Abstract
In response to cancer, AIDS, sepsis and other systemic diseases inducing muscle atrophy, the E3 ubiquitin ligase Atrogin1/MAFbx (MAFbx) is dramatically upregulated and this response is necessary for rapid atrophy. However, the precise function of MAFbx in muscle wasting has been questioned. Here, we present evidence that during muscle atrophy MAFbx targets the eukaryotic initiation factor 3 subunit 5 (eIF3-f) for ubiquitination and degradation by the proteasome. Ectopic expression of MAFbx in myotubes induces atrophy and degradation of eIF3-f. Conversely, blockade of MAFbx expression by small hairpin RNA interference prevents eIF3-f degradation in myotubes undergoing atrophy. Furthermore, genetic activation of eIF3-f is sufficient to cause hypertrophy and to block atrophy in myotubes, whereas genetic blockade of eIF3-f expression induces atrophy in myotubes. Finally, eIF3-f induces increasing expression of muscle structural proteins and hypertrophy in both myotubes and mouse skeletal muscle. We conclude that eIF3-f is a key target that accounts for MAFbx function during muscle atrophy and has a major role in skeletal muscle hypertrophy. Thus, eIF3-f seems to be an attractive therapeutic target.
Keywords: Atrogin/MAFbx, atrophy, eIF3-f, hypertrophy, ubiquitin–proteasome
Introduction
A dynamic balance between anabolic and catabolic processes controls skeletal muscle size. These two opposing phenomena are mechanically linked, in that either the activity or inactivity of a common set of molecules controlling a few cellular pathways determines whether the skeletal muscle tissue will respond to defined stimuli with increased protein synthesis and stimulation of cell growth (hypertrophy) or with increased protein breakdown and reduced cell proliferation (atrophy). Skeletal muscle atrophy is characterized by a decrease in the size of pre-existing muscle fibres and is observed in many physiological and pathological settings. Loss of muscle mass results primarily from accelerated protein degradation through the ubiquitin–proteasome pathway (Jagoe and Goldberg, 2001). Proteins degraded by this mechanism are first covalently linked to a chain of ubiquitin molecules, which marks them for rapid degradation by the 26S proteasome (Hershko and Ciechanover, 1998). The formation of ubiquitin–protein conjugates involves three components that participate in a cascade of ubiquitin transfer reactions: a ubiquitin-activation enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a ubiquitin ligase (E3) that acts at the last step of the cascade. The proteolysis of many cellular regulators is controlled by SCF (Skp1–Cul1–F-box protein) ubiquitin ligases (Guardavaccaro and Pagano, 2006). The F-box protein subunit provides specificity of SCF by recruiting the substrate (Winston et al, 1999). In multiple models of skeletal muscle atrophy, the muscle-specific F-box protein MAFbx/Atrogin1 (MAFbx) is upregulated and appears to be essential for accelerated muscle protein loss in a variety of experimental models of catabolisms, including diabetes, cancer, AIDS, fasting, renal failure, hind limb suspension, immobilization, oxidative stress and sepsis (Sacheck et al, 2007). Overexpression of MAFbx in skeletal myotubes leads to atrophy, and mice deficient in MAFbx are resistant to denervation atrophy of skeletal muscle (Bodine et al, 2001a). These observations suggest that MAFbx may regulate muscle size in association with a muscle-specific ubiquitin ligase complex but the substrates of this E3 are unknown.
The FOXO family of transcription factors was reported to regulate MAFbx transcription and to influence muscle atrophy. On the one hand, Akt negatively regulates FOXO transcription factors by phosphorylating them and promoting their nuclear export into the cytoplasm, where they are retained through association with the 14-3-3 proteins (Brunet et al, 1999). On the other hand, the IGF-1/Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo (Bodine et al, 2001b). Recently, it has been shown that in response to nutrients, energy sufficiency, hormones and mitogenic agents, the kinases mTOR and S6K1 mediate the assembly of the translation preinitiation complex by binding to the eIF3 complex through dynamic protein interchange and ordered phosphorylation events (Holz et al, 2005).
eIF3 is the largest (650 kDa) and one of the most complex initiation factors known (Hershey and Merrick, 2000). Mammalian eIF3 is composed of at least 12 individual subunits designated eIF3a-eIF3-k, in the order of decreasing size (from 166 500 to 25 100 Da) (Browning et al, 2001). The exact interactions and composition of the eIF3 subunits are poorly understood. eIF3 is a multifunctional initiation factor that has been shown to operate at different levels of the initiation pathway (Hinnebusch, 2006). Studies in Saccharomyces cerevisiae indicate that eIF3-a, eIF3-b, eIF3-c, eIF3-g and eIF3-i form an essential core, with the other subunits serving regulatory roles (Asano et al, 1998). Several eIF3 subunits have been known to be important in alternative, cap-independent mechanisms of translation that are still being elucidated (Bandyopadhyay and Maitra, 1999).
The MAFbx protein lacks common domains found in other F-box proteins that are known to interact with substrates. Almost all targets of F-box proteins identified to date interact with sequences located C-terminal to the F-box motif. Indeed, the COOH part of MAFbx interacts with cytoplasmic proteins such as calcineurin A and α-actinin-2 at the Z-disc in cardiomyocytes and inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex (Li et al, 2004). Furthermore the sequence located N-terminal to the F-box of MAFbx contains putative interacting protein domains such as a leucine zipper domain and a leucine-charged residue-rich domain (LCD), suggesting that potentially MAFbx could recognize different substrates. Indeed, we previously showed that the LCD domain of MAFbx interacts with MyoD, a critical transcription factor in muscle development, differentiation and regeneration. Ectopic expression of MAFbx in proliferating myoblasts antagonizes differentiation, inducing MyoD degradation and preventing muscle specific-gene activation (Tintignac et al, 2005).
To further determine substrates that account for MAFbx effect on muscle atrophy, we screened a human skeletal muscle library to identify proteins that interact with the NH2 domain of MAFbx using a yeast two-hybrid assay. Here, we report the identification of the eukaryotic initiation factor 3 subunit 5 (eIF3-f), which exhibited strong and specific interaction with MAFbx. eIF3-f is a subunit of the mammalian eIF3 multiprotein complex that binds to the 40S ribosome and promotes the binding of methionyl-tRNA and mRNA. Here, we demonstrate that eIF3-f is a target for MAFbx function in skeletal muscle atrophy. Further studies show that conditional repression of eIF3-f in differentiated myotubes causes a dramatic decrease in fibre size, whereas ectopic expression of eIF3-f promotes muscle hypertrophy by increasing muscle protein expression. Our surprising discovery that atrophy and hypertrophy pathways communicate with each other through eIF3-f should help in the search for the primary control mechanism mediating skeletal muscle atrophy and hypertrophy and to discover promising targets for developing novel therapies to combat muscle wasting.
Results
Evidence of interaction between eIF3-f and MAFbx
Stringent screening of human skeletal muscle yeast two-hybrid library with bait containing the N-terminal fragment of human MAFbx (aa 1–227) yielded repeatedly the same major hit: eIF3-f. The encoding region of the human eIF3-f cDNA is composed of 1231 bp and translates into a 47-kDa protein. To investigate the interaction between MAFbx and eIF3-f, we determined whether Myc-eIF3-f that had been overexpressed in C2C12 cells could be captured in vitro using glutathione-S-transferase (GST)–MAFbx bound to GSH–agarose (Figure 1A). As a control, we also assessed retention of Myc–eIF3-f by a resin prepared by binding GST to GSH–agarose. After incubating the two resins with total cell extracts, the samples were washed and the bound proteins were analysed by SDS–PAGE. The amount of Myc–eIF3-f recovered with the MAFbx affinity resin was much greater than that retained by the GST resin, indicating an interaction between MAFbx and eIF3-f. Next in transiently transfected C2C12 myoblasts, we determined whether an association could be detected between epitope-MAFbx and eIF3-f. Flag–MAFbx co-immunoprecipitated with Myc-eIF3-f and Myc-eIF3-f co-immunoprecipitated with Flag–MAFbx (Figure 1B) but did not associate with other ubiquitin ligases such as MuRF-1 (upregulated during atrophy) or Emi1 (G2/M transition) (Supplementary Figure S1A). Furthermore, the subunit eIF3-i did not co-immunoprecipitate with MAFbx in cotransfection experiments (Supplementary Figure S1B). Preliminary data indicated that eIF3-f is expressed in C2C12 myotubes (Supplementary Figure S2) and immunostaining experiments revealed that eIF3-f is localized to the cytoplasm (Figure 1C). MAFbx contains two nuclear localization signals and interacts with MyoD in the nucleus (Tintignac et al, 2005). Overexpression of MAFbx in myotubes led to atrophy (Bodine et al, 2001a) and triggered the translocation in part of eIF3-f from the cytoplasm to the nucleus (Figure 1C). In starvation-induced atrophy of C2C12 myotubes, we observed co-immunoprecipitation of endogenous eIF3-f with MAFbx (Supplementary Figure S1C) and an increase in nuclear endogenous eIF3-f and MAFbx (Figure 1D). Altogether, these results support the conclusion that MAFbx binds eIF3-f and suggest that MAFbx may control the function of eIF3-f during muscle atrophy.
Figure 1.
Interaction of the E3 ubiquitin ligase MAFbx with eIF3-f. (A) Interaction of MAFbx and eIF3-f. GST or GST–MAFbx beads (2 μg) were incubated with extracts (200 μg) of C2C12 myoblasts overexpressing Myc-tagged eIF3-f. After washing resins, the bound proteins were eluted, subjected to SDS–PAGE along with an extract sample and analysed by immunoblotting with anti-Myc antibody. (B) Co-immunoprecipitation of exogenously expressed MAFbx and eIF3-f. C2C12 myotubes were cotransfected with Myc-tagged eIF3-f and Flag-tagged MAFbx expression constructs. Total cellular extracts (250 μg) were subjected to immunoprecipitation with anti-Myc and/or anti-Flag, followed by immunoblotting analysis with anti-Flag and/or anti-Myc antibodies. (C) eIF3-f colocalizes in the nucleus in the presence of MAFbx. C2C12 myotubes were cotransfected with mammalian expression plasmids encoding GFP–MAFbx and Myc–eIF3-f and, after 24 h, the transfected cells were fixed and immunostained with anti-Myc antibodies and DAPI. Images of a representative field were obtained by indirect immunofluorescence microscopy. Scale bar, 50 μm. (D) Confocal fluorescence imaging of endogenous eIF3-f during starvation-induced atrophy in C2C12 myotubes. Myotubes were fixed 6 h after food deprivation and stained for DNA (blue), eIF3-f (green, upper panels) and MAFbx (green, lower panels). Scale bar, 50 μm. Arrows indicate nuclear localization of eIF3-f and MAFbx during starvation-induced atrophy.
Interaction domains of MAFbx and eIF3-f
To map the domains of eIF3-f and MAFbx that are implicated in their interaction, a series of MAFbx (Supplementary Figure S3A) and eIF3-f (Supplementary Figure S3B) mutants were generated and used in in vitro protein binding experiments. Full-length in vitro-translated eIF3-f efficiently bound to GST–MAFbx. Removing amino acids 1–140 from the N terminus of MAFbx did not affect the interaction with eIF3-f. In contrast, removing amino acids 1–222 completely abolished the association of GST–MAFbx with eIF3-f. Furthermore, a deletion of the F-box domain (ΔF-box) did not affect its binding to eIF3-f. On examination of additional overlapping deletion mutants, we found that the LCD domain mediated binding of MAFbx to eIF3-f. We performed reciprocal binding experiments with in vitro-translated MAFbx and GST–eIF3-f mutants. Full-length eIF3-f bound to MAFbx. Mutations that deleted amino acids 1–94 in the N terminus did not affect the interaction with MAFbx. A mutation that deleted amino acids 230–357 of the C terminus reduced association between eIF3-f and MAFbx. Both the N- and C-terminal regions of the molecule could be deleted and the resulting minimal central domain containing amino-acid residues 94–230 was sufficient for a high specific binding to MAFbx. The protein sequence of this minimal domain was found to correspond to the Mov-34 domain of eIF3-f. Thus, results from these experiments support the conclusion that the LCD domain of MAFbx physically interacts with the Mov34 domain of eIF3-f.
Degradation of eIF3-f in myotubes undergoing atrophy
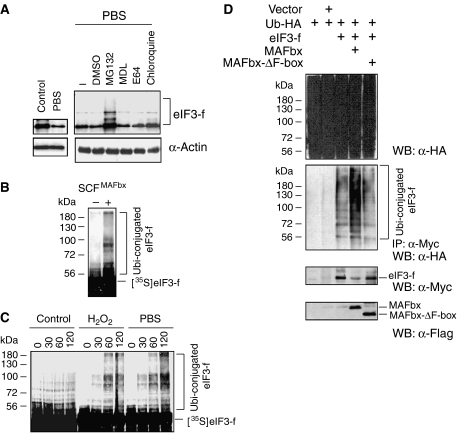
Because little is known about the expression of eIF3-f in muscle cells we initially characterized eIF-3-f expression during myogenic differentiation using the C2C12 cell line, a well-defined model for ex vivo differentiation. These cells proliferate as myoblasts in high serum concentrations and can be induced to differentiate by reducing the serum concentration from 20 to 2%. Under our conditions, C2C12 myoblasts fuse into myotubes within 48–60 h with an efficiency of 75–80%. Western blot analysis revealed that the eIF3-f protein is barely detectable in proliferating myoblasts, but is dramatically upregulated during terminal differentiation (Supplementary Figure S2) and is present in adult skeletal muscle (yeast two-hybrid library). This led us to investigate whether eIF3-f could be a substrate of MAFbx in experimental conditions previously shown to induce atrophy of C2C12 myotubes. In response to treatment with dexamethasone (DEX), food deprivation (PBS) and/or oxidative stress (H2O2), which all activated MAFbx RNA expression (Hasselgren, 1999; Li et al, 2003; Sandri et al, 2004; Stitt et al, 2004), C2C12 myotubes showed a distinct atrophic phenotype characterized by a decrease of ∼40–60% in myotube diameter and in the loss of myotube proteins (Supplementary Figure S4A–C). We also observed an increase of 2- to 4-fold in MAFbx protein levels and downregulation of eIF3-f protein expression during muscle atrophy (Figure 2A). Next, we asked whether eIF3-f regulation occurred at the RNA level. Semiquantitative RT–PCR results showed that food deprivation, increasing concentration of DEX or oxidative stress had no significant effect on the expression of eIF3-f mRNA (Figure 2B). We used small hairpin RNA interference (shRNAi) in atrophying C2C12 myotubes during fasting and oxidative stress to determine the role of MAFbx in the degradation of eIF3-f. Knockdown of MAFbx in these myotubes prevented by more than 50% the degradation of eIF3-f. By contrast, a control shRNAi did not affect the degradation of eIF3-f (Figure 2C). Subsequent studies examined whether MAFbx had similar effects on eIF3-f expression in adult mouse muscles. eIF3-f expression was reduced by 70% after induction of MAFbx in atrophying muscle during fasting. This change was completely reversed by supplying food to fasting mice for 2 days (Figure 2D). These findings indicate that MAFbx and eIF3-f showed a contrasting pattern of expression during skeletal muscle atrophy.
Figure 2.
Glucocorticoid treatment, starvation of cells and oxidative stress induced upregulation of MAFbx and degradation of eIF3-f. (A) Myotubes at 3 days of differentiation were treated with 225 μM H2O2 and 1, 10 and 50 μM DEX for 24 h or were starved by removal of growth medium, amino acids and glucose and incubated in PBS. Proteins were extracted and subjected to immunoblot analysis with specific antibodies against MAFbx and eIF3-f. α-Tubulin is shown as a loading control. (B) Expression of eIF3-f mRNA was analysed by semiquantitative RT–PCR. Myotubes were treated with increasing concentrations of DEX for 24 h (1, 10 and 50 μM), or incubated either in PBS for 6 h and medium was replaced in refed cultures for 12 h or in H2O2 (225 μM) for 4 and 9 h. GAPDH expression was used as an internal control. (C) Depletion of MAFbx affects eIF3-f degradation in C2C12 myotubes undergoing atrophy. C2C12 myoblasts were transfected with pTER+MAFbx or pTER+control. Myotubes at 2 days of differentiation were treated with H2O2 (225 μM) or were incubated in PBS and myotubes were harvested 6 h later. Cell lysates were prepared and total proteins were subjected to western blot with anti-eIF3-f and anti-MAFbx antibodies. (D) Opposite expression of MAFbx and eIF3-f in the skeletal muscle of mice during starvation and refeeding. Shown is an immunoblot analysis of protein extracts from hind limb muscles, using anti-eIF3-f, anti-MAFbx and anti-tropomyosin antibodies. Mice were starved for 48 h (starved) and refed for 48 h (Refed). Control mice were fed randomly.
Mediation of eIF3-f polyubiquitination by the SCFMAFbx
Given that many F-box proteins can target regulatory proteins for degradation by the ubiquitin–proteasome pathway, we hypothesized that MAFbx might target eIF3-f for SCF-mediated ubiquitination and thereby promote its turnover by the proteasome. To address this possibility, we first examined the effect of different protease inhibitors on the amount of eIF3-f in starved C2C12 myotubes. Addition of E64 (a cysteine protease inhibitor) E64, chloroquine (an inhibitor of lysosomal proteolysis), MDL (an inhibitor of calpains) and/or DMSO was ineffective in preventing the degradation of eIF3-f. In contrast, addition of the proteasome inhibitor MG132 induced an accumulation of eIF3-f protein in myotubes undergoing atrophy (Figure 3A). Thus, the proteasome appears to be the main pathway involved in the degradation of eIF3-f during muscle atrophy.
Figure 3.
Food deprivation- and oxidative stress-induced atrophy increases polyubiquitination and degradation of eIF3-f by the SCFMAFbx pathway. (A) C2C12 myotubes at day 4 of differentiation were incubated in PBS for 4 h in the presence of DMSO, MG132 (30 μM), MDL (50 μM), E64 (30 μM) and chloroquine (40 μM). Fifty micrograms of total proteins was subjected to western blotting with anti-eIF3-f antibodies. Anti-α-tubulin antibodies were used as an internal control. (B) The recombinant SCFMAFbx complex was assayed for the ability to mediate polyubiquitination of eIF3-f in the presence of E1, ATP, ubiquitin and cdc34 (E2). Reaction mixtures were separated on SDS–PAGE, followed by autoradiography. (C) Myotubes at 4 days of differentiation were treated with 225 μM H2O2 (9 h) and/or were starved by removal of growth medium and incubated in PBS (6 h). Cell lysates were prepared and 30 μg of total proteins was used for in vitro ubiquitination assay. 35S-labelled eIF3-f was subjected to in vitro ubiquitination reaction in the presence of normal and/or atrophic C2C12 myotube lysates. Reaction mixtures were separated on SDS–PAGE, followed by autoradiography. (D) MAFbx increases eIF3-f ubiquitination in vivo. C2C12 myotubes were transiently transfected with expression plasmids encoding Myc-eIF3-f, HA–ubiquitin, Flag–MAFbx and/or the F-box mutant Flag–MAFbx-ΔF-box. Transfected cells were treated for 2 h with 50 μM MG132 before harvesting. Cells were lysed in denaturation buffer containing 0.5% SDS. Aliquots (10%) were analysed by western blotting with anti-HA antibodies (upper panel). Cell lysates were then diluted in immunoprecipitation buffer, subjected to immunoprecipitation with anti-Myc antibodies (middle panel) and analysed by western blotting with anti-Flag, anti-Myc and anti-HA antibodies (lower panels).
The association of MAFbx with the essential Skp1, Roc 1 and Cul1 proteins, specific components of an E3 ubiquitin–protein ligase (SCFMAFbx), was previously described (Gomes et al, 2001; Li et al, 2004; Tintignac et al, 2005). In an in vitro ubiquitination assay, SCFMAFbx purified from recombinant baculoviruses produced in Sf9 cells (Tintignac et al, 2005) catalysed the polyubiquitination of eIF3-f (Figure 3B). As food deprivation and/or the oxidative stress lead to rapid myotube atrophy and a large increase in MAFbx (Figure 2A), we studied the effects of SCFMAFbx from these atrophic myotubes on eIF3-f ubiquitination. In an in vitro ubiquitination assay using total cellular extracts, the polyubiquitination of eIF3-f was dramatically increased in extracts from atrophic myotubes as compared with extracts from normal C2C12 myotubes (Figure 3C). Knockdown of MAFbx in atrophic myotubes prevented the polyubiquitination of eIF3-f. By contrast, a control shRNAi did not affect the polyubiquitination of eIF3-f (Supplementary Figure S5). Overexpression of MAFbx has been shown to induce atrophy in C2C12 myotubes (Figure 1C), suggesting that MAFbx-inducing C2C12 atrophy might increase eIF3-f polyubiquitination. To assess the effect of MAFbx overexpression on eIF3-f protein ubiquitination, expression vectors encoding MAFbx-wt or the mutant MAFbx-ΔF-box (deletion of the F-box domain required for Skp1 interaction) were cotransfected with Myc-eIF3-f and HA–ubiquitin into C2C12 cells. Cells were collected, lysed in SDS-containing buffer and eIF3-f was immunoprecipitated with anti-Myc antibody. Immunoprecipitates were then probed with anti-HA to detect ubiquitinated eIF3-f proteins. In the presence of MAFbx-wt, eIF3-f showed increasing amounts of polyubiquitination whereas the mutant MAFbx-ΔF-box was ineffective in the ubiquitination of eIF3-f (Figure 3D). Thus, MAFbx increases polyubiquitination of eIF3-f in atrophic myotubes.
Accelerated degradation of eIF3-f by MAFbx
To examine whether MAFbx increases the turnover of eIF3-f in vivo, we cotransfected eIF3-f with MAFbx or the mutant MAFbx-ΔF-box into C2C12 cells and then monitored eIF3-f protein stability after cycloheximide (CHX) treatment. As expected, the empty vector did not affect eIF3-f stability (half-life was ∼120 min). Overexpression of MAFbx resulted in enhanced degradation of eIF3-f by decreasing its half-life to ∼70–80 min. By contrast, addition of the proteasome inhibitor MG132 in cells compromised SCFMAFbx activity and eIF3-f degradation was affected (half-life >180 min). Moreover, overexpression of the mutant MAFbx-ΔF-box increased the half-life of eIF3-f (>180 min), a value similar to that observed in the presence of MG132 (Figure 4). These results indicate that MAFbx promotes polyubiquitination of eIF3-f and reduces its half-life.
Figure 4.
MAFbx increases eIF3-f turnover. C2C12 myoblasts were cotransfected using the expression vector encoding Myc–eIF3-f and Flag-tagged MAFbx-wt or the mutant MAFbx-ΔF-box as indicated. The cells were treated with CHX 36 h after transfection to block protein synthesis. Cell lysates were prepared at the indicated time and analysed by immunoblotting. Quantification of eIF3-f turnover following CHX treatment based on densitometric scanning of two experiments is shown. MG132 was added to control eIF3-f degradation by the proteasome.
Genetic repression of eIF3-f in differentiated myotubes induces atrophy whereas genetic activation of eIF3-f is sufficient to block starvation-induced atrophy
To develop a controllable protein knockout method in myogenic cells by using full-length antisense RNA, we constructed a bidirectional overexpression vector (pBI-eGFP vector, Clontech) for antisense DNA sequences of mouse eIF3-f cDNA. This vector was transiently cotransfected in C2C12 myoblasts with a vector expressing the tetracycline-controlled reverse transactivator (pRev-Tet-Off) in the presence of tetracycline (Tetra). Transfected cells were cultured in DM for 3 days in the presence of Tetra and for 3 more days in the absence of Tetra and then examined for myotube size and nuclei number per myotube (Figure 5A). Six days after differentiation, the control myotubes were of constant size and myoblast fusion no longer occurred (Rommel et al, 2001). Myotubes expressing antisense eIF3-f RNAs demonstrated that endogenous eIF3-f protein levels were efficiently and specifically reduced especially at 72 h after Tetra release as determined by western blot analysis (Figure 5C). Morphological examination of myotubes indicated an alteration in the number of nuclei and a 40% reduction in mean myotube diameter compared with those not transfected and/or expressing only GFP (vector control), which did not change diameter or the endogenous eIF3-f protein levels during this time (Figure 5B and C). Interestingly, in atrophy induced by starvation for 7 h, the diameter was of the same order in control myotubes and/or in those expressing GFP alone (45% of reduced myotube size). Myotubes expressing eIF3-f antisense showed a decrease of only 20% in mean myotube diameter, suggesting that eIF3-f is a major target for MAFbx function. To determine the role of eIF3-f in muscle atrophy, the C2C12 myoblasts were transiently transfected with the full-length sense DNA sequence of mouse eIF3-f and induced to differentiate. Myotubes expressing eIF3-f after 72 h of a Tetra release showed a 250% increase in mean diameter, a 300% increase in the number of nuclei and blocked the decrease in myotube diameter caused by starvation (Figure 5B–D). These findings demonstrate that, on the one hand, the downregulation of eIF3-f in myotubes induces atrophy and, on the other hand, conditional overexpression of eIF3-f can prevent muscle atrophy in vitro. Taken together, these findings implicate eIF3-f as a major target for MAFbx function.
Figure 5.
Genetic repression of eIF3-f in differentiated myotubes induces atrophy but genetic activation of eIF3-f is sufficient to block starvation-induced atrophy. (A) Scheme of the protocol applied to C2C12 cells to obtain transfected populations of myotubes. Myoblasts were transfected using Lipofectamin 2000 and cultured for 3 days in GM and then in DM for 6 days. During the first 5 days of culture, Tetra was added and then it was omitted until starvation-induced atrophy. (B) eIF3-f expression accounts for the atrophic and hypertrophic response of myotubes. Transfected cells were green fluorescent by virtue of GFP in the pBI bicistronic expression vector (images in the right column) in a field of myotubes (phase-contrast images in the left column). Scale bar, 50 μm. (C) Transiently transfected C2C12 myoblasts were induced to differentiate in the presence of Tetra for 3 days and then in the absence of Tetra for 3 more days. Myotubes were starved by removal of growth medium, amino acids and glucose and incubated in PBS for 7 h. Total cellular proteins (50 μg) were analysed by immunoblotting. Expression of eIF3-f sense constructs was confirmed by western blotting with anti-HA antibodies. The endogenous eIF3-f expression levels were analysed by western blotting with the polyclonal anti-eIF3-f. Anti-Cdk4 antibodies were used as loading controls. The graph represents averaged densitometric quantification of the data from three replicate experiments. *P<0.03. (D) Myotube diameter after food starvation for 4 and 7 h in the absence of Tetra. Experiments were repeated twice with similar results. Data represent the average±s.e. of at least 120 myotubes. *P<0.05. (E) Myotubes were stained with Hoechst 33258 to visualize the nuclei. The average number of nuclei per myotubes is indicated. Data are means±s.e.m. of at least 100 myotubes. *P<0.05.
eIF3-f induces hypertrophy in myotubes and in mouse skeletal muscle by increasing the muscle structural protein expression
The Akt/mTOR pathway has a prominent role in increased protein synthesis associated with muscle hypertrophy and is also thought to have a role, when chronically deactivated, in the progression of muscle atrophy (Bodine et al, 2001b). Recent data proved that the eIF3 complex acts as a scaffold to coordinate mTOR- and S6K1-mediated translation. Indeed, mTOR and S6K1, which are downstream targets of the IGF-1/Akt pathway and the phosphorylation (i.e. activation) of which is decreased with unloading atrophy, have just been characterized as directly interacting with eIF3-f (Holz et al, 2005). On the one hand, the eIF3-f protein is present during terminal differentiation (Supplementary Figure S2) and interacts with mTOR and S6K1 mediating the assembly of the translation preinitiation complex. On the other hand, MAFbx targets eIF3-f during muscle atrophy. Collectively, these data suggest that eIF3-f may have a pivotal role in the antagonism atrophy/hypertrophy of the skeletal muscle. Because overexpression of eIF3-f was recently shown to inhibit cell proliferation (Shi et al, 2006), we investigated its potential role in skeletal muscle hypertrophy by establishing stable cell lines that express conditional ectopic RNA of eIF3-f sense and/or antisense under the control of the muscle regulatory elements of muscle creatine kinase (MCK promoter) in C2C12 cells. Pools of stable cell lines were cultured in DM for 4 days and then examined for the formation of myotubes. Repression of eIF3-f by antisense eIF3-f RNAs driven by the MCK promoter in C2C12 myoblasts switched from GM to DM repressed the terminal differentiation of muscle, as evidenced by a decrease in the number and size of myotubes in vitro (Supplementary Figure S6). Conversely, overexpression of eIF3-f during skeletal muscle differentiation induced hypertrophy of myotubes, as observed by an increased diameter and a higher number of myonuclei in myotubes. Next, we examined the influence of ectopic expression of eIF3-f on proteins that are important for hypertrophy, such as sarcomeric proteins, in three independent stable cell lines (Figure 6A). The results showed that genetic activation of eIF3-f led to skeletal muscle hypertrophy characterized by a significant increase in muscle differentiation efficiency both at the level of muscle structural proteins such as troponin T, desmin and myosin heavy chains and at the level of the size of myotubes at 4 days of differentiation (Figure 6B–D). There was no positive effect of eIF3-f on the level of MyoD and myogenin proteins. To confirm whether eIF3-f promotes the hypertrophy programme in adult mouse muscles, we electroporated HA-tagged eIF3-f under the control of the MCK promoter into tibialis anterior (TA) muscle of the right hind leg and the empty vector into TA muscle of the left hind leg as control. We measured fibre size 14 days later. Muscle fibres expressing eIF3-f were bigger than the control fibres (Figure 7). At the same time, cross-sectional area determined in more than 1900 fibres from four muscles was increased by 11%. As overexpression of eIF3-f alone causes myotube and muscle hypertrophy, these data suggest that eIF3-f leads to enhanced translation of hypertrophy-related genes such as structural proteins necessary for such marked fibre enlargement.
Figure 6.
Overexpression of eIF3-f induces hypertrophy in muscle cells. (A) Clones of C2C12 myoblasts stably expressing conditional HA-tagged eIF3-f (MCK-eIF3-f) or empty vector (control) were differentiated for 3 days, fixed and stained with Hansen's hemalun. Images of a representative field were obtained by light microscopy. Scale bar, 100 μm. (B) Myotube diameter was measured at day 3 of differentiation. Histogram is a mean±s.e.m. of at least 125 myotubes. *P<0.05. (C) Clones of C2C12 myoblasts stably expressing conditional HA-tagged eIF3-f (MCK-eIF3-f) or empty vector (control) were cultured in GM (20% FCS) for 72 h and then in DM (2% FCS) for 96 h. Proteins were resolved on a 10% SDS–PAGE and HA–eIF3-f, endogenous eIF3-f and α-tubulin (as standard control) were detected by western blot analysis using specific antibodies. (D) Kinetics of differentiation of stably transfected C2C12 cells. Western blots with specific antibodies directed against myosin heavy chain, troponin T, desmin, MyoD and myogenin. * indicates a nonspecific band.
Figure 7.
Overexpression of eIF3-f in muscle fibres induces hypertrophy. (A) Adult TA muscles were electroporated with HA-tagged eIF3-f (pMCK-HA-eIF3-f-IRES-GFP, right leg) or the empty vector (left leg) and mice were killed after 14 days. Hypertrophic fibres expressing eIF3-f were detected in transverse sections stained with anti-HA (eIF3-f) (right panel). Scale bar, 20 and 50 μm for merge pictures. (B) Histogram shows the fibre size distribution of TA from four mice. Black bars, vector control; grey bars, HA–eIF3-f. (C) Mean cross-sectional area of TA fibre from four mice ±s.e.m. **P<0.03 between control and the eIF3-f electrotransferred fibres, by Student's t-test (right panel). NS: nonsignificant.
Discussion
In this work, we have provided several important new insights concerning the control of muscle fibre size and the mechanisms of atrophy induced by MAFbx. By using a stringent screening of a human adult skeletal muscle yeast two-hybrid library with bait containing the NH2 part of MAFbx, we identified eIF3-f, a regulatory subunit of the multisubunit eIF3 complex. eIF3 has an important role in translation initiation. Especially we provided direct biological and biochemical evidence that in response to various treatments inducing muscle atrophy, MAFbx physically interacts with eIF3-f and contributes to its ubiquitination and its degradation by the proteasome. We observed that blockade of MAFbx expression by shRNAi prevents eIF3-f degradation in myotubes undergoing atrophy. Furthermore, we showed that genetic repression of eIF3-f induces atrophy in normal myotubes whereas genetic activation of eIF3-f was sufficient to block atrophy induced by starvation of myotubes. Finally, eIF3-f can act as a ‘translational enhancer' that increases the efficiency of the structural muscle protein synthesis leading to muscle hypertrophy in vitro and in vivo. These data strongly suggest that eIF3-f is a substrate of MAFbx during skeletal muscle atrophy and that it is a key target that accounts for MAFbx function in atrophy by controlling translation of proteins important for hypertrophy. Such mechanism of regulation can be important for atrophy given that the atrophy associated with systemic catabolic states and following disuses involves the breakdown of myofibrillar proteins, the primary cause of the loss of fibre mass (Goldberg, 1969; Furuno et al, 1990).
In C2C12 myotubes that undergo atrophy, MAFbx is upregulated and accumulates in the nucleus (Figure 1D; manuscript in preparation). Ectopic expression of MAFbx in myotubes led to atrophy (Bodine et al, 2001a) and triggered the translocation of eIF3-f from the cytoplasm to the nucleus (Figure 1C). Although MAFbx and eIF3-f are still observed in the cytoplasm, a nuclear localization of eIF3-f was confirmed in atrophy induced by starvation of C2C12 myotubes. The translocation of the regulatory subunit eIF3-f can represent a particular response to the rapid downregulation of the synthesis of special proteins during atrophy. Changes in the composition of eIF3 represent a potential mechanism for controlling eIF3 function. The amount of the regulatory subunit eIF3j in eIF3 influences the amount of 40S subunit associated with eIF3 (Miyamoto et al, 2005). eIF3-f is one of two eIF3 subunits that contain a Mov34 motif, which is found in certain subunits in two macromolecular complexes that are homologous to eIF3: the COP9 signalosome and the lid of the 19S proteasome (Hofmann and Bucher, 1998). Whether MAFbx targets these other two complexes is unknown. We show that the Mov34 motif is implicated in the interaction with MAFbx and places MAFbx in a prime position to control the interaction between eIF3-f and other initiation factors.
The eIF3-f protein appears to be a Mov34 family member. Mov34 family members are involved in the regulation of proteasome, translational initiation and transcription (Asano et al, 1998; Aravind and Ponting, 1998; Pestova et al, 2001). Mov34 proteins act as the regulatory subunit of the 26 proteasome, which is involved in the ATP-dependent degradation of ubiquitinated proteins. The function of this domain is unclear, but it is found in the N-terminus of the proteasome regulatory subunits, eukaryotic initiation factor 3 (eIF3) subunits and regulators of transcription factors. These data are the first to implicate a specific regulatory subunit of the eIF3 translation initiation factor in the atrophic pathway regulated by MAFbx in muscle atrophy, suggesting an unexpected involvement of eIF3-f in the control of muscle cell size. However, the role of eIF3-f in the eIF3 complex has not been defined. eIF3-f is not found in S. cerevisiae, but in Schizosaccharomyces pombe eIF3-f is essential for viability, and depleting eIF3-f markedly decreases global protein synthesis (Zhou et al, 2005). Recent investigations on the biological function of eIF3-f in translation and apoptosis in tumour cells demonstrated that eIF3-f is downregulated in most human tumours and that overexpression of eIF3-f inhibited cell proliferation, suggesting a function associated with differentiation (Shi et al, 2006). Indeed, eIF3-f is upregulated during terminal differentiation of skeletal muscle and absent in the undifferentiated embryonic rhabdomyosarcoma (data not shown). Repression of eIF3-f by antisense RNA during terminal differentiation of skeletal muscle leads to sparse atrophic myotubes, suggesting that this protein can have a specific function in this tissue. We show that eIF3-f can increase the sarcomeric proteins expression but not the muscle regulatory factors such as MyoD and/or myogenin. These data suggest that eIF3-f may act as a ‘translational enhancer' driving specific mRNAs to polysomes and thus increasing the efficiency of protein synthesis. Our findings are consistent with previous suggestions that overload-induced hypertrophy is due to an increase in translational capacity and/or translational efficiency (Carson, 1997; Baar and Esser, 1999). The precise role of eIF3-f in terminal differentiation, at the level of satellite cell fusion and/or pre-existing fibre growth, remains to be elucidated.
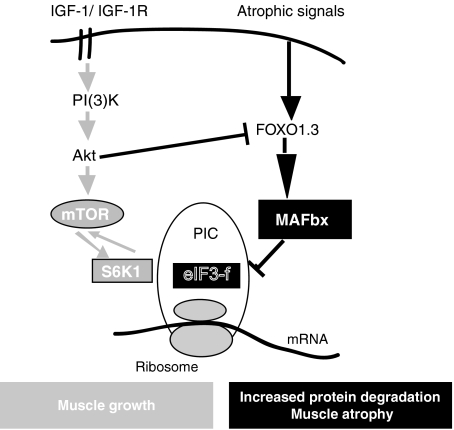
The IGF-1/PI3Akt/mTOR pathway mediates skeletal hypertrophy in C2C12 myotubes by an increase in protein synthesis by activation of S6K1 and inhibition of 4E-BP (Ohanna et al, 2005). The discovery that mTOR and S6K1 interact directly with eIF3-f and mediate the assembly of the translation preinitiation complex through dynamic protein/protein interchange provides important insights into the mechanisms of mTOR and S6K1 signalling and the control of protein synthesis (Holz et al, 2005). mTOR, S6K1 and eIF3 coordinate the assembly of a translation preinitiation complex with enhanced translational capacity in both nutrient- and hormone-dependent signalling pathways and combined with additional S6K1-activating inputs regulated by hormones such as insulin, IGF-1 (Tzatsos and Kandror, 2005; Harris et al, 2006). Under these conditions, this translation initiation complex will be particularly efficient in the translation of mRNA encoding proteins involved in muscle hypertrophy, as suggested by muscle hypertrophy induced by overexpression of eIF3-f. Thus, eIF3-f targeting could explain the decreased protein synthesis observed in multiple types of skeletal muscle atrophy through upregulation of MAFbx. Altogether, these data suggest that atrophy and hypertrophy pathways communicate with each other through eIF3-f (Figure 8). In the context of muscle growth, the mTOR pathway has recently been shown to positively regulate eIF3, whereas Akt is well known to negatively regulate MAFbx transcription. Targeting of subunit eIF3-f to the proteasome by MAFbx therefore represents a simple and powerful mechanism for inhibiting growth and stimulating muscle atrophy. This should help in the search for the primary control mechanism mediating skeletal muscle atrophy and hypertrophy and to discover promising targets for the development of new treatments for skeletal muscle disease.
Figure 8.
Model depicting the two signalling pathways converging on the eIF3-f factor in the antagonism atrophy/hypertrophy of the skeletal muscle.
Materials and methods
Reagents and descriptions of the yeast two-hybrid screen, plasmid constructs, preparation of recombinant proteins, immunoprecipitation, ex vivo ubiquitination, western blot, RNA extraction and RT–PCR analyses, antibodies, establishment of stable cell lines and GST pull-down assay have been included in Supplementary data.
Cell cultures
The mouse skeletal muscle cell line C2C12 was grown in DMEM 20% FCS. Myoblast fusion and differentiation was induced in subconfluent cells by replacing the medium with DMEM 2% FCS (differentiation medium). Atrophy was induced in cultured myotubes after 4 days in differentiation medium by switching the medium to PBS (100 mM NaCl, 5 mM KCl, 1.5 mM MgSO4, 50 mM NaHCO3, 1 mM NaH2PO4, 2 mM CaCl2). Myotubes were also treated with DEX at day 4. Myotubes were incubated with H2O2 in DMEM supplemented with 0.5% FCS. Myoblasts were cultured in 36-mm dishes and transfected using Lipofectamin 2000. For each dish, 3 μg of total plasmid was used as indicated in the legend of figures. High-level transfection efficiency of C2C12 myoblasts was achieved by using a modified protocol for Lipofectamin 2000 (Invitrogen). Freshly trypsinized myoblasts were transfected 30 min after plating (300 × 103 cells/36-mm dish) with a 2:1 ratio (μl/μg) of Lipofectamin 2000 (10 μl/36-mm dish) to plasmid DNA (5 μg/36-mm dish). CHX treatment (50 μg/ml) was carried out essentially as described (Tintignac et al, 2005).
Bright-field images of myotubes were randomly taken and analysed by the Axiovision 4.4 software (Zeiss). The diameters of at least 150 myotubes were measured in a region where myonuclei were absent and the diameter was constant. To count the number of nuclei per cell, myotubes were fixed in 3% paraformaldehyde and stained with DAPI. The number of nuclei in at least 100 myotubes was counted with the Perfect Image v5.5 software (Claravision, France).
Protein extraction and in vitro ubiquitination
C2C12 myotubes were suspended in 1.5 volume of ice-cold buffer consisting of 25 mM Tris–HCl (pH 7.4), 2 mM DTT, 0.25 mM EDTA, 10 μM/ml leupeptin and 10 μM/ml pepstatin and incubated on ice for 15 min. After addition of 0.2% Triton X-100 for 10 min, the lysate was transferred to an Eppendorf tube and centrifuged in a microcentrifuge at 10 000 g for 10 min. The supernatant was divided into smaller samples and frozen at −80°C. Ubiquitination assays were performed as described previously (Tintignac et al, 2005). In vitro-translated, 35S-labelled eIF3-f was incubated either with recombinant SCFMAFbx complexes produced in insect cells or with C2C12 myotube extracts (30 μg) in the presence of an ATP regenerating system (50 mM Tris pH 7.5, 5 mM MgCl2, 10 mM creatine phosphate, 5 U/ml phosphocreatine kinase, 5 mM ATP) with 1 μM ubiquitin aldehyde, 1 mg/ml ubiquitin, 1 mM DTT and 50 μM MG132. Reactions were carried out at 37°C and stopped with Laemmli sample buffer.
Immunofluorescence staining
Cells were cultured on coverslips and fixed in 3% paraformaldehyde for 30 min at 4°C and permeabilized with 0.25% Triton X-100 for 15 min at room temperature. The cells were treated with 5% normal goat serum and immunostained with mAb anti-Myc, the polyclonal anti-eIF3-f and the polyclonal anti-MAFbx. The Texas red-conjugated F(ab′)2 fragments of donkey anti-mouse IgG were used to visualize the mouse monoclonal antibodies and the FITC-conjugated F(ab′)2 fragment of goat anti-rabbit IgG to visualize the rabbit polyclonal antibodies.
Animal models of muscle atrophy and muscle electrotransfer
Fasted (48 h), refed (48 h) and control (fed) C57BL/6 mice were killed by cervical dislocation. Hind limb muscles from each animal were immediately removed and frozen in liquid nitrogen before storage at −80°C.
In vivo transfection experiments were carried out on four 8-week-old C57BL/6 females. Mice were first anaesthetized with isoflurane (0.75–1% in oxygen) and received a single injection of 0.4 U of bovine hyaluronidase (Sigma)/μl in 25 μl 0.9% NaCl into the TA muscle. After 2 h, a total of 15 μg of plasmid DNA in 0.9% NaCl was injected into the TA under conditions of ketamine (100 μg per g body weight) and xylazine (10 μg/g) anaesthesia. pMCK-IRES-GFP was used as a control and injected in the left leg whereas pMCK-3HA-eIF3-f-IRES-GFP was injected in the contralateral muscle. An electrical field was then applied to muscle with caliper rule electrodes coated with ultrasound transmission gel (Aquasonic 100, Parker) and placed on each side of the leg. Six square-wave 130 V/cm pulses, lasting 60 ms each with a 100 ms interval, were then applied with a BTX electrocell manipulator. Mice were killed by cervical dislocation and muscles were collected 14 days after electrotransfer.
Histological analysis and fibre size measurement
TA muscles were removed, embedded in cryomatrix and quickly frozen in isopentane cooled with liquid nitrogen. Muscles were then sectioned in a microtome cryostat (Leica). Transversal sections (14 μm) were fixed with 4% paraformaldehyde for 10 min on ice, rinsed three times in 1 × PBS, permeabilized with 0.1% Triton X-100 and left for 1 h in blocking solution (3% bovine serum albumin, 20% normal goat serum in 1 × PBS). Rat monoclonal anti-HA antibody (Roche, used at 1:1000 dilution) was then applied overnight to the treated sections. Bound primary antibodies were detected with cyanine 3-conjugated anti-rat IgG (Jackson Immunoresearch, used at 1:600 dilution). The indirect immunofluorescence steps were executed as much as possible in the dark to preserve GFP staining. Nuclei were stained with Hoechst. Fibre cross-sectional areas were measured using PerfetImage software and determined for two sections from four animals in each group.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary data
Acknowledgments
We thank Dr T Lorca for the Emi1 expression plasmid, Dr D Daegelen (Tissue Morphology Technology Facility, Cochin Institute Paris 5 University) for her valuable technical help, Dr Y Reynes and Dr B Chabi for microscopy assistance and Jonathan Levin for critically reading the manuscript. This work was supported by grants from the Association Française contre les Myopathies (AFM), the department PHASE from INRA and the ‘Institut National de la Santé et de la Recherche Médicale' (INSERM). Miss J Lagirand-Cantaloube held a graduate fellowship from the ‘Ministère de la Recherche et de la Technologie' (MRT) and an AFM fellowship. Mrs A Csibi holds a graduate AFM fellowship.
References
- Aravind L, Ponting CP (1998) Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci 7: 1250–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Phan L, Anderson J, Hinnebusch AG (1998) Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem 27: 18573–18585 [DOI] [PubMed] [Google Scholar]
- Baar K, Esser K (1999) Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol 276: C120–C127 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Maitra U (1999) Cloning and characterization of the p42 subunit of mammalian translation initiation factor 3 (eIF3): demonstration that eIF3 interacts with eIF5 in mammalian cells. Nucleic Acids Res 27: 1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ (2001a) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708 [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD (2001b) Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3: 1014–1019 [DOI] [PubMed] [Google Scholar]
- Browning KS, Gallie DR, Hershey JWB, Hinnebusch AG, Maitra U, Merrick WC, Norbury C (2001) Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci 26: 284. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Carson JA (1997) The regulation of gene expression in hypertrophying skeletal muscle. Exerc Sport Sci Rev 25: 301–320 [PubMed] [Google Scholar]
- Furuno K, Goodman MN, Goldberg AL (1990) Role of different proteolytic systems in the degradation of muscle proteins during denervation atrophy. J Biol Chem 265: 8550–8557 [PubMed] [Google Scholar]
- Goldberg AL (1969) Protein turnover in skeletal muscle. II. Effects of denervation and cortisone on protein catabolism in skeletal muscle. J Biol Chem 244: 3223–3229 [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL (2001) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA 98: 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardavaccaro D, Pagano M (2006) Stabilizers and destabilizers controlling cell cycle oscillators. Mol Cell 22: 1–4 [DOI] [PubMed] [Google Scholar]
- Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC Jr (2006) mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J 25: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselgren PO (1999) Glucocorticoids and muscle catabolism. Curr Opin Clin Nutr Care 2: 201–205 [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC (2000) Origins and principles of translational control. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds), pp 31–69. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2006) eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P (1998) The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci 23: 204–205 [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J (2005) mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123: 569–580 [DOI] [PubMed] [Google Scholar]
- Jagoe RT, Goldberg AL (2001) What do we really know about the ubiquitin–proteasome pathway in muscle atrophy? Curr Opin Clin Nutr Metab Care 4: 183–190 [DOI] [PubMed] [Google Scholar]
- Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, Patterson C (2004) Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest 114: 1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-P, Chen Y, Li AS, Reid MB (2003) Hydrogen peroxyde stimulates ubiquitin-conjugating activity and expression of genes for specific E2 and E3 proteins in skeletal muscle myotubes. Am J Physiol Cell Physiol 285: C806–C812 [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Patel P, Hershey JW (2005) Changes in ribosomal binding activity of eIF3 correlate with increased translation rates during activation of T lymphocytes. J Biol Chem 280: 28251–28264 [DOI] [PubMed] [Google Scholar]
- Ohanna M, Sobering AK, Lapointe T, Lorenzo L, Praud C, Petroulakis E, Sonenberg N, Kelly PA, Sotiropoulos A, Pende M (2005) Atrophy of S6K1(−/−) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat Cell Biol 7: 286–294 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98: 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ (2001) Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol 3: 1009–1013 [DOI] [PubMed] [Google Scholar]
- Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL (2007) Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J 21: 140–155 [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP, Nelson MA (2006) Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene 25: 4923–4936 [DOI] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ (2004) The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell 14: 395–403 [DOI] [PubMed] [Google Scholar]
- Tintignac LA, Lagirand J, Batonnet S, Sirri V, Leibovitch MP, Leibovitch SA (2005) Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J Biol Chem 280: 2847–2856 [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Kandror KV (2005) Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol Cell Biol 26: 63–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW (1999) A family of mammalian F-box proteins. Curr Biol 9: 1180–1182 [DOI] [PubMed] [Google Scholar]
- Zhou C, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA (2005) PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary data