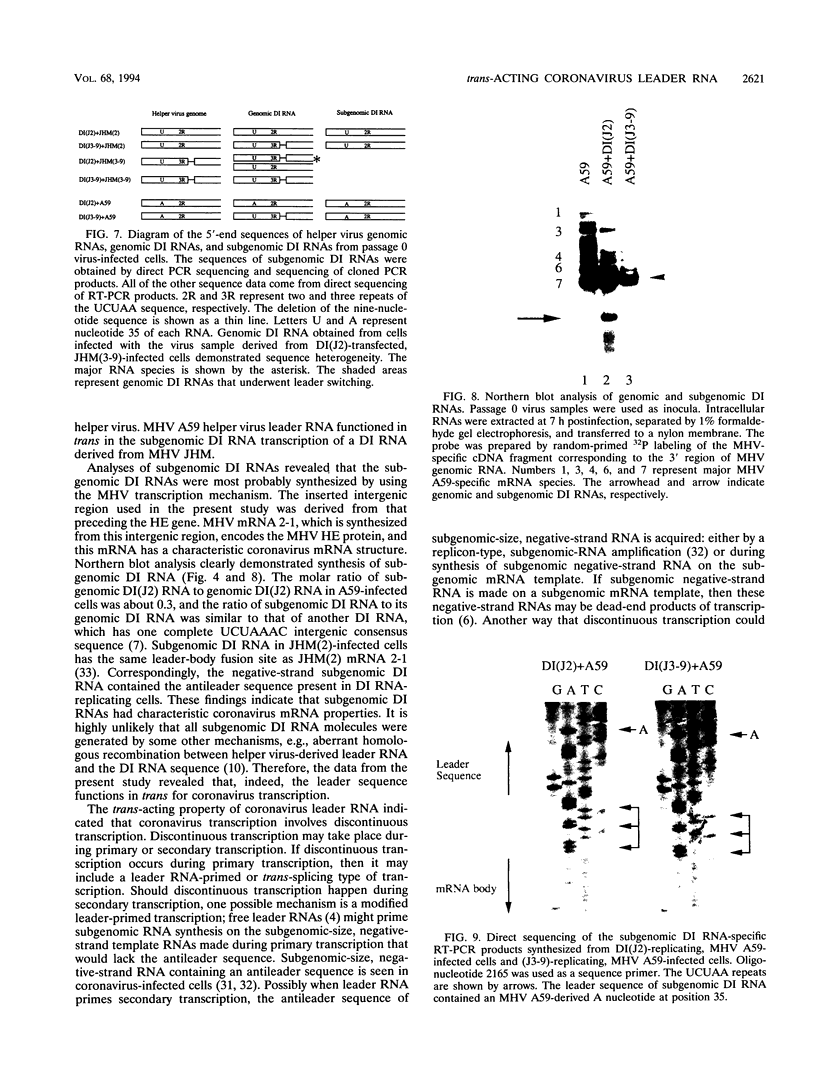
Coronavirus subgenomic mRNA possesses a 5'-end leader sequence which is derived from the 5' end of genomic RNA and is linked to the mRNA body sequence. This study examined whether coronavirus transcription involves a discontinuous transcription step; the possibility that a leader sequence from mouse hepatitis virus (MHV) genomic RNA could be used for MHV subgenomic defective interfering (DI) RNA transcription was examined. This was tested by using helper viruses and DI RNAs that were easily distinguishable. MHV JHM variant JHM(2), which synthesizes a subgenomic mRNA encoding the HE gene, and variant JHM(3-9), which does not synthesize this mRNA, were used. An MHV DI RNA, DI(J3-9), was constructed to contain a JHM(3-9)-derived leader sequence and an inserted intergenic region derived from the region preceding the MHV JHM HE gene. DI(J3-9) replicated efficiently in JHM(2)- or JHM(3-9)-infected cells, whereas synthesis of subgenomic DI RNAs was observed only in JHM(2)-infected cells. Sequence analyses demonstrated that the 5' regions of both helper virus genomic RNAs and genomic DI RNAs maintained their original sequences in DI RNA-replicating cells, indicating that the genomic leader sequences derived from JHM(2) functioned for subgenomic DI RNA transcription. Replication and transcription of DI(J3-9) were observed in cells infected with an MHV A59 strain whose leader sequence was similar to that of JHM(2), except for one nucleotide substitution within the leader sequence. The 5' region of the helper virus genomic RNA and that of the DI RNA were the same as their original structures in virus-infected cells, and the leader sequence of DI(J3-9) subgenomic DI RNA contained the MHV A59-derived leader sequence. The leader sequence of subgenomic DI RNA was derived from that of helper virus; therefore, the genomic leader sequence had a trans-acting property indicative of a discontinuous step in coronavirus transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker S. C., Lai M. M. An in vitro system for the leader-primed transcription of coronavirus mRNAs. EMBO J. 1990 Dec;9(12):4173–4179. doi: 10.1002/j.1460-2075.1990.tb07641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Fu K., Schaad M. C., Stohlman S. A. Establishing a genetic recombination map for murine coronavirus strain A59 complementation groups. Virology. 1990 Aug;177(2):646–656. doi: 10.1016/0042-6822(90)90530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Lai M. M. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983 Dec;48(3):633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Razavi M. K., Lai M. M. Characterization of leader-related small RNAs in coronavirus-infected cells: further evidence for leader-primed mechanism of transcription. Virus Res. 1985 Jul;3(1):19–33. doi: 10.1016/0168-1702(85)90038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N., Fujiwara K., Hino S., Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44(3):298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- Jeong Y. S., Makino S. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J Virol. 1992 Jun;66(6):3339–3346. doi: 10.1128/jvi.66.6.3339-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M., Makino S. Mutagenic analysis of the coronavirus intergenic consensus sequence. J Virol. 1992 Nov;66(11):6330–6337. doi: 10.1128/jvi.66.11.6330-6337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. N., Lai M. M., Makino S. Generation and selection of coronavirus defective interfering RNA with large open reading frame by RNA recombination and possible editing. Virology. 1993 May;194(1):244–253. doi: 10.1006/viro.1993.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Baric R. S., Brayton P. R., Stohlman S. A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Brayton P. R., Armen R. C., Patton C. D., Pugh C., Stohlman S. A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J Virol. 1981 Sep;39(3):823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Lai M. M., Patton C. D., Baric R. S., Stohlman S. A. Presence of leader sequences in the mRNA of mouse hepatitis virus. J Virol. 1983 Jun;46(3):1027–1033. doi: 10.1128/jvi.46.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. RNA recombination in animal and plant viruses. Microbiol Rev. 1992 Mar;56(1):61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Stohlman S. A. RNA of mouse hepatitis virus. J Virol. 1978 May;26(2):236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J. L., Wilhelmsen K. C., Bond C. W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology. 1981 Oct 15;114(1):39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J Virol. 1993 Jun;67(6):3304–3311. doi: 10.1128/jvi.67.6.3304-3311.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M., Makino J. K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991 Nov;65(11):6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Keck J. G., Stohlman S. A., Lai M. M. High-frequency RNA recombination of murine coronaviruses. J Virol. 1986 Mar;57(3):729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Lai M. M. Evolution of the 5'-end of genomic RNA of murine coronaviruses during passages in vitro. Virology. 1989 Mar;169(1):227–232. doi: 10.1016/0042-6822(89)90060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Lai M. M. High-frequency leader sequence switching during coronavirus defective interfering RNA replication. J Virol. 1989 Dec;63(12):5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Soe L. H., Shieh C. K., Lai M. M. Discontinuous transcription generates heterogeneity at the leader fusion sites of coronavirus mRNAs. J Virol. 1988 Oct;62(10):3870–3873. doi: 10.1128/jvi.62.10.3870-3873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Stohlman S. A., Lai M. M. Leader sequences of murine coronavirus mRNAs can be freely reassorted: evidence for the role of free leader RNA in transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4204–4208. doi: 10.1073/pnas.83.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Hirano N., Fujiwara K. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology. 1984 Nov;139(1):138–151. doi: 10.1016/0042-6822(84)90335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Yokomori K., Lai M. M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990 Dec;64(12):6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol. 1986 Jan;57(1):328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990 Mar;64(3):1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hofmann M. A., Brian D. A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991 Jan;65(1):320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hung S. L., Brian D. A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh C. K., Lee H. J., Yokomori K., La Monica N., Makino S., Lai M. M. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J Virol. 1989 Sep;63(9):3729–3736. doi: 10.1128/jvi.63.9.3729-3736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., Banner L. R., Lai M. M. Coronavirus mRNA transcription: UV light transcriptional mapping studies suggest an early requirement for a genomic-length template. J Virol. 1992 Aug;66(8):4671–4678. doi: 10.1128/jvi.66.8.4671-4678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]