Abstract
Nonenveloped animal viruses must disrupt or perforate a cell membrane during entry. Recent work with reovirus has shown formation of size-selective pores in RBC membranes in concert with structural changes in capsid protein μ1. Here, we demonstrate that μ1 fragments released from reovirus particles are sufficient for pore formation. Both myristoylated N-terminal fragment μ1N and C-terminal fragment φ are released from particles. Both also associate with RBC membranes and contribute to pore formation in the absence of particles, but μ1N has the primary and sufficient role. Particles with a mutant form of μ1, unable to release μ1N or form pores, lack the ability to associate with membranes. They are, however, recruited by pores preformed with peptides released from wild-type particles or with synthetic μ1N. The results provide evidence that docking to membrane pores by virus particles may be a next step in membrane penetration after pore formation by released peptides.
Keywords: cell entry, membrane penetration, membrane pore, nonenveloped virus, reoviridae
Introduction
Cell membranes compartmentalize biological processes, but pose a challenge for macromolecule translocation. The same impediment is confronted by foreign invaders, such as viruses, that must enter cells to replicate. The mechanisms by which nonenveloped animal viruses breach this barrier remain less well characterized than those of enveloped viruses (Smith and Helenius, 2004).
Studies of nonenveloped virus entry have exposed parallels with both enveloped viruses and bacterial toxins. Enveloped viruses express fusion proteins, which upon binding to a cell receptor or exposure to low pH in endosomes undergo conformational changes resulting in insertion of membrane-interacting sequences into target membranes. Subsequent rearrangements bring cell and virus membranes into proximity, favouring fusion and delivery of virus contents into the cell (Earp et al, 2005; Weissenhorn et al, 2007). Many bacterial toxins consist of two or more subunits or domains that include transmembrane and catalytic regions. They too respond to external cues, undergoing conformational changes that result in insertion of membrane-interacting sequences into target membranes. Transmembrane regions then serve as channels for translocation of catalytic regions (Geny and Popoff, 2006; Young and Collier, 2007). In addition to lacking the means to use membrane fusion similar to an enveloped virus, translocation of a large nonenveloped virus poses a different challenge than translocation of a polypeptide chain such as a bacterial toxin. In the case of mammalian orthoreovirus (reovirus), a subviral particle of 70–80 nm in diameter is delivered to the cytoplasm (Chandran and Nibert, 2003).
Nonenveloped virus entry involves stepwise capsid rearrangements and/or disassembly, leading to exposure or release of membrane-interacting peptides. For example, adenovirus releases its adhesion fibre soon after receptor binding at the cell surface (Nakano et al, 2000), followed by acid-dependent disassembly in endosomes (Greber et al, 1993). The latter process includes release of protein VI, which contains an amphipathic helix believed to disrupt endosomal membranes (Wiethoff et al, 2005). Similarly, receptor binding by poliovirus triggers structural changes in the capsid, allowing release of myristoylated peptide VP4 (De Sena and Mandel, 1977; Gromeier and Wetz, 1990; Tuthill et al, 2006) and exposure of the VP1 N terminus (Fricks and Hogle, 1990). Both released VP4 and particle-associated VP1 then contribute to membrane-pore formation, through which the virus genome is translocated to the cytoplasm (Hogle, 2002; Brandenburg et al, 2007).
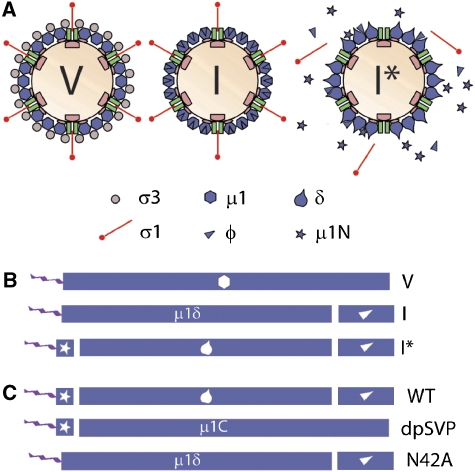
Reovirus also undergoes stepwise disassembly leading to membrane penetration (Figure 1A). Endosomal or intestinal proteases degrade outer capsid protein σ3, yielding a metastable particle, termed the infectious subvirion particle (ISVP) (Silverstein et al, 1970; Sturzenbecker et al, 1987; Bodkin et al, 1989; Nibert, 1993; Baer and Dermody, 1997). The ISVP converts to ISVP* in association with membrane disruption (Chandran et al, 2002, 2003). Several studies have implicated outer capsid protein μ1 in membrane penetration (Nibert et al, 1991; Lucia-Jandris et al, 1993; Nibert, 1993; Hooper and Fields, 1996a, 1996b; Chandran et al, 2002, 2003; Liemann et al, 2002; Odegard et al, 2004). μ1 undergoes a protease-induced C-terminal cleavage during ISVP generation (Nibert and Fields, 1992) and an autocatalytic N-terminal cleavage during the ISVP → ISVP* transition (Nibert et al, 2005; Zhang et al, 2006). These cleavages combined yield the following μ1 fragments: N-terminal myristoylated μ1N (4 kDa), central δ (59 kDa), and C-terminal φ (13 kDa) (Figure 1B). The μ1N cleavage, prevented by μ1 mutation N42A, is required for membrane penetration (Odegard et al, 2004), but the C-terminal cleavage, absent from ISVP-like particle form detergent-and-protease-treated subvirion particles (dpSVPs), is not required (Chandran and Nibert, 1998; Figure 1C; see Results for further descriptions of cleavage-deficient particles). Other hallmarks of the ISVP* transition include release of both μ1N and adhesion protein σ1 from particles (Chandran et al, 2002, 2003; Odegard et al, 2004; Agosto et al, 2006) and acquisition of protease sensitivity by the particle-associated δ fragment (Chandran et al, 2003; Figure 1A). Furthermore, the ISVP* transition results in the formation of size-selective pores, 4–9 nm in diameter, in RBC membranes (Agosto et al, 2006). Although the involvement of both particles and released μ1N in forming pores has been implied by their associations with RBC membranes (ibid), specific mediators of this pore formation have not been defined. Furthermore, events that follow formation of small pores, to allow translocation of a large virus particle across the membrane, remain unknown.
Figure 1.
Reovirus disassembly intermediates and μ1 cleavage products. (A) Reovirus particle-disassembly intermediates leading to membrane penetration: virion (V, left), ISVP (I, middle), and ISVP* (I*, right). (B) Linear representation of major products of μ1 processing in virions (top), ISVPs (middle), and ISVP*s (bottom). The N terminus of μ1 is myristoylated (wavy line). Exogenous protease cleaves μ1 near the C terminus generating μ1δ and φ (triangle). The N-terminal autocleavage during ISVP* transition generates μ1N (star), δ (teardrop), and φ. (C) Major products of μ1 processing in wild-type ISVP*s (WT, top), dpSVP*s (dpSVP, middle), and N42Aprc*s (N42A, bottom). See text for further descriptions of the cleavage-deficient particles.
In this report, we demonstrate that reovirus particles are dispensable for pore formation in RBC membranes and that this task is accomplished by peptides released from particles during the ISVP* transition. We identify myristoylated peptide μ1N as the primary and sufficient determinant of pore formation. These results are consistent with a paradigm in which peptides exposed or released from nonenveloped viruses have critical roles in membrane penetration. We also demonstrate that reovirus particles are recruited to preformed membrane pores and propose that particle docking to pores may be a next step in membrane penetration.
Results
Components released during ISVP* transition are sufficient for haemolysis
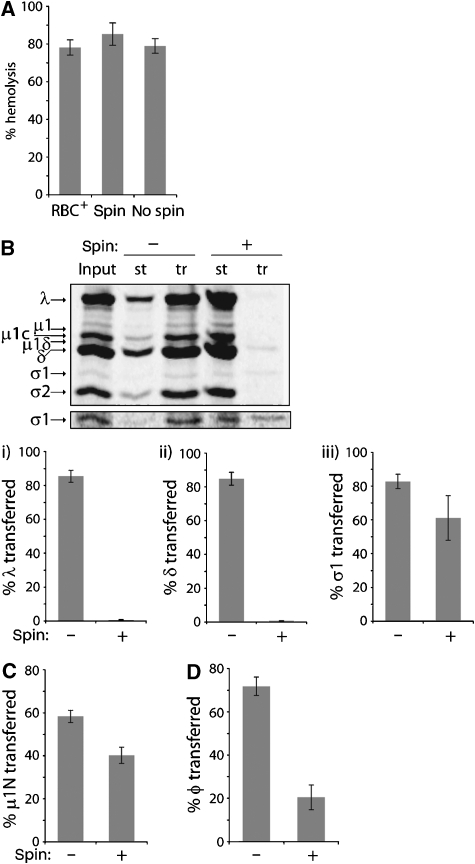
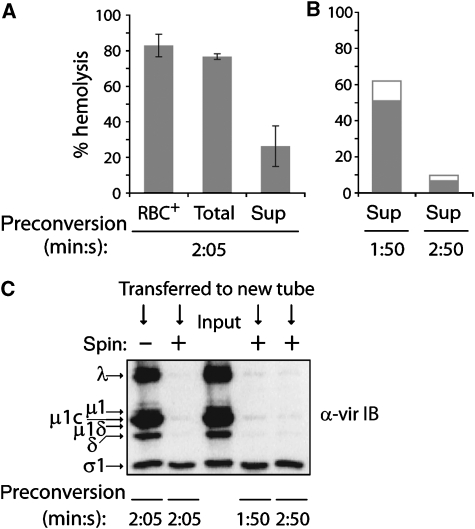
To determine whether reovirus particles are required for haemolysis, we developed two modifications of the original assay (Nibert, 1993; Chandran and Nibert, 1998; Chandran et al, 2002). In each case, nonpurified [35S]Met/Cys-labelled reovirus Type 1 Lang (T1L) ISVPs were preconverted to ISVP*s at 37°C in the absence of RBCs and then shifted to 4°C. We centrifuged one reaction to pellet particles (spin) or omitted the centrifugation step (no spin). We separately transferred aliquots of spin-reaction supernatant and no-spin reaction to tubes with RBCs at 37°C. The amount of RBC lysis was determined by measuring the absorbance of released haemoglobin. Spin, no-spin, and unmodified reactions performed in the presence of RBCs (RBC+ reactions), each yielded approximately 80% haemolysis (Figure 2A). These levels of haemolysis in modified reactions depended on careful monitoring of ISVP* preconversion times, as extending incubations at 37°C by several minutes resulted in loss of activity (data not shown).
Figure 2.
Haemolysis activity of ISVP* supernatant. (A) ISVP* transition was induced at 37°C either in the presence of RBCs (RBC+) or in their absence (spin + and spin −). One of the no-RBC reactions was centrifuged at 4°C to pellet virus particles (spin +), and the other was kept at 4°C without centrifugation (spin −). Aliquots of spin and no-spin reactions were then each separately transferred to tubes with RBCs at 37°C. Percentage haemolysis was expressed relative to reactions containing Triton X-100. (B–D) Spin and no-spin reactions were performed, and fractions staying in the tubes (st), as well as those transferred to new tubes (tr), were analysed. Percentage transfer was defined as the I(tr)/[I(tr)+I(st)], where I stands for intensity. For (B), nonpurified [35S]Met/Cys-labelled T1L ISVPs were used, and fractions were analysed by SDS–PAGE and phosphorimaging to quantify the amounts of λ (i), δ (ii), and σ1 (iii) protein transfer. A darker exposure of the σ1 region of this gel is also shown. For (C), purified [3H]myristate-labelled ISVPs containing the total of approximately 13 000 CPM were used, and μ1N protein transfer was measured by scintillation counting. For (D), purified unlabelled ISVPs were used, and φ transfer was quantified after SDS–PAGE, Coomassie Blue staining, and densitometry. For (A–D), means±s.d. of three independent experiments are shown.
To confirm that centrifugation cleared virus particles from the ISVP* supernatant, we analysed remaining and transferred fractions by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and phosphorimaging and quantified the transferred amounts of particle-associated δ and λ proteins and released σ1 protein (Figure 2B). Without centrifugation, we accounted for >80% transfer of each. After centrifugation, <1% of δ and λ, but approximately 60% of σ1, were found in the supernatant, confirming that centrifugation cleared particles and implicated released component(s) in haemolysis activity.
μ1-derived fragments μ1N and φ are enriched in supernatant of spin reactions
Which components of the ISVP* supernatant are candidates for haemolysis activity? To exclude a role for σ3 fragments formed during generation of ISVPs, we used purified ISVPs in all experiments described below. Previous work has shown the requirement for N-terminal autocleavage of μ1, generating the 4-kDa myristoylated fragment μ1N (Nibert et al, 1991, 2005), in both haemolysis and infection (Odegard et al, 2004). μ1N is released from ISVP*s (Odegard et al, 2004; Agosto et al, 2006) and recruited to membranes in concert with pore formation in RBC+ reactions (Agosto et al, 2006). These results suggest a direct role for released μ1N in membrane-pore formation, although an alternative function of μ1 autocleavage has not been ruled out, and neither has the possibility that μ1N is recruited to RBC membranes through ISVP*s, which also become membrane associated in RBC+ reactions (ibid). To quantify the amount of μ1N transfer in spin and no-spin reactions, we used purified [3H]myristate-labelled ISVPs and analysed the μ1N content of post-conversion pellet and supernatant fractions by scintillation counting. We observed approximately 40 and 60% of μ1N transfer in spin and no-spin reactions, respectively (Figure 2C). These data support prior evidence that μ1N is released from ISVP*s (Odegard et al, 2004; Agosto et al, 2006); moreover, its substantial presence in the ISVP* supernatant, which retains haemolysis activity, identifies μ1N as a candidate for this activity.
In contrast to prior evidence for μ1N, protease-mediated C-terminal cleavage of μ1, generating the C-terminal 13-kDa fragment φ (Nibert and Fields, 1992), is not required for haemolysis or infection (Chandran and Nibert, 1998; Chandran et al, 1999). As the φ region remains tethered to particles in the absence of cleavage, its role in these activities has not been excluded. We recently observed that φ is released from particles during the ISVP* transition in the presence of 0.1% Triton X-100 (data not shown). We have now asked whether φ is released under conditions of the current assays, which lack detergent. We used purified unlabelled ISVPs in spin and no-spin reactions and measured the amount of φ transfer by SDS–PAGE, Coomassie Blue staining, and densitometry. We observed approximately 20 and 70% of φ transfer in spin and no-spin reactions, respectively (Figure 2D). Although the amount of φ in spin-reaction supernatants was reduced relative to σ1 or μ1N, it was much larger than that of particle-associated δ and λ proteins (compare panel D to B and C in Figure 2). These results confirm that at least a portion of φ is released from ISVP*s and implicate φ as another candidate in haemolysis activity. We do not expect σ1 to contribute, because previous RBC+ reactions using recoated cores without σ1 showed normal levels of haemolysis (Chandran et al, 1999).
Supernatant of spin reactions retains capacity to form size-selective membrane pores
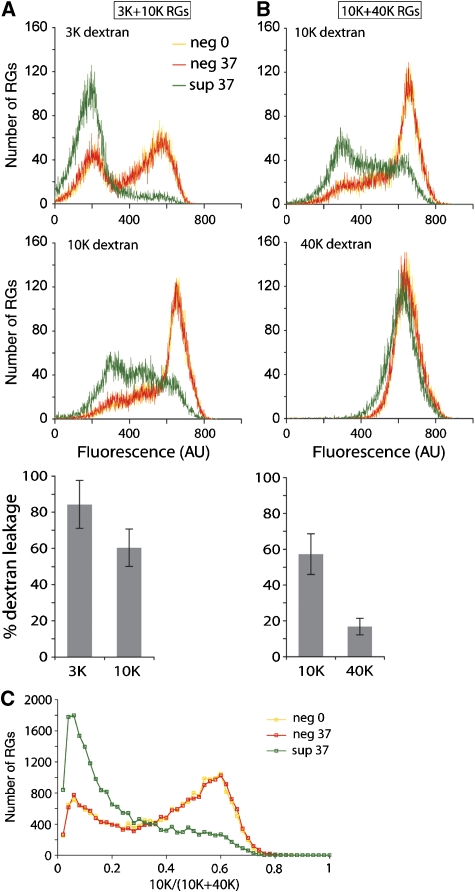
In RBC+ reactions, the ISVP* transition has been associated with osmotic lysis of RBCs, after formation of size-selective membrane pores, 4–9 nm in diameter (Agosto et al, 2006). We asked whether spin-reaction supernatant was sufficient to form similarly sized pores. We performed spin reactions and transferred supernatant to resealed RBC ghosts (RGs) loaded with fluorophore-conjugated dextrans of various sizes (Figure 3). To determine the efficiency of initial loading and the amount of nonspecific leakage, we incubated RGs in the same buffer, but lacking particle-derived components, on ice or at 37°C. RGs were doubly labelled with dextrans with mean molecular weights of either 3000 and 10 000 (3K/10K) (Figure 3A) or 10 000 and 40 000 (10K/40K) (Figure 3B), and the reactions were analysed by flow cytometry. Dextran leakage in the presence of supernatant strongly resembled that previously reported for RBC+ reactions (ibid). We observed approximately 80% leakage of 3K (Figure 3A) and approximately 60% leakage of 10K dextran (Figure 3A and B), and only a slight shift in the fluorescence signal of 40K dextran-labelled RGs, corresponding to <20% leakage. Frequency distributions of the relative intensities of 10K label over the total signal originating from 10K/40K doubly labelled RGs confirmed preferential leakage of 10K over 40K label in the presence of supernatant (Figure 3C). As in our previous determinations of pore size in RBC+ reactions (ibid), these results in combination with estimates of the hydrodynamic radii of dextrans (Scherrer and Gerhardt, 1971; Kuga, 1981) demonstrate that virus particles are dispensable for formation of size-selective membrane pores, 4–9 nm in diameter.
Figure 3.
Pore formation by ISVP* supernatant. Supernatants of spin reactions were transferred to RGs at 37°C (sup 37), containing either 3 and 10K dextrans (A) or 10 and 40K dextrans (B, C). A nonspecific dextran leakage control consisted of equivalent incubations lacking particle-derived components (neg 37). A loading control consisted of RGs kept at 0°C (neg 0). Representative histograms of data analysed by flow cytometry are shown (top). Mean percentages of dextran leakage±s.d. of three independent experiments are also shown (bottom). (C) Data represented by histograms in (B) are presented as frequency distributions of 10K dextran signal over the sum of 10 and 40K dextran signals for each sample.
μ1N and φ, but not σ1, associate with RGs after pore formation by spin-reaction supernatant
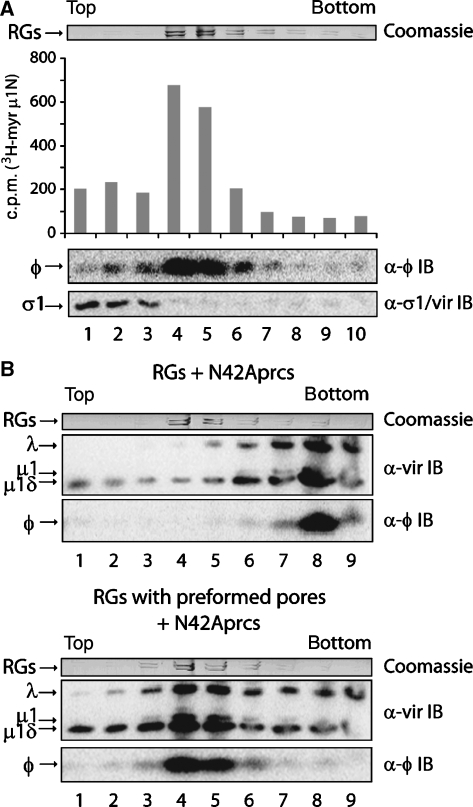
To distinguish among μ1N, φ, and σ1 as candidates for pore formation, we asked which of these proteins associate with RBC membranes after pore formation by the supernatant. We used [3H]myristate-labelled ISVPs in spin reactions with RGs and then fractionated the reactions by density sedimentation on Percoll gradients. A discrete RG band formed near the middle of the gradient. We analysed a portion of each fraction collected from the top by scintillation counting to detect μ1N, and the remainder by SDS–PAGE, and either Coomassie Blue staining to visualize RG protein bands or immunoblotting to visualize φ and σ1 (Figure 4A). μ1N and φ signals peaked in the same fractions as RG bands, while σ1 peaked near the top of the gradient. This pattern demonstrated that at least a portion of transferred μ1N and φ, but not σ1, had associated with RGs. The data are consistent with roles for μ1N and/or φ, but not σ1, in membrane-pore formation.
Figure 4.
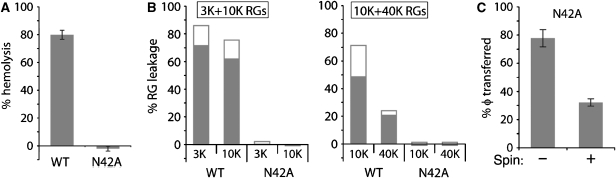
Membrane association of released components and virus particles. (A) Purified [3H]myristate-labelled ISVPs were used in spin reactions with RGs and fractionated on Percoll gradients. A portion of each fraction was analysed by scintillation counting to determine the amount of μ1N in each. The remainder of each fraction was analysed by SDS–PAGE and either Coomassie Blue staining to visualize RG protein bands or immunoblotting (IB) to detect φ or σ1. The same pattern of protein association (μ1N and φ) or its lack of association (σ1) with RGs was obtained from three additional experiments. (B) N42Aprc* transition was induced in the presence of RGs without pores (top) or those in which pores had been preformed using wild-type ISVP* supernatant (bottom). Reactions were fractionated as in (A) and analysed by SDS–PAGE and either Coomassie Blue staining to visualize RG protein bands or immunoblotting to detect virus particle-associated bands (vir) or φ. Representative results for one of three such experiments are shown.
Virus particles are recruited to pores formed by spin-reaction supernatant
Although ISVP*s associate with membranes in RBC+ reactions (Agosto et al, 2006), the data presented thus far suggest their dispensability for pore formation. It is possible that the previously reported association was a result, rather than a cause, of pore formation. To test whether virus particles can associate with membranes in the absence of pore formation, we used ISVP-like particles derived from cores recoated with a previously described, penetration-deficient μ1 autocleavage mutant, N42A, and wild-type σ3 (N42Aprcs) (Odegard et al, 2004) (see Figure 1C). We induced the N42Aprc* (ISVP*-like) transition in the presence of RGs and then fractionated the reactions on Percoll gradients. Fractions were analysed by SDS–PAGE. RG protein bands were visualized by Coomassie Blue staining and virus particle-associated bands and φ by immunoblotting. The N42Aprc* particles sedimented to the bottom of the gradient (Figure 4B, top), demonstrating their lack of association with RG membranes, which peaked near the middle. We confirmed that these particles had undergone the N42Aprc* transition by trypsin sensitivity of particle-associated δ (data not shown). We also verified by flow cytometry that N42Aprc* reactions did not form pores in target membranes (data not shown).
To determine whether the lack of N42Aprc* association with RG membranes was due to the absence of pore formation, rather than to an inability of these particles to associate with RGs, we performed equivalent reactions in the presence of RGs in which pores had been preformed using wild-type ISVP* supernatant. In this case, N42Aprc*s were effectively recruited to membranes, as indicated by their redistribution to the middle of the gradient, colocalizing with RGs (Figure 4B, bottom). We confirmed both pore formation by the supernatant and N42Aprc* transition as described above (data not shown). We also demonstrated that wild-type ISVP* supernatant did not alter the sedimentation behaviour of N42Aprc*s in the absence of RGs, which in this case sedimented near the bottom of the gradient (data not shown).
Distribution of N42Aprc*-derived φ fragment in these experiments mirrored particle distribution: φ sedimented near the bottom of the gradient in the presence of RGs without pores (Figure 4B, top), but near the middle in the presence of RGs with preformed pores (Figure 4B, bottom). These data provide strong evidence that neither virus particles nor φ can associate with RG membranes unless recruited to pores formed by released peptides. Interaction of φ with virus particles in the absence of pore formation may represent the way in which particles dock to membrane pores, but we cannot exclude a direct interaction of other particle-associated proteins with pore-associated μ1N. Particle docking to pores may be a discrete step after pore formation by released peptides during membrane penetration by reovirus.
μ1N is required for pore formation, φ is not sufficient, and σ1 is dispensable
To address directly whether μ1N is an active component of the ISVP* supernatant, we tested supernatant lacking μ1N for its ability to form membrane pores. We transferred N42Aprc* spin-reaction supernatants to tubes with either RBCs or doubly labelled 3K/10K or 10K/40K RGs (Figure 5A and B). We compared these supernatants with those generated from ISVP*-like particles derived from cores recoated with wild-type μ1 and σ3 (WTprc*s) (Chandran et al, 1999) (Figure 5A and B). Both the N42Aprcs and the WTprcs were recoated without σ1, so both types of supernatant lacked σ1. As μ1N remains particle-associated in N42Aprc*s, this supernatant also lacked μ1N. WTprc* supernatant induced normal levels of haemolysis as well as leakage of 3K and 10K dextrans from RGs. As in experiments with ISVP* supernatant, 40K dextran was largely retained. These results provide direct evidence that σ1 is dispensable. They further indicate that lack of an active component in the N42Aprc* supernatant is a likely cause of its failure to form pores. The observation that a normal amount of φ (∼30%) is transferred in the supernatant of N42Aprc*s argues for the requirement for μ1N (Figure 5C). Although φ may contribute to pore formation, it is not sufficient.
Figure 5.
μ1N is required for pore formation. WTprcs and/or N42Aprcs were used in spin reactions only (A, B) or both in no-spin and spin reactions (C). In (A), percentage haemolysis was measured as in Figure 2A (slightly negative values are due to a small variation in the amount of pipetted RBCs in individual reactions). Means±s.d. of three independent experiments are shown. In (B), percentage 3 and 10K dextran leakage (left) or 10 and 40K dextran leakage (right) from RGs was measured as in Figure 3A and B. Overlapping bars represent results of two independent experiments. In (C), the amount of φ transfer from N42Aprc*s was measured as in Figure 2D. Means±s.d. of three independent experiments are shown.
μ1N is sufficient for haemolysis, but φ is required for optimal activity
To address the role of φ in pore formation, we used dpSVPs (Chandran and Nibert, 1998). These particles are prepared similar to ISVPs, but under conditions that prevent the protease-mediated C-terminal cleavage that yields φ (see Figure 1C). RBC+ reactions using dpSVPs show normal levels of haemolysis (ibid). Can supernatant of dpSVP*s, which effectively lacks φ, mediate haemolysis? In initial experiments, dpSVP* supernatant appeared negative for haemolysis despite the transfer of approximately 30% of μ1N (data not shown). Shortening of 37°C incubation during preconversion by fractions of a minute ultimately yielded conditions that resulted in partial activity of the supernatant (Figure 6A and B), while still allowing dpSVP* transition (data not shown). No-spin dpSVP* reactions were not as sensitive to the length of 37°C incubation during preconversion, and they invariably yielded a higher level of haemolysis than spin dpSVP* reactions (Figure 6A). These results suggest that separation of μ1N from φ after preconversion by particle pelleting causes accelerated loss of μ1N activity. Using SDS–PAGE and immunoblotting, we also analysed the amount of dpSVP* particle transfer in these reactions. Figure 6C confirms that the level of particle, and thus also φ, clearance from the supernatant is not affected by altering the length of incubation at 37°C during preconversion. These results establish the sufficiency of μ1N in mediating haemolysis. Although the role of φ is not yet defined, the φ region contributes to the optimal level of haemolysis.
Figure 6.
μ1N is sufficient for haemolysis. (A) RBC+, no-spin, and spin reactions were performed using dpSVPs preconverted for 2 min and 5 s. Means±s.d. of three independent experiments are shown. (B) Spin reactions were performed using dpSVPs preconverted for 1 min and 50 s or 2 min and 50 s. Overlapping bars represent results of two independent experiments. (C) The amount of virus particle (vir) transfer in representative experiments shown in (A) and (B) was assessed by SDS–PAGE and immunoblotting (IB). Percentage haemolysis was measured as in Figure 2A.
Synthetic μ1N peptide mediates pore formation and particle recruitment to pores
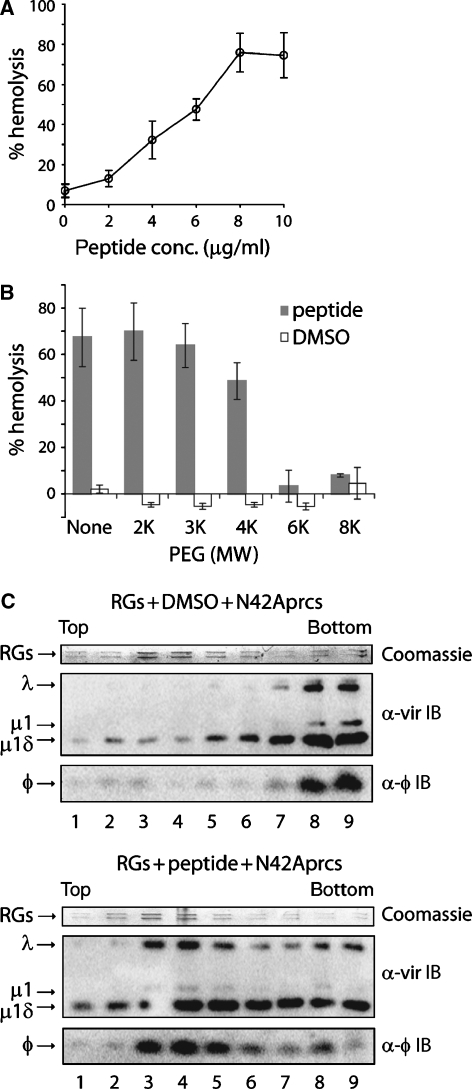
To demonstrate directly that μ1N is sufficient for pore formation, we performed haemolysis experiments with synthetic myristoylated μ1N. Reactions contained RBCs and μ1N peptide in a final concentration of 10% dimethyl sulphoxide (DMSO). DMSO alone caused up to approximately 10% haemolysis, whereas approximately 80% was observed with 10 μg/ml peptide (Figure 7A). DMSO, however, caused severe RG leakage, preventing pore-size determination using dextran-loaded RGs (data not shown). As an alternative, we performed osmotic-protection experiments with RBCs and polyethylene glycols (PEGs) of various sizes. This approach has allowed pore-size determination in a number of systems, including reovirus RBC+ reactions (Lang and Palmer, 2003; Agosto et al, 2006). Although PEGs of molecular weight 4000 and smaller had little or no effect, larger PEGs provided complete protection from haemolysis by synthetic μ1N (Figure 7B). These results resemble data obtained in RBC+ reactions and demonstrate formation of size-selective membrane pores, 4–6 nm in diameter (Agosto et al, 2006).
Figure 7.
Synthetic μ1N peptide mediates haemolysis, pore formation, and particle recruitment. (A) Haemolysis reactions were performed in the presence of 10% DMSO and a range of concentrations (0–10 μg/ml) of synthetic μ1N peptide. Means±s.d. of three independent experiments are shown. (B) Haemolysis reactions were performed in the presence of 10% DMSO with or without 10 μg/ml synthetic μ1N peptide. Some reactions also included 30 mM PEG of indicated molecular weights (MW). Means±s.d. of three independent experiments are shown. In (A) and (B), percentage haemolysis was measured as in Figure 2A. (C) The N42Aprc* transition was induced in the presence of RGs, treated with either DMSO alone (top) or synthetic μ1N peptide (bottom). Reactions were analysed as in Figure 4B (IB, immunoblotting). Representative results for one of three independent experiments are shown.
We next tested whether membrane pores preformed with synthetic μ1N can recruit virus particles. The N42Aprc* transition was induced in the presence of RGs preincubated with DMSO alone or μ1N peptide. The reactions were then fractionated on Percoll gradients and analysed by SDS–PAGE and either Coomassie Blue staining to visualize RG protein bands or immunoblotting to detect particle-associated proteins or φ. Particles and φ cosedimented near the bottom of the gradient in the absence of pore formation, showing their lack of association with RGs, peaking near the middle (Figure 7C, top). RGs containing μ1N-induced pores effectively recruited particles and particle-associated φ, which peaked along with RG protein bands near the middle of the gradient (Figure 7C, bottom). μ1N peptide did not alter the sedimentation behaviour of N42Aprc*s in the absence of RGs, which in this case sedimented near the bottom of the gradient (data not shown). These experiments confirm the sufficiency of μ1N in both membrane-pore formation and particle recruitment to pores.
Discussion
In this report, we demonstrate that myristoylated peptide μ1N, released from reovirus particles during the ISVP* transition, is sufficient for pore formation in RBC membranes. Both particle-associated components and released φ proved dispensable, but released φ contributed to optimal activity, apparently by keeping μ1N in active form for longer times. One possible role for φ is in helping to keep μ1N soluble until membrane insertion. Another possibility is that φ might prevent premature or erroneous interactions between μ1N molecules. Future experiments will explore the role of φ as a μ1N chaperone.
Synthetic μ1N peptide forms similarly sized pores to those previously reported for RBC+ reactions (Agosto et al, 2006) or those formed with spin-reaction supernatants shown here. In contrast to RBC+ reactions, in which pore size varied to a limited extent with particle concentration (ibid), altering synthetic μ1N concentration showed no clear effect on pore size (data not shown). This difference could reflect a role for other virus-derived components, such as φ or particle-associated proteins, or it could be due to limiting solubility of μ1N in 10% DMSO. Maximal haemolysis occurred with synthetic μ1N concentrations above 1.5 μM (Figure 7A), comparable to the efficiency of melittin (Asthana et al, 2004).
Both released μ1N and φ associate with RBC membranes in the absence of virus particles (Figure 4A and B). From the total amount of μ1N transferred in spin reactions, only approximately 40% associated with RG membranes in these experiments (data not shown). This result suggests that the portion of μ1N found in association with membranes is the active fraction that makes pores, but we cannot exclude the possibility that some portion of the association is nonproductive.
As μ1N mediates pore formation, and N42Aprc*-derived φ does not associate with RG membranes lacking pores, the association of φ with membranes upon pore formation might represent recruitment by μ1N. A substantial amount of N42Aprc*-derived φ remains particle associated as demonstrated by copelleting on Percoll gradients (see Figures 4B, top, and 7C, top). As a normal amount of φ is transferred in spin reactions using N42Aprc*s, the remaining association might represent normal interaction, rather than defects in φ release from N42Aprc*s. These observations raise the possibility that if φ can interact with both μ1N pores and particles, it might in fact mediate particle recruitment to pores. The possibility that particles interact with pore-associated μ1N through the δ fragment of μ1 and/or even through some other protein has not been excluded.
The pore formed by μ1N is approximately one-tenth the diameter of the reovirus particle. What subsequent steps allow particle translocation to the cytoplasm? Proposed scenarios include widening of the pore by host factors and osmotic lysis of the endosome (Agosto et al, 2006). The results described here provide novel evidence that a next step in membrane penetration may be recruitment of virus particles to these pores (Figure 5). As endosome rupture as a direct result of pore formation would seem to eliminate the need for particle recruitment, this observation may favour a penetration mechanism by which pore formation leads to a more regulated process of particle translocation. Docking of particles to the pores might allow particle-associated sequences to access cytoplasmic host factors, which could then mediate subsequent steps, including pore widening and particle translocation (e.g., see recent evidence for a possible role of Hsc70 (Ivanovic et al, 2007)).
We have obtained evidence both for and against the involvement of host factors in particle translocation across the membrane. Docking of N42Aprc*s to RGs containing μ1N-formed pores does not cause further increase in pore size (data not shown), suggesting that our in vitro system does not support translocation of docked particles and thus that host factors might be required. Arguing to the contrary is that treatment of docked N42Aprc*s with trypsin degrades particle-associated δ, but dissociates only a fraction of the resulting core-like particles (data not shown). Future studies will address this question more directly.
Our estimates indicate that the μ1N concentration that may be reached in an endosome containing a single reovirus ISVP that undergoes the ISVP* transition is similar to or even much higher than that needed for pore formation in RBC membranes. It is thus possible that the size of pores formed in endosomal membranes might be larger than those obtained in RBC membranes. It is also possible that events after particle recruitment result in endosome rupture. These questions will be better addressed through studies of infected cells. For example, size-selective pores have been demonstrated in endosomes of cells infected with rhinovirus and proposed to mediate genome release into cells (Brabec et al, 2005). Other evidence supports endosome rupture by adenovirus (FitzGerald et al, 1983; Meier et al, 2002; Brabec et al, 2005; Farr et al, 2005). Pore formation followed by particle docking and translocation of virus genome, nucleoprotein complex, or subviral particle into the cell interior, may be a common series of steps during membrane penetration by nonenveloped animal viruses.
Our current findings lend further support to models in which nonenveloped viruses penetrate cell membranes during entry by deploying membrane-interacting peptides, leading to pore formation. Although in some cases these membrane-interacting peptides may remain particle associated, there are a number of cases in which they seem to be released. In addition to protein VI of adenovirus (Greber et al, 1993; Wiethoff et al, 2005) and myristoylated peptide VP4 of poliovirus (De Sena and Mandel, 1977; Gromeier and Wetz, 1990; Tuthill et al, 2006), the membrane-interacting γ peptide dissociates from an entry intermediate of flock house virus (Walukiewicz et al, 2006). The release of membrane-interacting pep46 from infectious bursal disease virus in association with membrane-pore formation in vitro has also been suggested (Galloux et al, 2007). Thus, entry mechanisms of many distinct nonenveloped animal viruses, known to differ with respect to the ‘payload' delivered to the cytoplasm, may include mechanistically similar processes centred on pore formation by released peptides.
Strong evidence that peptides released from nonenveloped virus particles participate directly in membrane penetration raises a number of interesting questions. How does the virus ensure sufficient concentration(s) of the released peptides? How are the released peptides targeted to the membrane? How does the virus particle then ‘find' the membrane pore(s)? One simple way a virus may overcome these challenges is by sequestration in membrane-bounded compartments (e.g., endosomes or constricted cell-surface invaginations). In addition to the possibility that released peptides may become diluted or misdirected on the cell surface, or that contacts between virus particles and membranes may be lost, formation of pores on the cell surface may be detrimental to the cell. Indeed, it appears that internalization of virus particles prior to membrane penetration is a common requirement for nonenveloped viruses that release active peptides.
Materials and methods
Virus particles
Virions of reovirus T1L were purified by standard protocol (Furlong et al, 1988) and stored in virion buffer (VB) (150 mM NaCl, 10 mM MgCl2, 10 mM Tris–HCl, pH 7.5). For [35S]Met/Cys- or [3H]myristate-labelled virions, 7 mCi Tran35S label (ICN) or 10 mCi [3H]myristic acid (Perkin Elmer) was added at the start of infection. Using the latter protocol, Nibert et al (1991, 2005) have demonstrated that μ1, μ1δ, and μ1N are the only viral products that incorporate the [3H]myristate label. Recoated cores lacking σ1 were generated as described previously (Agosto et al, 2007) using baculovirus-expressed wild-type (Chandran et al, 1999) or N42A (Odegard et al, 2004) μ1. Nonpurified ISVPs and protease-treated recoated cores (WTprcs, N42Aprcs, [35S]Met/Cys-labelled ISVPs) were obtained by digesting virions in VB at 1.2 × 1013 particles/ml with 200 μg/ml chymotrypsin (Sigma-Aldrich) for 10–15 min at 37°C. Digestion was stopped with 2–5 mM phenylmethyl sulphonyl fluoride (Sigma-Aldrich) on ice. For purified ISVPs (unlabelled ISVPs, [3H]myristate-labelled ISVPs), chymotrypsin digestions were performed at 5 × 1012 particles/ml, and particles were purified on CsCl step gradients (1.3–1.5 g/cm3), which in some experiments included a 20% sucrose layer. Purified dpSVPs were prepared using the protocol for ISVPs, except that 1 mM sodium tetradecyl sulphate was included during chymotrypsin digestions, which was then precipitated on ice for 7 min and removed by two 1-min centrifugations at 16 000 g before particle purification.
RBCs and RGs
RBCs (Colorado Serum) and both unlabelled and dextran-loaded RGs were prepared as described previously (Agosto et al, 2006), and approximately 20% stock solutions were made in phosphate-buffered saline (PBS) supplemented with 2 mM MgCl2 (PBS-Mg). RBCs were always prepared freshly for the experiments, and RGs were stored for up to 2 weeks at 4°C.
RBC+, no-spin, and spin reactions
All types of reactions were performed in siliconized low-retention microtubes (Fisher Scientific). RBC+ reactions contained 4 × 1012 particles (ISVPs, dpSVPs, WTprcs, or N42Aprcs), 200 mM CsCl, 33% VB, 53% 10 mM Tris–HCl, pH 7.4, and approximately 7% RBC stock in a final volume of 30 μl. RBCs were omitted from no-spin and spin reactions. Times of preconversion for ISVPs, dpSVPs, and WTprcs were measured by incubating RBC+ reactions at 37°C and checking for RBC lysis every 30–45 s. Time of preconversion for N42Aprcs was measured by taking aliquots of RBC+ reactions every minute for several minutes to tubes with trypsin (Sigma-Aldrich) (final concentration 200 μg/ml in VB), incubating on ice for 30–40 min, and analysing by SDS–PAGE and Coomassie Blue staining. Preconversion times were typically <6 min. All post-conversion steps were performed quickly, and no more than six transfer reactions were performed in parallel at any time point. After preconversion, no-spin reactions were incubated at 4°C, while spin reactions were centrifuged at 16 000 g for 10 min also at 4°C.
Haemolysis. RBC+ reactions were incubated at 37°C and moved to ice at preconversion times. In no-spin or spin reactions, the top 25 μl were transferred to new tubes containing 2 μl of RBC stock and incubated at 37°C either for 1 min (ISVP supernatants) or 5 min (dpISVP, WTprc, and N42Aprc supernatants). Unlysed RBCs were removed by centrifugation at 380 g for 1 min, and 10 or 15 μl of the supernatant were added to 40 or 45 μl of VB. Absorbance at 415 nm (A415) was then measured in a microplate reader (Molecular Devices). Percentage haemolysis was defined as [(A415(sample)−A415(blank))/(A415(detergent)−A415(blank))] × 100, where blank lacked virus particles and detergent sample additionally contained 0.3% Triton X-100.
RG leakage. A 25 μl volume of spin supernatants were transferred to new tubes containing 3 μl RG stock and incubated at 37°C for 5 min. Three or 4 μl of each reaction were analysed by flow cytometry as described previously (Agosto et al, 2006). Data were analysed with CellQuest (BD Biosciences) and MFI softwares (E Martz, University of Massachusetts–Amherst, USA). Percentage dextran leakage was defined as {1-[(MI of sup 37-Bkgnd)/(MI of neg 37-Bkgnd)]} × 100, where MI stands for mean intensity, Bkgnd is the mean intensity of the unlabelled population in the sample kept on ice, and neg 37 is a sample incubated at 37°C in a mixture lacking virus-derived components.
Measurements of protein transfer
After removal of spin and no-spin aliquots for absorbance measurements, the fractions left in tubes and equivalent amounts of pellet fractions were boiled in gel sample buffer, and λ, δ, and σ1 protein contents were analysed by SDS–PAGE and either phosphorimaging ([35S]Met/Cys-labelled T1L ISVPs) or immunoblotting (purified unlabelled dpSVPs). For μ1N transfer, we used purified [3H]myristate-labelled ISVPs, performed each spin and no-spin reaction in duplicate, and analysed both supernatant and pellet fractions of one set not used in haemolysis by scintillation counting in a Beckman LS 600IC. For φ transfer using purified unlabelled ISVPs or nonpurified unlabelled N42Aprcs, we performed each reaction in triplicate, pooled and boiled in gel sample buffer two reactions that were not used in haemolysis and analysed them by SDS–PAGE, Coomassie Blue staining, and densitometry.
Associations with RGs
Association of released components with RGs. Spin reactions with RGs using [3H]myristate-labelled ISVPs were layered atop 400-μl Percoll gradients (25% Percoll (GE Healthcare), 1 × VB, and 400 mM CsCl) and centrifuged at 30 000 g for 15 min. 45-μl fractions were collected from the top as described previously (Agosto et al, 2006). Five-sixth of each fraction was analysed by scintillation counting, and the remainder by SDS–PAGE and either Coomassie Blue staining or immunoblotting.
Association of virus particles with RGs. Spin reactions were performed with RGs using either wild-type ISVP* supernatants or omitting particles altogether. Pore formation or its absence was verified by flow cytometry in each case (data not shown). N42Aprcs were then added at 2.5–4 × 1012 particles/ml and reactions were incubated at 37°C for an additional 30 min. They were then fractionated and analysed as described above for released components. Using SDS–PAGE and immunoblotting with anti-virion serum antibodies, the wild-type ISVP* particle clearance from initial spin reactions was verified in a portion of sample not incubated with N42A particles (data not shown).
Synthetic μ1N peptide
Haemolysis. Myristoylated μ1N peptide, consisting of aa 2–42 of T1L μ1, N-terminally N-myristoylated, was synthesized as will be described (LZ and SCH, submitted manuscript). Haemolysis reactions contained approximately 3% RBCs, 77% VB, 10% PBS-Mg, 400 mM CsCl, and 10% DMSO. Peptide was added last, from a 10 × stock solution made in 100% DMSO. Reactions were incubated at 37°C for 30 min, unlysed cells were pelleted by centrifugation at 380 g for 1–3 min, and 10 μl of supernatant was added to 90 μl of VB. Haemoglobin release was measured by A405, and percentage haemolysis was calculated as described above. Osmotic protection experiments were identical, except that reactions additionally contained 30 mM PEG (Sigma-Aldrich).
Particle docking. We used synthetic μ1N peptide or DMSO alone in similar reactions to those outlined above under Haemolysis, except that reactions contained approximately 2% unlabelled RGs in place of RBCs and had 200 mM CsCl. Following 30-min incubation at 37°C, N42Aprcs were added to each reaction at 2.5 × 1012 particles/ml and incubated at 37°C for additional 30 min. Reactions were fractionated on Percoll gradients and analysed as described above under Association of virus particles with RGs.
Anti-φ serum antibodies
DNA encoding T1L φ (μ1 aa 582–708) was cloned into pGEX-2T (GE Healthcare) in frame with glutathione-S-transferase (GST) (GST at the C terminus) and expressed in Escherichia coli strain BL21 DE3. Two hundred millilitre of cells were grown to an optical density of 0.6–0.9 at 600 nm, induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside for 4 h, pelleted, washed, and frozen. Bacterial pellets were lysed in Laemmli sample buffer. Total lysates were resolved by SDS–PAGE and stained with KCl. The φ/GST fusion protein band was excised and macerated. Protein was electroeluted and used to generate polyclonal antisera in rats. Two boosts were performed followed by a terminal bleed. Injections and bleeds were carried out at the Polyclonal Antibody Service at the University of Wisconsin-Madison Medical School, USA.
SDS–PAGE and immunoblotting
For detection of RG protein or particle-associated bands, we used 10% Tris/glycine gels. In experiments in which φ was also detected, step-gradient 10–16% Tris/tricine gels with 4% stacker were used. For detection of particle-associated bands, proteins were electrophoretically transferred to 0.45 μm nitrocellulose membranes (BioRad) for 1 h at 100 V. In experiments in which φ was also detected, small proteins were electrophoretically transferred to 0.2 μm polyvinylidene fluoride membranes (Whatman) overnight at 10 mA. After transfer of proteins for φ detection, membranes were fixed with 0.2% glutaraldehyde in TBS for 30 min and then washed extensively with TBS. For detection of particle-associated bands, antibody incubations were performed in 20 mM Tris–HCl (pH 7.5), 500 mM NaCl, 5% milk, 0.05% Tween-20. No Tween-20 was used in blots for φ detection. T1L virion-specific serum (anti-virion) (Virgin et al, 1988), φ/GST-specific serum (anti-φ), and T1L σ1-specific serum (anti-σ1) (Chandran et al, 2002) antibodies were all used at 1:1000 dilution. Rabbit-specific donkey IgG or rat-specific IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch) were used in secondary detection at 1:5000 dilution. Antibody binding was detected with Western Lightning chemiluminescence reagents and scanned on a Typhoon imager (GE Healthcare).
Acknowledgments
We thank Elaine Freimont for technical support, Aaron Engel for aid in making anti-φ serum, and Fernando Cruz-Guilloty, Gregg Espiritu Santo, and Amal Rahmeh for helpful discussions and comments on the paper. We are also very grateful to David King at the Howard Hughes Medical Institute, University of California-Berkeley, for μ1N peptide. We thank Branislav Ivanovic for drawings. This work was supported, in part, by NIH grants R01 AI46440 to MLN, R37 CA13202 to SCH, and F31 AI064142 to MAA. TI and MAA received earlier support from NIH training grants T32 AI07245 and T32 GM07226, respectively. SCH is an investigator of the Howard Hughes Medical Institute.
References
- Agosto MA, Ivanovic T, Nibert ML (2006) Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc Natl Acad Sci USA 103: 16496–16501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto MA, Middleton JK, Freimont EC, Yin J, Nibert ML (2007) Thermolabilizing pseudoreversions in reovirus outer-capsid protein μ1 rescue the entry defect conferred by a thermostabilizing mutation. J Virol 81: 7400–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana N, Yadav SP, Ghosh JK (2004) Dissection of antibacterial and toxic activity of melittin: a leucine zipper motif plays a crucial role in determining its hemolytic activity but not antibacterial activity. J Biol Chem 279: 55042–55050 [DOI] [PubMed] [Google Scholar]
- Baer GS, Dermody TS (1997) Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J Virol 71: 4921–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodkin DK, Nibert ML, Fields BN (1989) Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J Virol 63: 4676–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabec M, Schober D, Wagner E, Bayer N, Murphy RF, Blaas D, Fuchs R (2005) Opening of size-selective pores in endosomes during human rhinovirus serotype 2 in vivo uncoating monitored by single-organelle flow analysis. J Virol 79: 1008–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM (2007) Imaging poliovirus entry in live cells. PLoS Biol 5: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Farsetta DL, Nibert ML (2002) Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J Virol 76: 9920–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Nibert ML (1998) Protease cleavage of reovirus capsid protein μ1/μ1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J Virol 72: 467–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Nibert ML (2003) Animal cell invasion by a large nonenveloped virus: reovirus delivers the goods. Trends Microbiol 11: 374–382 [DOI] [PubMed] [Google Scholar]
- Chandran K, Parker JSL, Ehrlich M, Kirchhausen T, Nibert ML (2003) The δ region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J Virol 77: 13361–13375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Walker SB, Chen Y, Contreras CM, Schiff LA, Baker TS, Nibert ML (1999) In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J Virol 73: 3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sena J, Mandel B (1977) Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology 78: 554–566 [DOI] [PubMed] [Google Scholar]
- Earp LJ, Delos SE, Park HE, White JM (2005) The many mechanisms of viral membrane fusion proteins. Curr Top Microbiol Immnuol 285: 25–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GA, Zhang LG, Tattersall P (2005) Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc Natl Acad Sci USA 102: 17148–17153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald DJ, Trowbridge IS, Pastan I, Willingham MC (1983) Enhancement of toxicity of antitransferrin receptor antibody-Pseudomonas exotoxin conjugates by adenovirus. Proc Natl Acad Sci USA 80: 4134–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricks CE, Hogle JM (1990) Cell-induced conformational change in poliovirus: externalization of the N-terminus of VP1 is responsible for liposome binding. J Virol 64: 1934–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong DB, Nibert ML, Fields BN (1988) Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol 62: 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloux M, Libersou S, Morellet N, Bouaziz S, Da Costan B, Ouldali M, Lepault J, Delmas B (2007) Infectious bursal disease virus, a non-enveloped virus, possesses a capsid-associated peptide that deforms and perforates biological membranes. J Biol Chem 282: 20774–20784 [DOI] [PubMed] [Google Scholar]
- Geny B, Popoff MR (2006) Bacterial protein toxin and lipids: pore formation or toxin entry into cells. Biol Cell 98: 667–678 [DOI] [PubMed] [Google Scholar]
- Greber UF, Willetts M, Webster P, Helenius A (1993) Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75: 477–486 [DOI] [PubMed] [Google Scholar]
- Gromeier M, Wetz K (1990) Kinetics of poliovirus uncoating in HeLa cells in a nonacidic environment. J Virol 64: 3590–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle JM (2002) Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu Rev Microbiol 56: 677–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper J, Fields BN (1996a) Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J Virol 70: 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JW, Fields BN (1996b) Monoclonal antibodies to reovirus σ1 and μ1 proteins inhibit chromium release from mouse L cells. J Virol 70: 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovic T, Agosto MA, Chandran K, Nibert ML (2007) A role for molecular chaperone Hsc70 in reovirus outer capsid disassembly. J Biol Chem 282: 12210–12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuga S (1981) Pore size distribution analysis of gel substances by size exclusion chromatography. J Chromatogr 206: 449–461 [Google Scholar]
- Liemann S, Chandran K, Nibert ML, Harrison SC (2002) Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108: 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucia-Jandris P, Hooper JW, Fields BN (1993) Reovirus M2 gene is associated with chromium release from mouse L cells. J Virol 67: 5339–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier O, Boucke K, Hammer SV, Keller S, Stidwill RP, Hemmi S, Greber UF (2002) Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J Cell Biol 158: 1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MY, Boucke K, Suomalainen M, Stidwill RP, Greber UF (2000) The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J Virol 74: 7085–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert ML (1993) Structure and function of reovirus outer capsid proteins as they relate to early steps in infection. PhD thesis. Cambridge, MA, USA: Harvard University
- Nibert ML, Fields BN (1992) A carboxy-terminal fragment of protein μ1/μ1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol 66: 6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert ML, Odegard AL, Agosto MA, Chandran K, Schiff LA (2005) Putative autocleavage of reovirus μ1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J Mol Biol 345: 461–474 [DOI] [PubMed] [Google Scholar]
- Nibert ML, Schiff LA, Fields BN (1991) Mammalian reoviruses contain a myristoylated structural protein. J Virol 65: 1960–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard AL, Chandran K, Zhang X, Parker JSL, Baker TS, Nibert ML (2004) Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J Virol 78: 8732–8745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer R, Gerhardt P (1971) Molecular sieving by the Bacillus megaterium cell wall and protoplast. J Bacteriol 107: 718–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S, Palmer M (2003) Characterization of Streptococcus agalactiae CAMP factor as a pore-forming toxin. J Biol Chem 278: 38167–38173 [DOI] [PubMed] [Google Scholar]
- Silverstein SC, Schonberg M, Levin DH, Acs G (1970) The reovirus replicative cycle: conservation of parental RNA and protein. Proc Natl Acad Sci USA 67: 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Helenius A (2004) How viruses enter cells. Science 304: 237–241 [DOI] [PubMed] [Google Scholar]
- Sturzenbecker LJ, Nibert M, Furlong D, Fields BN (1987) Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol 61: 2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill TJ, Bubeck D, Rowlands DJ, Hogle JM (2006) Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J Virol 80: 172–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW IV, Bassel-Duby R, Fields BN, Tyler KL (1988) Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J Virol 62: 4594–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walukiewicz HE, Johnson JE, Scheemann A (2006) Morphological changes in the T=3 capsid of Flock House virus during cell entry. J Virol 80: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Hinz A, Gaudin Y (2007) Virus membrane fusion. FEBS Lett 581: 2150–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiethoff CM, Wodrich H, Gerace L, Nemerow GR (2005) Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol 79: 1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JAT, Collier RJ (2007) Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem 76: 243–265 [DOI] [PubMed] [Google Scholar]
- Zhang L, Chandran K, Nibert ML, Harrison SC (2006) Reovirus μ1 structural rearrangements that mediate membrane penetration. J Virol 80: 12367–12376 [DOI] [PMC free article] [PubMed] [Google Scholar]