Abstract
Using Caenorhabditis elegans genetic screens, we identified receptor-mediated endocytosis (RME)-4 and RME-5/RAB-35 as important regulators of yolk endocytosis in vivo. In rme-4 and rab-35 mutants, yolk receptors do not accumulate on the plasma membrane as would be expected in an internalization mutant, rather the receptors are lost from cortical endosomes and accumulate in dispersed small vesicles, suggesting a defect in receptor recycling. Consistent with this, genetic tests indicate the RME-4 and RAB-35 function downstream of clathrin, upstream of RAB-7, and act synergistically with recycling regulators RAB-11 and RME-1. We find that RME-4 is a conserved DENN domain protein that binds to RAB-35 in its GDP-loaded conformation. GFP–RME-4 also physically interacts with AP-2, is enriched on clathrin-coated pits, and requires clathrin but not RAB-5 for cortical association. GFP–RAB-35 localizes to the plasma membrane and early endocytic compartments but is lost from endosomes in rme-4 mutants. We propose that RME-4 functions on coated pits and/or vesicles to recruit RAB-35, which in turn functions in the endosome to promote receptor recycling.
Keywords: C. elegans, clathrin-coated pits, endocytic recycling, Rab GTPase
Introduction
The endocytic system of eukaryotic cells consists of a complex network of membrane-bound compartments. Internalization of many receptor-ligand complexes from the plasma membrane is clathrin mediated (Brodsky et al, 2001). Clathrin and cargo molecules are assembled into coated pits on the plasma membrane together with adaptor proteins that link clathrin and transmembrane receptors, concluding in the formation of mature clathrin-coated vesicles. Clathrin-coated vesicles are then actively uncoated and transported to early endosomes. After delivery to early endosomes, sorting occurs, determining whether the cargo is degraded or recycled (Maxfield and McGraw, 2004). Transport from early endosomes to late endosomes, and ultimately to lysosomes, leads to degradation. Cargo to be recycled from early endosomes, such as the transferrin receptor, are sorted into carriers that take them to recycling endosomes, or directly to the plasma membrane. These processes are highly regulated, but the precise molecular mechanisms controlling these steps are not fully understood.
Rab GTPases have emerged as critical regulators of intracellular transport and have been proposed to define functional domains within the endomembrane system (Zerial and McBride, 2001). Rab function is primarily controlled via nucleotide binding, by which the protein cycles between the active GTP-bound state, in which it physically interacts with effectors on membranes, and the inactive GDP-bound state, where it is thought to be quiescent in the cytoplasm complexed with its chaperone Rab-GDI. The nucleotide cycle is locally controlled by specific nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). GTP-bound Rabs associate with specific membrane compartments and, through their effectors, influence critical steps in membrane traffic such as vesicle tethering/fusion, vesicle budding, cargo sorting, and membrane-cytoskeleton interaction. These Rab-regulated processes are not acting all at once, but rather are spatially and temporally controlled.
In the genomes of higher eukaryotes, Rabs represent the largest group within the Ras superfamily (Pereira-Leal and Seabra, 2001). The Caenorhabditis elegans genome contains 30 Rabs, whereas the human genome contains over 60 Rab genes. A number of Rab proteins are known to function in the endocytic pathway. For instance, Rab5 is a master regulator of early endocytic events (Christoforidis et al, 1999; McBride et al, 1999; Rink et al, 2005). Rab7 functions directly downstream of Rab5 and is required for transport/maturation from early endosomes to late endosomes (Bucci et al, 2000; Rink et al, 2005). Rab11 regulates endocytic recycling through the recycling endosomes, whereas Rab4 is thought to direct recycling from the early endosomes to the plasma membrane (van der Sluijs et al, 1992; Ullrich et al, 1996). Some Rabs are expressed and function only in the more elaborate context of polarized cell endomembranes, such as those of epithelia and neurons (Fischer von Mollard et al, 1994; Chen et al, 2006; Sato et al, 2007). Although a few Rabs have been studied in great detail, the role of many Rabs still remains to be established, along with their regulators and effectors.
To further elucidate the molecular mechanisms mediating the endocytic pathway of higher eukaryotes, we have applied C. elegans genetics. We previously described a sensitive in vivo assay for the receptor-mediated endocytosis and trafficking the yolk protein YP170, a ligand related to the mammalian cholesterol carrier ApoB-100 (Grant and Hirsh, 1999). A YP170–GFP fusion protein, like endogenous YP170, is synthesized in the intestine and secreted basolaterally into the body cavity from which it is endocytosed by the receptor-mediated endocytosis (RME)-2 yolk receptors expressed in the oocytes, in a clathrin- and adaptor complex AP-2-dependent manner (Grant and Hirsh, 1999). This assay has allowed us to isolate a large number of rme mutants showing defects in endocytosis (Grant and Hirsh, 1999; Grant et al, 2001; Zhang et al, 2001; Sato et al, 2005, 2006).
Here, we report molecular cloning and functional characterization of rme-4 and rme-5, previously uncharacterized complementation groups showing very similar phenotypes. We found that RME-4 is a DENN domain protein with a proline-rich domain and consensus AP-2 and clathrin-binding sequences, most similar to the mammalian coated-pit protein Connecdenn (Allaire et al, 2006). RME-5 is the worm Rab35 orthologue, a protein recently implicated in the regulation of endocytic recycling in cultured mammalian cells (Kouranti et al, 2006). We provide genetic and biochemical evidence that RME-4 functions with RAB-35, likely recruiting or activating it on endocytic vesicles. In turn, RAB-35 accumulates on early endosomes where it is required for the recycling of receptors to the plasma membrane. In the absence of RME-4 or RME-5/RAB-35, yolk receptors are lost from the early endocytic pathway and accumulate aberrantly in small cytoplasmic vesicles, indicating a defect in receptor recycling. These results identify an unexpected connection between coated-pit biogenesis and the downstream recycling of endocytosed cargo.
Results
rme-4 and rme-5 are required for yolk uptake by oocytes
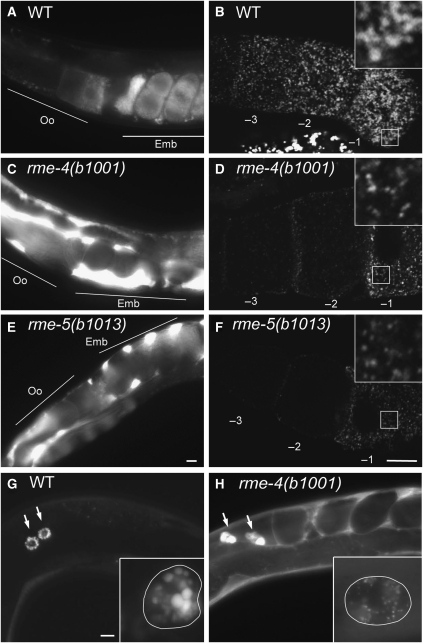
We isolated one allele of rme-4(b1001) and two alleles of rme-5(b1013 and b1034) in our previously described genetic screen for mutants defective in endocytosis of YP170–GFP (Grant and Hirsh, 1999). In both rme-4 and rme-5 mutants, yolk uptake by oocytes was strongly reduced, resulting in high level accumulation of YP170–GFP in the body cavity (Rme phenotype; Figure 1C and E). Similar to other endocytosis mutants such as rme-1 and rme-6 (Grant et al, 2001; Sato et al, 2005), the block in uptake in rme-4 and rme-5 mutants was not absolute, as some YP170–GFP could still be detected in mutant oocytes (Figure 1D and F). YP170–GFP that was taken up localized to vesicles that were fewer and smaller than in wild-type oocytes (average yolk granule area; WT, 0.915±0.192 μm2; rme-4(b1001), 0.283±0.086 μm2; rme-5(b1013), 0.320±0.086 μm2). These results indicate that RME-4 and RME-5 are important regulators of receptor-mediated endocytosis in oocytes.
Figure 1.
rme-4 and rme-5 mutants display endocytosis defects. (A, C, E) YP170–GFP endocytosis by oocytes of adult hermaphrodites. In wild-type, YP170–GFP is efficiently endocytosed by oocytes (A). In the rme-4(b1001) and rme-5(b1013) mutants, endocytosis of YP170–GFP by oocytes is greatly reduced and YP170–GFP accumulation in the body cavity is greatly increased (C, E). Positions of oocytes (Oo) and embryos (Emb) are indicated. (B, D, F) Gonads from wild-type or mutants expressing YP170–GFP were dissected and fixed before imaging. In the oocytes of mutant animals, the overall intensity of YP170–GFP fluorescence was reduced, and YP170–GFP that was internalized was observed in abnormally small vesicles. Insets show enlargements ( × 3) of the boxed area. Oocytes proximal to the spermatheca are numbered as −1. (G, H) Coelomocyte endocytosis. GFP secreted from body-wall muscle cells (ssGFP) is taken up by coelomocytes and accumulates in endosomes/lysosomes of wild-type coelomocytes (G). In rme-4(b1001), increased accumulation of ssGFP in the body cavity was observed, indicating poor endocytosis by coelomocytes (H). Arrows indicate each coelomocyte. Enlarged images of coelomocytes are shown in the insets. Cell boundaries of coelomocytes are outlined for clarity. Bars, 10 μm.
Another well-developed assay for endocytosis in C. elegans is based upon the uptake of foreign compounds by scavenger cells called coelomocytes. This is most easily followed in strains in which the muscle cells have been engineered to secrete a signal sequence modified form of GFP (ssGFP) (Fares and Greenwald, 2001). In wild-type animals, ssGFP is efficiently cleared from the body cavity and accumulates in the endosomes and lysosomes of coelomocytes before its degradation (Figure 1G). However, in rme-4 mutants, an abnormally high level of ssGFP remains in the body cavity, indicating that endocytosis by coelomocytes is impaired (Figure 1H). Some ssGFP was taken up by rme-4 coelomocytes, but the size and morphology of the GFP-positive endocytic compartments were abnormal. On the other hand, rme-5 mutants showed normal coelomocyte endocytosis (data not shown). These results suggest that rme-4 is generally required for endocytosis in multiple cell types, whereas rme-5 may be cell type or cargo specific.
RME-2 is lost from cortical endosomes in rme-4 and rme-5 mutants
Several types of endocytic defects can result in failure to efficiently internalize ligands. These include defects in the uptake or transport of cognate receptors to the early endosome or the failure to recycle such receptors back to the plasma membrane for multiple rounds of uptake. Results of epistasis tests suggested that RME-4 and RME-5 function upstream of RAB-7 (Supplementary Figure 1A). To better understand the defect in yolk endocytosis displayed by rme-4 and rme-5 mutants, we determined the fate of the yolk receptor (RME-2) in these mutants. We have previously shown, by immunofluorescence and immunoelectron microscopy, that at steady state, endogenous RME-2 yolk receptors are found sparsely distributed on the oocyte plasma membrane and are enriched in cortical vesicles and tubules representing early and recycling endosomes (Figure 2A and G; Supplementary Figure 2A) (Grant and Hirsh, 1999; Sato et al, 2005). Little or no yolk receptor can be detected in the late endosomal yolk granules in wild-type oocytes, indicating efficient sorting of yolk from yolk receptors during endocytic transport. The late endosomal yolk granules are thought to be storage organelles with relatively little degradative capacity, only acquiring high-level proteolytic activity during embryogenesis, probably via fusion with lysosomes later in development.
Figure 2.
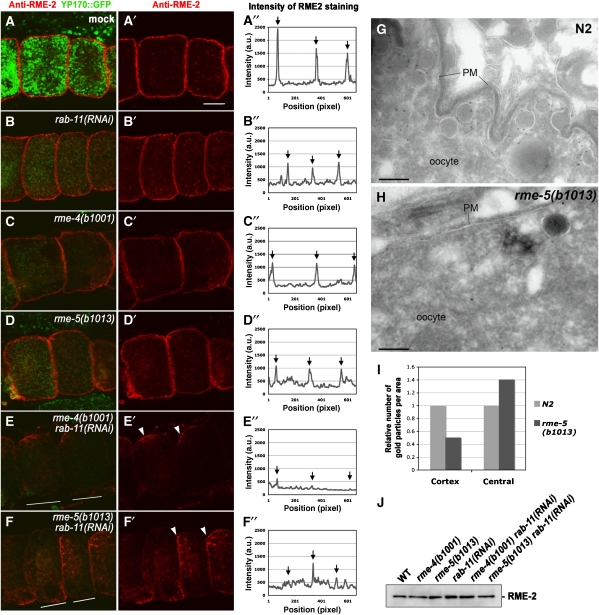
RME-2 is lost from cortical endosomes in rme-4 and rme-5 mutants. (A–A″, B–B″, C–C″, D–D″, E–E″, F–F″) Dissected gonads were immunostained with anti-RME-2 antibody. (A–A″) wild-type; (B–B″) rab-11(RNAi); (C–C″) rme-4(b1001); (D–D″) rme-5(b1013); (E–E″) rme-4(b1001) rab-11(RNAi); (F–F″) rme-5(b1013) rab-11(RNAi). All strains expressed YP170–GFP. Middle focal planes of oocytes are shown. Arrowheads in (E′) and (F′) indicate tubular structures aggregated in small patches of the oocyte cortex (see text). Bar, 10 μm. Fluorescence intensity of RME-2 staining as a function of position was graphed along a line through the centre of the oocytes as indicated in the paired panels (A″–F″). Arrows indicate the relative position of the oocyte plasma membrane. At least 10 animals for each strain were analysed and representatives are shown. (G, H) Immunoelectron microscopy of RME-2 was performed in wild-type N2 (G) and rme-5(b1013) mutants (H). In the cortical area of wild-type oocytes, RME-2 localizes to the plasma membrane and tubular and vesicular structures (endosomes). In rme-5 mutants, the RME-2 signal, and the number of yolk-containing vesicles were significantly reduced. Bars, 200 nm. (I) Quantitative analysis of RME-2 localization in oocytes. The number of gold particles in the cortical or central regions of oocytes per unit area were counted, and the ratio as compared with wild-type controls is shown (see Materials and methods). (J) RME-2 protein level is not reduced in rme-4 and rme-5 mutant animals. Total lysates were prepared from the strains indicated and were probed with anti-RME-2 antibody in western blots.
In rme-4 or rme-5 mutant oocytes, the intensity of cortical RME-2 detected by immunofluorescence was reduced by about twofold (Figure 2C and D; Supplementary Figure 2C and D). Conversely, the diffuse RME-2 signal deeper in the cytoplasm was increased in rme-4 or rme-5 mutants. The remaining cortical RME-2 was found in small punctate structures lacking YP170–GFP, suggesting residence in early endosomal compartments (Supplementary Figure 2C and D).
By immunoelectron microscopy, the loss of RME-2 from cortical endosomal vesicles in rme-5 mutants was also clear (Figure 2G–I). In the cortex of rme-5(b1013) oocytes, we found a twofold reduction in anti-RME-2 labelling. The vesicles containing RME-2 appeared smaller and more sparsely distributed than in wild-type oocytes. In accordance with the immunofluorescence results, we also observed a measurable increase in anti-RME-2 labelling of vesicles deeper in the cytoplasm. In mutant oocytes, mislocalized RME-2 was found in fine vesicles and tubules smaller than 30 nm in diameter. There was no increase in plasma membrane RME-2 labelling, suggesting that uptake of receptors from the plasma membrane was not impaired. These results suggest that, in rme-4 and rme-5 mutants, sorting and/or recycling of RME-2 is impaired such that RME-2 is not maintained in the cortical endosomes. This is different than the phenotype in rme-6 mutants, where yolk receptors accumulate in small cortical vesicles, failing to fuse with early endosomes (Sato et al, 2005). This is also different than rme-1 mutants that are impaired in a late recycling event, leading to yolk receptor accumulation in enlarged cortical vesicles (W. Liou and B.D. Grant, unpublished data; Grant et al, 2001). The overall level of RME-2 protein in rme-4 and rme-5 mutants as determined by immunoblotting was unchanged, indicating that the altered RME-2 receptor distribution does not reflect a change in receptor synthesis or degradation (Figure 2J).
rme-4 and rme-5 mutations strongly enhance rab-11(RNAi)-associated defects
As rme-4 and rme-5 appeared to be required for sorting/recycling of RME-2 after endocytosis, we investigated their relationship with RAB-11, a well-known regulator of endocytic recycling in many eukaryotic species. As we have reported previously, in C. elegans oocytes, RNAi of rab-11 causes a yolk uptake defect and results in accumulation of RME-2 yolk receptors in a tubular endosomal compartment in the cortex and deep in the cytoplasm (Figure 2B; Supplementary Figure 2B) (Grant and Hirsh, 1999). We measured an overall reduction of cortical RME-2 under these conditions (Figure 2B). This phenotype is somewhat different from that of rme-4 or rme-5 mutants in which non-cortical RME-2 showed a fine punctate and diffuse staining rather than tubular accumulation (Figure 2C and D; Supplementary Figure 2).
Our results suggested that loss of any of these proteins affects yolk receptor recycling. Using genetics, we asked whether RME-4/RME-5 and RAB-11 function sequentially in the recycling pathway or in parallel pathways. If RME-4/5 and RAB-11 function in the same pathway, simultaneous knockdown would be expected to give a phenotype similar to single mutants of either gene. However, when RNAi of rab-11 was performed in rme-4(b1001) or rme-5(b1013) mutants, the phenotype was dramatically enhanced. The punctate RME-2 staining disappeared completely from most cortical areas. The remaining RME-2 signal accumulated in enlarged tubular structures (Figure 2E and F). These tubular structures were often observed aggregated in small patches of the oocyte cortex (Figure 2E′ and F′, arrowheads). In some double-mutant animals, the gonad was severely disorganized (data not shown). These strongly enhanced phenotypes resulting from rme-4/rme-5 mutations combined with rab-11(RNAi) depletion suggest that RME-4/5 and RAB-11 function in parallel pathways rather than sequentially. We observed a similar synthetic effect on RME-2 localization when another known recycling endosome regulator rme-1 was depleted in rme-4/5 mutant backgrounds (Supplementary Figure 1B).
rme-4 encodes a DENN domain protein
We identified the wild-type rme-4 gene (F46F6.1) by standard methods (see Materials and methods). It encodes an uncharacterized protein of 661 amino acids containing an N-terminal DENN domain (Figure 3A). The DENN domain occurs in several proteins involved in Rab-mediated processes as well as mitogen-activated protein kinase (MAPK) signalling pathways (Levivier et al, 2001). The molecular function of the DENN domain remains unclear. Human DENN (differentially expressed in neoplastic versus normal cells) is also called MADD (MAPK activating protein containing death domain) and functions in signalling pathways that regulate neuronal cell death (Chow and Lee, 1996; Schievella et al, 1997; Miyoshi and Takai, 2004). Human DENN/MADD is orthologous to rat Rab3GEP and C. elegans AEX-3, guanine nucleotide exchange factors for Rab3/RAB-3 (and RAB-27 in C. elegans), GTPases that regulate neurotransmitter release at synapses (Iwasaki et al, 1997; Wada et al, 1997; Mahoney et al, 2006). The DENN domain is also found in Rab6ip1, a protein that physically interacts with Rab6 and Rab11 (Miserey-Lenkei et al, 2007). In addition to the DENN domain, RME-4 contains a cluster of SH3-binding motifs (PXXP) in the C-terminal region. This organization of domains within RME-4 is most similar overall to mouse Connecdenn, a neuronal protein associated with clathrin-coated pits (Allaire et al, 2006).
Figure 3.
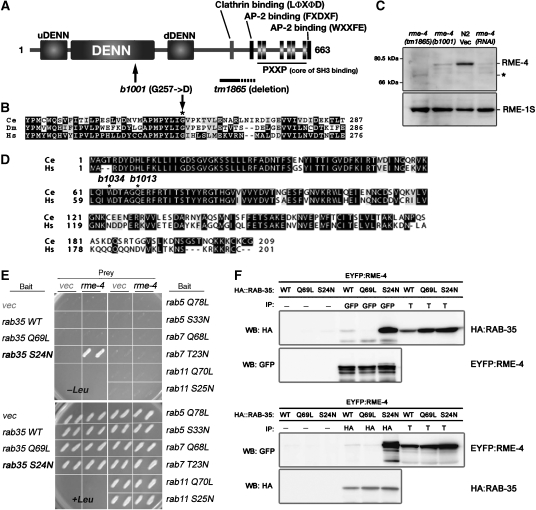
RME-4 is a novel protein containing a DENN domain and physically interacts with RME-5/RAB-35. (A) Domain structure of RME-4, showing the N-terminal DENN domain accompanied by the uDENN and dDENN domains. In addition, RME-4 contains AP-2-binding motifs, a clathrin-binding motif, and a cluster of SH3-binding motifs all located in the C-terminal half of the protein. Mutations identified in rme-4 alleles are also shown. (B) Amino-acid alignment of the highly conserved region of the DENN domain from the human (Hs), C. elegans (Ce), and fly (Dm) RME-4 homologues. In rme-4(b1001), glycine 257 (marked with an asterisk) is mutated. (C) Detection of RME-4 protein. Total lysates were prepared from rme-4 mutant worms and N2 wild-type worms treated with RNAi of either control vector or rme-4. They were examined by immunoblotting using anti-RME-4 antibody. The same membrane was also probed with anti-RME-1 antibody. The asterisk indicates the mutant RME-4(tm1865) protein containing a partial deletion. (D) RME-5 is the worm Rab35 orthologue. Amino-acid alignment of worm RME-5/RAB-35 (Ce) and human Rab35 (Hs). Two alleles of rme-5 contain nonsense mutations at the indicated positions. (E) Yeast two-hybrid interaction between RME-4 and RAB-35. Full-length RME-4 was expressed in a yeast reporter strain as a fusion with the transcriptional activation domain of B42 (prey). C. elegans RAB-35, RAB-5, RAB-7, RAB-11, and their mutant forms were expressed in the same yeast cells as fusions with the DNA binding domain of LexA (bait). Interaction between bait and prey was tested using LEU2 (shown) and β-galactosidase (not shown) reporter assays. (F) Co-immunoprecipitation of EYFP–RME-4 and mRFPCherryHA–RAB-35 or their mutant forms. Total lysates were prepared from worms co-expressing EYFP–RME-4 and mRFPCherryHA–RAB-35 or its mutant forms and subjected to immunoprecipitation with anti-GFP antibody (upper panel) or anti-HA antibody (lower panel). Precipitants were probed on immunoblots with anti-HA and anti-GFP antibodies. T (total lysate) equals approximately 4% of the total input into the assay.
The rme-4(b1001) mutation results in a predicted single amino-acid change (G257D) in a highly conserved residue of the RME-4 DENN domain (Figure 3B). We obtained another allele of rme-4, tm1865, from the Japanese National Bioresource Project for nematodes. This deletion mutant removes part of the RME-4 protein (at least residues 465–529, Figure 3A). rme-4(tm1865) mutants display a yolk-endocytosis defect identical to that of rme-4(b1001) (data not shown).
Using an antibody that we developed against RME-4 in western blot analysis, we detected a single band of the predicted size for RME-4 (74 kDa; Figure 3C). The intensity of the band was greatly reduced after RNAi of rme-4, confirming the specificity of the antibody. Consistent with the predicted partial deletion, rme-4(tm1865) blots displayed an apparent reduction in molecular weight for the anti-RME-4 reactive species and strongly reduced band intensity. The missense mutant rme-4(b1001) displayed an RME-4 reactive band of normal mobility but with strongly reduced intensity, suggesting that the integrity of the DENN domain is important for RME-4 folding or stability.
rme-5 encodes C. elegans Rab35
We cloned the wild-type rme-5 gene using standard methods and found that it encodes C. elegans Rab35 (RAB-35), a member of the Rab GTPase family recently implicated in recycling from early endosomes (see Materials and methods) (Kouranti et al, 2006). Nonsense mutations were associated with each rme-5 allele (b1013 and b1034) and are predicted to truncate the protein at amino acid 69 and 64, respectively (Figure 3D). Both alleles are thus predicted null alleles of rab-35.
RME-4 specifically interacts with the GDP form of RAB-35
Given the similarity in phenotypes of rme-4 and rme-5/rab-35 mutants, and the RME-4 DENN domain that suggests association with a Rab protein, we hypothesized that RME-4 and RAB-35 might function together. As a first test of this possibility, we examined physical interaction in a yeast two-hybrid assay (Figure 3E). Full length of RME-4 did not interact with the wild type or a GTPase-defective mutant form (Q69L) of RAB-35. However, RME-4 strongly interacted with a mutant form of RAB-35 predicted to be locked in the GDP-bound conformation (S24N). RME-4 did not interact with similar GTP- or GDP-locked forms of other endocytic Rab proteins (RAB-5, -7, and -11), indicating the specificity of the interaction. To verify this physical interaction, we also performed co-immunoprecipitation experiments testing the interaction of RME-4 and RAB-35 in worm lysates. As predicted by the two-hybrid interactions, YFP–RME-4 co-precipitated with mRFPCherryHA–RAB-35(S24N) (Figure 3F). Little or no interaction was observed between YFP–RME-4 and the wild-type or the Q69L mutant of mRFPCherryHA–RAB-35. These results show that RME-4 preferentially interacts with RAB-35(S24N), suggesting that RME-4 normally interacts with GDP-bound RAB-35.
RME-4 regulates the association of RAB-35 with cortical endosomes
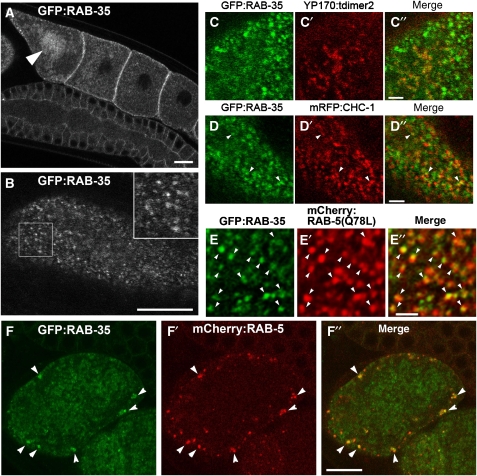
To determine the subcellular localization of RAB-35 in oocytes, we expressed GFP–RAB-35 using a germline-specific promoter or its own promoter. GFP–RAB-35 driven by its own promoter was ubiquitously expressed with a punctate distribution in many cell types (Supplementary Figure 3). We confirmed that expression of GFP–RAB-35 in the germ line of the rme-5(b1013) mutants fully rescued the yolk-uptake defect, indicating that GFP–RAB-35 is functional and very likely properly localized (Supplementary Figure 4A and C). In oocytes, GFP–RAB-35 was mainly found on small punctate structures in the cortex (Figure 4A and B). In addition, some GFP–RAB-35 was also observed on larger vesicles deeper within the cytoplasm (Figure 4A). The latter large vesicles contained high levels of YP170, indicating that they are yolk granules, which are late endosome-like compartments in these cells (Figure 4C–C″). During ovulation, residence of GFP–RAB-35 on yolk granules became more prominent (Figure 4A, arrowhead). In fertilized embryos, GFP–RAB-35 was found on yolk granules, smaller vesicles, and on or near the cell surface (Figure 4F; Supplementary Figure 3E). Human Rab35 (HsRab35), expressed as a GFP fusion, also rescued yolk endocytosis in rme-5(b1013) mutant worms, indicating that the worm and human proteins are true orthologues (Supplementary Figure 4B and D). The subcellular localization of GFP–HsRab35 in oocytes was very similar to that of worm RAB-35.
Figure 4.
Subcellular localization of rescuing GFP–RAB-35 in the germ line. (A, B) GFP–RAB-35 was expressed under germline-specific promoter control and observed in live animals. Middle (A) and top (B) focal planes are shown. In developing oocytes, GFP–RAB-35 mainly localizes to small puncta in the cortex. Localization of GFP–RAB-35 to the yolk granules deeper in the cytoplasm becomes prominent in oocytes close to the spermatheca (an arrowhead in panel A). An enlarged ( × 2) image of the boxed area is shown in the inset (B). Bars, 10 μm. (C–C″, D–D″, E–E″) Colocalization of GFP–RAB-35 with endocytic markers in oocytes. GFP–RAB-35 and either YP170–tdimer2 (yolk protein; C–C″), mRFP–CHC-1 (D–D″) or mRFPCherry–RAB-5(Q78L) (E–E″) were co-expressed in the germ line and their subcellular localization was observed in live animals. Bars, 2 μm. (F–F″) Colocalization of GFP–RAB-35 and mRFPCherry–RAB-5(WT) in one-cell-stage embryos. Bar, 10 μm.
Rab35 has recently been shown to function in recycling from early endosomes to the plasma membrane in mammalian cells (Kouranti et al, 2006). To determine if the cortical GFP–RAB-35 signal that we observed corresponds to early endosomes in C. elegans, we tested for colocalization with early endosome marker mRFPCherry–RAB-5 in oocytes and early embryos. We observed partial overlap, suggesting residence of RAB-35 on early endosomes (Figure 4F–F″; data not shown). Colocalization of GFP–RAB-35 and the early endosome became more evident on the enlarged endosomes present in oocytes expressing low levels of activated mRFPCherry–RAB-5(Q78L) (Figure 4E–E″). Most often, the RAB-35 and RAB-5 signals appeared as partially overlapping puncta, suggesting that the two Rab proteins may segregate into distinct subdomains within the endosome. We also observed occasional GFP–RAB-35 puncta associated with clathrin-coated pit markers mRFP–clathrin heavy chain-1 (CHC-1) or mRFP–α-adaptin (APT-4) (Figure 4D–D″; data not shown). Our results indicate that most punctate GFP–RAB-35 signal represents cortical early endosomes, whereas the diffuse cortical GFP–RAB-35 signal represents the plasma membrane and perhaps occasional clathrin-coated pits.
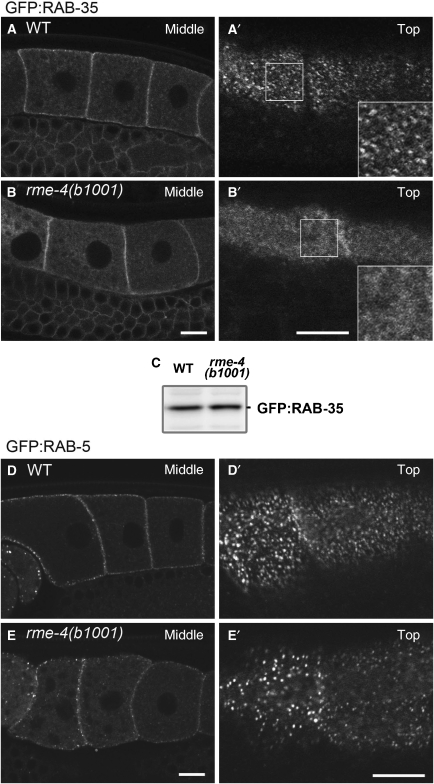
If RME-4 regulates RAB-35 recruitment or activity, then loss of RME-4 should adversely affect the subcellular localization of RAB-35. In rme-4 mutants, the large cortical GFP–RAB-35 puncta were lost, although a weaker, fuzzy-appearing GFP–RAB-35 signal persisted in the cell periphery on or very near the plasma membrane (Figure 5B and B′). By contrast, GFP–RAB-5 (Figure 5E) or GFP–CHC-1 (data not shown) labelling of cortical endosomes and pits was not greatly altered in rme-4 mutants, although the number of GFP–RAB-5-positive endosomes were reduced by about one-third. The level of GFP–RAB-35 protein was normal in rme-4 mutants as judged by anti-GFP western blot, indicating that RME-4 affects the localization of RAB-35 but not its stability (Figure 5C). Taken together, these results suggest that RME-4 functions as a regulator of RAB-35, important for its association with endosomes.
Figure 5.
RME-4 is required for correct subcellular localization of GFP–RAB-35 in the oocytes. (A–B′) Subcellular localization of GFP–RAB-35 was observed in oocytes of wild-type (A, A′) or rme-4(b1001) mutants (B, B′). Middle planes (A, B) and cortical planes (A′, B′) of oocytes are shown. (C) Detection of GFP–RAB-35 by immunoblotting using an anti-GFP antibody. GFP–RAB-35 protein levels were not affected by the rme-4(b1001) mutation. (D–E′) Subcellular localization of GFP–RAB-5 in the oocytes of wild-type (D, D′) or rme-4(b1001) mutant (E, E′) animals. Middle planes (D, E) and cortical planes (D′, E′) of oocytes are shown. Bars, 10 μm.
RME-4 localizes to the clathrin-coated pit and physically interacts with AP-2
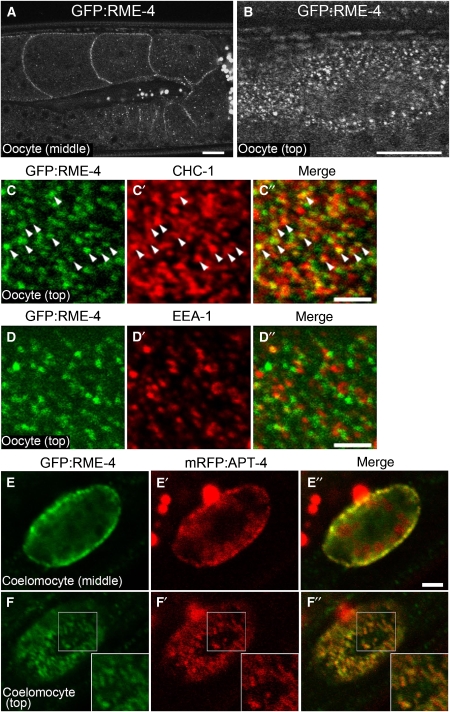
To further understand how RME-4 regulates the endocytic pathway, we determined its subcellular localization. This was achieved by analysis of a GFP–RME-4 fusion protein expressed in oocytes that we found rescued the yolk-endocytosis defect of rme-4 mutants, showing that it is functional and therefore is very likely to faithfully represent RME-4 localization (Supplementary Figure 4A and C). Unlike RAB-35, we found that, at steady state, GFP–RME-4 colocalized well with coated pit markers mRFP–CHC-1 (Supplementary Figure 5G–G″ and H–H″) and mRFP–APT-4 (data not shown), suggesting that GFP–RME-4 is mainly found in clathrin-coated pits and/or vesicles. GFP–RME-4 also colocalized with endogenous CHC-1 on the oocyte surface, whereas GFP–RME-4 failed to colocalize with endogenous EEA-1, an early endosome marker (Figure 6C–C″ and D–D″). GFP–RME-4 expressed under the control of its own promoter was broadly expressed with notably high levels in coelomocytes, neurons, pharynx, and intestine (Supplementary Figure 5). In coelomocytes, GFP–RME-4 localized to puncta on or very close to the plasma membrane. In agreement with our results in oocytes, GFP–RME-4 in coelomocytes also colocalized with coated-pit marker mRFP–APT-4 (Figure 6E–E″ and F–F″).
Figure 6.
GFP–RME-4 colocalizes with clathrin in the oocyte cortex. (A, B) Rescuing GFP–RME-4 was expressed under germline-specific promoter control and observed in the oocytes. Middle (A) and top (B) focal planes are shown. Bars, 10 μm. (C–C″, D–D″) GFP–RME-4 colocalizes with endogenous CHC-1. Dissected gonads of animals expressing GFP–RME-4 were stained with anti-GFP antibody and either anti-CHC-1 (C–C″) or anti-EEA-1 (D–D″) antibodies. Cortical focal planes of oocytes are shown. Bars, 2.5 μm. (E–E″, F–F″) Colocalization of GFP–RME-4 and mRFP–APT-4 in coelomocytes. Middle (E–E″) and top (F–F″) focal planes of coelomocytes are shown. Insets show enlargements ( × 1.5) of the boxed area. Bars, 2.5 μm.
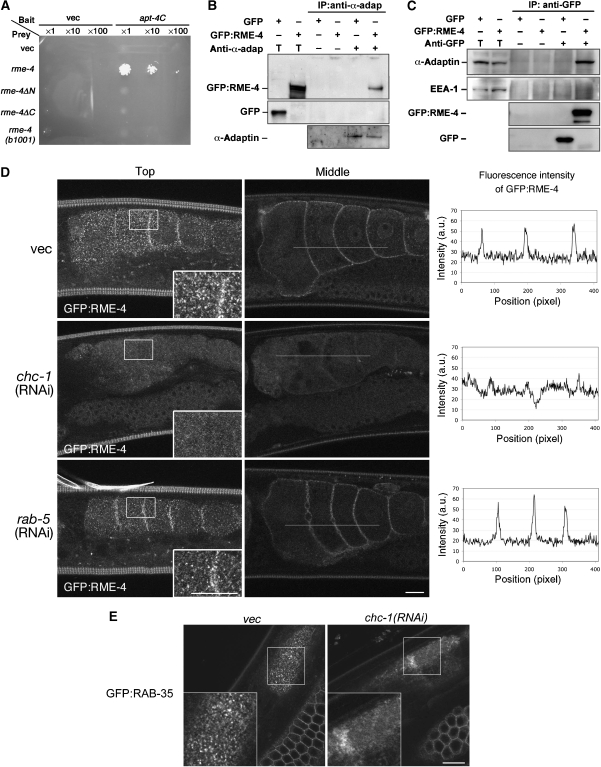
Given that RME-4 localizes to clathrin-coated pits and contains consensus binding sites for coated-pit components CHC-1 and AP-2 (Figure 3A) (ter Haar et al, 2000; Brett et al, 2002; Mishra et al, 2004), we sought to test the ability of RME-4 to physically interact with these components. We found that full-length RME-4 interacted with the C-terminal ear-domain of APT-4 in a yeast two-hybrid assay (Figure 7A). N- or C-terminal fragments of RME-4, or the mutant form of RME-4(G257D), failed to interact with APT-4 in this assay. We confirmed the physical interaction in lysates from C. elegans expressing GFP–RME-4 under its own promoter control. We found that immunoprecipitation of endogenous APT-4 co-precipitated GFP–RME-4 (Figure 7B and C). The converse was also true: immunoprecipitation of GFP–RME-4 co-precipitated endogenous APT-4. In contrast, GFP–RME-4 did not co-immunoprecipitate with EEA-1, an early endosome protein, under the same conditions. Collectively, these results indicate that RME-4 physically interacts with AP-2.
Figure 7.
RME-4 physically interacts with AP-2. (A) RME-4 interacts with the C-terminal ear region of APT-4 in a yeast two-hybrid assay. The ear domain of APT-4 (682–925 residues) was expressed in a yeast reporter strain as a fusion with the DNA-binding domain (bait). Full-length RME-4 or mutant forms of RME-4 were expressed in the same yeast cells as fusions with the transcriptional activation domain (prey). Interaction between bait and prey was tested using the LEU2 growth assay. Cell were serially diluted 1-, 10-, or 100-fold and spotted on growth medium. (B, C) Co-immunoprecipitation of GFP–RME-4 and endogenous APT-4. Total lysates were prepared from worms expressing GFP–RME-4 or GFP alone and subjected to immunoprecipitation with anti-APT-4 antibody (B) or anti-GFP antibody (C). Precipitants were probed on immunoblots with anti-APT-4, anti-GFP, and anti-EEA-1 antibodies. T (total lysate) equals approximately 1.5% of the total input into the assay. (D) RNAi-mediated knockdown of chc-1 but not rab-5 disrupts the cortical localization of GFP–RME-4 in oocytes. RNAi of chc-1 or rab-5 was performed in worms expressing GFP–RME-4 in the germ line. P0 worms were grown on RNAi bacteria for 24 h at 25°C. Top focal planes (projections of three images with 1 μm intervals; left panels) and middle focal planes (middle panels) of oocytes are shown. The insets show enlargements ( × 2) of the boxed area. Fluorescence intensity of GFP–RME-4 as a function of position was graphed along the lines indicated in the middle panels and are shown in paired panels. (E) RNAi-mediated knockdown of chc-1 disrupts the punctate localization of GFP–RAB-35. RNAi of chc-1 was performed in worms expressing GFP–RAB-35 in the germ line. Top focal planes of oocytes are shown. The insets show enlargements ( × 2) of the boxed area. Bars, 10 μm.
We further examined whether the cortical localization of GFP–RME-4 depends on clathrin-coated pit structures. Indeed, we found that RNAi-mediated knockdown of chc-1, but not rab-5, disrupts the cortical localization of GFP–RME-4 in oocytes (Figure 7D). Under the conditions used, RNAi of either chc-1 or rab-5 blocked yolk endocytosis and resulted in embryonic lethality, indicating that the RNAi-mediated depletions were efficient. Thus, the cortical localization of GFP–RME-4 depends upon clathrin. We also examined the effect of chc-1(RNAi) on the subcellular localization of GFP–RAB-35 (Figure 7E). RNAi-mediated knockdown of chc-1 impaired accumulation of GFP–RAB-35 on cortical endosomes, similar to the effect observed in rme-4(b1001) mutants, further suggesting that the coated-pit localization of RME-4 is important for downstream regulation of RAB-35.
Discussion
Here, we genetically identified C. elegans RME-4 and RAB-35 as regulators of endocytic transport and provide in vivo evidence that they function to promote receptor recycling.
RME-4 and RAB-35 are required for recycling of yolk receptors at early endosomes
Our detailed phenotypic analyses indicate that, in rme-4 and rab-35 mutants, RME-2 yolk receptors are internalized from the plasma membrane, but are not efficiently retained in cortical endosomes. In the case of rab-35, we showed directly that many of the mislocalized receptors are found in small vesicles, approximately 30 nm in diameter, suggesting that the receptors are accumulating in transport intermediates. The synthetic phenotype produced by combining loss of rme-4 or rab-35 with depletion of rab-11 or rme-1 suggests that RME-4 and RAB-35 function in a recycling pathway in parallel to that controlled by RAB-11 and RME-1 on recycling endosomes. Given our finding that GFP–RAB-35 strongly localizes to early endosomes in developing oocytes, our results suggest that RAB-35 functions in a direct recycling route from early endosomes to the plasma membrane. Consistent with the results from C. elegans null-mutant analysis, Rab35 has also recently shown to associate with early endosomes in mammalian tissue culture cells, and kinetic tests suggested a requirement for Rab35 in rapid recycling of transferrin from early endosomes to the plasma membrane (Kouranti et al, 2006).
Knockdown of Rab35 in HeLa cells causes a defect in the abscission step of cytokinesis (Kouranti et al, 2006). As C. elegans rab-35 null mutants are viable and fertile, we conclude that RAB-35 is not essential for cytokinesis in the nematode. On the other hand, rab-35 mutants do show a slightly reduced brood size (N2, 329±30; rab-35(b1013), 250±12 at 20°C). In addition, we observed severely disorganized oocytes in animals lacking rab-35 and rab-11, suggesting a redundant function for these Rabs in oogenesis (data not shown). Cytokinetic events producing individual oocytes from the germline syncytium might be impaired under these conditions.
RME-4 is an upstream regulator of RAB-35
To date, nothing has been reported on regulators or direct effectors of Rab35 in any species. We genetically identified rme-4 and rab-35 as functionally related genes. The most notable feature of RME-4 is the DENN domain at its N-terminus. DENN domains appear in several proteins involved in Rab function and are also associated with signalling pathways (Levivier et al, 2001; Miyoshi and Takai, 2004; Miserey-Lenkei et al, 2007). However, the molecular function of the DENN domain has remained elusive. Importantly, this domain was found in rat Rab3GEP and worm AEX-3, guanine nucleotide exchange factors for Rab3/RAB-3 (Iwasaki et al, 1997; Wada et al, 1997; Mahoney et al, 2006). The DENN domain of Rab3GEP is necessary for its biochemical GEF activity (Miyoshi and Takai, 2004), suggesting that the DENN domain may contribute part of the catalytic function. The preferential interaction of RME-4 with RAB-35(S24N), and the RME-4-dependent early endosomal localization of GFP–RAB-35, suggests that RME-4 functions to recruit RAB-35 to the early endosome, possibly leading to nucleotide exchange and Rab activation. Alternatively, RME-4 could act to recruit RAB-35 to membranes where it is activated by other regulators. Further investigation will be required to address these possibilities.
It is also clear from our studies that RME-4 has additional functions independent of its regulation of RAB-35, as rme-4 mutants show clear defects in endocytosis by coelomocytes, whereas rab-35 mutants have no such defect. One likely explanation is that RME-4 regulates at least one additional Rab GTPase, one that is required in coelomocyte cells. A similar situation has been well documented for AEX-3, which acts as a GEF for both RAB-3 and RAB-27 and displays mutant phenotypes similar to rab-3; rab-27 double mutants (Mahoney et al, 2006).
Regulation of RAB-35 by a clathrin-coated pit component RME-4
The colocalization of GFP–RME-4 with clathrin-coated pit markers and its physical interaction with AP-2 suggest that RME-4 is a clathrin-coated pit component. RME-4 is likely to bind directly to AP-2, and possibly CHC-1, through its consensus AP-2- and clathrin-binding motifs. It should be noted that RME-4 also has several SH3-binding motifs. Many components of clathrin-coated pits contain SH3 domains, or SH3-binding motifs, and are known to form complex networks of interacting proteins (Brett and Traub, 2006). Thus the proline-rich motifs present in RME-4 could also contribute to coated-pit association. Three human proteins are similar in overall domain structure to RME-4 (DENN/MADD-containing protein 1A, 1B, and 1C). Recently, mouse Connecdenn, which is an orthologue of human DENN/MADD-containing protein 1A, was shown to localize to clathrin-coated pits in neurons and was shown to interact with endophilin A1 and intersectin, SH3 domain proteins involved in synaptic clathrin-mediated endocytosis (Allaire et al, 2006). These results suggest that RME-4 is a conserved component of clathrin-coated pits, and further suggest potential interaction with additional clathrin-coated pit components. Knockdown of Connecdenn in cultured neurons partially reduced FM4-64 uptake by synaptic boutons after chemically stimulated depolarization (Allaire et al, 2006). Connecdenn function was not tested in non-neuronal cells, and connecdenn was not tested for effects on recycling that might indirectly affect uptake. We provide evidence that, in C. elegans, RME-4 primarily functions in receptor recycling rather than receptor uptake per se and thus suggest that a similar situation may exist for RME-4 homologues in other species such as mammalian connecdenn. This unexpected connection between coated-pit biogenesis and the downstream recycling of endocytosed cargo indicated by our work represents a new paradigm that will need to be taken into consideration.
We have shown that the coated-pit localization of RME-4 is disrupted by depletion of CHC-1 and that the endosomal localization of GFP–RAB-35 is impaired under these same conditions. This suggests that recruitment of RME-4 to the coated pit is important for downstream control of RAB-35, linking clathrin-coated vesicle formation with subsequent recycling processes. These results suggest that receptor recycling is primed during vesicle formation, ensuring that this important downstream event occurs efficiently. Our results with RME-4 and RAB-35 are highly reminiscent of the previously demonstrated recruitment of exocyst components to the coated pits of Drosophila oocytes, a process that is also not required for yolk receptor internalization but rather is required for the subsequent recycling of yolk receptors (Sommer et al, 2005). Our results with RME-4 and RAB-35 are also reminiscent of our previous results with RME-6, a RAB-5 GEF that is recruited to coated pits and is required to activate RAB-5 for downstream fusion events (Sato et al, 2005). Taken together, these studies suggest that formation of clathrin-coated vesicles includes the loading and activation of multiple components not required for uptake per se, but are rather required to prepare the cargo carrier for multiple downstream events, including fusion with early endosomes and rapid recycling of receptors from endosomes back to the plasma membrane.
Materials and methods
General methods
Worm cultures, genetic crosses, and other C. elegans methods were performed according to standard protocols (Brenner, 1974). Strains expressing transgenes in germline cells were grown at 25°C. Other strains were grown at 20°C. RNA interference (RNAi) was performed by the feeding method (Kamath and Ahringer, 2003). cDNAs were prepared from EST clone provided by Yuji Kohara (National Institute of Genetics, Japan) and subcloned into RNAi vector L4440 (Kamath and Ahringer, 2003). For RNAi knockdown of rab-5, rab-11, or chc-1, L4 worms were placed onto RNAi plates, and P0 adults were subsequently scored for phenotype after 24 h. For RNAi of rme-4, rme-5, or rme-1, F1 progeny were scored for phenotype.
Strains
Transgenic strain bIs1 (vit-2:GFP), arIs37 (pmyo-3:ssGFP), pwIs53 (ppie-1:mRFP:chc-1) and pwIs177 (punc-122:mRFP:apt-4) were described previously (Grant and Hirsh, 1999; Fares and Greenwald, 2001; Sato et al, 2005). A deletion allele of rme-4(tm1865) was provided by Shohei Mitani of the Japanese National Bioresource Project for nematodes. rab-7(ok551)/mIs1 and unc-119(ed3) were provided by the Caenorhabditis Genetics Center.
Genetic mapping and molecular cloning
rme-4(b1001) and rme-5(b1013 and b1034) were isolated in a screen described previously (Grant and Hirsh, 1999). The rme-4 mutation was mapped between unc-115 and egl-15 of LGX by classical three-point mapping. To identify a candidate for rme-4, we surveyed predicted genes in the C. elegans genomic sequence in this region by RNAi for the Rme phenotype and found that RNAi for one of genes, F46F6.1, caused a strong Rme phenotype. All exons from this gene were amplified from the rme-4(b1001) mutant by PCR and sequenced. Comparison of these sequences with the wild-type gene revealed a single change in the coding region (G770A). To determine the precise coding region of rme-4, the apparently full-length cDNA yk629b9 was sequenced. The rme-5 mutation was mapped between vab-7 and dpy-18 on LGIII by three-point mapping. The map position was further refined by complementation test with deficiencies; tDf5 failed to complement rme-5 but ctDf3 complemented. Similarly, predicted genes in this region were surveyed by RNAi, and the candidate gene of rme-5, Y47D3A.25/rab-35, was identified. All exons of this gene amplified from the rme-5 mutants were sequenced and identified a single mutation for each allele (b1013; C205T, b1034; G191A). Expression of F46F6.1a or rab-35 as GFP fusions under the pie-1 promoter rescued the Rme phenotype of the rme-4(b1001) or rme-5(b1013) mutants, respectively. RNAi of F46F6.1 and rab-35 by the feeding method caused the Rme phenotype only in the F1 and subsequent generations.
Plasmids and transgenic strains
To express GFP fusions in the germ line, rme-4 genomic DNA or cDNAs of C. elegans rab-35 and human Rab35 (Guthrie cDNA Resource Center) were cloned into pDONR221 and then transferred into pID3.01B (Pellettieri et al, 2003) by Gateway cloning (Invitrogen, CA). To express RFPmChery–RAB-5 and RFPmChery–RAB-5(Q78L), rab-5 and its mutant form were cloned into AZ132_mCherry_Gtwy (a gift from A Audhya and K Oegema). To express EYFP–RME-4 in the intestine, the coding region of GFP in pvha-6-GFP (Chen et al, 2006) was replaced with that of EYFP and a genomic rme-4 fragment was cloned downstream of EYFP. To express mRFPCherryHA fusions, the act-5 promoter (2005 bp) and the coding sequence of mRFPCherryHA was subcloned into pPD117.01 (a gift of A Fire). A Gateway cassette (Invitrogen, CA) and the unc-119 gene from C. briggsae was further subcloned into the vector. The coding sequence of mRFPCherry was amplified from AZ132_mCherry_Gtwy by PCR, and a linker fragment coding an HA epitope (YPYDVPDYA) was inserted just after the mRFPCherry coding sequence. cDNA fragments of rab-35, and its mutant forms generated by site-directed mutagenesis, were cloned into this destination vector by Gateway cloning.
To construct GFP–rme-4 driven by its own promoter control, the entire genomic rme-4 fragment including the upstream (2369 bp) and downstream (2140 bp) regions was cloned into pBluescriptII SK(+) (Stratagene), and then a GFP fragment was inserted in frame into the intrinsic NdeI site at the start codon of the rme-4 gene (pBSSK–GFP–rme-4). As we could not subclone the entire genomic fragment of rab-35, a genomic fragment containing the 1st to 3rd exons were subcloned into pDONR221-rab35(cDNA), resulting a genomic-cDNA hybrid minigene. This rab-35 minigene was cloned into pvha-6-GFP, and then the genomic fragment upstream of rab-35 (6049 bp) was subcloned upstream of GFP.
All integrated transgenic lines were obtained by the microparticle bombardment method (Praitis et al, 2001). pBSSK–GFP–rme-4 was cobombarded with plasmid MM016B encoding the wild-type unc-119 gene into unc-119(ed3) mutant worms. A list of transgenic lines constructed for this study is provided in Supplementary data.
Immunoblotting and antibodies
For immunoblotting of whole worm lysates, lysates were prepared from 50 adults (24 h after L4) by boiling in Laemmli sampling buffer and subjected to immunoblotting using rabbit anti-RME-4 or anti-RME-2 antibodies or goat anti-GFP antibody.
The entire coding sequence of rme-4 was subcloned into pEXP1–DEST (Invitrogen). cDNA sequences encoding the hub domain of CHC-1 (aa 1176–1681) were subcloned into the pET15 vector (Novagen). His-tagged RME-4 and CHC-1hub were expressed in Escherichia coli and purified under denaturing conditions and outsourced for injection into rabbits (Advanced Technology Planner, Japan). This RME-4 antibody worked well on western blots but could not detect RME-4 in situ after aldehyde or methanol fixation. The CHC-1 antibody worked well on both western blots and in situ after aldehyde fixation. Anti-RME-2 and anti-EEA-1 antibodies was described previously (Grant and Hirsh, 1999; Shaye and Greenwald, 2002).
Two-hybrid assay
The DupLEX-A two-hybrid system (OriGene Technologies Inc., MD) was used according to the manufacturer's instructions. A full-length cDNA of rme-4 was cloned into pJG4-5. A cDNA fragment corresponding to the C-terminal region of APT-4 (aa 682–925) was cloned into pEG202. cDNA of rab-35 lacking the last four residues and its mutant forms (S24N and Q69L) were cloned into pEG202. C. elegans rab-5, rab-7, rab-11, and their mutant forms were also cloned into pEG202. These plasmids were introduced into a reporter strain EGY48 included with the system. To assess the expression of the LEU2 reporter, transformants were grown on plates lacking leucine, histidine, tryptophan, and uracil, containing 2% galactose/1% raffinose at 30°C for 3 days.
Immunoprecipitation
To detect physical interaction between GFP–RME-4 and endogenous APT-4, a transgenic line expressing GFP–RME-4 (pwIs268) was used to prepare whole worm lysates. Co-immunoprecipitation experiments between GFP–RME-4 and APT-4 were performed as described previously (Sato et al, 2005). To detect physical interaction between RME-4 and RAB-35, transgenic worms strains were constructed expressing EYFP–RME-4 (dkIs163) and either mRFPCherryHA–RAB-35 (dkIs187) or with its mutant forms (dkIs191 or dkIs194). Approximate 0.3 ml of young adults harbouring the two transgenes were harvested and washed with M9 buffer. They were resuspendend in 1 ml of 25 mM HEPES–KOH (pH 7.4), 50 mM potassium acetate, 5 mM magnesium acetate, and 1 mM dithiothreitol containing 0.1% Triton X-100 and protease inhibitors (Complete EDTA-free, Roche Diagnostics GmbH; IP buffer) and homogenized in a stainless steel homogenizer (Wheaton Science Products, NJ). Homogenates were incubated on ice for 30 min and cleared twice by centrifugation for 10 min at 16 000 g at 4°C. For immunoprecipitation, extracts were incubated with mouse anti-GFP monoclonal antibody (3E6, Q-BIOgene Inc., CA) or mouse anti-HA monoclonal antibody (16B12, Covance Research Products Inc., CA) for 2 h at 4°C followed by incubation with protein G-Sepharose (Sigma) for 2 h at 4°C. Beads were washed with IP buffer three times and IP buffer without Triton X-100 once. Precipitants were eluted with Laemmli sampling buffer and subjected to immunoblotting using goat anti-GFP polyclonal antibody (Research Diagnostics Inc., NJ) and rat anti-HA monoclonal antibody (3F10, Roche Diagnostics GmbH).
Microscopy
To observe live worms expressing transgenes, worms were mounted on agarose pads with 10 mM levamisol in M9 buffer. In some cases, when two germline-expressed transgenes were crossed together, one or both transgenes became silenced. In such cases, transgene expression was restored before imaging by blocking the silencing mechanism, using rde-2 feeding RNAi (Kim et al, 2005). Immunostaining of dissected gonads were performed as described previously (Grant and Hirsh, 1999). Confocal images were obtained using an Olympus confocal microscope system FV1000 with × 60, 1.35 NA UPlanSApo oil-objective lens (Olympus Corp., Japan). Quantification of images was performed with software included in the FV1000 system. Fluorescence images were obtained using an Axiovert 200 M (Carl Zeiss MicroImaging Inc., Germany) microscope equipped with a digital CCD camera (C4742-95-12ER, Hamamatsu Photonics, Japan) and Metamorph software (Universal Imaging Corp., PA) and then deconvolved with AutoDeblur software (AutoQuant Imaging Inc., NY).
For immunoelectron microscopy, adult N2 and rme-5(b1013) were anaesthetized in M9 containing 20 mM sodium azide, transferred to a drop of fixative (2% glutaraldehyde in 0.1 M sodium cacodylate buffer), and cut immediately at the level of the pharynx. After 2 h of fixation at room temperature, animals were embedded in 12% gelatin. Ultrathin cryosectioning and immunogold labelling with anti-RME-2 INT antibody were performed sequentially as described (Sato et al, 2005). To quantify the RME-2 labelling density, sampling was taken from a block that harboured N2 and rme-5 gonads side by side, so that both could be subject to precisely the same preparation and labelling conditions. Electron micrographs were taken from 11 section pairs from five different grids. The cortical zone was defined as within 500 nm of the cell surface. Gold particle number per unit area was estimated by the point counting method (Griffiths, 1993). Total areas quantified were 20 μm2 (N2) and 22 μm2 (rme-5) for the central zone and 82 μm2 (N2) and 127 μm2 (rme-5) for the cortical zone, respectively.
Supplementary Material
Supplementary Figures 1
Supplementary Figures 2
Supplementary Figures 3
Supplementary Figures 4
Supplementary Figures 5
Supplementary data
Acknowledgments
We thank Laura Pedraza and Yinhua Zhang for performing the initial mapping of rme-4 and rme-5 mutants, and David Hirsh for supporting the early stages of this work. We thank Anjon Audhya, Karen Oegema, and Geraldine Seydoux for provided important reagents, Yuji Kohara for EST clones, Shohei Mitani for rme-4(tm1865), the Caenorhabditis Genetic Center for strains, and Ken Sato for suggestions on biochemical assays. We also thank Peter Schweinsberg and Katsuya Sato for expert technical assistance. MS was supported by JSPS Research Fellowships for Young Scientists. This work was supported by the Ministry of Education, Science, Sports and Culture, Grant-in-Aid for Young Scientists (A), 2005, Scientific Research on Priority Areas 2005, Scientific Research (B), 2005, and Bioarchitect Research Projects of RIKEN to KS and grants-in-aid and the Global Center of Excellence Program from the Japanese Ministry of Education, Culture, Sports, Science and Technology to MS, KS, and AH. This work was also supported by an NIH Grant GM067237 to BG. WL was supported by a Grant EMRPD 160251 from the Ministry of Education in Taiwan.
References
- Allaire PD, Ritter B, Thomas S, Burman JL, Denisov AY, Legendre-Guillemin V, Harper SQ, Davidson BL, Gehring K, McPherson PS (2006) Connecdenn, a novel DENN domain-containing protein of neuronal clathrin-coated vesicles functioning in synaptic vesicle endocytosis. J Neurosci 26: 13202–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett TJ, Traub LM (2006) Molecular structures of coat and coat-associated proteins: function follows form. Curr Opin Cell Biol 18: 395–406 [DOI] [PubMed] [Google Scholar]
- Brett TJ, Traub LM, Fremont DH (2002) Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure 10: 797–809 [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE (2001) Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol 17: 517–568 [DOI] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B (2000) Rab7: a key to lysosome biogenesis. Mol Biol Cell 11: 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Schweinsberg PJ, Vashist S, Mareiniss DP, Lambie EJ, Grant BD (2006) RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol Biol Cell 17: 1286–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow VT, Lee SS (1996) DENN, a novel human gene differentially expressed in normal and neoplastic cells. DNA Seq 6: 263–273 [DOI] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M (1999) The Rab5 effector EEA1 is a core component of endosome docking. Nature 397: 621–625 [DOI] [PubMed] [Google Scholar]
- Fares H, Greenwald I (2001) Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stahl B, Li C, Sudhof TC, Jahn R (1994) Rab proteins in regulated exocytosis. Trends Biochem Sci 19: 164–168 [DOI] [PubMed] [Google Scholar]
- Grant B, Hirsh D (1999) Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell 10: 4311–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D (2001) Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol 3: 573–579 [DOI] [PubMed] [Google Scholar]
- Griffiths G (1993) Fine Structure Immunocytochemistry. Heidelberg, Germany: Springer-Verlag [Google Scholar]
- Iwasaki K, Staunton J, Saifee O, Nonet M, Thomas JH (1997) aex-3 encodes a novel regulator of presynaptic activity in C. elegans. Neuron 18: 613–622 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321 [DOI] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, Vidal M, Ruvkun G (2005) Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167 [DOI] [PubMed] [Google Scholar]
- Kouranti I, Sachse M, Arouche N, Goud B, Echard A (2006) Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol 16: 1719–1725 [DOI] [PubMed] [Google Scholar]
- Levivier E, Goud B, Souchet M, Calmels TP, Mornon JP, Callebaut I (2001) uDENN, DENN, and dDENN: indissociable domains in Rab and MAP kinases signaling pathways. Biochem Biophys Res Commun 287: 688–695 [DOI] [PubMed] [Google Scholar]
- Mahoney TR, Liu Q, Itoh T, Luo S, Hadwiger G, Vincent R, Wang ZW, Fukuda M, Nonet ML (2006) Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol Biol Cell 17: 2617–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE (2004) Endocytic recycling. Nat Rev Mol Cell Biol 5: 121–132 [DOI] [PubMed] [Google Scholar]
- McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M (1999) Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell 98: 377–386 [DOI] [PubMed] [Google Scholar]
- Miserey-Lenkei S, Waharte F, Boulet A, Cuif MH, Tenza D, El Marjou A, Raposo G, Salamero J, Heliot L, Goud B, Monier S (2007) Rab6-interacting protein 1 Links Rab6 and Rab11 function. Traffic 8: 1385–1403 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Hawryluk MJ, Brett TJ, Keyel PA, Dupin AL, Jha A, Heuser JE, Fremont DH, Traub LM (2004) Dual-engagement regulation of protein interactions with the AP-2 adaptor alpha appendage. J Biol Chem 279 (44): 46191–46203 [DOI] [PubMed] [Google Scholar]
- Miyoshi J, Takai Y (2004) Dual role of DENN/MADD (Rab3GEP) in neurotransmission and neuroprotection. Trends Mol Med 10: 476–480 [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Reinke V, Kim SK, Seydoux G (2003) Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C.elegans. Dev Cell 5: 451–462 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC (2001) Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313: 889–901 [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J (2001) Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122: 735–749 [DOI] [PubMed] [Google Scholar]
- Sato K, Sato M, Audhya A, Oegema K, Schweinsberg P, Grant BD (2006) Dynamic regulation of caveolin-1 trafficking in the germ line and embryo of Caenorhabditis elegans. Mol Biol Cell 17: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Sato K, Fonarev P, Huang CJ, Liou W, Grant BD (2005) Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat Cell Biol 7: 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Mushiake S, Kato Y, Sato K, Sato M, Takeda N, Ozono K, Miki K, Kubo Y, Tsuji A, Harada R, Harada A (2007) The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature 448: 366–369 [DOI] [PubMed] [Google Scholar]
- Schievella AR, Chen JH, Graham JR, Lin LL (1997) MADD, a novel death domain protein that interacts with the type 1 tumor necrosis factor receptor and activates mitogen-activated protein kinase. J Biol Chem 272: 12069–12075 [DOI] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I (2002) Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature 420: 686–690 [DOI] [PubMed] [Google Scholar]
- Sommer B, Oprins A, Rabouille C, Munro S (2005) The exocyst component Sec5 is present on endocytic vesicles in the oocyte of Drosophila melanogaster. J Cell Biol 169: 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T (2000) Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci USA 97: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG (1996) Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol 135: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs P, Hull M, Webster P, Male P, Goud B, Mellman I (1992) The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70: 729–740 [DOI] [PubMed] [Google Scholar]
- Wada M, Nakanishi H, Satoh A, Hirano H, Obaishi H, Matsuura Y, Takai Y (1997) Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J Biol Chem 272: 3875–3878 [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2: 107–117 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Grant B, Hirsh D (2001) RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol Biol Cell 12: 2011–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1
Supplementary Figures 2
Supplementary Figures 3
Supplementary Figures 4
Supplementary Figures 5
Supplementary data