Abstract
Heteroplasmic mitochondrial DNA (mtDNA) mutations (mutations present only in a subset of cellular mtDNA copies) arise de novo during the normal ageing process or may be maternally inherited in pedigrees with mitochondrial disease syndromes. A pathogenic mtDNA mutation causes respiratory chain deficiency only if the fraction of mutated mtDNA exceeds a certain threshold level. These mutations often undergo apparently random mitotic segregation and the levels of normal and mutated mtDNA can vary considerably between cells of the same tissue. In human ageing, segregation of somatic mtDNA mutations leads to mosaic respiratory chain deficiency in a variety of tissues, such as brain, heart and skeletal muscle. A similar pattern of mutation segregation with mosaic respiratory chain deficiency is seen in patients with mitochondrial disease syndromes caused by inherited pathogenic mtDNA mutations. We have experimentally addressed the role of mosaic respiratory chain deficiency in ageing and mitochondrial disease by creating mouse chimeras with a mixture of normal and respiratory chain-deficient neurons in cerebral cortex. We report here that a low proportion (>20%) of respiratory chain-deficient neurons in the forebrain are sufficient to cause symptoms, whereas premature death of the animal occurs only if the proportion is high (>60–80%). The presence of neurons with normal respiratory chain function does not only prevent mortality but also delays the age at which onset of disease symptoms occur. Unexpectedly, respiratory chain-deficient neurons have adverse effect on normal adjacent neurons and induce trans-neuronal degeneration. In summary, our study defines the minimal threshold level of respiratory chain-deficient neurons needed to cause symptoms and also demonstrate that neurons with normal respiratory chain function ameliorate disease progression. Finally, we show that respiratory chain-deficient neurons induce death of normal neurons by a trans-neuronal degeneration mechanism. These findings provide novel insights into the pathogenesis of mosaic respiratory chain deficiency in ageing and mitochondrial disease.
INTRODUCTION
Mutations of mitochondrial DNA (mtDNA) are common causes of human genetic disease (1,2) and are also heavily implicated in the ageing process (3,4). Heteroplasmic mutations of mtDNA typically undergo an apparently random segregation in somatic tissues and they will impair respiratory chain function if they are present above a certain threshold level (5). This uneven distribution of mutated mtDNA among cells in a tissue often creates a mosaic pattern of respiratory chain deficiency in patients with mitochondrial diseases caused by inherited pathogenic mtDNA mutations.
It has been recognized for almost two decades that acquired mtDNA mutations (6–11) and decreased respiratory chain capacity (12–14) are associated with mammalian ageing. Experimental studies of mtDNA mutator mice expressing a proof-reading-deficient mtDNA polymerase have shown that high levels of somatic mtDNA mutations in the mouse impair respiratory chain function and cause a variety of premature ageing phenotypes, such as osteoporosis, hair loss, alopecia, lean-ness, weight loss, reduced fertility and heart enlargement (15). Somatic mtDNA mutations thus have the capacity to cause premature ageing phenotypes, but it has been questioned whether the levels observed in normal ageing are sufficient to impair respiratory chain function (16). When discussing this issue, it is important to recognize that somatic mtDNA mutations are subject to the same apparently random mitotic segregation as inherited mtDNA mutations (5). Ageing humans invariably develop focal respiratory chain deficiency (14,17–19) in a variety of tissues, such as heart (20), skeletal muscle (21), colon (19) and brain (18, 22,23). There is good evidence to support the hypothesis that clonal expansion of somatic mtDNA mutations is the main factor explaining this phenomenon (14,17–19).
The pathogenesis of focal respiratory chain deficiency is poorly understood and the fraction of respiratory chain-deficient cells that need to be present in an organ in order to cause symptoms has not been previously defined. In this study, we generated chimeras between normal mice and mitochondrial late-onset neurodegeneration (MILON) mice (24) to create mice with varying proportions of focal respiratory chain deficiency in forebrain neurons. The MILON mice have a conditional knockout for mitochondrial transcription factor A (Tfam) in neocortex and medial parts of hippocampus, which leads to adult onset of progressive respiratory chain deficiency in large forebrain neurons, neurodegeneration and premature death (24). Characterization of a large number of mouse chimeras show that a rather low proportion (>20%) of respiratory chain-deficient neurons in the forebrain is sufficient to cause clinical symptoms, whereas premature death of the animal occurs only if the proportion is high (>60–80%). The presence of normal neurons leads to later onset of disease and reduced mortality. Unexpectedly, respiratory chain-deficient neurons induce degeneration of normal adjacent neurons by a trans-neuronal mechanism. These findings shed new light on the pathogenesis of focal respiratory chain deficiency in mitochondrial disease and ageing.
RESULTS
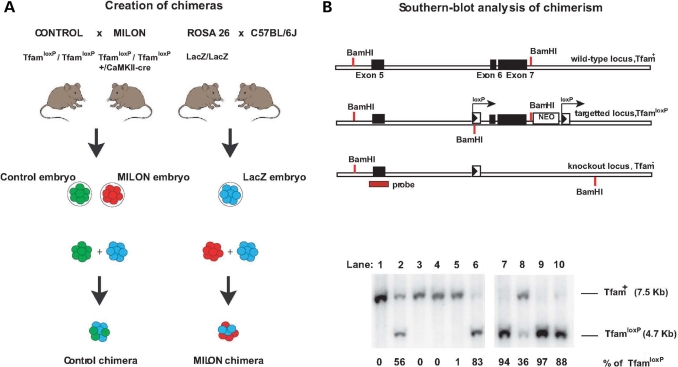
We used an embryo aggregation procedure to generate mouse chimeras between LacZ embryos and MILON embryos (Fig. 1A). The LacZ embryos are heterozygous offspring from the ROSA26 mice and express LacZ ubiquitously (25,26). The MILON mice are conditional knockouts with postnatal inactivation of the gene encoding mitochondrial transcription factor A (Tfam) in neurons of neocortex and medial parts of the hippocampus (24). Conditional knockout of TFAM causes mtDNA depletion, severely reduced mtDNA expression and a profound respiratory chain deficiency (24,27,28), because the TFAM protein is critical for maintenance (29,30) and transcription of mtDNA (31). We isolated a total of ∼103 LacZ embryos and aggregated them with ∼103 embryos that were either control embryos or MILON embryos (Fig. 1A). We obtained 781 fused embryos and 501 of these developed to at least the eight-cell stage in vitro and were subsequently implanted into pseudopregnant foster mothers. A total of 216 pups were born and genotyped by polymerase chain reaction (PCR) analysis of tail DNA. We excluded 84 animals because of non-chimeric genotype (n = 71) or because of inconsistent results from repeated genotyping (n = 13). We thus obtained a total of 132 chimeric mice, which were either MILON chimeras (n = 66) or control chimeras (n = 66), and these chimeras were used for the subsequent studies.
Figure 1.
Creation of mouse chimeras. (A) Control mice were crossed with mitochondrial late-onset neurodegeneration (MILON) mice to generate control embryos (genotype TfamloxP /TfamloxP, indicated by green color) and MILON embryos (genotype TfamloxP/TfamloxP, +/CaMKII-cre, indicated by red color). Homozygous ROSA26 mice (LacZ/LacZ) were crossed with C57BL/6J mice to generate LacZ embryos (genotype +/LacZ, indicated by blue color). Embryo aggregations were performed by mixing control embryos and LacZ embryos or MILON embryos and LacZ embryos to generate control chimeras and MILON chimeras. (B) Genotyping of mouse chimeras using Southern blot analysis. DNA was digested with the restriction endonuclease BamHI and Southern blots were hybridized with a radioactively labeled probe covering the genomic regions surrounding exon 5 of the Tfam gene. The wild-type allele (Tfam+) corresponds to a fragment of 7.5 kb, the non-recombined loxP-flanked allele (TfamloxP) corresponds to a fragment of 4.7 kb and the knock-out allele (Tfam−) corresponds to a fragment of 9.5 kb (upper panel). Genotyping of cerebellar DNA from chimeras was performed to assess levels of chimerism (lower panel). The wild-type Tfam allele (Tfam+) corresponds to cells derived from the LacZ embryo. The loxP-flanked Tfam allele (TfamloxP) corresponds to cells derived from either control embryos or MILON embryos. Additional genotyping to determine the presence of the CaMKII-cre transgene was performed to distinguish between chimeras containing control or MILON cells. The knockout Tfam allele (Tfam−) is not present in cerebellum as cells derived from MILON embryos only express cre-recombinase in neocortex and hippocampus. The ratio between Tfam+ and TfamloxP alleles in cerebellum of individual chimeras thus corresponds to the level of chimerism, expressed as the percentage of TfamloxP alleles.
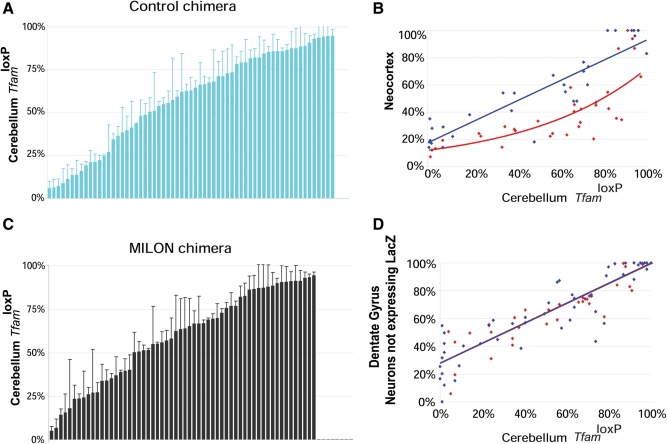
We used a Southern blot assay to quantify chimera composition based on the fact that cells derived from control (genotype TfamloxP/TfamloxP) or MILON embryos (genotype TfamloxP/TfamloxP, +/CamKII-cre) are homozygous for the TfamloxP allele, whereas cells derived from LacZ embryos are homozygous for the wild-type (Tfam+) allele (Fig. 1B and C). The relative amount of TfamloxP alleles thus reflects the contribution of control or MILON cells to the chimeras (Fig. 1B). The composition of the chimeras was assessed by Southern blot analyses of cerebellum as this region of the brain is unaffected by the neuron-specific Tfam knockout and the subsequent nerve cell loss occurring in MILON mice (24). Chimeras had an even distribution of various levels of control (Fig. 2A) or MILON cells (Fig. 2B) in cerebellum. Next, we investigated whether the grade of chimerism in cerebellum, as determined by Southern blot analyses, was representative of the grade of chimerism, as determined by histochemical staining for LacZ enzyme activity, in neurons of neocortex and the dentate gyrus (Fig. 2C and D). The control or MILON cell contribution in neocortex and the dentate gyrus was defined as the proportion of neurons that did not express LacZ. In control chimeras there was a linear correlation between the grade of chimerism in cerebellum and neocortex (Fig. 2C) or the dentate gyrus (Fig. 2D). We thus conclude that the observed chimerism in cerebellum is a good indicator of the grade of chimerism in neocortex and the dentate gyrus of controls.
Figure 2.
Level of chimerism in cerebellum, neocortex and dentate gyrus of chimeras as assessed by Southern blot analysis (proportion of TfamloxP alleles) or by enzyme histochemistry (proportion of LacZ-expressing cells). (A, B) Level of chimerism of control chimeras (blue bars; n = 63) and mitochondrial late-onset neurodegeneration (MILON) chimeras (black bars; n = 58) assessed by Southern blot analysis of DNA from cerebellum. Each bar represents one animal. Bars indicate mean levels ± 1 SD. (C) Correlation between levels of chimerism in neocortex (proportion of neurons not expressing LacZ) and cerebellum (proportion of TfamloxP alleles) of different control chimeras (n = 32; blue dots) and MILON chimeras (n = 32; red dots). Neurons with nuclear staining for Hoechst 33342 but lacking LacZ enzyme activity were classified as non-LacZ-expressing neurons. There is a linear correlation (blue line; r2 = 0.81; P = 2.5 × 10−12) between levels of chimerism in neocortex and cerebellum of control chimeras. The level of chimerism in neocortex in comparison with cerebellum is generally lower in MILON chimeras (best fitted red line shown) indicating cell loss. (D) The correlation between levels of chimerism in the dentate gyrus (proportion of neurons not expressing LacZ) and cerebellum (proportion of TfamloxP alleles) of different control chimeras (n = 50; blue dots) and MILON chimeras (n = 35; red dots). The levels of chimerism in neocortex and cerebellum show a linear correlation in control chimeras (blue line; r2 = 0.80; P = 1.7 × 10−18) and MILON chimeras (red line; r2 = 0.81; P = 1.8 × 10−13).
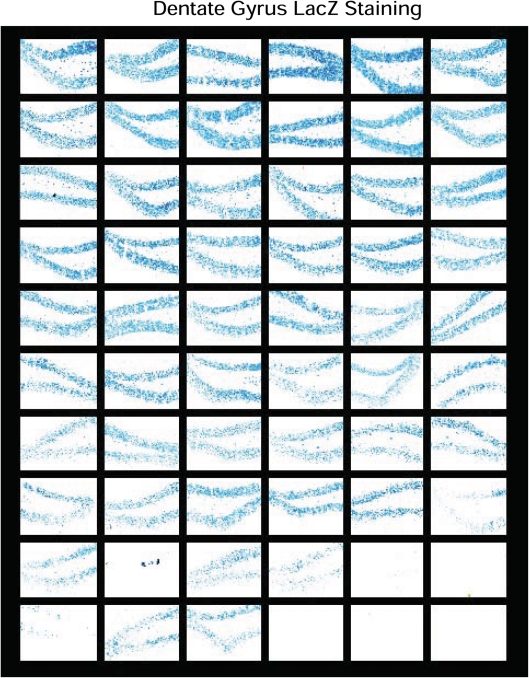
We also performed histochemical stainings to determine the distribution of LacZ-expressing cells in the dentate gyrus (Fig. 3) and neocortex (not shown) of MILON and control chimeras with widely varying levels of chimerism according to Southern blot analysis of DNA from cerebellum. We found that the LacZ-expressing cells were evenly distributed throughout the dentate gyrus (Fig. 3). The aggregation protocol we used for generating chimeras thus leads to a finely dispersed mixture of the two cell types, consistent with the observations in other types of mouse chimeras (32).
Figure 3.
Analysis of chimerism in the dentate gyrus. LacZ staining of the dentate gyrus was performed in series (n = 60) of different control and mitochondrial late-onset neurodegeneration chimeras. The sections are ordered according to increasing mean levels of chimerism (proportion of Tfamlox alleles) in cerebellum as follows (starting at the top from left to right): row 1, 0–6%; row 2, 7–19%; row 3, 21–27%; row 4, 32–39%; row 5, 40–49%; row 6, 51–58%; row 7, 62–68%; row 8, 71–79%; row 9, 82–89%; row 10, 91–100%.
We have previously demonstrated that MILON mice develop a massive nerve cell loss in different regions of neocortex and medial hippocampus at the end of their life (24). As expected, MILON cells were encountered at less than the expected frequency in neocortex of MILON chimeras (Fig. 2C), indicating a substantial loss of respiratory chain-deficient neurons in this region. There was no clear loss of neurons in the dentate gyrus as demonstrated by the very good correlation between levels of chimerism in cerebellum and the dentate gyrus in MILON chimeras (Fig. 2D). The reason for the preservation of nerve cells in the dentate gyrus of MILON chimeras can be that this region is affected late in the neurodegenerative process (24) and that animal death therefore may occur before extensive cell death becomes apparent in this region. It is also possible that compensatory neurogenesis in the dentate gyrus may mask an ongoing cell loss, consistent with our previous observation of PCNA-positive nerve cell clusters indicative of neurogenesis in the dentate gyrus of end-stage MILON mice (24).
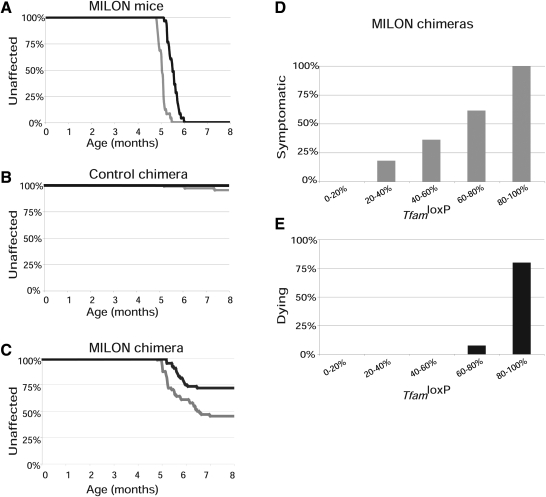
We used a clinical assessment scale to determine age at onset of symptoms and followed the lifespan (Fig. 4A–C) in cohorts of pure MILON mice (n = 25), control chimeras (n = 57) and MILON chimeras (n = 64). The genetically determined neurodegeneration in MILON mice invariably caused symptoms and premature death (Fig. 4A), consistent with our previous results from studies of these mice (24). Only three of the control chimeras developed symptoms and no animals in this cohort died during the 8-month-long follow-up period (Fig. 4B). These results demonstrate that chimerism per se does not cause clinical symptoms or substantially reduced lifespan.
Figure 4.
Observation of clinical symptoms and determination of lifespan in chimeras. Age at onset of symptoms was scored by a clinical assessment scale and is indicated by gray lines. Age at death (or age at onset grave symptoms requiring euthanasia) was determined and is indicated by black lines. (A) Symptoms and survival in pure mitochondrial late-onset neurodegeneration (MILON) mice (n = 25). (B) Symptoms and survival in control chimeras (n = 57). (C) Symptoms and survival in MILON chimeras (n = 63). (D) Relationship between grade of chimerism and occurrence of symptoms in MILON chimeras. (E) Relationship between grade of chimerism and occurrence of death in MILON chimeras.
The subset of MILON chimeras that developed clinical symptoms had a significantly (P < 5.0 × 10−5) higher mean age at onset of symptoms (mean ± 1SD = 5.6 ± 0.6 months, range, r = 4.8–7.0 months) (Fig. 4C) than MILON mice (5.1 ± 0.2 months, r = 4.8–5.5 months) (Fig. 4A). In addition, only about half of the symptomatic chimeras died (Fig. 4C). We also studied the correlation between MILON cell contribution in cerebellum and occurrence of symptoms or death in chimeras. Symptoms started to appear in chimeras with low contribution of MILON cells (>20%) and became much more prevalent as the MILON contribution increased (Fig. 4D). Death only occurred in chimeras with a high (>60%) contribution of MILON cells (Fig. 4E). All of the chimeras with a MILON cell contribution >80% developed symptoms and most of these animals died. The MILON chimeras that died had a lifespan (5.6 ± 0.3 months, r = 5.2–6.4 months), which was not significantly different (P = 0.32) from that of MILON mice (5.6 ± 0.2 months; r = 5.2–6.0 months).
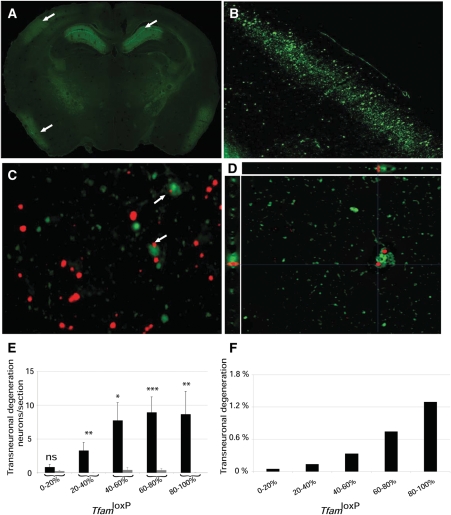
We performed FluoroJade staining (33,34) of control and MILON chimera brain sections and found evidence for neurodegeneration only in MILON chimeras. Chimeras with a high contribution of MILON cells showed massive neurodegeneration in medial hippocampus and neocortex (Fig. 5A and B). The pattern of neurodegeneration observed in MILON chimeras of this study (Fig. 5A and B) is very similar to the previously described pattern in MILON mice (24). As shown above, the presence of neurons with normal respiratory chain function delayed the onset of symptoms and prevented death in MILON chimeras (Fig. 4C). However, we also investigated whether respiratory chain-deficient neurons may exert a negative influence on neurons with normal respiratory chain function (LacZ-expressing neurons) in neocortex of MILON chimeras. We detected few LacZ-expressing and FluoroJade-positive neurons in control chimeras, whereas we found an increased number of such neurons in the neocortex of MILON chimeras (Fig. 5C and D). Respiratory chain-deficient neurons thus induce degeneration of neurons with a normal respiratory chain function. These trans-neuronal degeneration events increased if the contribution of MILON cells was high in the chimeras (Fig. 5E). In chimeras with >80% contribution of MILON cells ∼1% of total nerve cell degeneration in cortex was trans-neuronal (Fig. 5F). There is a low amount (<20%) of cortical neurons with normal respiratory chain function in chimeras with high contribution (>80%) of MILON cells and therefore a substantial proportion (∼1:20) of these neurons degenerate by this trans-neuronal mechanism.
Figure 5.
Neurodegeneration and trans-neuronal degeneration in mitochondrial late-onset neurodegeneration (MILON) chimeras. (A) Fluorescence microscopy to detect FluoroJade staining in a brain section of a MILON chimera with 90% TfamloxP alleles in cerebellum. Massive neurodegeneration is evident in the medial hippocampus (upper right arrow), the somatosensory cortex (upper left arrow) and the piriform cortex (lower left arrow). (B) Higher magnification of area containing the neocortex region indicated by the upper left arrow in (A). (C) Combined image obtained by fluorescence microscopy to detect FluoroJade staining (green) and bright field microscopy for immunohistochemical detection of LacZ expression (red) in the neocortex of a MILON chimera with 56% TfamloxP alleles in cerebellum. Arrows indicate LacZ-expressing neurons undergoing trans-neuronal degeneration. (D) Deconvolution microscopy image demonstrating the presence of FluoroJade (green) and LacZ (red) in a single neuron undergoing trans-neuronal degeneration in the neocortex of a MILON chimera with 65% TfamloxP alleles in cerebellum. The xz and yz projections (0.3 µm thick, 32 stacks) are shown. (E) The correlation between levels of chimerism (proportion of TfamloxP alleles in cerebellum) and trans-neuronal degeneration (FluoroJade-positive neurons expressing LacZ) in control chimeras (gray bars) and MILON chimeras (black bars). (F) The proportion of degenerating neurons (FluoroJade-positive neurons) that show evidence of trans-neuronal degeneration (FluoroJade-positive neurons expressing LacZ) at different levels of chimerism (proportion of TfamloxP alleles in cerebellum).
DISCUSSION
Pathogenic mutations of mtDNA are often heteroplasmic and undergo mitotic segregation to create a mosaic pattern of respiratory chain deficiency in various organs. The role of this mosaicism in the pathogenesis of mitochondrial dysfunction in disease and ageing has been difficult to assess experimentally. We have now addressed this issue by analyzing mouse chimeras with mosaic respiratory chain deficiency in cerebral forebrain. We have previously demonstrated that respiratory chain-deficient neurons are prone to undergo apoptosis (24,35) and consistent with this we found clear evidence of cell loss in the forebrain of high-grade MILON chimeras. Relatively low levels (>20%) of respiratory chain-deficient large forebrain neurons are sufficient to cause clinical symptoms, whereas animal death only occurs if a large proportion of the neurons (>60%) are affected. All animals with very high levels (>80%) of respiratory chain-deficient neurons in neocortex and hippocampus develop symptoms and most of them die from neurodegeneration.
The proportion of respiratory chain-deficient neurons in the ageing human hippocampus (∼1%) is well below the threshold level (∼20%) needed to cause clinical symptoms in chimeric mice (22). However, it should be emphasized that studies of ageing human brains only provide a ‘snap-shot’ of a continuously ongoing process (22), whereas the MILON mice have a genetically defined narrow temporal onset of neurodegeneration (24). It is quite possible that the continuously ongoing formation of respiratory chain-deficient neurons in the ageing human brain may lead to a substantial cell loss over time (22). In addition, there is focal respiratory chain deficiency in a variety of organs in aged humans and the combined cell loss in all tissues may be substantial and impact lifespan.
We observed a significant loss of healthy neurons because of a trans-neuronal degeneration mechanism in the chimeric mice. It has been suggested that neurons are connected in trophic units (36), which may explain the observed trans-neuronal degeneration. The ageing process is likely multi-factorial and determined by accumulated damage to a variety of cellular maintenance systems (37). Therefore, such trans-neuronal degeneration may be even more pronounced in the normal ageing process when the respiratory chain-deficient neurons are surrounded by neurons with other types of acquired age-associated defects (37). In mitochondrial disease, the nerve cell loss may be additionally enhanced by the presence of respiratory chain deficiency in non-neuronal cells such as astrocytes and other types of glial cells. Our experimental results may also be of relevance for a variety of degenerative processes causing cell death in the nervous system (38), e.g. ischemic stroke and Alzheimer disease, because trans-neuronal degeneration may enhance the consequence of various types of primary lesions causing nerve cell death.
In summary, we have experimentally defined the minimal threshold level of respiratory chain-deficient neurons in cerebral cortex that are needed to cause clinical symptoms or death. We demonstrate that the presence of normal neurons delay onset of disease and prevent death. Finally, we identify trans-neuronal degeneration as a novel mechanism creating additional nerve cell loss. These findings shed new light on the pathogenesis of mosaic respiratory chain deficiency in ageing and mitochondrial disease.
MATERIALS AND METHODS
Creation of chimeras
Matings to obtain embryos for aggregation were performed after superovulation of female mice at age 8–12 weeks. The mice were first injected i.p. with 7.5 IU pregnant mare serum (Intervet, The Netherlands) and 48 h later injected i.p. with 7.5 IU chorionic gonadotrophin hormone (HCG; Intervet). Males were added to the cages after the second injection. Female or male mice of the genotype TfamloxP/TfamloxP were mated to mice of the opposite sex with the genotype TfamloxP/TfamloxP, +/CaMKII-cre. This cross is expected to give 50% TfamloxP/TfamloxP embryos and 50% TfamloxP/TfamloxP, +/CaMKII-cre embryos. We used the same protocol to superovulate and mate ROSA26 mice, which are homozygous for LacZ gene (LacZ/LacZ), and C57BL/6 mice to obtain embryos heterozygous for LacZ (+/LacZ). Pregnant females were killed 55 h after the HCG injection and the uterus was removed. The oviducts and uterus were dissected and flushed with M2 (Sigma) medium and embryos at the eight-cell stage up to the early blastocyst stage were recovered and incubated in KSOM (Molecular Biotechnologies) media. We obtained a total of 1094 embryos of the genotype TfamloxP/TfamloxP or TfamloxP/TfamloxP, +/CaMKII-cre and 1003 embryos of the genotype +/LacZ. The zona pellucida was removed from the embryos by exposing them to acidic Tyrodes for 30–120 s followed by several washes with M2 medium. Embryos of the genotype TfamloxP/TfamloxP or TfamloxP/TfamloxP, +/CaMKII-cre were aggregated with +/LacZ embryos in microwells with KSOM (Molecular Biotechnologies) medium covered with OVOIL (Vitolife). Aggregations were performed at 37°C for 28 h in an incubator with 5% CO2 and saturated humidity. Fused embryos that had developed to the blastocyst stage were implanted into 2.5-day pseudopregnant foster females to obtain chimeric mice.
Genotyping and southern blot analyses
Chimeras were first genotyped using PCR assays to detect the Tfam+, TfamloxP, Tfam− and cre genotypes in tail DNA, as previously described (29). Southern blot analyses were performed to assess levels of chimerism in the cerebellum of all chimeras. Half of the cerebellum was homogenized and total DNA was extracted. An aliquot of 3 µg DNA was digested with BamHI, separated in 0.8% agarose, blotted onto a nylon membrane and probed with a DNA fragment corresponding to the region surrounding exon 5 of the Tfam gene. The exon 5 probe was obtained by PCR amplification of genomic DNA with the primer pair E5A (5′-acaagcatacttcatttacaaatccc-3′) and E5B (5′-gtagaaaatgttctctctcataagag-3′). The probe was labeled using the Prime-It II kit (Stratagene, USA). Hybridized blots were analyzed with the Personal Molecular Imager FX (BioRad, USA) and the software Quantity One 4.6 was used to determine the proportion of Tfam+ and TfamloxP alleles. At least three independent analyses were performed on each chimeric mouse.
Clinical scoring of symptoms and analysis of locomotion and rearing
MILON mice reproducibly and invariably develop all five criteria we used for clinical scoring of chimeras at 5 months of age (Fig. 4A), i.e. decreased motor activity, reduced grooming, characteristic posture with anxiety and social avoidance, and aggressive behavior and hyperactivity in response to stress. We developed a scoring system based on five parameters to determine age-of-onset symptoms in cohorts of MILON mice, control chimeras and MILON chimeras. We scored a mouse as symptomatic when three of these five criteria were present: (i) decreased spontaneous motor activity, (ii) unattended, poorly groomed fur, (iii) kyphosis of spine, (iv) gait disturbance with wobbly walking, (v) aggressive behavior and/or hyperactivity in response to stress. Mice were scored on a weekly basis from 4 months of age. We first observed the mouse in its normal environment (the cage). We then repeated the observation after transferring the mouse to a novel environment (a laboratory bench).
Immunohistochemistry and microscopy
Chimeras were subjected to transcardiac perfusion with Ca2+-free Tyrode’s solution with heparin to remove blood and thereafter fixed by perfusion with a solution containing 4% paraformaldehyde and 0.4% picric acid in 0.16 m phosphate buffer, pH 7.0. The brains were removed and post-fixed overnight in the same solution at 4°C. Next, brains were equilibrated in 10% sucrose containing 0.1% sodium azide and stored in this solution at +4°C. Brains were frozen on CO2 ice and sectioned with a cryostat to obtain 14 µm sections. LacZ enzyme activity was detected by incubating brain sections with substrate at 37°C overnight using a kit (Sigma). Cresyl violet (Sigma) and FluoroJade (Chemicon) stainings (33,34) were performed as suggested by the manufacturer. Sections were analyzed (NIKON Eclipse E600) and images collected using appropriate software (Spot v.4.5). Fluorescence images corresponding to entire coronal sections of the brain were digitally constructed (Photoshop to combine four smaller 5X lens images). Bright field images were adjusted to contain the same background intensity and the same average signal intensity per object to correct for the variation of LacZ staining. Assessment of chimerism in neocortex and the dentate gyrus was performed by cell counting of sections stained to detect nuclear DNA (Hoechst 33342) and LacZ enzyme activity. Images of double-stained sections were obtained with a deconvolution microscope (LEICA ASMDW). Image deconvolution was performed (Leica software) and co-localization was digitally analyzed (Volocity 3D Software). Results from deconvolution microscopy were confirmed by confocal microscopy (LSM 510 Meta, Zeiss, Germany) in selected cases. Assessment of chimerism in neocortex was obtained by counting a total of 99 cortical neurons of each animal at three defined locations, i.e. layer III of the retrospinal granular cortex (33 neurons), the somatosensory one-barrel field (33 neurons) and the piriform cortex (33 neurons). Hoescht staining was used to identify neurons (based on location, size of the nucleus and the presence of several nucleoli) prior to bright-field counting of the proportion of neurons expressing LacZ.
Statistical analyses
Variations in the measurements are shown as standard deviation. Regression analyses were performed using appropriate software (Excel regression tool of the Analysis Toolpack Microsoft Office).
Conflict of Interest statement. The authors have no commercial interests in this study.
FUNDING
This study was supported by the Swedish Research Council, Torsten and Ragnar Söderbergs Stiftelse, the Swedish Heart and Lung Foundation, Swedish Brain Power, Vinnova, AFA, the Swedish Brain Foundation and USPH grants. Funding to pay the Open Access publication charges for this article was provided by The Swedish Research Council.
REFERENCES
- 1.Smeitink J., van Den Heuvel L., DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2001;2:342–352. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- 2.Schaefer A.M., McFarland R., Blakely E.L., He L., Whittaker R.G., Taylor R.W., Chinnery P.F., Turnbull D.M. Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 3.Wallace D.C. A mitochondrial paradigm for degenerative diseases and ageing. Novartis Found. Symp. 2001;235:247–263. doi: 10.1002/0470868694.ch20. [DOI] [PubMed] [Google Scholar]
- 4.Trifunovic A., Larsson N.G. Mitochondrial dysfunction as a cause of ageing. J. Int. Med. 2008;263:167–178. doi: 10.1111/j.1365-2796.2007.01905.x. [DOI] [PubMed] [Google Scholar]
- 5.Larsson N.G., Clayton D.A. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 6.Piko L., Hougham A.J., Bulpitt K.J. Studies of sequence heterogeneity of mitochondrial DNA from rat and mouse tissues: evidence for an increased frequency of deletions/additions with aging. Mech. Ageing Dev. 1988;43:279–293. doi: 10.1016/0047-6374(88)90037-1. [DOI] [PubMed] [Google Scholar]
- 7.Sato W., Tanaka M., Ohno K., Yamamoto T., Takada G., Ozawa T. Multiple populations of deleted mitochondrial DNA detected by a novel gene amplification method. Biochem. Biophys. Res. Commun. 1989;162:664–672. doi: 10.1016/0006-291x(89)92362-0. [DOI] [PubMed] [Google Scholar]
- 8.Soong N.W., Hinton D.R., Cortopassi G., Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat. Genet. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 9.Corral-Debrinski M., Horton T., Lott M.T., Shoffner J.M., Beal M.F., Wallace D.C. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 10.Schwarze S.R., Lee C.M., Chung S.S., Roecker E.B., Weindruch R., Aiken J.M. High levels of mitochondrial DNA deletions in skeletal muscle of old rhesus monkeys. Mech. Age. Dev. 1995;83:91–101. doi: 10.1016/0047-6374(95)01611-3. [DOI] [PubMed] [Google Scholar]
- 11.Khaidakov M., Heflich R.H., Manjanatha M.G., Myers M.B., Aidoo A. Accumulation of point mutations in mitochondrial DNA of aging mice. Mutat. Res. 2003;526:1–7. doi: 10.1016/s0027-5107(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 12.Trounce I., Byrne E., Marzuki S. Decline in skeletal muscle mitochondrial respiratory chain function: possible factor in ageing. Lancet. 1989;8639:637–639. doi: 10.1016/s0140-6736(89)92143-0. [DOI] [PubMed] [Google Scholar]
- 13.Yen T.-C., Chen Y.-S., King K.-L., Yeh S.-H., Wei H. Liver mitochondrial respiratory functions decline with age. Biochem. Biophys. Res. Commun. 1989;165:994–1003. doi: 10.1016/0006-291x(89)92701-0. [DOI] [PubMed] [Google Scholar]
- 14.Cottrell D.A., Turnbull D.M. Mitochondria and ageing. Curr. Opin. Clin. Nutr. Metab. Care. 2000;3:473–478. doi: 10.1097/00075197-200011000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly Y.M., Gidlöf S., Oldfors A., Wibom R., et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 16.Krishnan K.J., Greaves L.C., Reeve A.K., Turnbull D. The ageing mitochondrial genome. Nucleic Acids Res. 2007;35:7399–7405. doi: 10.1093/nar/gkm635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fayet G., Jansson M., Sternberg D., Moslemi A.R., Blondy P., Lombes A., Fardeau M., Oldfors A. Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul. Disord. 2002;12:484–493. doi: 10.1016/s0960-8966(01)00332-7. [DOI] [PubMed] [Google Scholar]
- 18.Bender A., Krishnan K.J., Morris C.M., Taylor G.A., Reeve A.K., Perry R.H., Jaros E., Hersheson J.S., Betts J., Klopstock T., et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 19.Taylor R.W., Barron M.J., Borthwick G.M., Gospel A., Chinnery P.F., Samuels D.C., Taylor G.A., Plusa S.M., Needham S.J., Greaves L.C., et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller-Hocker J. Cytochrome c oxidase deficient cardiomyocytes in the human heart: an age-related phenomenon. Am. J. Pathol. 1989;134:1167–1173. [PMC free article] [PubMed] [Google Scholar]
- 21.Müller-Höcker J. Cytochrome c oxidase deficient fibres in the limb muscle and diaphragm of man without muscular disease: an age related alteration. J. Neurol. Sci. 1990;100:14–21. doi: 10.1016/0022-510x(90)90006-9. [DOI] [PubMed] [Google Scholar]
- 22.Cottrell D.A., Blakely E.L., Johnson M.A., Ince P.G., Borthwick G.M., Turnbull D.M. Cytochrome c oxidase deficient cells accumulate in the hippocampus and choroid plexus with age. Neurobiol. Age. 2001;22:265–272. doi: 10.1016/s0197-4580(00)00234-7. [DOI] [PubMed] [Google Scholar]
- 23.Cottrell D.A., Borthwick G.M., Johnson M.A., Ince P.G., Turnbull D.M. The role of cytochrome c oxidase deficient hippocampal neurones in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2002;28:390–396. doi: 10.1046/j.1365-2990.2002.00414.x. [DOI] [PubMed] [Google Scholar]
- 24.Sorensen L., Ekstrand M., Silva J.P., Lindqvist E., Xu B., Rustin P., Olson L., Larsson N.G. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J. Neurosci. 2001;21:8082–8090. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich G., Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Gen. Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 26.Zambrowicz B.P., Imamoto A., Fiering S., Herzenberg L.A., Kerr W.G., Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wredenberg A., Wibom R., Wilhelmsson H., Graff C., Wiener H.H., Burden S.J., Oldfors A., Westerblad H., Larsson N.G. Increased mitochondrial mass in mitochondrial myopathy mice. Proc. Natl Acad. Sci. USA. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva J.P., Kohler M., Graff C., Oldfors A., Magnuson M.A., Berggren P.O., Larsson N.G. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat. Genet. 2000;26:336–340. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- 29.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 30.Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 31.Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N.G., Gustafsson C.M. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 32.Low-Zeddies S.S., Takahashi J.S. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmued L.C., Albertson C., Slikker W., Jr Fluoro-Jade: a novel fluorochrome for the sensitive and reliable histochemical localization of neuronal degeneration. Brain. Res. 1997;751:37–46. doi: 10.1016/s0006-8993(96)01387-x. [DOI] [PubMed] [Google Scholar]
- 34.Schmued L.C., Hopkins K.J. Fluoro-Jade B: a high affinity fluorescent marker for the localization of neuronal degeneration. Brain. Res. 2000;874:123–130. doi: 10.1016/s0006-8993(00)02513-0. [DOI] [PubMed] [Google Scholar]
- 35.Wang J., Silva J., Gustafsson C.M., Rustin P., Larsson N.G. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc. Natl Acad. Sci. USA. 2001;98:4038–4043. doi: 10.1073/pnas.061038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agnati L.F., Cortelli P., Pettersson R., Fuxe K. The concept of trophic units in the central nervous system. Prog. Neurobiol. 1995;46:561–574. doi: 10.1016/0301-0082(95)00017-p. [DOI] [PubMed] [Google Scholar]
- 37.Kirkwood T.B. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 38.Bredesen D.E., Rao R.V., Mehlen P. Cell death in the nervous system. Nature. 2006;443:796–802. doi: 10.1038/nature05293. [DOI] [PMC free article] [PubMed] [Google Scholar]