Abstract
Ribosomal RNA synthesis is the eukaryotic cell's main transcriptional activity, but little is known about the chromatin domain organization and epigenetics of actively transcribed rRNA genes. Here, we show epigenetic and spatial organization of mouse rRNA genes at the molecular level. TTF-I-binding sites subdivide the rRNA transcription unit into functional chromatin domains and sharply delimit transcription factor occupancy. H2A.Z-containing nucleosomes occupy the spacer promoter next to a newly characterized TTF-I-binding site. The spacer and the promoter proximal TTF-I-binding sites demarcate the enhancer. DNA from both the enhancer and the coding region is hypomethylated in actively transcribed repeats. 3C analysis revealed an interaction between promoter and terminator regions, which brings the beginning and end of active rRNA genes into close contact. Reporter assays show that TTF-I mediates this interaction, thereby linking topology and epigenetic regulation of the rRNA genes.
Keywords: chromatin, looping, nucleolus, transcription
Introduction
In most cells, more than half of the RNA synthesis is rRNA transcription, which is essential to maintain protein synthesis capacity. This massive energy-consuming process needs tight and economic regulation. The nucleolus is the site of the 47S precursor rRNA synthesis, which is transcribed by RNA polymerase I and processed into 5.8S, 18S and 28S rRNA (Russell and Zomerdijk, 2005).
Eukaryotic rRNA genes are flanked on both sides by terminator elements. In mouse, the terminator sequence motif is recognized by the transcription termination factor TTF-I. Binding of TTF-I to the terminators (T1–T10) downstream of the transcription unit stops elongating RNA polymerase I and mediates transcription termination (Grummt et al, 1986b). A similar sequence element, defined as T0, is located immediately upstream of the ribosomal gene promoter (Grummt et al, 1986a). Binding of TTF-I to the promoter proximal terminator is a key event that facilitates transcription initiation on rDNA reconstituted into chromatin (Längst et al, 1997, 1998).
The fact that binding sites for the rDNA-specific transcription factor TTF-I are located at the 5′ and the 3′ end of each rRNA gene suggests a functional link between transcription initiation and termination. Homologous sites of the yeast rRNA genes have been proposed to serve as the base for DNA loops juxtaposing the promoter and terminator regions. According to this ‘ribomotor' model, RNA polymerase I could be directly transferred from the termination site to the initiation site, thereby increasing the re-initiation rate (Kempers-Veenstra et al, 1986). However, experimental support for the ribomotor model has so far been lacking. The observation that TTF-I is capable to form oligomers and bridges separate DNA molecules in vitro (Sander and Grummt, 1997) tempts speculation that TTF-I may be an active player in the formation of looped rRNA gene domains in mammals.
A typical eukaryotic cell contains several hundred rRNA genes, most of them arranged as tandem repeats, but even in metabolically active cells only a proportion of these genes are transcriptionally active. Active and inactive copies are marked by different chromatin structures. The role of chromatin remodelling, histone modifications and DNA methylation in the silencing process has been intensively studied (Grummt and Pikaard, 2003). However, the epigenetics of actively transcribed copies and the establishment and maintenance of active rRNA gene status are poorly understood. We analysed the chromatin domain organization of active and inactive rRNA genes in mammalian cells using the combination of ChIP, chromosome conformation capture (3C), DNA methylation sensitivity and functional assays. These assays revealed that rRNA genes exist in different topological conformations depending on gene activity and epigenetic modifications. Furthermore, we show that TTF-I has a central function in defining the topology of the rRNA gene and separates three functional chromatin domains; the promoter/enhancer, the coding and the intergenic spacer domains.
Results
Mapping factor occupancies at the rDNA transcription unit
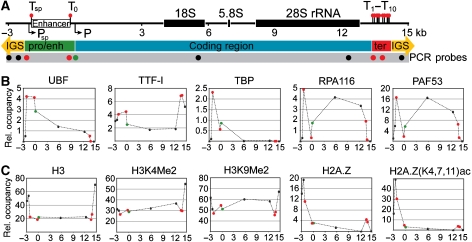
To characterize protein occupancies at the rRNA genes, ChIP experiments were performed. Previous ChIP studies of the mammalian rDNA system were restricted either to the promoter region (Santoro et al, 2002; Philimonenko et al, 2004; Mayer et al, 2006) or to a single factor (O'Sullivan et al, 2002). The most comprehensive ChIP analysis of rRNA genes so far showed occupancies of 4 features (transcription factors and histone modifications) at 10 locations in human cells (Grandori et al, 2005). In this study, we mapped 10 features at 14 locations within the mouse rDNA transcription unit (Figure 1A), including known cis-regulatory elements, the coding region and non-repetitive sequence elements of the intergenic spacer (Figure 1 and Supplementary Figure S1).
Figure 1.
Transcription factor and histone modification occupancies of mouse rRNA genes. (A) Scheme of regulatory sequence elements from −3000 to +15 000 relative to the transcription start site. Lollipops show binding sites for TTF-I (Tsp, T0 and T1−10); arrows indicate promoter regions (Psp: spacer promoter; P: core promoter element, primary transcript start site); white rectangle marks the enhancer element; black rectangles label processed transcript coding regions (18S, 5.8S and 28S rRNA). Circles below the ruler show the positions of the PCR amplicons. Red circles indicate TTF-I-binding sites, the green circle marks the core promoter. Numbers indicate kilo basepairs. (IGS: intergenic spacer; pro/enh: promoter/enhancer; ter: terminator element). (B) Chromatin immunoprecipitation (ChIP) analysis of transcription factor occupancy. Relative occupancies were estimated by real-time quantitative PCR as described in Materials and methods, and labelled as dots at each analysed position. ChIP experiments were performed using polyclonal antibodies against proteins indicated above the graphs. (C) ChIP analysis of histone modification patterns. Relative occupancies of histone modifications were normalized to nucleosome occupancies of the corresponding region. ChIP experiments were performed using polyclonal antibodies against epitopes indicated above each plot. Dashed lines are present only to aid visualization of differences between relative occupancies; they do not represent factor occupancies.
Figure 1B and Supplementary Figure S1 show binding of the multifunctional protein TTF-I to its suggested target sites at the terminator (T1–T10) and the promoter (T0). Moreover, TTF-I binds to an additional site, the newly characterized spacer terminator (Tsp) site, which is located about 2 kb upstream of the gene promoter and 63 bp downstream of the spacer promoter.
Binding of the architectural transcription factor UBF is restricted to the enhancer and coding regions of the rDNA (Figure 1B and Supplementary Figure S1). The factor is not uniformly distributed within these regions, but it is more abundant close to the enhancer and at the gene promoter, than within the coding region. Protein occupancy gradually decreases downstream of the transcription start site, indicating the presence of high-affinity binding sites for UBF at the promoter and enhancer.
The relative TBP occupancy, diagnostic of the basal transcription factor complex TIF-IB (Eberhard et al, 1993), was found to be higher at the spacer promoter than at the core promoter. Similar to TBP, RNA polymerase I, as revealed by the occupancy pattern of its RPA116 and RPA53 subunits, is more enriched at the spacer promoter than at the core promoter, suggesting that the spacer promoter functions as a high-affinity binding site or an entry site for the enzyme. As expected, RNA polymerase I is highly enriched in the coding region, but absent from the TTF-I-dependent transcription termination sites (Figure 1B and Supplementary Figure S1).
The epigenetic status of the rRNA genes was characterized by measuring the relative occupancies of five different chromatin features throughout the rDNA repeat. First, we analysed nucleosome occupancies by using an antibody that recognizes all forms of histone H3. Compared with the IGS sequences we detect reduced nucleosome levels (about 50%) at the enhancer and coding regions, inversely correlating with the occupancy of the transcription machinery (Figure 1C and Supplementary Figure S1). Using an antibody recognizing H3K4Me2, a modification that marks transcriptionally active sites (Schneider et al, 2004; Mayer et al, 2006), revealed a relative increase of this modification within the coding region as compared with H3 levels. We were also able to detect high levels of nucleosomes carrying the repressive mark H3K9Me2 throughout the whole coding region, with reduced occupancies at both ends of the coding region, that is, the promoter/enhancer and terminator (Figure 1C and Supplementary Figure S1).
In contrast, H2A.Z and its acetylated form H2A.Z (K4,7,11)ac were found primarily 500–600 bp upstream of the spacer promoter and to a lesser extent at the rRNA gene terminator (Figure 1C and Supplementary Figure S1). H2A.Z was recently described as a marker of active and poised promoters, marking a switch in local chromatin structure, separating different structures of neighbouring chromatin domains (Guillemette and Gaudreau, 2006). Two remarkable aspects of our H2A.Z results can be highlighted: first, the primary peak is located close to the spacer promoter and not at the core promoter, suggesting that this region serves specific functions and in addition marks the boundary between the IGS and the functional rRNA gene domains. Second, we observed the appearance of a minor peak at the terminator site that—similar to TTF-I-, marks the 3′ boundary of the functional rRNA gene. This signal could reflect an indirect crosslinking effect, suggesting that the terminator is in close proximity to the promoter in the three-dimensional nucleolar space. However, ChIP assays cannot discriminate between H2A.Z occupancy at the terminators and interactions in three-dimensional nucleolar space.
Characterization of the TTF-I-binding site at the spacer promoter
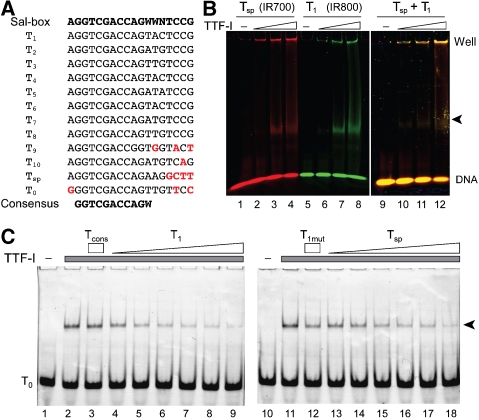
As described above, ChIP analysis of mouse rRNA genes revealed a new TTF-I-binding site close to the spacer promoter. We compared the nucleotide sequence of the binding site with the previously described Sal-boxes (Grummt et al, 1986a). Apparently, TTF-I requires a smaller recognition sequence than described before (Figure 2A). TTF-I binding to the Tsp site was confirmed by electromobility shift assays, exhibiting comparable binding affinities as for T1, the first terminator site (Figure 2B). To compare TTF-I-binding affinities more accurately, we performed competitive binding assays. Competition of TTF-I binding to the T0 site was similar for T1 and Tsp, whereas the mutant T1mut oligonucleotide and the minimal consensus binding site Tcons were not able to interfere with TTF-I binding (Figure 2C). Our results demonstrate that TTF-I binds with comparable affinity to the spacer terminator and to the T1 site in vitro, whereas the 10-bp-long consensus sequence is not sufficient to mediate high-affinity binding of TTF-I. Our data suggest that additional nucleotides flanking the core sequence increase the binding affinity, but not the sequence specificity of TTF-I.
Figure 2.
Characterization of the Tsp-binding site of TTF-I. (A) Sequence comparison of murine TTF-I-binding sites. Differences from the ‘Sal-box' sequence are highlighted in red. The consensus sequence is shown at the bottom. (B) Electrophoretic mobility shift assay. Increasing amounts of TTFdN209 (25–100 fmol) were incubated with the IR700-labelled Tsp (lanes 1–4), IR800-labelled T1 (lanes 5–8) or an equimolar mixture of both TTF-I-binding sites (lanes 9–12). TTF-I–DNA (marked by an arrow) complexes were analysed on 4.5% native polyacrylamide gels. (C) Binding competition assay. Increasing amounts (62.5–2000 fmol) of T1 (lanes 4–9) and Tsp (lanes 13–18), 2000 fmol of the minimal consensus sequence Tcons (lane 3), or the binding site mutant T1mut (lane 12) oligos were added to Alexa555-labelled T0 oligo (250 fmol) and TTFdN348 (400 fmol). TTF-I–DNA complexes (arrow) were resolved as in (B).
CpG domains of mammalian rRNA genes
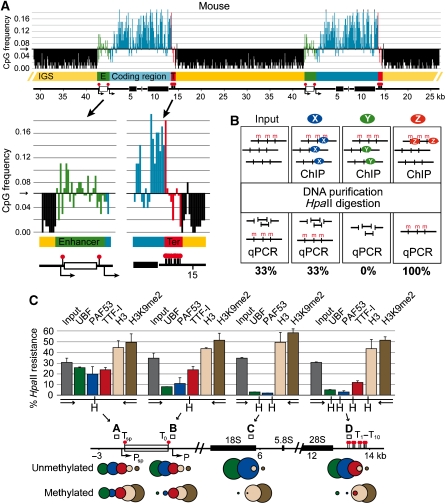
Biological systems relying on DNA methylation as a regulatory mechanism should maintain a significant proportion of CpG dinucleotides, whereas the average level of this dinucleotide should decrease with time by cytosine deamination and other mutagenic mechanisms (Pfeifer, 2006). The pattern of DNA methylation in human rRNA genes and the %CpG content throughout the gene has been studied before (Brock and Bird, 1997). We calculated the absolute dinucleotide occurrence of rDNA across different species with a resolution of 100 bp (Figure 3A and Supplementary Figure S2). The frequency of CpG dinucleotides should be 0.0625 at random distribution. In mouse rDNA (GenBank acc. no. BK000964), CpG dinucleotides are enriched in the primary transcript coding region and depleted in the IGS between the TTF-I-binding sites T10 and Tsp. A local enrichment of CpG content is observed around the spacer promoter, followed by levels that correlate with random distribution at the enhancer (Figure 3A). The human rDNA shows a comparable distribution of CpG along the locus (Supplementary Figure S2; GenBank acc. no. U13369). Indeed, the CpG domain transitions at the ends of the intergenic spacer are even more defined in the human rDNA locus. The picture was different in yeast (GenBank acc. no. U53879) and Arabidopsis (GenBank acc. no. X52322) rDNA (Supplementary Figure S2). Thus, neither eukaryotes lacking DNA methylation (yeast) or those possessing multiple DNA methylating activities modifying the rDNA promoter (Arabidopsis; Lawrence et al, 2004) have CpG distributions similar to those seen in mammals.
Figure 3.
CpG domains coexist with functional chromatin domains in mammalian rDNA. (A) CpG domains of mouse rDNA (GenBank acc. no. BK000964). Two repeats of the tandem rDNA array are shown on the scheme, and different DNA elements are colour coded and labelled as in Figure 1A. CpG dinucleotide frequency distribution is shown at 100-bp resolution. CpG domain transitions are shown enlarged. The threshold is set at 0.0625 (1:16=random distribution). (B) Scheme of the methylation-sensitive ChIP-chop assay. X, Y and Z indicate hypothetical proteins that bind DNA independent of DNA methylation, bind to unmethylated or methylated DNA, respectively. The expected outcome of the precipitation reactions is shown in the scheme. (C) Differential methylation of mouse rDNA domains. The rDNA ruler is labelled as in Figure 1A. Rectangles and letters A–D mark PCR amplicons for the methylation-sensitive ChIP analysis. HpaII cleavage sites are labelled with H. Circle diagrams aid visualization of unmethylated/methylated DNA ratio in different ChIP samples.
The DNA methylation status from the promoter to the terminators marks the activity status of the rRNA genes
ChIP assays investigate chromatin features in a population-averaged manner, not selective for the activity status of the genes. In a subset of species—including mouse—DNA methylation at CpG sites is frequently coupled to transcriptional regulation through different mediator mechanisms (Klose and Bird, 2006). To reveal relationships between transcription factor occupancy, DNA methylation and transcriptional activity, we analysed factor occupancies on the methylated and non-methylated rRNA genes by using the ChIP-chop assay (Figure 3B) (Preuss and Pikaard, 2007). Immunoprecipitated DNA was digested with the methylation-sensitive enzyme HpaII and its isochizomer MspI that cleaves independent of cytosine methylation. The proportion of HpaII-sensitive DNA, present in the input DNA and the immunoprecipitated material was quantified by real-time quantitative PCR, using DNA primer pairs that encompass a DNA fragment containing HpaII/MspI sites.
Low-resolution Southern blot analysis revealed that DNA methylation does not exist as isolated patches on the transcription units, but the rRNA coding region is either fully methylated or non-methylated (Brock and Bird, 1997) (data not shown). Accordingly, the DNA methylation levels measured at the spacer promoter, promoter, transcribed region and terminator were similar (between 31 and 35%), representing the DNA methylation status of the whole rDNA transcription unit. About one-third of the rRNA genes are methylated in the mouse 3T3 cell line (Figure 3C, input).
Analysis of the histone H3 content and the H3K9me2 modification revealed their association with methylated DNA, therefore correlating with inactive rRNA genes. The predominant association of histone H3 with inactive rRNA genes suggests a decreased amount of crosslinkable histones on active genes (Figure 3C). Next, we analysed the distribution of transcription factors that should be associated with the active rRNA genes. UBF and PAF53 bind to the promoter, coding and terminator regions of non-methylated genes, correlating with active transcription. Surprisingly, a different result was obtained for the spacer promoter: The methylation levels of the DNA precipitated with UBF and PAF53 antibodies were similar to the methylation level of the input DNA. These factors bind to the spacer promoter sequences irrespective of the DNA methylation status. This suggests that rRNA spacer and gene promoters are functionally different. Only active gene promoters are loaded with the transcription machinery, whereas all spacer promoters are loaded with the transcription factors (Figure 3C). Our data suggest that each rRNA gene—active or inactive—is preceded by a spacer promoter fully loaded with RNA polymerase I factors.
TTF-I, exhibiting sequence-specific binding sites near to the promoters and at the terminator, showed different occupancies at the promoters compared with the terminator. TTF-I is present at the spacer promoter and promoter irrespective of DNA methylation. However, the protein is mainly associated with non-methylated DNA at the terminators (Figure 3C). Our data indicate that TTF-I binds to the spacer promoter and promoter irrespective of DNA methylation and binds selectively to the non-methylated rDNA terminator. This suggests that the presence of TTF-I at the gene terminator is a hallmark of active rRNA genes.
The promoter and terminator regions of active rRNA genes do physically interact
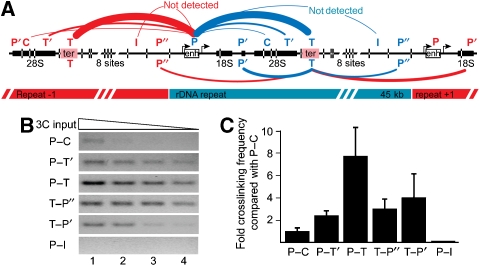
To analyse the three-dimensional organization of mouse rDNA more directly, we performed 3C analysis (Dekker et al, 2002; Dekker, 2006) in neonatal liver cells (Figure 4A and Supplementary Figure S3). The 3C technology involves quantitative PCR analysis of crosslinking frequencies between two DNA restriction fragments, which gives a measure of their proximity in nuclear space (Dekker et al, 2002). A schematic representation of our 3C experiment is given in Figure 4A. As TTF-I was our candidate for DNA bridging, we focused on the determination of relative crosslinking frequencies of DNA pairs where at least one of the two fragments contains a binding site for TTF-I. The systematic analysis of the rDNA revealed close proximity between the promoter and terminator regions, as shown by the quantification of the specific PCR products (Figure 4C). We observed an eight-fold increase in crosslinking frequency between the promoter and the terminator regions and reduced interaction frequencies within the coding and intergenic regions. The levels of crosslinking frequencies are presented schematically in Figure 4A.
Figure 4.
Promoter and terminator sequences of mouse rDNA are juxtaposed in vivo. (A) Scheme of the 3C assay shows the analysed fragment pairs (P—promoter, C—coding region, T′—terminator neighbouring, T—terminator; I—intergenic spacer; P′, P″—promoter neighbouring). Restriction enzyme sites are marked with lines crossing the rDNA ruler. Thickness of the connecting line between different DNA fragments indicates relative crosslinking frequency. Possible intragenic interactions are labelled with blue lines and the intergenic ones with red. (B) 3C results of neonatal mouse liver cells. Agarose gel pictures showing the PCR amplicons of a 3C experiment. (C) Average values and standard deviations of three 3C biological replicate samples are shown on the plot as fold relative crosslinking frequency compared to the P–C interaction.
The results clearly show that the promoter and terminator regions, bearing the TTF-I-binding sites, are in close proximity in the nucleolar space. As described by Dekker and colleagues, the crosslinking frequency gradually decreases as the distance between the DNA fragments and the interacting sites increases (see interaction frequencies at the globin locus, measured by 3C and 5C; Dostie et al, 2006; Figures 2, 3 and 4). Most importantly, the interacting sites show a peak in crosslinking frequency between promoter–terminator (P–T) fragments (Figure 4). However, the assay does not reveal whether these interactions are formed between the promoter and the terminator of one transcription unit, neighbouring or even distant rRNA genes. In addition, the assay does not discriminate between active and inactive genes.
Active rRNA genes form DNA loops
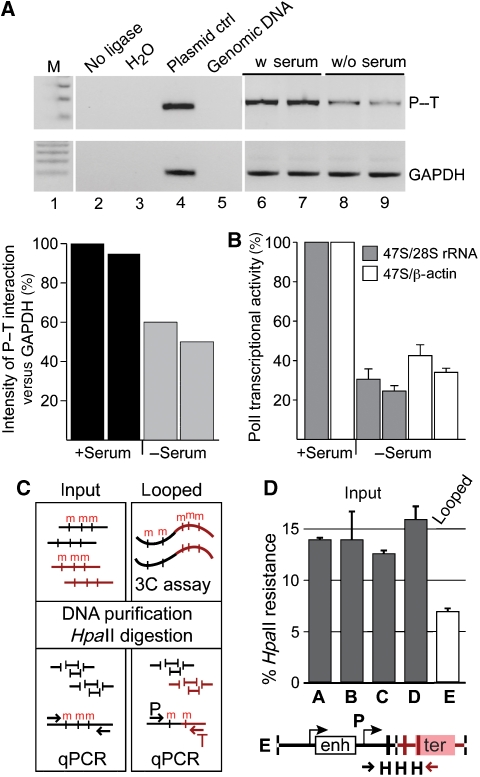
To address the latter question, we performed 3C analysis in mouse fibroblasts after serum starvation (Figure 5A and B) and quantified DNA methylation levels of the promoter–terminator ligation products of mouse neonatal liver cells in a 3C-chop assay (Figure 5C and D), as described before for the ChIP-chop assay.
Figure 5.
Promoter–terminator interactions of rRNA genes correlate with transcriptional activity. (A) Analysis of promoter–terminator interactions in mouse 3T3 fibroblasts upon serum starvation. Control PCR reactions (lanes 2–5) and 3C results of two biological replicate experiments (lanes 6–9) are shown and interaction frequencies are plotted on the graph. (B) The results of the qRT–PCR reaction, quantifying the newly synthesized pre-rRNA upon serum starvation, are shown in the graph. (C) Scheme of the methylation-sensitive 3C-chop assay. Long-range chromatin interactions are trapped by the 3C method (looped). The ligated DNA and the genomic DNA (input) are digested with methylation-sensitive restriction enzymes and levels of DNA methylation are quantified by real-time PCR. Different DNA regions are marked (black and red) and DNA methylation (m) is indicated. (D) 3C-chop analysis of mouse rDNA from neonatal liver cells. The overall methylation level of the rDNA coding region was measured as described above: letters A–D mark the same PCR amplicons as in Figure 3C. Letter E marks the 3C ligation product of the promoter and the terminator (scheme is shown below). HpaII cleavage sites are indicated (H).
Serum starvation of 3T3 mouse fibroblast cells, accompanied by the repression of rRNA transcription, resulted in decreased crosslinking frequencies between promoter and terminator fragments as revealed by the 3C assay (Figure 5A and B). This suggests that DNA looping correlates with gene activity, and should therefore be established within non-methylated rDNA copies.
To verify the results obtained with tissue culture cells, we used native tissue to analyse the impact of DNA methylation on the formation of promoter–terminator interactions. Analysis of the DNA methylation levels in mouse neonatal liver cells, at the spacer promoter, gene promoter, coding region and the terminator, revealed that about 14% of the rRNA genes were methylated and therefore inactive. Quantification of the DNA methylation levels in the 3C ligated material revealed more than two-fold reduced levels of DNA methylation. This result shows that promoter–terminator interactions occur mainly at and within active gene copies (Figure 5C and D), as suggested by the results of the serum starvation experiment.
TTF-I oligomerization and transactivation domains are required for rRNA gene activation
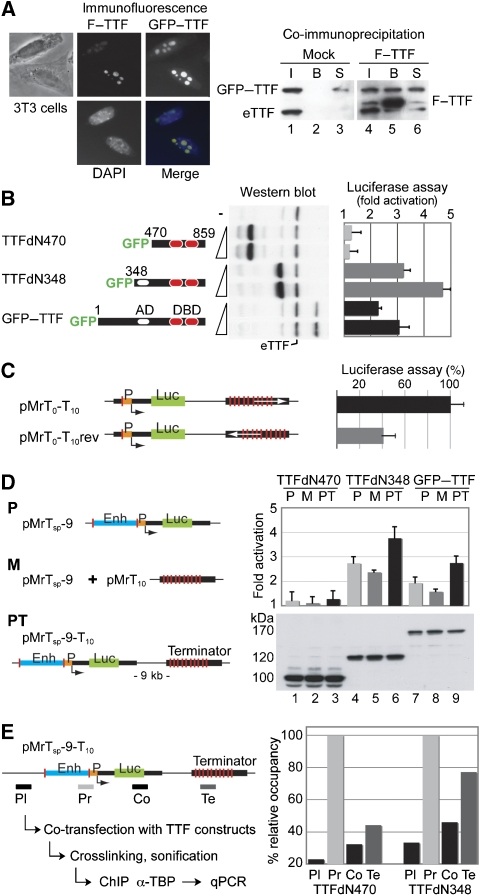
TTF-I was shown to form oligomers in vitro, requiring the N-terminal 470 amino acids for this function (Sander and Grummt, 1997). To analyse the subcellular localization of TTF-I mutants and to test its ability to form multimers in vivo, Flag-tagged full-length TTF-I (F–TTF) and N-terminal truncated GFP–TTF-I plasmids were co-transfected into mouse 3T3 fibroblasts and CHO cells. Similar to the endogenous TTF-I protein, the Flag–TTF-I and the full-length GFP–TTF-I construct, all GFP-tagged TTF-I deletion mutants, either lacking 348 (TTFdN348) or 470 (TTFdN470) N-terminal amino acids, showed predominantly nucleolar localization (Figure 6A and Supplementary Figure S4). Co-immunoprecipitation experiments, using the Flag-tagged, full-length TTF-I reveals the capability of this protein to multimerize in vivo (Figure 6A). Flag–TTF-I was capable of co-immunoprecipitating the endogenous TTF-I (eTTF) and the GFP-tagged full-length TTF-I (GFP–TTF).
Figure 6.
Coupling of initiation and termination is required for full transcriptional activity of rDNA reporters. (A) GFP– and Flag–TTF-I (F–TTF) do physically interact. Immunofluorescence and phase-contrast micrographs of cells, transiently transfected with GFP– and Flag–TTF-I (left panel). In co-immunoprecipitation experiments (right panel), cells were co-transfected with GFP-tagged TTF-I in combination with pcDNA–Flag (lanes 1–3) or pcDNA–Flag–TTF-I plasmids (lanes 4–6). Western blot shows 5% of the input (I, lanes 1 and 4), 50% of the bound (B, lanes 2 and 5) and 5% of the unbound supernatant (S, lanes 3 and 6) fractions of the co-immunoprecipitations. The proteins were detected using an α-TTF-I antibody. The co-immunoprecipitation of the endogenous TTF-I is marked (eTTF). (B) Transcriptional activation of TTF-I depends on the transactivation domain. RNA polymerase I reporter plasmids were co-transfected with GFP–TTF-I expression vectors, and the transcriptional activation was estimated in comparison with co-transfected GFP. TTF-I construct overviews are shown on the left. Numbers indicate amino-acid positions, empty ovals label the transactivation domain (AD) and red ovals show the DNA-binding domain (DBD). The cartoon of the pMrTsp-9-T10 reporter plasmid is shown below (D). Expression levels of different GFP–TTF-I mutants are shown on the western blot. eTTF marks the endogenous TTF-I protein. (C) Transcriptional activation of TTF-I depends on the orientation of terminator-binding sites. The indicated reporter plasmids were co-transfected with Renilla luciferase control plasmids and transcriptional activities were compared. The transcriptional activity of the ‘wild-type' reporter—harbouring T0 and T1−10 TTF-I-binding sites in the proper orientation—was set as 100%. Thin black lines label mouse rDNA sequences and the transcriptional start is marked by an arrow. Green boxes mark the luciferase coding region and orange boxes label the gene promoter. Red lines indicate TTF-I-binding sites and white arrows show the orientation of the terminator. Abbreviations: Mr—mouse rDNA; T—TTF-I-binding site; rev—reverse orientation. (D) Transcriptional activation of TTF-I depends on the cis configuration of the promoter proximal and terminator-binding sites. RNA polymerase I reporter plasmids were co-transfected with GFP–TTF-I expression vectors, and the transcriptional activation was estimated in comparison with GFP–TTFdN470. The terminator-less construct is labelled with letter P, whereas M labels the mixture of terminator-less reporter with terminator containing plasmids (trans configuration of TTF-I-binding sites) and PT marks the complete rRNA minigene reporter (cis configuration of TTF-I-binding sites). Blue boxes label enhancer (Enh) sequences. The numbers in the plasmid name indicate the distance between the transcriptional start and terminator sites in kilo basepairs. Expression levels of different GFP–TTF-I mutants are shown on the western blot (lanes 1–3: GFP–TTFdN470; lanes 4–6: GFP–TTFdN348; lanes 7–9: GFP–TTF-I). (E) TTF-I mediates promoter–terminator interactions. Scheme of the experimental procedure is shown on the left side. The pMrTsp-9-T10 reporter plasmid was either co-transfected with TTFdN470 or with the TTFdN348 mutant. Four DNA fragments of the reporter plasmid—shown as grey or black rectangles—were analysed by quantitative real-time PCR after TBP-ChIP. The amounts of precipitated DNA are shown relative to the TBP-binding site bearing promoter fragment. Abbreviations: Pl—plasmid backbone; Pr—promoter; Co—transcribed coding region; Te—terminator element.
To address the role of TTF-I in the formation of the higher order chromosome structure and its function in gene regulation, we performed functional rRNA gene reporter assays. To visualize RNA polymerase I activity, we constructed a series of rRNA minigene constructs. rRNA minigenes contained an internal ribosome entry site (IRES) downstream of the mouse rRNA promoter, followed by the coding sequence for the Luciferase protein (Ghoshal et al, 2004; Németh et al, 2004). Transcription of the newly constructed mouse rRNA minigene was α-amanitin resistant, indicating that it transcribed by RNA polymerase I (Supplementary Figure S5).
The different TTF-I constructs were co-transfected with the rRNA minigene pMrTsp-9-T10 (scheme shown in Figure 6D), TTF-I expression was monitored by western blot and gene activity was quantified with the Luciferase assay. In these assays, co-transfected TTF-I was competing with the endogenous protein for minigene binding. Efficient expression was achieved for TTFdN470 and TTFdN348 constructs, whereas expression of the full-length TTF-I construct was limited (Figure 6B). We observed dosage-dependent activation of the rRNA minigene by the full-length and TTFdN348 constructs. The lower activation levels of full-length TTF-I are most likely due to the reduced expression levels that compare to the levels of endogenous TTF-I. In contrast, we did not observe transcriptional activation by TTFdN470, suggesting that the N-terminal 470 amino acids encompassing the transactivation (Längst et al, 1998) and oligomerization (Sander and Grummt, 1997) domains are required for efficient transcriptional activation.
A correct spatial arrangement of the promoter and terminator is required for full rRNA gene activity
Initial experiments addressed the effects of the rRNA gene terminator on the overall rate of transcription. Therefore, minigenes were constructed, carrying the rRNA terminator in the correct and inverse orientation. The transcription rates of these constructs were quantified in the absence of exogenous TTF-I. Placing the terminator in inverse orientation resulted in reduced transcriptional activity, suggesting synergistic effects between the TTF-I-bound initiation and termination sites (Figure 6C).
To exclude the possibility that the transcribed rDNA sequences would influence TTF-I-dependent transcriptional activation, reporter plasmids with rDNA versus non-rDNA inserts were compared in our test system. The TTF-I-dependent activation of RNA polymerase I transcription was similar using reporter plasmids with different coding regions, showing that the regulatory effect of TTF-I does not depend on the sequence composition of the transcribed region (Supplementary Figure S5).
To dissect the role of the gene terminator, we analysed the effects of the different TTF-I mutants on intact rRNA minigenes (pMrTsp-9-T10; PT), minigenes either lacking the terminator sequences (pMrTsp-9; P) or terminator-less constructs supplemented with the terminator sequences on an extra plasmid (pMrTsp-9+pMrT10; M). Constructs containing the terminator sequences in cis, lacking the intact termination sequences, or co-transfections of the terminator sequences located on an extra plasmid in trans were activated by TTF-I and TTFdN348. However, the presence of terminator sequences in trans reduced, but in cis configuration further stimulated the activation. TTFdN470 failed to stimulate transcription on the rRNA minigenes (Figure 6D). These observations indicate that the functional integrity of terminator and promoter sequences is required for efficient transcriptional activation.
TTF-I mediates promoter–terminator interactions
To assess the role of TTF-I in the establishment of promoter–terminator interactions, we analysed the dependence of DNA looping on the two TTF-I constructs TTFdN348 and TTFdN470. The analysis of DNA looping was performed by TBP-ChIP experiments, a factor present in the promoter bound TIF-IB complex. TTF-I constructs were co-transfected with the rRNA minigene and the experiment was divided into three samples. First, we monitored TTFdN348 and TTFdN470 expression by western blot analysis (not shown). Second, transcription was quantified with the luciferase assay, and the activation by TTFdN348 was three-fold higher than with TTFdN470. Third, the sample was crosslinked, sonified and used for immunoprecipitation with α-TBP antibodies. Co-precipitation of different regions of the rRNA minigene was monitored by quantitative real-time PCR. Amplicons comprised the promoter and termination sequences, with a further two placed in the coding region and vector backbone serving as controls. Relative DNA quantification revealed that only TTFdN348, harbouring the oligomerization domain and efficiently activating the rRNA minigene transcription, was capable to co-precipitate similar amounts of promoter and terminator DNA (Figure 6E). These results are in good agreement with the 3C data and support the model that transcriptionally active rRNA genes form DNA loops that are mediated by the transcription termination factor TTF-I.
Discussion
Epigenetic control of rRNA gene transcription
Previous studies have shown that the chromatin architecture and the epigenetic modifications of DNA and histones are major players in the establishment and maintenance of transcriptionally active and inactive rRNA genes. The results of the present study support this view and demonstrate that epigenetic modifications correlate with the three-dimensional topology of the rRNA genes, bringing distant DNA elements into close proximity. We could show that the termination factor TTF-I mediates the formation of these rRNA gene loops, allowing efficient rRNA transcription.
We performed an extensive ChIP analysis combined with real-time quantitative PCR on mouse rRNA genes. The highest relative occupancies of TBP, RNA polymerase I, UBF and H2A.Z were found around the spacer promoter. Our results therefore suggest that the spacer promoter functions as an entry and assembly site for the RNA polymerase I transcriptional machinery.
In an attempt to link factor occupancies to transcriptional activity, DNA methylation-sensitive ChIP analysis was performed. We could show that transcription factors of rRNA genes are associated with non-methylated, active genes, whereas histone H3 and the repressive mark H3K9Me2 are enriched on inactive, methylated DNA fragments. Surprisingly, factor binding at the spacer promoter did not correlate with DNA methylation status. This suggests that only part of the rRNA gene locus—from the promoter to the terminators—is the target of regulatory DNA methylation. The fact that this region has a higher frequency of CpG dinucleotides only in organisms with CpG methylation supports this hypothesis.
Our finding about the binding of the architectural RNA polymerase I transcription factor UBF is in contrast with the previously reported UBF binding characteristic, which showed non-restrictive binding of this transcription factor throughout the vertebrate rDNA repeat by Southern blot and semiquantitative PCR analysis of the immunoprecipitated DNA (O'Sullivan et al, 2002). In turn, it is in good agreement with the results of the ChIP-qPCR analysis of the human rDNA repeat (Grandori et al, 2005). In our experience, ChIP analysis shows clear results only in combination with real-time qPCR and with the selection of unique, non-repetitive sequences of the rDNA transcription unit. However, it is likely that in lower vertebrates such as Xenopus, in which the whole intergenic spacer functions as enhancer, the binding of UBF is not restricted to the coding region.
We characterized an additional TTF-I-binding site downstream of the spacer promoter and observed that binding sites for this protein demarcate a functional rRNA gene unit. This unit is characterized by high levels of CpG dinucleotides that are absent in the intergenic spacer, suggesting it may be subject to regulation by DNA methylation. In addition, this domain is flanked by H2A.Z-containing nucleosomes that were recently shown to separate functional chromatin domains (Meneghini et al, 2003). A second, weak binding site of H2A.Z was observed at the rRNA gene terminator. It is tempting to speculate that looping of the rRNA genes cause this phenomenon.
Promoter–terminator interactions of mammalian rRNA genes
In the present study, topological features of mouse rRNA genes were analysed in vivo. In summary, we were able to provide compelling support for the ribomotor model (Kempers-Veenstra et al, 1986) that suggested promoter–terminator interactions in the rRNA gene. We showed the existence and functional importance of this structure in the mammalian system. In addition, we were able to identify TTF-I as a factor that mediates the formation of these rRNA gene loops, allowing efficient rRNA transcription. By using psoralen-crosslinked rRNA reporter constructs it was shown for Xenopus that recycling of the RNA polymerase I complex from the upstream terminator to the neighbouring promoter was not necessary for maintaining the high rate of transcription (Lucchini and Reeder, 1989). The results of this study argue against a read-through transcription between individual repeats, which is in good agreement with a looping-based polymerase recycling model. However, the existence of the promoter–terminator interaction of rRNA genes in yeast, Xenopus and other organisms still remain to be shown.
Transcriptional activity correlates with epigenetic status and topology of mammalian rRNA genes
Analysis of the methylation status of the looped mouse rRNA genes showed that the majority of looped genes are devoid of DNA methylation, suggesting a model in which promoter–terminator interactions are present on active rRNA genes, but not on inactive genes. However, we detected a significant, but minor fraction of methylated DNA forming DNA loops. In addition, serum starvation reduced the level of promoter–terminator interactions in the cells, but did not abolish all contacts. This implies that some transcriptionally inactive and methylated rRNA genes retain the topology of active genes. We therefore propose that rRNA genes can be grouped into three species: (a) the transcriptionally inactive, methylated and non-looped rRNA genes; (b) the active, non-methylated and looped genes and (c) a transcriptionally inactive, methylated and looped rRNA gene species that may represent poised rRNA genes, being in the process of inactivation. However, further experiments are necessary to confirm the existence and clarify the role of the latter species. In summary, our rRNA gene studies confirm the view that interactions between promoter and termination sites are present in a large number of actively transcribed genes, as shown for active mRNA and mitochondrial rRNA coding genes (O'Sullivan et al, 2004; Martin et al, 2005).
One can imagine that linking the terminator of active genes to active gene promoters augments transcription rates by increasing the efficiency of transcription re-initiation, as described by the ribomotor model (Kempers-Veenstra et al, 1986). Our 3C assays cannot rule out that promoter–terminator interactions may occur between distant rRNA genes and not within the same gene. As mainly active genes form DNA loops, the ribomotor model would be functional in such a trans-loop scenario as well. In the trans-loop scenario, an uninterrupted chain of promoter–terminator interactions of active genes would be formed, resulting in the establishment of an extended ribomotor. However, we favor the view of gene-specific DNA loops, but testing this model has to await the detailed knowledge of rRNA gene sequence variations and the use of high-resolution chromosome capturing assays.
The rRNA gene promoter, the coding and the terminator regions follow the classical rules of the epigenetic code, in which active genes are loaded with transcription factors, exhibit increased levels of nucleosomes carrying active marks and on which the DNA is not methylated. Respectively, inactive genes are devoid of the transcription machinery, the histones are marked by H3K9 methylation and the DNA is methylated. The spacer promoter complicates this rule as it is loaded with UBF, PolI and TBP irrespectively of the methylation status of the DNA (Figure 3C). Our result suggests that all spacer promoters in the cell are decorated with these factors. This surprising outcome may be confirmed by the observation that ectopically introduced enhancer sequences form binding sites for transcription factors in the absence of an active promoter (Mais et al, 2005). Thus, spacer promoters could serve as entry sites for these factors, providing sufficient initiation-competent RNA polymerases for active gene promoters. However, this working model requires functional testing.
TTF-I mediates promoter–terminator interactions of mammalian rRNA genes
TTF-I is a multifunctional protein with roles in transcription termination, replication fork arrest and transcriptional activation in chromatin. We add another function by showing that TTF-I mediates contacts between the rRNA gene promoter and the terminator in vivo.
The topological difference between active and inactive genes may result from the presence or absence of TTF-I, or the activity status of this protein, which could be regulated by post-translational modifications. In summary, we envision that TTF-I bound to the terminator and promoter bridges the DNA elements by direct TTF–TTF interactions. The mechanism regulating TTF-I binding to the rRNA terminators may be a major determinant of rRNA gene activity. However, the order of events, that is, epigenetic alterations and TTF-I regulation, remain to be established. To analyse the role of TTF-I in loss-of-function experiments, we have tested several siRNA constructs. However, none were successful in knocking down TTF-I, although we could efficiently knock down other targets. Given the multiple functions of TTF-I in the regulation of transcription termination, transcription initiation, replication and gene looping, we think that the gain-of-function analysis using the reporter constructs and TTF-I mutants is not only technically easier, but also provides a more direct readout system for the characterization of TTF-I function in the regulation of promoter–terminator interaction.
Our results give the first insight into the epigenetics and regulation of higher order rDNA organization at the molecular level. Future studies will clarify the details of genomic interactions within active and inactive rRNA genes using 4C and/or 5C assays (Dostie et al, 2006; Ling et al, 2006; Simonis et al, 2006; Zhao et al, 2006) and describe the role of the nucleolus in the spatial organization of eukaryotic genomes.
Materials and methods
Plasmids
Mouse and human ribosomal DNA sequences (GenBank acc. no. BK000964 and U13369, respectively) were numbered relative to the transcription start site (+1). Mouse rDNA reporter plasmids were prepared as described in the Supplementary data. TTF-I expression plasmids were cloned by inserting previously described fragments (Németh et al, 2004) into pEGFP (Clontech), resulting in pEGFP–TTF, pEGFP–TTFdN348 and pEGFP–TTFdN470. As a normalization control, Renilla luciferase expressing phRL-TK plasmid was co-transfected in the reporter assays.
Chromatin immunoprecipitations
Exponentially growing NIH3T3 mouse fibroblast cells were crosslinked using 0.25% or 1% formaldehyde for 10 min. Alternatively, cells were crosslinked with 10 or 25 mM dimethyl-adipimidate (DMA, dissolved in 1 × PBS, pH 8.1) prior to 1% formaldehyde treatment. Chromatin immunoprecipitations were performed according to standard protocols (Supplementary data). Oligonucleotide sequences and quantitative PCR assay characteristics are shown in Supplementary Table 1. Relative quantifications of occupancies at the regions of interest (Oa—relative occupancy of a given ChIP sample) were calculated as % of input after background (normal rabbit IgG ChIP) subtraction and averaging duplicate or triplicate PCR measurements:
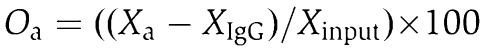 |
where X is calculated from the Ct values:
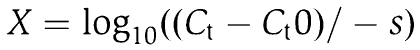 |
where s is the slope of the corresponding calibration curve and Ct0 is the mean threshold cycle (see Supplementary Table 1).
Histone modification and variant occupancies were normalized to H3 occupancy at each region and shown relative to histone H3 (i.e. nucleosome) occupancy:
Electrophoretic mobility shift assays
Reactions (10 μl) contained 50 fmol of IR700- or IR800-labelled oligonucleotides, 20 mM Tris–HCl, pH 8.0; 80 mM KCl; 5 mM MgCl2; 0.2 mM EDTA; 2 mM TCEP; 10% glycerol; 0.2 μg poly(dI–dC) and 25–100 fmol of purified TTFdN209. After incubation for 10 min at 30°C, protein–DNA complexes were separated by electrophoresis on native 4.8% polyacrylamide gels and visualized with the Odyssey® Infrared Imaging System. In competition experiments, 400 fmol of purified TTFdN348 was added to 250 fmol of Alexa555-labelled T0-binding site mixed with 62.5–2000 fmol of T1-, T1mut-, Tsp- or the consensus Tcons-binding site. The signals were visualized using the FUJI FLA-5000 Fluorescent Imaging System. Oligonucleotide sequences are listed in Supplementary Table 1.
3C analysis
The 3C assays on liver cells from C57BL/6*SD7 mice were carried out essentially as has been described before (de Laat and Grosveld, 2003) with minor modifications. The PCR analysis of the 3C material was carried out using the quality control procedure as we described before (see Supplementary data) (Kurukuti et al, 2006). The calreticulin locus, proposed to adopt a similar spatial conformation in all tissues (Tolhuis et al, 2002) was used as a positive internal control for intramolecular ligation and to normalize crosslinking efficiency among various samples. In serum starvation experiments, mouse 3T3 fibroblasts after 48 h serum deprivation were crosslinked and analysed for rDNA promoter–terminator interactions as described above. The gapd locus was used as internal control in these experiments as described before (Spilianakis and Flavell, 2004). The transcriptional activity of rRNA genes was measured by monitoring the level of the short half-life 47S rRNA precursor in comparison with the stable 28S rRNA species and with β-actin mRNA using qRT–PCR. Oligonucleotide sequences are listed in Supplementary Table 1.
Methylation-sensitive ChIP-chop and 3C-chop assays
ChIP and 3C samples were subjected to HpaII or MspI enzyme digestions prior to quantitative PCR analysis. The PCR reactions were performed using SybrGreen detection. Oligonucleotide sequences and quantitative PCR assay characteristics are shown in Supplementary Table 1. MspI restriction enzyme sensitivity of the ChIP input and 3C samples was measured at the spacer promoter, promoter, 18S rRNA coding regions and at the T1−3 and T6−7 sites of the terminator. In 3C samples, the promoter–terminator ligation product was also analysed by using the MeP–MeT primers. In all samples, 20–30% of the rDNA genes were resistant to MspI digestion at the T6−7 terminators, suggesting either alterations from the GenBank reference sequence or CpNpG methylation at this locus. This site was omitted from the HpaII analysis. Methylation of a promoter CpG site at position −143 served as a marker for transcriptional status (Santoro and Grummt, 2001; Santoro et al, 2002). For normalization between samples, a promoter fragment was used containing no HpaII restriction site (Santoro et al, 2002). Quantification of % HpaII resistance was calculated as follows:
X, analysed fragment and C, control promoter fragment.
Fluorescence microscopy
Cells were grown on cover slips, washed in PBS, fixed in 2% paraformaldehyde in PBS for 15 min on ice, and permeabilized with 0.25%. Triton X-100 in the presence of 1% paraformaldehyde in PBS for 10 min on ice. All following steps were carried out at room temperature. After washing with PBS, cells were blocked for 1 h in PBS, 2% BSA and 5% goat serum. Cells were then incubated for 1 h with mouse monoclonal anti-GFP (Dianova) and rabbit polyclonal anti-Flag (Sigma) antibodies in blocking solution. After washing, bound antibodies were visualized by incubation for 1 h with Texas Red-conjugated goat anti-rabbit and Cy3-conjugated goat anti-mouse F(ab)2 fragments (Dianova) diluted in blocking solution. Cover slips were then washed three times for 5 min in PBS. The second wash contained 80 ng/ml 4,6-diamidino-2-phenylindole to stain genomic DNA. In vivo images were taken by direct visualization of GFP-tagged fluorescent proteins. Fluorescence imaging was achieved using a Zeiss Axiovert 200 inverse microscope.
Co-immunoprecipitation assays
CHO cells were co-transfected with pcDNA–Flag–TTF-I and GFP–TTF-I. As control, pcDNA–Flag plasmid was co-transfected with GFP–TTF-I. Transfected cells (4 × 105) were harvested after 48 h, resuspended in 0.1 ml of lysis buffer (50 mM Tris, pH 8, 0.2% Nonidet P-40, 300 mM NaCl, 10 mM MgCl2, 5 mM TCEP, supplemented with protease inhibitors Complete®; Roche Diagnostics) and incubated for 15 min on ice. Samples were cleared by centrifugation (15 min, 13 000 r.p.m., 4°C), and supernatants were collected and diluted with 0.1 ml of dilution buffer (50 mM Tris, pH 8, 0.2% Nonidet P-40). Immunoprecipitation was performed by adding 20 μl of packed Flag-M2 beads (Sigma) for 2 h at 4°C under gentle rotation. Beads were then washed three times with 0.2 ml washing buffer (50 mM Tris, pH 8, 300 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P-40), and bound proteins were subjected to western blot analysis.
RNA polymerase I reporter assays
NIH3T3 mouse fibroblasts and CHO cells were grown in D-MEM medium supplemented with 10% fetal calf serum and 1% penicillin–streptomycin. Luciferase reporter assays were performed in 12-well plate format. After 48 h, luciferase activity was measured using the Dual Luciferase Assay kit (Promega). Renilla luciferase (transcribed by RNA polymerase II) was co-transfected in each experiment to normalize for differences in transfection efficiency. The firefly luciferase counts (F) of the RNA polymerase I reporter constructs were divided by the Renilla luciferase counts (R) and compared with GFP control transfections. Average and standard deviation values of 3–4 biological replicates—each performed in triplicate—are shown on the plots. Protein expression levels were estimated in western blots.
Supplementary Material
Supplementary Data
Acknowledgments
We thank K Dachauer and R Strohner for technical help. A Villar Garea, R Strohner, J Hochstatter, A Brehm, A Hochheimer and T Straub for helpful discussions. L Karagyozov for mouse rDNA plasmids. ST Jacob for the pHr–IRES–Luc plasmid. I Grummt, Y Isogai and R Tjian for antibodies. G Gilfillan and A Brehm for critical reading of the paper. This research was supported by the DFG, Fonds der Chemischen Industrie and the EMBO Young Investigator Program to GL.
References
- Brock GJ, Bird A (1997) Mosaic methylation of the repeat unit of the human ribosomal RNA genes. Hum Mol Genet 6: 451–456 [DOI] [PubMed] [Google Scholar]
- de Laat W, Grosveld F (2003) Spatial organization of gene expression: the active chromatin hub. Chromosome Res 11: 447–459 [DOI] [PubMed] [Google Scholar]
- Dekker J (2006) The three ‘C's of chromosome conformation capture: controls, controls, controls. Nat Methods 3: 17–21 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J (2006) Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res 16: 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D, Tora L, Egly JM, Grummt I (1993) A TBP-containing multiprotein complex (TIF-IB) mediates transcription specificity of murine RNA polymerase I. Nucleic Acids Res 21: 4180–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST (2004) Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem 279: 6783–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ (2005) c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Grummt I, Kuhn A, Bartsch I, Rosenbauer H (1986a) A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell 47: 901–911 [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS (2003) Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4: 641–649 [DOI] [PubMed] [Google Scholar]
- Grummt I, Rosenbauer H, Niedermeyer I, Maier U, Öhrlein A (1986b) A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell 45: 837–846 [DOI] [PubMed] [Google Scholar]
- Guillemette B, Gaudreau L (2006) Reuniting the contrasting functions of H2A.Z. Biochem Cell Biol 84: 528–535 [DOI] [PubMed] [Google Scholar]
- Kempers-Veenstra AE, Oliemans J, Offenberg H, Dekker AF, Piper PW, Planta RJ, Klootwijk J (1986) 3′-End formation of transcripts from the yeast rRNA operon. EMBO J 5: 2703–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP (2006) Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G, Becker PB, Grummt I (1998) TTF-I determines the chromatin architecture of the active rDNA promoter. EMBO J 17: 3135–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Längst G, Blank TA, Becker PB, Grummt I (1997) RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J 16: 760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS (2004) A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR (2006) CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312: 269–272 [DOI] [PubMed] [Google Scholar]
- Lucchini R, Reeder RH (1989) A test of ‘polymerase handover' as a mechanism for stimulating initiation by RNA polymerase I. Nucleic Acids Res 17: 373–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B (2005) UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev 19: 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G (2005) Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell 123: 1227–1240 [DOI] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Németh A, Strohner R, Grummt I, Längst G (2004) The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res 32: 4091–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan AC, Sullivan GJ, McStay B (2002) UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 22: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ (2004) Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Pfeifer GP (2006) Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol 301: 259–281 [DOI] [PubMed] [Google Scholar]
- Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozak P, Grummt I (2004) Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol 6: 1165–1172 [DOI] [PubMed] [Google Scholar]
- Preuss S, Pikaard CS (2007) rRNA gene silencing and nucleolar dominance: insights into a chromosome-scale epigenetic on/off switch. Biochim Biophys Acta 1769: 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC (2005) RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci 30: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander EE, Grummt I (1997) Oligomerization of the transcription termination factor TTF-I: implications for the structural organization of ribosomal transcription units. Nucleic Acids Res 25: 1142–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I (2001) Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell 8: 719–725 [DOI] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32: 393–396 [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2004) Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 6: 73–77 [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA (2004) Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Singh Sandhu K, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R (2006) Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data