Abstract
We explore the role of differential compartmentalization of Rhomboid (Rho) proteases that process the Drosophila EGF receptor ligands, in modulating the amount of secreted ligand and consequently the level of EGF receptor (EGFR) activation. The mSpitz ligand precursor is retained in the ER, and is trafficked by the chaperone Star to a late compartment of the secretory pathway, where Rho-1 resides. This work demonstrates that two other Rho proteins, Rho-2 and Rho-3, which are expressed in the germ line and in the developing eye, respectively, cleave the Spitz precursor and Star already in the ER, in addition to their activity in the late compartment. This property attenuates EGFR activation, primarily by compromising the amount of chaperone that can productively traffic the ligand precursor to the late compartment, where cleavage and subsequent secretion take place. These observations identify changes in intracellular compartment localization of Rho proteins as a basis for signal attenuation, in tissues where EGFR activation must be highly restricted in space and time.
Keywords: EGF receptor, intramembrane proteolysis, Rhomboid, Spitz, Star
Introduction
EGF receptor (EGFR) signalling has an important function in the differentiation of a wide variety of tissues, during multiple stages of Drosophila development (Shilo, 2003). Pathway activation is initiated upon binding of the receptor to secreted, TGF-α-like ligands, which are produced in source cells from inactive, transmembranal precursors. A key step in ligand maturation involves precursor cleavage by members of the evolutionarily conserved Rhomboid (Rho) family of serine proteases (Urban et al, 2001). Of the seven Rho-family proteins encoded in the Drosophila genome, Rho-1 is responsible for EGFR pathway activation in most tissues. Other family members are utilized for EGFR activation in more restricted settings, such as Rho-2 in the germ line and Rho-3 in the developing eye (Wasserman et al, 2000; Schulz et al, 2002).
Intracellular compartmentalization and trafficking of ligands within the cells producing the signal were revealed as crucial determinants in EGFR pathway activation. The major ligand, Spitz, is retained in its precursor form in the endoplasmic reticulum (ER), through COPI-dependent retrograde trafficking (Lee et al, 2001; Tsruya et al, 2002; Schlesinger et al, 2004). This retention is essential to prevent fortuitous generation of secreted ligand on the plasma membrane by metalloproteases (Reich and Shilo, 2002). In contrast, Rho-1 localizes to late compartments of the secretory pathway (Lee et al, 2001; Tsruya et al, 2002, 2007). The separation between the intracellular locations of ligand precursor and protease is overcome through the activity of the chaperone Star, which associates with Spitz in the ER and traffics it to the late compartment, where ligand processing ensues (Lee et al, 2001; Tsruya et al, 2002). Interestingly, Star, a type II transmembrane protein, was also shown to be a substrate for Rhos and, in contrast to Spitz, is inactivated by cleavage (Tsruya et al, 2007). Thus, the number of ligand trafficking cycles that a Star molecule can accomplish is limited in cells where Rho is active.
The EGFR pathway commonly employs short-range signalling. The rate-limiting event for activation of the pathway is the expression of rho-1, which is highly dynamic, and prefigures the sites of EGFR activation (Bier et al, 1990; Gabay et al, 1997). Restrictions on the spatial range of pathway activation are achieved by negative-feedback regulators such as Argos and Sprouty, whose expression is induced by the pathway in signal-receiving cells (Golembo et al, 1996; Kramer et al, 1999; Reich et al, 1999). However, attenuation of EGFR signalling by restricting the level of active ligand that is produced and secreted has not been reported. This work will focus on the role of the signal-producing cells in modulating the level of EGFR activation.
The development of ommatidia within the Drosophila compound eye represents a particularly revealing scenario with regard to spatial restriction of EGFR activation. Following establishment of founder R8 photoreceptor cells by an EGFR-independent mechanism, reiterative rounds of pathway activation are employed to recruit and specify all other photoreceptors, as well as accessory cone and pigment cells (Freeman, 1996). Tight restrictions must be placed on the range of pathway activation in each signalling cycle, due to the limited pool of undifferentiated precursor cells. The EGFR signalling cassette employed during eye development displays unique ligand trafficking features, which may contribute to pathway regulation. EGFR activation in the eye is highly sensitive to levels of the chaperone Star, in contrast to all other tissues in which Star is required (Bridges and Morgan, 1919; Heberlein and Rubin, 1991; Hsiung et al, 2001; Tsruya et al, 2002; Brown and Freeman, 2003). Furthermore, loss of the small wing (sl) gene, which encodes Drosophila PLCγ and is expressed ubiquitously, leads to EGFR hyperactivation only in the eye (Thackeray et al, 1998). Recent work has demonstrated that Sl is required for ER retention of the cleaved form of Spitz, implying eye-specific cleavage of the Spi precursor in the ER (Schlesinger et al, 2004).
Utilization of Rho-3, in addition to Rho-1, for cleavage of ligand precursors during eye development (Wasserman et al, 2000), raises the possibility that specific attributes of Rho-3 contribute to the unique features of ligand trafficking in this tissue. Rho-1 and Rho-3 show similar substrate specificity with respect to their ability to cleave the precursors of the EGFR ligands Spitz, Keren and Gurken in cell culture (Ghiglione et al, 2002; Urban et al, 2002), as well as Star (Tsruya et al, 2007). However, whereas cleavage by Rho-1 in cell culture is fully dependent on the presence of Star, Rho-2 and Rho-3 are able to cleave Spitz and Gurken in the absence of the chaperone (Ghiglione et al, 2002; Urban et al, 2002).
This work explores the basis for these distinct behaviours and demonstrates that Rho-3 is partially localized in vivo to the ER, where it cleaves both the Spi precursor and the chaperone Star. This property attenuates EGFR activation, primarily by compromising the amount of chaperone that can productively traffic the ligand precursor to the late compartment, where cleavage (by both Rho-1 and Rho-3) and subsequent secretion take place. Rho-based cleavage thus has a dual role during EGFR pathway activation in the developing eye: late-compartment activity is responsible for production of active ligand, whereas ER activity imparts a parallel, modulating effect on signalling.
Processing of Spitz in germ cells is carried out by Rho-2/Brho/Stet (Guichard et al, 2000; Schulz et al, 2002; Gilboa and Lehmann, 2006), which we now show to be partially located and active in the ER as well, raising the possibility that EGFR-based signalling in this tissue is modulated in a similar fashion to the developing eye. Consistent with this scenario, we have observed Star dosage sensitivity, as well as EGFR hyperactivation in sl mutants in larval female gonads. Taken together, these observations identify changes in intracellular compartment localization of Rho proteins as a basis for signal attenuation, in tissues where the range and level of EGFR activation must be highly restricted.
Results
Rho-1 and -3 exhibit distinct intracellular localization in the eye
Recurrent rounds of EGFR activation lead to the induction of cell fates in the developing-eye imaginal disc (Freeman, 1996). In this tissue, both Rho-1 and Rho-3 participate in the proteolytic processing that generates the active form of the Spi ligand. Rho-3, however, is the prominant protease during eye development: whereas adult eyes lacking rho-3 show extensive loss of photoreceptors and cone cells (Wasserman et al, 2000), loss of Rho-1 in the eye does not lead to detectable phenotypes, indicating that Rho-3 activity is sufficient.
As discussed above, EGFR signalling in the eye displays several unique features, including exceptional sensitivity to the levels of the Star chaperone, and EGFR hyperactivation in a sl mutant background, indicative of ER-based ligand cleavage (Schlesinger et al, 2004). One possibility is that novel features of Rho-3, which is expressed mainly in the developing eye, could account for these features. To address this issue, we compared the subcellular localizations of Rho-1 and Rho-3, following eye-disc-specific expression of tagged variants of these proteins. Distinctions in the localization patterns of the two Rho proteases were readily apparent, regardless of the tag used (Figure 1, and data not shown). Rho-1 localizes to discrete punctate structures that are enriched on the apical side of eye-disc cells (Figure 1B) and do not co-localize with markers for the ER, Golgi or endosomes (Supplementary Figure S1). Although Rho-3 can also be found in the same apical puncta as Rho-1, Rho-3 localization displays a specific, perinuclear distribution (Figure 1C–E). Co-localization of Rho-3 with mSpi (Figure 1E) or the ER marker BiP (not shown) in this ring-like pattern indicates that Rho-3 localizes to the ER.
Figure 1.
Subcellular localization of Rho-1 and Rho-3 in the eye imaginal disc (A–E) GMR-Gal4 drives expression of the indicated UAS constructs in all cells posterior to the morphogenetic furrow. ElaV (Blue) marks the differentiated photoreceptors, FasIII (red) stains plasma membranes and BiP marks the ER. (A) mSpi–GFP (green) localizes exclusively to the perinuclear ER within photoreceptor cells, where it co-localizes with BiP (red). (B) Optical sections through the apical (top) and middle (bottom) portions of ommatidia reveal that Rho-1-HA (red) does not co-localize with mSpi but is found within more apical punctate structures. (C) Rho-3–GFP (green) is detected both around the nuclei, in a pattern reminiscent of mSpi–GFP, and in punctate structures. (D) Rho-3–GFP (green) co-localizes with Rho-1–HA (red) in apical punctate structures. (E) Rho-3–GFP (green) and mSpi–HRP (red) co-localize in the peri-nuclear ER. A full-colour version of this figure is available at the EMBO Journal Online.
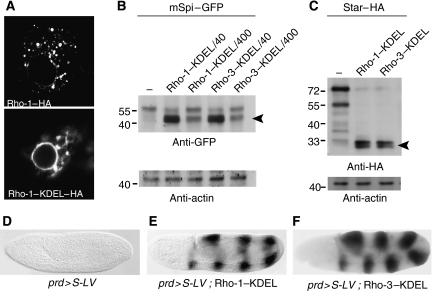
These observations raised the possibility that ER-based Rho activity influences EGFR signalling in the developing eye. Are Rho proteins functional when localized to the ER? To address this issue, we appended the ER retrieval sequence KDEL (Munro and Pelham, 1987) to the C-terminus (lumen side) of Rho-1 or Rho-3, to form Rho-1–KDEL or Rho-3–KDEL. Transfection of Rho-1–KDEL in S2R+ cells shows that the punctate distribution of Rho-1 has been replaced with a perinuclear one (Figure 2A). mSpi is typically localized to the ER when expressed in these cells (Tsruya et al, 2002, 2007). Co-expression of Rho-1–KDEL or Rho-3–KDEL with mSpi–GFP demonstrated efficient ER cleavage, even upon 400-fold dilutions of the proteases relative to mSpi–GFP, indicating that their catalytic activity is maintained in the ER (Figure 2B). Furthermore, both Rhos showed equivalent proteolytic potencies in the ER, suggesting that there is no intrinsic difference in their enzymatic properties.
Figure 2.
Rhos can cleave both Spitz and Star in the ER. (A) Addition of a KDEL ER-retrieval sequence to HA-tagged Rho-1 re-localizes it in S2R+ cells, to the perinuclear ER. (B) Cell lysates from Schneider cells transfected with the indicated constructs and probed with an anti-GFP antibody. Even when diluted 400-fold compared with mSpi–GFP, Rho–KDEL constructs efficiently produce the faster migrating band, which corresponds to the cleaved ligand (arrowhead). The membrane was reblotted with an anti-actin antibody to ensure equal loading for all lanes. (C) Star–HA is also efficiently cleaved by Rho-1–KDEL and Rho-3–KDEL, such that most of the precursor is converted to a faster-migrating band (arrowhead). The cleaved C-terminal, extracellular domain of Star is detected by anti-HA. (D–F) To monitor specifically the capacity of Rho proteins to cleave Star in the ER in embryos, a Star protein tagged at the cytoplasmic domain with a LexA VP16 transcriptional activator and a LexA-lacZ reporter were used. Upon expression of Rho-1–KDEL or Rho-3–KDEL by the prd-Gal4 driver, prominent cleavage of Star was detected by X-Gal staining.
To determine if Rhos can also cleave Star in the ER, the same Rho–KDEL constructs were co-transfected with Star, which normally shuttles between the ER and the Rho-1 compartment (Tsruya et al, 2007). Indeed, prominent bands of cleaved Star were generated (Figure 2C).
We have previously described the utilization of a fusion construct between Star and LexA VP16, to demonstrate Rho-dependent cleavage of Star in embryos (Tsruya et al, 2007). However, as the LexA VP16 domain also contains a nuclear-localization signal, this assay could not distinguish between cleavage of Star in the ER or in a late compartment. To specifically test the capacity of Rho proteins to cleave Star in the ER, we expressed in ectodermal stripes using prd-Gal4, the Star construct together with Rho-1–KDEL or Rho-3–KDEL and monitored the ability of cleaved Star–LexA VP16 to trigger the expression of a lacZ reporter. Indeed, either Rho-1–KDEL or Rho-3–KDEL could efficiently cleave Star, as detected by the expression of lacZ in prd stripes (Figure 2D–F).
Taken together, the results above indicate that when Rhos are present in the ER, they can efficiently cleave the Spitz ligand precursor, as well as the chaperone Star. One may therefore expect the partial presence of Rho-3 in the ER to impinge on Spi processing and trafficking, and thus on EGFR activation in vivo.
ER-resident Rhos induce lower levels of EGFR signalling
To estimate the potency of ER-resident Rhos in generating active secreted ligand in vivo, we monitored the level of EGFR activation following expression of the proteases in developing embryos. We compared Rho-1-, Rho-3- and Rho-1–KDEL-induced activation, representing an increasing level of ER residency. Under these conditions, endogenous, ubiquitously expressed mSpi and Star are utilized, and the extent of activation depends solely on the protease. We also verified that the tags used had no effect on activity by comparing phenotypes of tagged and non-tagged Rhos expressed in wing discs (not shown).
We first ascertained that the subcellular localization of the tagged Rhos in embryos mirrored the pattern previously observed in the developing eye (Figure 3). Indeed, when expressed in stripes of cells within the embryonic ectoderm, HA-tagged Rho-1 localizes to discrete punctate structures (Figure 3A), whereas GFP-tagged Rho-3 was found both around the nuclei, in a pattern indicative of ER localization, and in punctate structures (Figure 3B).
Figure 3.
ER cleavage by Rhos induces low levels of EGFR activation. (A, A′) Lateral view of a stage-14 embryo. UAS–Rho-1–HA (green) expressed under the control of en-Gal4 localizes to discrete punctate structures, reminiscent of its localization in the eye imaginal disc. (B, B′) Similarly expressed Rho-3–GFP (green) localizes in the embryo around the nuclei (consistent with ER localization), as well as in punctate structures. (C) Ventral view of a stage 9–10 WT embryo (anterior to the left). Yan protein (red) is degraded in the 1–2 cell rows bordering the ventral midline (vm), which expresses rho-1. (D) Ectopic expression of UAS–Rho-1–KDEL driven by prd-Gal4. The EGFR pathway does not show signs of ectopic activation within the prd stripes (defined by co-expression of UAS–GFP, marked by white lines), as assayed by Yan degradation. (E) Following similar expression of Rho-1, Yan is fully degraded throughout the prd stripe, as well as in adjacent cells. (F, F′) Intermediate levels of EGFR activation are observed upon ectopic expression of Rho-3. Many nuclei within the prd stripe still display Yan, and the non-autonomous effect is hardly detected. (G) Co-expression of Rho-1 and Rho-3. The partial Yan degradation pattern in prd stripes resembles that observed upon expression of Rho-3 alone (F). (H–K) Ventral views of stage 9–10 embryos expressing the indicated Rho in the ventral midline (arrowheads), where Rho-1 is endogenously expressed. (H) Yan is degraded in 1–2 cells bordering the ventral midline in WT embryos. (I) Expression of Rho-1 in the midline by btl-Gal4 does not modify the endogenous Yan degradation pattern. (J) Rho-1–KDEL expression is epistatic to the endogenous Rho-1 and leads to a reduced range of EGFR activation, manifested by Yan presence in cells bordering the midline. (K) Expression of Rho-3 is also epistatic to that of Rho-1. A full-colour version of this figure is available at the EMBO Journal Online.
As a marker for EGFR activation, we followed the degradation of Yan, a ubiquitous nuclear protein that is exported to the cytoplasm and degraded upon EGFR-induced MAPK activation (Rebay and Rubin, 1995; Melen et al, 2005). At stages 9–10 of embryonic development, Yan protein is found in nuclei throughout the ectoderm and is normally degraded only in cells adjacent to the ventral midline, which express endogenous rho-1 (Figure 3C). Ectopic expression of Rho-1 by prd-Gal4 led to degradation of Yan throughout the prd stripes, as well as in the adjacent 1–2 rows of ectodermal cells, consistent with the generation of a secreted ligand with a non-cell autonomous activity (Figure 3E). In contrast, expression of Rho-1–KDEL did not significantly alter the normal pattern of Yan protein (Figure 3D). Clear distinctions between strong EGFR hyperactivation phenotypes resulting from ectopic expression of the late-compartment resident Rho-1 and very subtle phenotypes induced by Rho-1–KDEL or Rho-3 KDEL are readily apparent at additional developmental stages and tissues, such as the developing eye or the wing (Supplementary Figure S2).
The consequences of expressing Rho-3, which localizes to both the ER and cytoplasmic puncta, differed significantly from those of Rho-1 expression (Figure 3F). Using several independent transgenic lines, expression of Rho-3 resulted in markedly lower levels of EGFR activation than Rho-1. Many cells within the prd stripes still contained Yan, and the non-autonomous effect on adjacent cells was hardly detectable (Figure 3F). A possible interpretation of these observations is that combining ER and late-compartment Rho activities within the same cell attenuates the level of EGFR activation achieved by late-compartment activity alone.
To test this notion further, we monitored the outcome following co-expression of Rho-1 and Rho-3. We observed the milder, Rho-3-like EGFR activation phenotype (Figure 3G), suggesting that the ER-based activity of Rho-3 is epistatic and prevents full EGFR activation by Rho-1. This effect is not due to the presence of multiple UAS constructs, as co-expression of UAS–LacZ and UAS–CD8–GFP with UAS–Rho-1 did not alter its phenotype (data not shown). The same conclusions were reached when the capacity to induce the expression of the EGFR-target gene pointedP1 non-autonomously was monitored (Supplementary Figure S3).
In a complementary experiment, the endogenous activity of Rho-1 in the midline-glial cells was supplemented by co-expression of Rho proteins, driven by btl-Gal4. Normally, Yan is degraded in 1–2 cell rows adjacent to the midline, with a sharp boundary (Melen et al, 2005). We found that the level of EGFR activation was compromised upon expression of either Rho-1–KDEL or Rho-3, as monitored by the appearance of Yan in nuclei that are adjacent to the midline, and a jagged border of Yan degradation, whereas co-expression of Rho-1 did not alter the normal Yan degradation pattern (Figure 3H–K). ER-active Rhos can thus attenuate the activity of endogenous Rho-1, in a normal developmental setting.
We conclude that both Rho-1 and Rho-3 localize and are active in a late compartment of the secretory pathway; however, Rho-3 is also present and active in the ER. We propose that a high output of EGFR pathway activation is obtained when Rho cleavage generates active ligand exclusively in late compartments (e.g. by native Rho-1), whereas confining Rho activity to the ER results in very limited activation. Intermediate levels of EGFR pathway activation are observed in situations where the functional Rho (e.g. Rho-3) is active in both the ER and the late compartment.
ER cleavage sensitizes EGFR activation to the levels of Star
ER activity of Rho-3 may compromise its potency to activate the EGFR pathway because of premature cleavage of either mSpi or Star. Cleavage of mSpi in the ER could interfere with its correct folding, or prime it for ER retention via a Small wing-dependent mechanism (Schlesinger et al, 2004). ER cleavage would also prematurely inactivate Star, before trafficking of mSpi for processing in a late compartment (Tsruya et al, 2007). To address this issue, we tested if the reduced EGFR activation observed upon expression of Rho-3 is sensitive to the gene dosage of spi or Star. The embryonic prd expression assay was repeated in embryos heterozygous for null spi or Star alleles, thereby halving spi or Star functional gene dosage. Whereas Rho-3-induced Yan degradation remained unaltered in spi heterozygous embryos (Figure 4A), a corresponding reduction in Star levels suppressed the Rho-3 induced phenotype (Figure 4B). This effect was even more dramatic when we examined EGFR-induced phosphorylation of MAPK, which occurs under a lower threshold of receptor activation (Supplementary Figure S4).
Figure 4.
ER cleavage sensitizes EGFR signalling to the level of Star. UAS–Rho-3–GFP and UAS–mCD8–GFP were jointly expressed under the control of prd-Gal4. (A) Degradation of Yan (red) following Rho-3 expression in spi heterozygous embryos displays a similar pattern to the one observed upon Rho-3 expression in WT embryos. (B) Star heterozygosity largely suppresses the EGFR hyperactivation caused by Rho-3 expression. (C) Simultaneous expression of mSpi with Rho-3 does not significantly alter the Yan degradation pattern. (D) Co-expression of Star and Rho-3 significantly elevate EGFR activation levels. The degradation of Yan reaches the extent observed upon expression of Rho-1 (Figure 3E). A full-colour version of this figure is available at the EMBO Journal Online.
In a complimentary assay, Spi or Star levels were elevated by co-expression of UAS-based constructs with UAS–Rho-3 in the prd stripes. Again, the system shows sensitivity only to levels of Star: co-expression of mSpi and Rho-3 yielded the same outcome as Rho-3 expression alone, whereas co-expression of Star and Rho-3 raised the Rho-3-induced activation levels to a Rho-1-like profile (Figure 4C and D). Unlike Rho-3 dependent activation, the Rho-1-induced phenotype was insensitive to either spi or Star dosage (Supplementary Figure S5).
Finally, the interplay between ER activity of Rhos and the gene dosage of Star or spi was assayed in the wing imaginal disc, where the EGFR pathway governs vein patterning following restricted expression of rho-1 (Sturtevant et al, 1993). Ectopic expression of either Rho-1 or Rho-3 in the developing wing disc led to appearance of ectopic veins in the mature wing, an established consequence of EGFR pathway hyperactivation (Sturtevant et al, 1993). The Rho-3 phenotype is suppressed by halving the dose of Star but not of spi, whereas the Rho-1 phenotype is insensitive to reduced gene dosage of either Star or spi (Figure 5C and E).
Figure 5.
ER-active Rhos display mild EGFR activation in the wing, which is sensitive to the levels of Star. Rho-proteases were expressed in female wing imaginal discs using MS1096-Gal4 in the genetic backgrounds indicated. (A) WT wings. (B) Rho-1–KDEL expression yields very subtle phenotypes, except when it is co-expressed with Star, where EGFR activation becomes comparable to that seen with Rho-1. (C) Rho-1 expression leads to very pronounced EGFR hyperactivation phenotypes. There is no sensitivity to Star gene dosage. (D) Expression of Rho-2 leads to intermediate phenotypes, namely vein expansion. This phenotype is suppressed upon reduction in Star gene dosage and becomes more severe when Star is co-expressed with the protease. (E) Rho-3 expression produces intermediate phenotypes. Importantly, Star heterozygosity or overexpression strongly suppresses or enhances the phenotype, respectively. A full-colour version of this figure is available at the EMBO Journal Online.
In accordance with the highly restricted ability of Rho-1–KDEL to induce EGFR activation, very few ectopic veins were observed upon its expression in the wing disc. In this tissue, the expression of ER-active proteases did not interfere with endogenous Rho-1 signalling, presumably reflecting a requirement for lower levels of EGFR activation. However, when Star was co-expressed with Rho-1–KDEL, the ectopic vein phenotype was significantly enhanced, whereas co-expression of mSpi did not have an effect (Figure 5B). Taken together, these observations indicate that Rho ER cleavage activity reduces EGFR activation mostly due to Star inactivation, and thus sensitizes the system to Star gene dosage. Furthermore, these results recapitulate the pronounced sensitivity to Star levels that is observed in the developing eye, a tissue in which EGFR signalling is controlled by the ER-active Rho-3.
Rho-3 attenuates EGFR activation during eye development
The endogenous setting in which the ER cleavage activity of Rho-3 is expected to have an important function in EGFR activation is the developing eye. Although both Rho-1 and Rho-3 are expressed in this tissue, mutant analysis indicated that the latter is the prominent protease, whereas Rho-1 has a redundant function (Wasserman et al, 2000).
To elucidate the differences in the function of Rhos during eye development, we first examined the expression pattern of rho-1 and rho-3 by using enhancer trap lines. rho-1 transcription is detected in R8, R2 and R5, the first three photoreceptor cells to be determined in each ommatidium, and is maintained in these cells throughout eye disc development (Figure 6A and Sturtevant et al, 1996; Baonza et al, 2001). rho-3 expression is more dynamic, starting in R8 immediately after the furrow, rapidly expanding to the outer photoreceptors, and then to precursors of the other cell types (Figure 6B). Thus, rho-3 transcription follows the expanding activation profile of the EGFR (Freeman, 1996) and supports a model in which activation of EGFR leads to expression of the protease, to expand the source of ligand processing. Indeed, ectopic EGFR activation gave rise to an expanded pattern of rho-3 expression (Supplementary Figure S6). Furthermore, the expression patterns of the two proteases account for the more prominent role of rho-3 in eye development compared with rho-1.
Figure 6.
Expression pattern of Rho-1 and Rho-3 and hyperactivation of the EGFR pathway in rho-3 mutant eyes. (A) Expression of the rho-1 enhancer trap X81 is detected by anti-β-galactosidase staining (red) in R8, R2 and R5 photoreceptor cells, in ommatidia throughout the eye disc. Photoreceptors, visualized with the neuronal nuclear marker ElaV (blue), form discrete eight-cell clusters . The morphogenetic furrow (mf) is indicated. (B) Expression of the rho-3 enhancer trap Inga (anti-β-galactosidase, red) is observed in the R8 cells of photoreceptor clusters near the mf, and expands to the other cell types in the ommatidium. ElaV (blue) and FasIII (green) mark neuronal nuclei and outlines of cells in the eye disc, respectively. Insets in (A) and (B) show magnified views of marked regions. (C) Scheme representing the expression pattern of both rho genes in the eye disc. Rho-1 and Rho-3 are co-expressed in R8, R2 and R5, whereas Rho-3 is expressed alone in all other cells. (D–E) The R4 photoreceptor cell fate, monitored in eye imaginal discs by expression of mδ0.5-LacZ. β-gal (red), is detected in only one cell per ommatidium in WT discs (D; and Cooper and Bray, 1999), but is often found in several cells of a single ommatidium in rho-3-null (ruPLLb) mutant discs (E). FasIII (green). (F–G) The R7 cell fate is monitored by the co-expression of Prospero (red) and the neuronal marker ElaV (blue). Cells that express Prospero but not ElaV are cone cell precursors. Only one nucleus in each WT ommatidium shows co-expression of the two markers (F), whereas induction of ectopic R7 cell fates is observed in rho-3– (arrows in G). (H) Mutant clones of rho-3 were generated (detected by absence of GFP), and expression of the cone cell marker, Cut, was followed. A decrease in Cut expression is observed within the mutant clones. Arrow shows the position posterior to the morphogenetic furrow (mf, arrowhead), at which Cut begins to be expressed in the WT cells. (I–K′) Sections through the retinas of adult flies of the indicated genotypes. In bottom schemes of the data, trapezoids indicate a full complement of photoreceptors and their spatial orientation, whereas red and green circles indicate ommatidia with missing or extra photoreceptors, respectively. (H) Expression of the apoptosis inhibitor p35 under control of GMR-Gal4 does not alter photoreceptor number and ommatidial patterns. (I) In rho-3− retinas, only 17% of the ommatidia show a full wild-type complement of photoreceptors. (J) Bypassing apoptosis by expressing p35 in rho-3− eyes reveals hyperactivation of EGFR. Most ommatidia (54%) now show the WT array of seven photoreceptors, and in some cases (22%) extra photoreceptors are detected. (J′) Examples of single ommatidia from GMR>p35; rho-3− eyes, showing missing photoreceptors (a) or extra photoreceptors (b) and (c). (L) Quantification of photoreceptor numbers in adult ommatidia from the indicated genotypes. Note: the death of photoreceptor cells in eye imaginal discs expressing only Rho-1, which was uncovered by co-expression of p35, may stem either from insufficient phosphorylation of HID by MAPK, or from death of mis-specified photoreceptor cells. A full-colour version of this figure is available at the EMBO Journal Online.
In the eye imaginal disc, newly recruited photoreceptors elicit the next round of EGFR activation by expression of Rho protein(s) (Figure 6A and B; Freeman, 1996). In rho-3-null mutant eye discs, ommatidial patterning via the EGFR pathway is triggered exclusively by Rho-1, which is expressed only in photoreceptors R8, R2 and R5. Remarkably, examination of a series of photoreceptor markers in rho-3− eye imaginal discs reveals an overall normal ommatidial array (not shown), with a number of mutant ommatidia displaying an excess of various photoreceptor cell types. These include extra R4 cells (Figure 6D and E) ectopic R7 specification (Figure 6F and G) and ommatidia in which the svp LacZ marker (which marks R1, R6, R3 and R4; Mlodzik et al, 1990) was expressed in more than four cells (not shown). Following the expanded recruitment of photoreceptor cells, a reduction in the subsequent recruitment of cone cells (marked by Cut) was observed in rho-3 mutant clones (Figure 6H), due to the depletion of the precursor pool. A similar reduction in Cut expression was observed when the entire disc was mutant for rho-3 (Wasserman et al, 2000). Thus, in the absence of ER cleavage activity of Rho-3, late-compartment cleavage by Rho-1 appears to allow a broader spatial range of EGFR activation, so that production of active ligand in only three photoreceptor cells is sufficient to induce an almost normal array of photoreceptors, with an excess of several cell types.
The surprisingly normal patterning of rho-3 mutant eye discs contrasts with the rho-3 adult eye phenotype, in which most ommatidia do not show a full complement of photoreceptors. This phenotype, however, may be due to the requirement of the EGFR pathway for cell viability during the ensuing pupal stages of development (Bergmann et al, 1998). To examine this possibility, and obtain a quantitative assessment of photoreceptor recruitment in the absence of ER cleavage by Rho-3, we analysed adult rho-3 mutant eyes in which cell death was circumvented by the expression of the apoptosis inhibitor p35. In rho-3 mutant eyes, only 17% of ommatidia possess a full complement of photoreceptors (Figure 6J). Following expression of p35, 54% of the ommatidia displayed a wild-type number of photoreceptors, with the remaining 24% showing photoreceptor loss and 22% exhibiting extra photoreceptors (Figure 6K and K′). Histograms displaying percentage of ommatidia with the different photoreceptor cell numbers are shown in Figure 6L.
The developing eye disc thus presents a developmental setting in which Rho cleavage activity is simultaneously present in both the ER and late compartments, a situation that could lead to attenuated activation of the EGFR pathway. The potency of the EGFR pathway to induce ectopic cell fates upon hyperactivation, along with its recurrent usage, and the limited number of precursor cells in the eye disc provide a plausible advantage for the utilization of a Rho protease with an attenuated activating capacity in this particular setting.
The results described above indicate that ER-based activity reduces the levels of active secreted ligand that can be generated by a Rho protease. This is attributed mainly to cleavage of Star and leads to the sensitivity to Star gene dosage, which is manifest during eye development.
Rho-2-dependent EGFR activation in the female larval gonad is attenuated by ER cleavage of Star
Rho-2 represents a second Rho-family member that can cleave the mSpi and Star substrates in the ER in cell culture (data not shown). In addition, we found that Rho-2 displays an ER localization when expressed in embryos, similar to Rho-3 (Supplementary Figure S7). It also shows a perinuclear distribution when expressed in the larval female gonad, a tissue in which it is normally active (Supplementary Figure S7). Furthermore, ectopic expression of Rho-2 results in relatively weak activation of the EGFR pathway in both embryos and wing imaginal discs and is suppressed by reduced levels of Star (Figure 5D; Supplementary Figure S5), a phenotypic profile closely resembling ectopically expressed Rho-3. Induction of EGFR activation in the wing by Rho-2 is enhanced by co-expression of Star (Figure 5D, and Urban et al, 2002). We were, therefore, motivated to determine whether Rho-2-dependent patterning of developing gonads would show similar features to those observed in the developing eye.
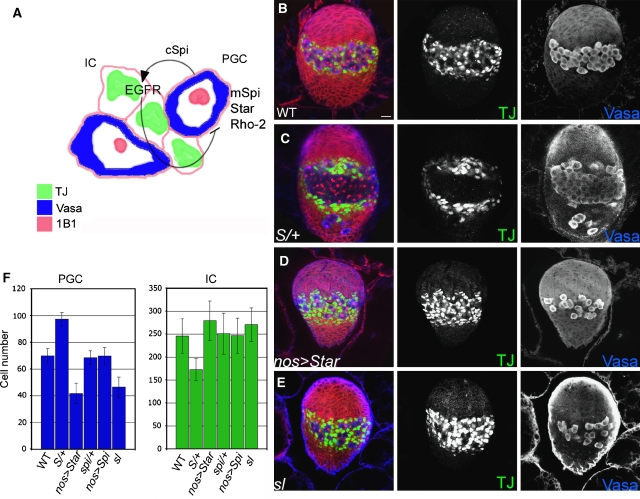
Recent work on the female larval gonad has implicated Rho-2 in the processing of Spi, which mediates germline–soma interactions. Spi, secreted from the primordial germ cells (PGCs), promotes the survival of somatic intermingled cells (IC), which in turn inhibit the proliferation of the germ line (Gilboa and Lehmann, 2006), as schematized in Figure 7A. The ability to quantitatively follow the levels of EGFR activation, by monitoring the number of PGCs and ICs, renders this system particularly suitable for detecting perturbations in the level of EGFR signalling.
Figure 7.
ER localization of Rho-2 in the female larval gonad attenuates EGFR signalling. (A) Cleaved Spi is produced by Rho-2 in the primordial germ cells (PGC-blue), leading to activation of EGFR in the somatic intermingled cells (IC-green), promoting their survival and preventing an unknown signal that inhibits PGC proliferation (Gilboa and Lehmann, 2006). (B) In WT gonads, ICs, marked by anti-Traffic-Jam (TJ, green) are found between and around germ cells (anti-Vasa, blue). Membranes of somatic cells, as well as the fusome, a germline-specific organelle, are labelled with 1B1 antibody (red). (C) Star heterozygous gonads are characterized by increased number of Vasa-positive cells, which are tightly clustered, and fewer intermingled cells, which are no longer spatially mixed with the germ line. (D) Overexpression of UAS-Star in the germ line using nanos-Gal4-VP16 leads to an increase in the number of ICs, with a concomitant reduction in PGC numbers. (E) sl-null mutant female gonads are small, with a lower number of germ cells, and elevated numbers of ICs, indicative of EGFR hyperactivation. (F) Quantification of germ cell (Vasa-positive, blue) and IC (TJ-positive, green) mean numbers in gonads of the indicated genotypes. Eight specimens were examined per genotype. Error bars represent standard deviation. A full-colour version of this figure is available at the EMBO Journal Online.
Wild-type gonads from late third instar larvae contain an average of 70 (±5, n=8) PGCs and 246 ICs (±37, n=8) (Figure 7B and F). These numbers did not significantly change in spi heterozygotes or upon overexpression of Spi in either the germ line or soma (Figure 7F). On the other hand, Star heterozygous gonads displayed a marked elevation in PGC number (98±5.1, n=8) and a concomitant reduction in IC numbers (175±24, n=8) (Figure 7C and F). Increasing Star levels in the germ line by driving UAS–Star with the nanos-Gal4 driver led to the opposite phenotype (Figure 7D and F), with PGC number decreasing to 41 (±8, n=8) and IC numbers rising to 280 (±43, n=8). Star overexpression in somatic cells did not lead to significant alterations in IC or PGC numbers (not shown), confirming that Star dosage sensitivity is a feature of the ligand-processing cells. The restricted Spi signal generated as a consequence of ER activity of Rho-2 may thus contribute to the tight and short-range interactions between germ line and somatic cells in the gonad.
Finally, the hallmark of ER cleavage activity by Rhos is the generation of cleaved Spi in the ER, and thus a requirement for the retention activity of Small wing (Sl) (Schlesinger et al, 2004). In view of the apparent ER cleavage by Rho-2 in the larval female germ line, we tested if the level of EGFR activation is altered in sl-null mutants (Figure 7E). Indeed, the PGC number decreased to 45 (±8, n=8), whereas the IC number increased to 270 (±37, n=8), consistent with the release of excess ligand, leading to EGFR hyperactivation. The sl mutant phenotype in this tissue uncovers the presence of cleaved Spi in the ER, but it does not imply that Spi levels are normally limiting in this setting.
Taken together, these observations identify changes in intracellular compartment localization of Rho proteins as a basis for signal attenuation, in the tissues where the range and level of EGFR activation must be highly restricted.
Discussion
Rho proteases are active in distinct intracellular compartments
Processing of EGFR ligands, in particular Spi, provides the basis for spatial and temporal regulation of EGFR activation throughout Drosophila development.
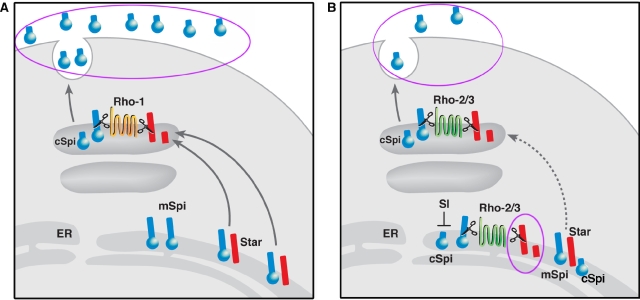
Intracellular compartmentalization of the components comprising the Spi processing machinery is critical. This work has demonstrated that two Rho proteins, Rho-2 and Rho-3, show partial localization to the ER in vivo (Figures 1 and 3; Supplementary Figure S5) and have the capacity to cleave substrates in the ER, in addition to the ability to cleave Spi in the late compartment. These features contrast with those of the major Drosophila protease, Rho-1, restricted to a late compartment, whose ability to cleave the ligand precursor mSpi is completely dependent on the Star chaperone ((Urban et al, 2002, and data not shown). Rho-2 and Rho-3 are thus capable of both signal attenuation through their ER-based activity and productive ligand processing in the late compartment (schematized in Figure 8). We find that this mode of Rho activity is utilized in developmental settings that require tight restrictions over EGFR pathway activation levels.
Figure 8.
ER targeting of Rhos reduces active ligand secretion. (A) The canonical Spitz processing by Rho-1. Cleavage in the late compartment occurs after the chaperone, Star, has accomplished at least one round of Spitz trafficking and results in high levels of secreted ligand. (B) ER activity of Rho-2 or Rho-3 reduces the amounts of active, secreted Spitz. Star cleavage in the ER (circled) prevents the chaperone from trafficking Spitz to the late compartment. ER-cleaved Spitz is retained by a specific, Sl-dependent mechanism. Thus, only the residual Star molecules, which ‘escaped' cleavage by the protease in the ER, are capable of trafficking Spitz precursor (and possibly also cleaved Spitz) to the late compartment, where the activity of Rho-2 or Rho-3 produces the secreted ligand, at reduced levels. A full-colour version of this figure is available at the EMBO Journal Online.
What structural features are responsible for the difference between the activities of Rho proteins in distinct intracellular compartments? The conservation of sequences comprising the enzymatic site and the ER activity of Rho-1–KDEL (Figure 2), suggest that Rho proteins are constitutively active towards their specific substrates, regardless of the compartment in which they reside. Furthermore, when both Rho-1 and Rho-3 were targeted to the ER by a KDEL sequence, they displayed equivalent levels of enzymatic activity (Figure 2). The difference is thus likely to reside in sequences responsible for intracellular trafficking of the Rho proteins. Rho-2 and Rho-3 may possess residues that allow retrograde trafficking or facilitate retention in the ER, in contrast to Rho-1.
The significance of Star cleavage in the ER
Rho proteins appear to have a highly restricted set of proteolytic targets. The molecular basis for this specificity is not clear, as the crystal structure of a bacterial Rho protein suggests that the only essential feature for a Rho substrate is the presence of helix-breaking residues, which allow to denature part of its transmembrane domain within the active site of the protease (Wang et al, 2006; Wu et al, 2006; Ben-Shem et al, 2007).
ER cleavage of the endogenous mSpi precursor produces only traces of an active ligand. This was demonstrated following expression of Rho-1–KDEL in the embryo, eye and wing discs, which yielded very marginal EGFR activation (Figures 3; Supplementary Figure S2). Generation of an active Spi ligand following ER cleavage is likely prevented in part by the novel ER retention mechanism of cleaved Spi, mediated by the PLCγ Small wing (Schlesinger et al, 2004). Inactivation of Star by ER-active Rhos is consistent with previous studies, which showed that Star can function as a chaperone for mSpi only in its full-length form, and is rendered inactive by cleavage (Tsruya et al, 2007). Cleavage of Star in the ER may provide a more effective attenuating mechanism, as chaperone molecules can be inactivated even before they perform a single round of ligand transport.
A priori, ER cleavage by Rhos could compromise EGFR activation by reducing either the levels of biologically active Spi or Star, or both. However, in the eye disc where Rho-3 is expressed, dosage sensitivity is observed only for Star and not for Spi, suggesting that Star becomes the limiting factor. This notion was corroborated by demonstrating that, in every tissue where Rho-3 was expressed ectopically, Star-dosage sensitivity ensued (Figures 4 and 5). Furthermore, in the female gonad where Rho-2 is active, we were also able to demonstrate Star dosage sensitivity, in accordance with the ER activity of this protease (Figure 7). The exclusive dosage sensitivity of Star may result from higher endogenous levels of expression of mSpi or a higher affinity of Star for the protease. In this context, we note that elevation of Star levels strongly enhanced EGFR activation by Rho-1–KDEL (Figure 5B), suggesting that intact Star is also capable of trafficking the cleaved form of Spi out of the ER, and therefore underscoring the significance of ER-based cleavage of Star.
Even when ER-active Rhos were expressed at high levels, they were incapable of completely eliminating trafficking of ligand out of the ER. It is possible that when the ligand and its chaperone are loaded onto secretory COPII vesicles immediately after synthesis, they are refractive to cleavage in the ER, whereas the pool of ligand and chaperone that awaits loading onto these vesicles is readily cleaved under a broad range of concentrations of ER-localized Rhos. The net amount of secreted ligand that is normally generated by Rho-2 or Rho-3 may thus be independent of the dosage of these proteins and rely only upon the intrinsic kinetic probability of ‘escaping' the ER before cleavage.
Rho ER activity restricts EGFR activation range
The ability of Rho proteins to cleave in the ER has a profound effect on the level of active, secreted ligand they produce, and hence on the range of EGFR signalling.
This was demonstrated in the embryo with different markers of EGFR activation (Figure 3; Supplementary Figure S3). In addition, as both Rho-1 and Rho-3 are normally expressed in the eye disc (Figure 6), the complete removal of Rho-3 allowed to monitor quantitatively the resulting activity of Rho-1 alone. Indeed, although Rho-1 is expressed in only three photoreceptor cells, the signal it generated was sufficient to pattern a full eight-cell array, and even recruit supernumerary photoreceptor cells (Figure 6). Thus, the absence of Rho-3 results in hyperactivation of Rho-1-based EGFR signalling, implying that Rho-3 normally attenuates the production of active ligand in the developing eye disc. In a complementary experiment, we showed that co-expressing Rho-3 in the embryonic midline-glial cells, where Rho-1 is normally expressed, attenuated the normal level of EGFR activation (Figure 3).
The biological significance of restricting EGFR signalling is highlighted in tissues such as the eye disc, where multiple discrete rounds of EGFR activation are responsible for recruiting photoreceptor cells, and the number of undifferentiated precursor cells is limited (Freeman, 1996). In this scenario, only the cells immediately adjacent to the Rho-expressing cells are normally recruited, presumably due to the limited amount of ligand that is secreted. When this tight control is compromised, for example, in the rho-3 mutant background, more distant cells can be recruited, and more than one cell of each type can be induced in every round.
The male and female gonads, where Rho-2 is expressed, may also represent a scenario where tight and local activation of the EGFR is required. In the male gonad, production of cleaved Spi by the germ cells, leads to activation of EGFR in the somatic support cells which ensheath the germ cells and physically isolate them from neighbouring cells (Schulz et al, 2002; Sarkar et al, 2007). In the female larval gonad, the Spi signal activating EGFR is likely to exert a local effect to provide the intermingled cells with an accurate mechanism to monitor the number of germ cells producing the activating signal (Gilboa and Lehmann, 2006). We suggest that the ER cleavage ability of Rho-2, which is expressed in the germ cells, leads to the restricted signal that is produced.
The Rho-2 protease is also expressed in the adult female ovary, specifically within the oocyte (Guichard et al, 2000). In this tissue, the mRNA encoding the EGFR ligand Grk is targeted to the vicinity of the oocyte nucleus (Neuman-Silberberg and Schupbach, 1993). Upon translation in the rough ER, the Grk protein is cleaved and secreted, leading to EGFR activation in the neighbouring follicle cells. The tight localization of grk RNA and protein is responsible for establishing the dorso-ventral axis of the egg chamber (Neuman-Silberberg and Schupbach, 1996). The ER activity of Rho-2 in the adult ovary may impose a problem, as cleaved Grk would be uniformly distributed within the ER, and asymmetry would be lost. This problem appears to be circumvented by the Cornichon (Cni) protein (Roth et al, 1995). Cni was shown to associate with the precursor form of Grk in the ER, and facilitate its inclusion within CopII-secretory vesicles, before cleavage (Bokel et al, 2006). Thus, the asymmetric distribution of Grk is maintained.
In conclusion, our studies suggest that the ability to partially target Rho proteases to the ER provides a mechanism for attenuating the production of active EGFR ligands. This is achieved by limiting the amount of intact Star chaperone that is capable of trafficking the ligand precursor (and possibly also the cleaved ligand) to a late compartment, where it is cleaved and secreted. Two modes of EGFR activation are thus employed, while still utilizing the canonical signalling cassette: higher-level activation driven by Rho-1 and limited activation facilitated by Rho-2 or Rho-3.
Materials and methods
DNA constructs
The following constructs were generated by PCR, sequenced and subcloned into UAS-based vectors. Star–HA, Rho–1HA and Rho–3HA (Tsruya et al, 2002) were tagged at their C-termini with a KDEL sequence for ER targeting. Rho-1–GFP, Rho-2–GFP and Rho-3–GFP were constructed by cloning the appropriate cDNAs into pTWR or pTWG (T Murhpy, Carnegie Institution of Washington) using the Gateway cloning system (Invitrogen). mSpi–GFP and mSpi–HRP were described previously (Schlesinger et al, 2004).
Fly strains
The following lines were used: prd-Gal4, en-Gal4, btl-Gal4, MS1096-Gal4, GMR-Gal4, nanos-Gal4-VP16 (Van Doren et al, 1998), C587-Gal4 (Zhu and Xie, 2003; obtained from C Schultz), UAS–p35, UAS–CD8–GFP, UAS–Rho-2 (obtained from M Freeman), UAS–cSpiGFP, UAS–S–LexA VP16, LexOp-lacZ (Tsruya et al, 2007), mδ0.5-LacZ (obtained from S Bray), SIIN23, spi1, ruPLLb, ruPLLbFRT2A, sl9, rhoX81 and ruInga (obtained from M Gallio), ey-flp; ubi–GFP FRT 2A. UAS–mSpiGFP, UAS–mSpi–HRP, UAS–Rho–1HA and UAS–Star were generated in our lab and described previously (Tsruya et al, 2002; Schlesinger et al, 2004). UAS–Rho-3–GFP, UAS–Rho-2–GFP, UAS–Rho-1-GFP, UAS–Rho-1–KDEL and UAS–Rho-3–KDEL were generated by standard fly transformation protocols.
Cell culture and western blots
Drosophila Schneider cells (S2 or S2R+) were transiently transfected using ESCORT IV (Sigma). Expression of UAS-based vectors was achieved by co-transfection of an actin-Gal4 plasmid. Efficiency of the transfection was monitored by GFP expression. Cells were harvested 72 h after transfection, lysed in RIPA buffer and equal amounts of proteins were loaded onto SDS-PAGE gels. For staining, transfected cells were plated on poly-L-Lysine 90 min before fixation.
Immunohistochemistry and histology
Primary antibodies used in this study were the following: anti-GFP (Rabbit, 1:2000; Molecular Probes), anti-HA (mouse, 1:1000; Roche), anti-actin (mouse 1:2000; Sigma), anti–FasIII (mouse, 1:20), anti-Vasa (rabbit, 1:400; Lasko and Ashburner, 1990; obtained from P Lasko), anti dGMAP (rabbit, 1:1000; obtained from P Therond), anti Rab 5, anti Rab7, anti Rab11 (rabbit, 1:50; obtained from M Gonzalez-Gaitan), anti-Traffic-Jam (guinea pig, 1:3000; obtained from D Godt; Li et al, 2003), anti-PntP1 (rabbit, 1:200; obtained from J Skeath), anti–Troponin H to detect BiP (rat, 1:100; Babraham Bioscience Technologies), anti dpERK (mouse, 1:5000; Sigma), anti-Drosophila Golgi (mouse, 1:200; Calbiochem) and anti-β galactosidase (rabbit, 1:1000; Cappel). Anti-Yan (mouse, 1:10), anti-1B1 (mouse, 1:10), anti ElaV (rat, 1:100), anti-Prospero (1:100) and anti-Cut (1:50) were obtained from the Developmental Studies Hybridoma Bank, University of Iowa. Cy-2 conjugated goat anti-HRP, as well as Cy-2, Cy-3 and Cy-5 conjugated secondary antibodies (1:200) were obtained from Jackson ImmunoResearch.
Staining for all antigens was after fixation in 4% formaldehyde and washes with 0.1% Triton X-100, except for Yan and dpERK staining, which were performed as described by Melen et al (2005). Eye sections and scanning electron microscopy were done as described by Schlesinger et al (2004).
Supplementary Material
Supplementary Figures 1–7
Acknowledgments
We thank S Bray, M Freeman, M Gallio, D Godt, M Gonzalez-Gaitan, P Lasko, C Schulz, J Skeath and P Therond for generously providing reagents. We are grateful to E Arama, E Bibi and A Schlesinger for critical reading of the paper, and to the Shilo lab members for support and discussions. We thank Tamar Shapira-Cohen for help in eye sections. This work was supported by grants from the Israel Science Foundation, the US-Israel BSF and Nissim Foundation for Life Sciences Research to BS, who is an incumbent of the Hilda and Cecil Lewis chair for Molecular Genetics.
References
- Baonza A, Casci T, Freeman M (2001) A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol 11: 396–404 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Fass D, Bibi E (2007) Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci USA 104: 462–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H (1998) The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95: 331–341 [DOI] [PubMed] [Google Scholar]
- Bier E, Jan LY, Jan YN (1990) rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev 4: 190–203 [DOI] [PubMed] [Google Scholar]
- Bokel C, Dass S, Wilsch-Brauninger M, Roth S (2006) Drosophila Cornichon acts as cargo receptor for ER export of the TGFalpha-like growth factor Gurken. Development 133: 459–470 [DOI] [PubMed] [Google Scholar]
- Bridges C, Morgan TH (1919) The Drosophila ‘Red Book' of Lindsley and Zimm. Carnegie Institution. Washington Publ 278: 279 [Google Scholar]
- Brown KE, Freeman M (2003) Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development 130: 5401–5412 [DOI] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ (1999) Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature 397: 526–530 [DOI] [PubMed] [Google Scholar]
- Freeman M (1996) Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87: 651–660 [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ (1997) In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Bach EA, Paraiso Y, Carraway KL III, Noselli S, Perrimon N (2002) Mechanism of activation of the Drosophila EGF Receptor by the TGFalpha ligand Gurken during oogenesis. Development 129: 175–186 [DOI] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R (2006) Soma-germline interactions coordinate homeostasis and growth in the Drosophila gonad. Nature 443: 97–100 [DOI] [PubMed] [Google Scholar]
- Golembo M, Schweitzer R, Freeman M, Shilo BZ (1996) Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development 122: 223–230 [DOI] [PubMed] [Google Scholar]
- Guichard A, Roark M, Ronshaugen M, Bier E (2000) brother of rhomboid, a rhomboid-related gene expressed during early Drosophila oogenesis, promotes EGF-R/MAPK signaling. Dev Biol 226: 255–266 [DOI] [PubMed] [Google Scholar]
- Heberlein U, Rubin GM (1991) Star is required in a subset of photoreceptor cells in the developing Drosophila retina and displays dosage sensitive interactions with rough. Dev Biol 144: 353–361 [DOI] [PubMed] [Google Scholar]
- Hsiung F, Griffis ER, Pickup A, Powers MA, Moses K (2001) Function of the Drosophila TGF-alpha homolog Spitz is controlled by Star and interacts directly with Star. Mech Dev 107: 13–23 [DOI] [PubMed] [Google Scholar]
- Kramer S, Okabe M, Hacohen N, Krasnow MA, Hiromi Y (1999) Sprouty: a common antagonist of FGF and EGF signaling pathways in Drosophila. Development 126: 2515–2525 [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M (1990) Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev 4: 905–921 [DOI] [PubMed] [Google Scholar]
- Lee JR, Urban S, Garvey CF, Freeman M (2001) Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107: 161–171 [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D (2003) The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol 5: 994–1000 [DOI] [PubMed] [Google Scholar]
- Melen GJ, Levy S, Barkai N, Shilo BZ (2005) Threshold responses to morphogen gradients by zero-order ultrasensitivity. Mol Syst Biol 1: 2005 0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y, Weber U, Goodman CS, Rubin GM (1990) The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell 60: 211–224 [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HR (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48: 899–907 [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T (1993) The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell 75: 165–174 [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T (1996) The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech Dev 59: 105–113 [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM (1995) Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81: 857–866 [DOI] [PubMed] [Google Scholar]
- Reich A, Sapir A, Shilo B (1999) Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development 126: 4139–4147 [DOI] [PubMed] [Google Scholar]
- Reich A, Shilo BZ (2002) Keren, a new ligand of the Drosophila epidermal growth factor receptor, undergoes two modes of cleavage. EMBO J 21: 4287–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schupbach T (1995) Cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell 81: 967–978 [DOI] [PubMed] [Google Scholar]
- Sarkar A, Parikh N, Hearn SA, Fuller MT, Tazuke SI, Schulz C (2007) Antagonistic roles of rac and rho in organizing the germ cell microenvironment. Curr Biol 17: 1253–1258 [DOI] [PubMed] [Google Scholar]
- Schlesinger A, Kiger A, Perrimon N, Shilo BZ (2004) Small wing PLCgamma is required for ER retention of cleaved Spitz during eye development in Drosophila. Dev Cell 7: 535–545 [DOI] [PubMed] [Google Scholar]
- Schulz C, Wood CG, Jones DL, Tazuke SI, Fuller MT (2002) Signaling from germ cells mediated by the rhomboid homolog stet organizes encapsulation by somatic support cells. Development 129: 4523–4534 [DOI] [PubMed] [Google Scholar]
- Shilo BZ (2003) Signaling by the Drosophila epidermal growth factor receptor pathway during development. Exp Cell Res 284: 140–149 [DOI] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, Bier E (1993) The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes Dev 7: 961–973 [DOI] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, O'Neill JW, Biehs B, Colley N, Bier E (1996) The Drosophila rhomboid protein is concentrated in patches at the apical cell surface. Dev Biol 174: 298–309 [DOI] [PubMed] [Google Scholar]
- Thackeray JR, Gaines PC, Ebert P, Carlson JR (1998) Small wing encodes a phospholipase C-(gamma) that acts as a negative regulator of R7 development in Drosophila. Development 125: 5033–5042 [DOI] [PubMed] [Google Scholar]
- Tsruya R, Schlesinger A, Reich A, Gabay L, Sapir A, Shilo BZ (2002) Intracellular trafficking by Star regulates cleavage of the Drosophila EGF receptor ligand Spitz. Genes Dev 16: 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsruya R, Wojtalla A, Carmon S, Yogev S, Reich A, Bibi E, Merdes G, Schejter E, Shilo BZ (2007) Rhomboid cleaves Star to regulate the levels of secreted Spitz. EMBO J 26: 1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M (2001) Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107: 173–182 [DOI] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M (2002) A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J 21: 4277–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R (1998) Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol 8: 243–246 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Ha Y (2006) Crystal structure of a rhomboid family intramembrane protease. Nature 444: 179–180 [DOI] [PubMed] [Google Scholar]
- Wasserman JD, Urban S, Freeman M (2000) A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes Dev 14: 1651–1663 [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y (2006) Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol 13: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Zhu CH, Xie T (2003) Clonal expansion of ovarian germline stem cells during niche formation in Drosophila. Development 130: 2579–2588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–7