Abstract
Signal transduction pathways that mediate activation of serum response factor (SRF) by heterotrimeric G protein α subunits were characterized in transfection systems. Gαq, Gα12, and Gα13, but not Gαi, activate SRF through RhoA. When Gαq, α12, or α13 were coexpressed with a Rho-specific guanine nucleotide exchange factor GEF115, Gα13, but not Gαq or Gα12, showed synergistic activation of SRF with GEF115. The synergy between Gα13 and GEF115 depends on the N-terminal part of GEF115, and there was no synergistic effect between Gα13 and another Rho-specific exchange factor Lbc. In addition, the Dbl-homology (DH)-domain-deletion mutant of GEF115 inhibited Gα13- and Gα12-induced, but not GEF115 itself- or Gαq-induced, SRF activation. The DH-domain-deletion mutant also suppressed thrombin- and lysophosphatidic acid-induced SRF activation in NIH 3T3 cells, probably by inhibition of Gα12/13. The N-terminal part of GEF115 contains a sequence motif that is homologous to the regulator of G protein signaling (RGS) domain of RGS12. RGS12 can inhibit both Gα12 and Gα13. Thus, the inhibition of Gα12/13 by the DH-deletion mutant may be due to the RGS activity of the mutant. The synergism between Gα13 and GEF115 indicates that GEF115 mediates Gα13-induced activation of Rho and SRF.
Four classes of G protein α subunits—Gαs, Gαi, Gαq, and Gα12 (1)—are involved in signal transduction of various hormones, neurotransmitters, and many other biologically active molecules such as lysophosphatidic acid (LPA) and thrombin (2–4). The Gαs subunits and Gαi subunits regulate adenyl cyclase activities, and the Gαq subunits regulate phospholipase C activities (5, 6). However, the direct effectors for the Gα12 class of G proteins, which includes Gα12 and Gα13 (7), remains to be elucidated. Activated forms of Gα12 and Gα13 were shown to induce transformation phenotypes when transfected into fibroblasts, suggesting that they are involved in regulation of cell growth (8–10). In addition, Gα12 and Gα13 were shown to induce formation of stress fibers in fibroblast cells and apoptosis through the small G protein RhoA (11, 12). This observation was supported by the report that Gα12 activated serum response factor (SRF) through RhoA (13). Moreover, a study using mice that lack Gα13 indicates that Gα13 is involved in the function of endothelial cells because mice lacking Gα13 are embryonic lethal apparently due to the failure to develop vasculature (14). In this study, thrombin-mediated chemotaxis of fibroblasts lacking Gα13 was blocked, indicating that the thrombin receptor couples to Gα13. This is consistent with the observation that thrombin could stimulate the binding of a photoaffinity GTP analog to Gα13 (15).
In this report, we describe the involvement of a Rho-specific guanine nucleotide exchange factor, GEF115 (16), in Gα13- but not Gα12- or Gαq-mediated SRF activation. We found that the N-terminal portion of GEF115, which contains a region homologous to the regulator of G protein signaling (RGS) domain of the RGS12 protein, is required for mediating Gα13-induced SRF activation. Moreover, both RGS12 and a GEF115 mutant lacking the Dbl-homology (DH) domain were able to inhibit Gα13 and Gα12 but not Gαq function.
METHODS
Cell Culture and Transfection.
COS-7 and NIH 3T3 cells were maintained in DMEM containing 10% fetal calf serum at 37°C under 5% CO2/95% air. For transfection, cells (5 × 104 cells per well) were seeded into 24-well plates the day before transfection. Cells were transfected with 0.25 μg of DNA per well for COS-7 cells and 0.5 μg of DNA per well for NIH 3T3 cells by using Lipofectamine Plus (Life Technologies), as suggested by the manufacturer. The transfection was stopped after 3 hr by switching to culture medium containing 0.5% fetal bovine serum. Cell extracts were collected 24 hr later for luciferase assays, kinase assays, and Western blot analysis.
Construct.
All of the G protein subunits and receptors were in pCMV expression vectors as described (8, 16, 17). The SRE.L-luciferase reporter plasmid was constructed as described (18), except the luciferase gene was used as the reporter instead of the chloramphenicol acetyltransferase gene.
Luciferase Assay.
Luciferase assays were performed with Boehringer Mannheim Constant Light luciferase assay kit as instructed. Transfection efficiency was normalized by quantifying the fluorescence intensity emitted by cotransfected green fluorescence protein (GFP) using a Wallac multi-counter. The Wallac counter (Wallac, Gaithersburg, MD) is capable of counting both fluorescence and luminescence. The luciferase substrate was then added to the cell lysates, and luciferase activities were determined by measuring luminescence intensity using the same counter. Luminescence intensities were normalized against fluorescence intensities. DNA concentrations were adjusted if transfection of any of the cDNAs resulted in significant differences between normalized and nonnormalized data.
RESULTS AND DISCUSSION
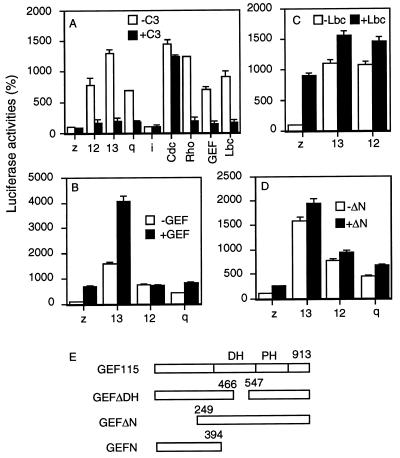
To define the components that mediate Gα13-induced RhoA activation, a system was established for assaying Gα12/13-induced Rho-dependent activation of SRF. SRF activation was evaluated by determining SRF-dependent transcriptional activation of a luciferase reporter gene, the transcription of which is controlled by a transcription regulatory sequence element, called SRE.L. SRE.L is a derivative of c-fos serum response element (SRE), to which SRF but not tertiary complex factor binds (18). Thus, SRE.L-mediated production of luciferase mainly depends on the activity of SRF. Expression of activated forms of Gα12, Gα13, and Gαq markedly stimulated the production of the reporter luciferase when G proteins were cotransfected with the reporter gene construct (Fig. 1A). G protein-mediated SRF activation was sensitive to coexpression of Clostridium butulinum C3 transferase (C3), suggesting that the SRF activation is mediated by the small GTP-binding protein RhoA (Fig. 1). C3 is a specific RhoA inactivator that ADP ribosylates RhoA (18). This result is consistent with previous findings that Gα12, Gα13, and Gαq activate RhoA (11, 13, 19–21). Expression of GEF115 (16) and Lbc (22, 23) also stimulated SRE.L-dependent transcription, and C3 was able to block the stimulation (Fig. 1A). These results are consistent with the idea that GEF and Lbc are Rho-specific exchange factors (16, 22, 23). Furthermore, C3 did not significantly inhibit Cdc42-mediated activation of SRF but clearly inhibited Rho-mediated SRF activation (Fig. 1A), confirming that C3 acts specifically in our system.
Figure 1.
Regulation of SRF-mediated gene transcription by G proteins and guanine nucleotide exchange factors. (A) NIH 3T3 cells were cotransfected with 0.1 μg of SRE.L-luciferase reporter plasmid, 0.1 μg of GFP expression construct, and 0.1 μg of activated Cdc42V12 or RhoV14, Lbc, GEF115, or 0.01 μg of various activated forms of G protein subunits in the presence (solid bar) or absence (open bar) of 0.02 μg of C3 expression plasmid. (B) NIH 3T3 cells were cotransfected with 0.1 μg of SRE.L-luciferase reporter plasmid, 0.1 μg of GFP expression construct, and 0.01 μg of activated Gαq, Gα12 or Gα13 in the presence (solid bar) or absence (open bar) of 0.05 μg of GEF115 (GEF). (C) NIH 3T3 cells were cotransfected with 0.1 μg of SRE.L-luciferase reporter plasmid, 0.1 μg of GFP expression construct, and 0.01 μg of activated Gα12 or Gα13 in the presence (solid bar) or absence (open bar) of 0.05 μg of Lbc. (D) NIH 3T3 cells were cotransfected with 0.1 μg of SRE.L-luciferase reporter plasmid, 0.1 μg of GFP expression construct, and 0.01 μg of activated Gαq, Gα12, or Gα13 in the presence (solid bar) or absence (open bar) of 0.05 μg of GEFΔN. LacZ (z) expression plasmid was used to make the total amount of DNA equal (0.5 μg per well) in all transfection. One day later, cells were lysed, and GFP levels and luciferase activity were determined. The luciferase activities presented are normalized against the levels of GFP expression. Data show similar tendencies with or without normalization. Experiments were carried out in triplicate and repeated at least twice. The representative experiments are shown, and error bars represent standard derivations. (E) Schematic representation of GEF115 and its mutants.
To test whether G protein α subunits can activate GEF115 and Lbc, NIH 3T3 cells were cotransfected with cDNA encoding activated Gα12, Gα13, or Gαq and cDNA encoding GEF115 or Lbc in the presence of the SRE.L reporter gene plasmid. Cells coexpressing Gα13 and GEF115 produced significantly more reporter gene products than the sum of those produced by cells separately transfected with the G protein and GEF115 cDNA (Fig. 1B); i.e., there is a synergistic effect between Gα13 and GEF115 in activation of SRF. This synergistic effect between Gα13 and GEF115 is not due to any increase in the expression levels of Gα13 or GEF115 during the cotransfection, because cells expressing Gα13 alone or coexpressing Gα13 and GEF115 showed similar levels of Gα13 (Fig. 2C) or GEF115 (data not shown). Such synergistic effects do not appear to exist between Gα12 or Gαq and GEF115 (Fig. 1B). The luciferase activities in cells cotransfected with Gαq/α12 and GEF115 are approximately equal or less than the sum produced by cells transfected with Gαq/α12 and GEF115 separately. Synergistic effects were also not observed between Lbc and Gα13 or Gα12 (Fig. 1C). All these results indicate that Gα13 but not Gα12 or Gαq can lead to specific activation of GEF115.
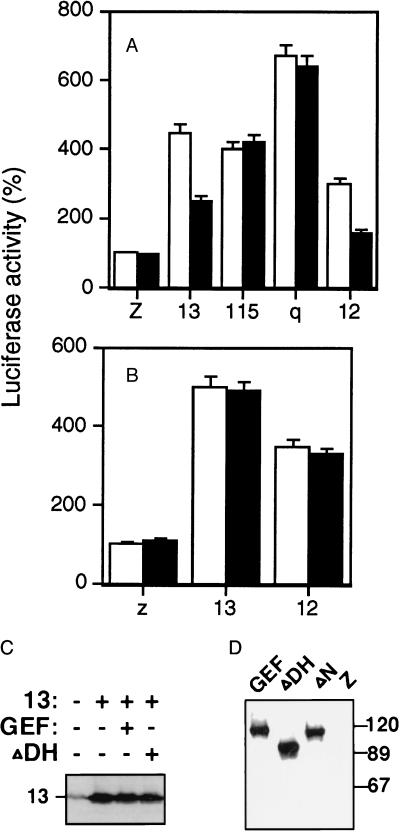
Figure 2.
Effect of GEFΔDH on G protein-mediated SRF activation. NIH 3T3 cells were cotransfected with 0.1 μg of SRE.L-luciferase reporter plasmid, 0.1 μg of GFP expression construct, and 0.05 μg of various wild-type forms of G protein subunits in the presence (solid bar) or absence (open bar) of 0.2 μg of GEFΔDH (A) or LbcΔDH (B). LacZ (z) expression plasmid was used to make the total amount of DNA equal (0.5 μg per well) in all transfections. Assays were carried out as described in Fig. 1. The expression of Gα13 was detected by an antibody specific to Gα13 (C), and GEF115 and its mutants were detected with an anti-myc antibody that recognizes the myc-tag in these proteins (D).
To investigate which structural domains of GEF115 are required for SRF activation, two deletion mutants of GEF115 were tested. GEFΔN contains an N-terminal deletion of residues 1–248, and GEFDΔH contains a deletion in its DH domain from residues 466 to 547 (16). The expression of the wild-type GEF115 and its mutants are shown in Fig. 2D. When GEFΔN was cotransfected with the SRE.L reporter gene plasmid into NIH 3T3 cells, GEFΔN showed about 2-fold stimulation of luciferase activity. However, when GEFΔN was coexpressed with Gα13, there were no synergistic effects between GEFΔN and Gα13 (Fig. 1D). Neither was there any synergism between GEFΔN and Gα12 or Gαq in activation of SRF (Fig. 1D). Therefore, the N-terminal part of GEF115 is required for activation by Gα13.
When GEFΔDH was expressed with the reporter gene plasmid, this mutant lost its ability to stimulate SRF-dependent transcription (Fig. 2A). This result is consistent with previous findings that the DH domain is involved in promoting guanine nucleotide exchange activity of small G proteins (16, 23, 24). The requirement of the N-terminal part of the GEF115 molecule for activation by Gα13 suggests that the DH-deletion mutant may function as a dominant-negative mutant, because GEFΔDH lacks the effector activation ability but seems to be able to interact with upstream regulators. To test whether GEFΔDH is capable of inhibiting Gα13-mediated SRF activation, GEFΔDH was coexpressed with the wild-type Gα13 in 3T3 cells. Cells coexpressing Gα13 and GEFΔDH showed approximately 50% of the luciferase activity compared with those expressing Gα13 alone (Fig. 2A). The expression of GEFΔDH did not change the expression level of Gα13 (Fig. 2C). Thus, the inhibition by GEFΔDH cannot be attributed to changes in the expression levels of Gα13. GEFΔDH showed no inhibitory effect on GEF115- or Gαq-mediated SRF activation (Fig. 2A), suggesting that GEFΔDH does not inhibit GEF115 downstream proteins. This result also confirms the idea that Gαq may act through pathways other than GEF115. Thus, the observations that Gα13 activates SRF synergistically with GEF115 and that GEFΔDH inhibits Gα13-mediated SRF activation indicate that GEF115 is involved in activation of SRF by Gα13 in NIH 3T3 cells. The inability of the Lbc mutant that lacks its DH domain (deletion of residues 76–295) to inhibit SRF activation by Gα13 or Gα12 (Fig. 2B) further confirms the findings that Lbc is not involved in SRF activation by Gα13 or Gα12. Surprisingly, GEFΔDH seems to be a potent inhibitor of Gα12 (Fig. 2A). Because Gα12 showed no synergistic effect with GEF115 in activation of SRF when they were coexpressed, it is unlikely that Gα12 acts through GEF115 in activation of RhoA and SRF.
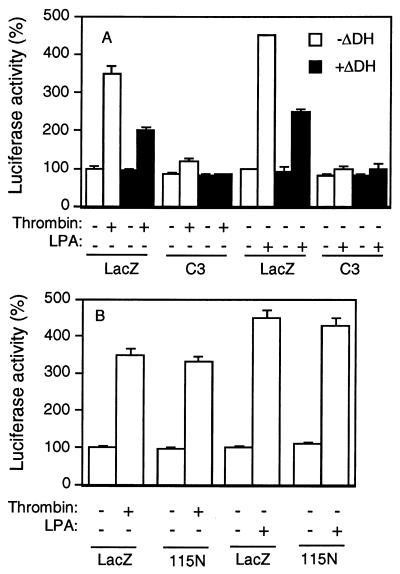
The effect of GEFΔDH on receptor-mediated activation of SRF was also investigated. Both the thrombin receptor and the lysophosphatidic acid (LPA) receptor have been shown to couple to Gα12 and Gα13 (14, 15, 25). The thrombin-receptor-mediated biological effect was also shown to depend on Gα13, because fibroblasts derived from Gα13-deficient mice showed diminished ability to respond to thrombin (14). NIH 3T3 cells presumably contain endogenous thrombin receptors and LPA receptors because LPA or thrombin can induce SRE.L-mediated transcription in cells transfected with the reporter plasmid. The fact that LPA- and thrombin-mediated SRF activation is sensitive to C3 suggests that these receptors act through RhoA. Cells cotransfected with the GEFΔDH cDNA and the reporter plasmid showed less luciferase activity in response to thrombin or LPA than those transfected with lacZ and the reporter gene (Fig. 3A). Expression of the N-terminal part of GEF115, termed GEF115N, did not inhibit thrombin- or LPA-induced SRF activation (Fig. 3B), suggesting that the sequences downstream of the DH domain are important for the inhibitory function of GEFΔDH. GEF115N did not inhibit Gα12- or Gα13-mediated SRF activation as well (data not shown). Both thrombin and LPA receptors were able to stimulate inositol phosphate accumulation, suggesting possible coupling of these receptors to Gq in addition to G12/13. The observation that GEFΔDH could not inhibit Gαq-mediated SRF activation (Fig. 2A) implies that GEFΔDH is likely to specifically inhibit Gα12/13 in these cells.
Figure 3.
Inhibition of receptor-mediated SRF activation by GEFΔDH. NIH 3T3 cells were cotransfected with 0.1 μg of SRE.L-luciferase reporter plasmid, 0.1 μg of GFP expression construct, and 0.2 μg of GEFΔDH (ΔDH) (A) or GEF115N (B) with or without 0.02 μg of C3 expression plasmid as indicated. LacZ (z) expression plasmid was used to make the total amount of DNA equal (0.5 μg per well) in all transfections. The next day, cells were lysed 6 hr after the addition of ligands of thrombin (10 units/ml) or 5 μM LPA. Data are processed and presented as described in Fig. 1.
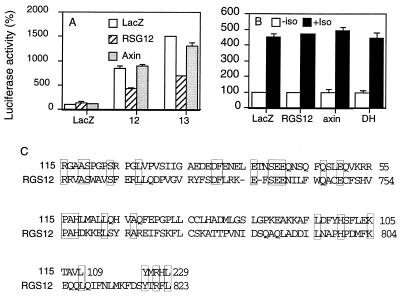
To provide an explanation for inhibition of Gα12/13 by GEFΔDH, we compared the amino acid sequences of RGS12 and GEF115. RGS proteins are a group of proteins that share homologous sequence motifs, the RGS domains, and inhibit G protein function by promoting GTPase activity of the Gα subunits (26, 27). RGS12 (28) inhibits both Gα12 and Gα13 (Fig. 4A) whereas it did not inhibit Gαs-mediated effects (Fig. 4B). Another RGS-domain-containing protein, Axin, did not inhibit either Gα12 or Gα13 (Fig. 4A). Thus, it appears that GEFΔDH behaved similarly to RGS12 because GEFΔDH also inhibited both Gα12 and Gα13 but did not inhibit Gαs-mediated effects (Fig. 4B). The sequence comparison between GEF115 and RGS12 revealed that the N-terminal region of GEF115 contains a motif that is homologous with the RGS domain of RGS12 (Fig. 4C). Some of the amino acids identical between RGS12 and GEF115 are also conserved in other RGS proteins (Fig. 4C). Therefore, inhibition of Gα12/13 by GEF115ΔDH may be attributed to the RGS motif of GEF115. This idea is supported by the observation that GEFΔN showed no inhibitory effects on Gα12 or Gα13 (Fig. 1D).
Figure 4.
N-terminal part of GEF115 contains a RGS motif. (A) NIH 3T3 cells were cotransfected with 0.15 μg of SRE.L-luciferase reporter plasmid, 0.15 μg of GFP expression construct, and 0.2 μg of LacZ, Axin or RGS12 in the presence or absence of 0.05 μg of wild-type Gα12 or Gα13 subunits. One day later cells were lysed, and GFP levels and luciferase activity were determined as in Fig. 1. (B) NIH 3T3 cells were cotransfected with 0.15 μg of CRE-luciferase reporter plasmid, 0.15 μg of GFP expression construct, and 0.2 μg of LacZ, GEF115ΔDH or RGS expression plasmids. Isoprenaline (10 μM) was added the next day for 6 hr. Then GFP levels and luciferase activity were determined as in Fig. 1. (C) The amino acid sequences of GEF115 and RGS12 were compared by using a computer program macaw. Homologous regions were shown. Identical amino acids between RGS12 and GEF115 are shown in boxes.
In this report, we describe the discovery of a Rho-specific guanine nucleotide exchange factor GEF115 to mediate Gα13-induced activation of Rho and SRF in NIH 3T3 cells. The evidence shows that GEF115 acted synergistically with Gα13 in activation of SRF in a C3-sensitive manner and that a GEF115 mutant lacking the DH domain inhibited Gα13-induced SRF activation. The mechanism by which Gα13 regulates GEF115 remains unclear. However, the activity requires the N-terminal part of the GEF115 molecule. GEF115 did not act synergistically with Gα12 in activation of SRF although GEFΔDH inhibited Gα12-induced SRF activation. The inhibition may be due to the RGS activity of GEFΔDH. Thus, with the facts that GEF115 did not act synergistically with Gαq in SRF activation and that GEFΔDH showed no inhibitory effect on Gαq-mediated SRF activation, GEF115 is unlikely to be involved in regulation of RhoA and SRF by Gα12 and Gαq. These two G proteins may act through different guanine nucleotide exchange factors or different pathways. Recent studies indicate that Gα12 and Gαq may act through the Tec family of nonreceptor tyrosine kinases to regulate RhoA and SRF (20, 29).
Our data are largely consistent with two recent reports demonstrating that GEF115 acts as GTPase-activating protein for both Gα12 and Gα13 but that only Gα13 stimulates the nucleotide exchange activity of GEF115 (30, 31). Similarity between RGS proteins and the N-terminal portion of GEF115 was also noted. The sequence comparison was made based on secondary structure predictions in these reports (30, 31), whereas we made comparisons based on the primary sequences. Thus, different sequence alignments are produced although both methods point at a similar regions of GEF115 and RGS12 molecules. We are investigating whether our prediction of the involvement of the RGS homologous sequences of GEF115 in regulating Gα13 activation of RhoA is correct by using the site-directed mutagenesis approach.
Acknowledgments
We thank Lisa Estey and Huiping Jiang for reading the manuscript. We also thank Alan Hall, Solvio Gutkind, Dennis Toksoz, and Matthrew Hart for providing cDNAs. This work is supported by grants to D.W. from NIH (GM53162 and GM54167) and from National Heart Association.
ABBREVIATIONS
- DH
Dbl homology
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescence protein
- LPA
lysophosphatidic acid
- RGS
regulator of G protein signaling
- SRE
serum response element
- SRF
serum response factor
References
- 1. Simon M I, Strathman M P, Gautum M. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaumer L, Birnbaumer M. J Recept Signal Transduct Res. 1995;15:213–252. doi: 10.3109/10799899509045218. [DOI] [PubMed] [Google Scholar]
- 3.Gilman A G. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 4.Bourne H R. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 5.Hepler J R, Gilman A G. Trends Biochem Sci. 1992;17:383–387. doi: 10.1016/0968-0004(92)90005-t. [DOI] [PubMed] [Google Scholar]
- 6.Neer E J. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 7.Strathmann M, Simon M I. Proc Natl Acad Sci USA. 1990;87:9113–9117. doi: 10.1073/pnas.87.23.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H, Wu D, Simon M I. FEBS Lett. 1993;330:319–322. doi: 10.1016/0014-5793(93)80896-3. [DOI] [PubMed] [Google Scholar]
- 9.Xu N, Voyno-Yasenetskaya T, Gutkind J S. Biochem Biophys Res Commun. 1994;201:603–609. doi: 10.1006/bbrc.1994.1744. [DOI] [PubMed] [Google Scholar]
- 10.Voyno-Yasenetskaya T A, Pace A M, Bourne H R. Oncogene. 1994;9:2559–2565. [PubMed] [Google Scholar]
- 11.Buhl A M, Johnson N L, Dhanasekaran N, Johnson G L. J Biol Chem. 1995;270:24631–24634. doi: 10.1074/jbc.270.42.24631. [DOI] [PubMed] [Google Scholar]
- 12.Althoefer H, Eversole-Cire P, Simon M I. J Biol Chem. 1997;272:24380–24386. doi: 10.1074/jbc.272.39.24380. [DOI] [PubMed] [Google Scholar]
- 13.Fromm C, Coso O A, Montaner S, Xu N, Gutkind J S. Proc Natl Acad Sci USA. 1997;94:10098–10103. doi: 10.1073/pnas.94.19.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Offermanns S, Mancino V, Revel J P, Simon M I. Science. 1997;275:533–536. doi: 10.1126/science.275.5299.533. [DOI] [PubMed] [Google Scholar]
- 15.Offermanns S, Laugwitz K L, Spicher K, Schultz G. Proc Natl Acad Sci USA. 1994;91:504–508. doi: 10.1073/pnas.91.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hart M J, Sharma S, elMasry N, Qiu R G, McCabe P, Polakis P, Bollag G. J Biol Chem. 1996;271:25452–25458. doi: 10.1074/jbc.271.41.25452. [DOI] [PubMed] [Google Scholar]
- 17.Wu D, Lee C-H, Rhee S G, Simon M I. J Biol Chem. 1992;267:1811–1817. [PubMed] [Google Scholar]
- 18.Hill C S, Wynne J, Treisman R. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 19.Sah V P, Hoshijima M, Chien K R, Brown J H. J Biol Chem. 1996;271:31185–311890. doi: 10.1074/jbc.271.49.31185. [DOI] [PubMed] [Google Scholar]
- 20.Mao, J., Xie, W., Yuan, H., Simon, M. I., Mano, H. & Wu, D. (1998) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 21.Mao, J., Yuan, H., Xie, W. & Wu, D. (1998) J. Biol. Chem., in press. [DOI] [PubMed]
- 22.Toksoz D, Williams D A. Oncogene. 1994;9:621–628. [PubMed] [Google Scholar]
- 23.Zheng Y, Olson M F, Hall A, Cerione R A, Toksoz D. J Biol Chem. 1995;270:9031–9034. doi: 10.1074/jbc.270.16.9031. [DOI] [PubMed] [Google Scholar]
- 24.Olson M F, Sterpetti P, Nagata K, Toksoz D, Hall A. Oncogene. 1997;15:2827–2831. doi: 10.1038/sj.onc.1201594. [DOI] [PubMed] [Google Scholar]
- 25.Gohla A, Harhammer R, Schultz G. J Biol Chem. 1998;273:4653–4659. doi: 10.1074/jbc.273.8.4653. [DOI] [PubMed] [Google Scholar]
- 26.Dohlman H G, Thorner J. J Biol Chem. 1997;272:3871–3874. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 27.Koelle M R. Curr Opin Cell Biol. 1997;9:143–147. doi: 10.1016/s0955-0674(97)80055-5. [DOI] [PubMed] [Google Scholar]
- 28.Snow B E, Antonio L, Suggs S, Gutstein H B, Siderovski D P. Biochem Biophys Res Commun. 1997;233:770–777. doi: 10.1006/bbrc.1997.6537. [DOI] [PubMed] [Google Scholar]
- 29.Bence K, Ma W, Kozasa T, Huang X Y. Nature (London) 1997;389:296–299. doi: 10.1038/38520. [DOI] [PubMed] [Google Scholar]
- 30.Kozasa T, Jiang X J, Hart M J, Sternweis P M, Singer W D, Gilman A G, Bollag G, Sternweis P C. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 31.Hart M J, Jiang X J, Kozasa T, Roscoe W, Singer W D, Gilman A G, Sternweis P C, Bollag G. Science. 1998;280:2112–2114. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]