Abstract
The presynaptic protein α-synuclein is involved in several neurodegenerative diseases, including Parkinson's disease (PD). In rare familial forms of PD, causal mutations (PARK1) as well as multiplications (PARK4) of the α-synuclein gene have been identified. In sporadic, idiopathic PD, abnormal accumulation and deposition of α-synuclein might also cause degeneration of dopaminergic midbrain neurons, the clinically most relevant neuronal population in PD. Thus, cell-specific quantification of α-synuclein expression-levels in dopaminergic neurons from idiopathic PD patients in comparison to controls would provide essential information about contributions of α-synuclein to the etiology of PD. However, a number of previous studies addressing this question at the tissue-level yielded varying results regarding α-synuclein expression. To increase specificity, we developed a cell-specific approach for mRNA quantification that also took into account the important issue of variable RNA integrities of the individual human postmortem brain samples. We demonstrate that PCR –amplicon size can confound quantitative gene-expression analysis, in particular of partly degraded RNA. By combining optimized UV-laser microdissection- and quantitative RT–PCR-techniques with suitable PCR assays, we detected significantly elevated α-synuclein mRNA levels in individual, surviving neuromelanin- and tyrosine hydroxylase-positive substantia nigra dopaminergic neurons from idiopathic PD brains compared to controls. These results strengthen the pathophysiologic role of transcriptional dysregulation of the α-synuclein gene in sporadic PD.
INTRODUCTION
The progressive degeneration of dopaminergic (DA) midbrain neurons, in particular within the substantia nigra (SN), and in consequence the dramatic reduction of DA innervation in the striatal target areas is the clinically most relevant, pathological hallmark in Parkinson's disease (PD) and related neurodegenerative disorders (1,2). The etiology for most forms of PD is still unclear (sporadic or idiopathic PD, iPD), however, for some rare familial forms of PD, several underlying causal gene-mutations have been identified (3,4). Alpha-synuclein (α-SYN) has been identified as the first causative gene (PARK1) in familial forms of Parkinson's disease, harboring dominant gain-of-function mutations (5). Human α-SYN is coded by the SNCA-gene (=NACP; 4q21; 6 exons) and exists in three distinct splice variants, 140 amino acids (full length), 126 amino acids (no exon 3) and 112 amino acids (no exon 5) (6–8). Mutations in SNCA lead to a range of neuropathologic phenotypes, from PD to diffuse Lewy-body disease or dementia with Lewy-bodies (DLB) (9). Lewy-bodies—neuronal protein-inclusions—are a hallmark of iPD, and other neurodegenerative diseases (10), and α-SYN is one major constituent of Lewy-bodies (11–13). Point-mutations found in SNCA reduce α-SYN protein degradation by lysosomal and/or proteasomal pathways (14), and lead to accumulation and aggregation of α-synuclein in the cell (15). Furthermore, increased expression of wild-type α-SYN due to SNCA gene duplications or triplications has also been identified as causes for Parkinsonism (PARK4) (16–19). These findings led to the view that elevated levels of α-synuclein expression might be sufficient to cause PD in a dose-dependent manner (20–22). This assumption is supported by the findings that transcriptional dysregulation of the SNCA gene (23) as well as posttranslational processing of wild-type α-SYN (7) might contribute to the neurodegenerative process of PD. Furthermore, overexpression of wild-type SNCA is sufficient to kill dopaminergic neurons in several animal models (20,24, 25), further substantiating the importance of transcriptional control of α-SYN-levels. Importantly, the Parkinsonism-inducing toxin MPTP and other forms of neuronal injury increase α-SYN expression in rodent DA neurons, thus suggesting an α-SYN-dependent final pathway of DA neurodegeneration (26,27). Accordingly, DA neurons from α-SYN KO mice are resistant to MPTP-induced neurotoxicity (28,29). Surprisingly, single α-SYN KO-, or double α-, β- and/or γ-synuclein KO-mice are viable, fertile and display no major phenotype (30–32). These findings suggest that α-synuclein is not fundamentally important for the cell, and its spectrum of physiological roles for synaptic neuronal functions are still not clear (33,34). Dopamine-dependent selective neurotoxicity of α-SYN has been described (35). However, a neuroprotective role of presynaptic α-SYN acting as co-chaperone for the formation of SNAP/SNARE complexes has also been demonstrated in a mouse model of neurodegeneration (32), arguing that transcriptional control of the SNCA-gene might be important in both directions. Given these important—but mechanistically still unclear—roles of α-SYN in context of neurodegeneration and PD (36), it is mandatory to conclusively answer the question whether α-SYN gene-expression is up- or down-regulated, or unchanged in dopaminergic neurons from iPD brains, in comparison to age-matched controls. Up to now, evidence for changes of α-SYN expression in human iPD midbrain-tissues in both directions have been reported (37–42). However, tissue-based studies cannot directly compare α-SYN expression in dopaminergic midbrain neurons from control and PD brains, as these approaches only report averaged expression-levels across a complex multitude of neuronal and nonneuronal cell-types present in midbrain. Even more, the relative number of DA neurons in substantia nigra-/midbrain-tissue differs dramatically among individual PD brains as well as between PD brains and controls, which renders tissue-based approaches in this case even more problematic. Another crucial factor that will also seriously affect gene-expression studies using human postmortem material is caused by case-to-case variations in mRNA quality/integrity of the individual brain samples, due to e.g. clinical differences in end-stage disease (e.g. degree of cerebral ischemia, age and differences of medication) as well as significant differences in postmortem factors like the delay between death and tissue collection, tissue-pH and the protocol for preservation/freezing of the human tissue, respectively (37,39,41,43–45). Thus, in order to overcome the combination of these confounding factors of tissue-based approaches and variable RNA qualities, gene-expression studies should combine cell specificity with a rigorous assessment and consideration of distinct and suboptimal mRNA integrities. Here, we describe, validate and utilize such an optimized target-cell-specific quantitative approach that combines contact-free UV-laser-microdissection-based selective sampling of individual DA SN neurons from human postmortem iPD midbrain-tissue and respective controls, and subsequent quantitative real-time RT–PCR gene-expression analysis. We detail and discuss our specific experimental design and interpretation of results, given that RNA integrities of different individual human brain samples were not homogeneous. By applying this protocol, we detected about 6-fold higher α-SYN mRNA-levels in surviving neuromelanin- and tyrosine-hydroxylase-positive DA SN neurons from PD patients compared to those of unaffected controls. These experimental findings support the hypothesis that transcriptional upregulation of α-SYN does indeed exist in the DA SN neurons, the essential target population in iPD.
METHODS
Preparation and cryosectioning of fresh mouse brains for UV-LMD
For data shown in Figure 2, 1-month-old male C57Bl/6 mice from our breeding colony were deeply anesthetized with isoflurane (Abbot) and decapitated. Brains were quickly removed, and a coronal tissue-block containing the midbrain was cut and mounted with tissue freezing medium (Jung), and immediately frozen by insertion into the snap-freeze holder of a cryostat (Leica CM1850). After equilibration at −18°C for 30 min, 12 μm serial coronal cryosections of mouse midbrains were cut and mounted on RNAse-free UV-C-treated membrane slides (1 mm PEN-membrane, Microdissect). A fresh, ethanol-cleaned single-use blade (Leica Type 819) was used for each brain. All animal-procedures were in accordance with the German guidelines (approved by the Regierungspräsidium Giessen, Germany).
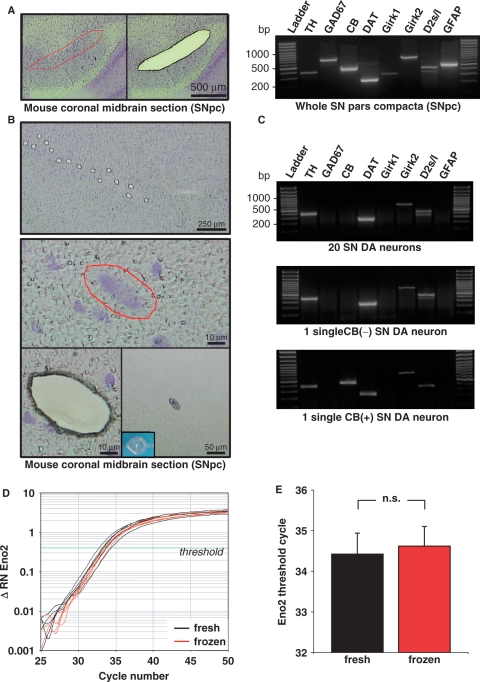
Figure 2.
UV-LMD of individual neurons. (A) Left panel: cresylviolet (CV)-stained mouse coronal midbrain section indicating the area of the substantia nigra pars compacta (SNpc) before (left) and after (right) UV-laser-microdissection of the entire area. Right panel: gel electrophoresis results of qualitative reverse transcription (RT) multiplex-nested PCR products for a whole LMD SNpc. RT–PCR signals for all eight different dopaminergic (TH, DAT, Girk2, D2s/l) and nondopaminergic (GAD, Girk1, GFAP, CB) marker-genes, detected in the heterogeneous cellular mixture of dopaminergic, GABAergic and glial cells. (DNA-ladder: 100-bp marker). (B) Top: CV-stained coronal midbrain section after UV-laser-microdissection of 15 individual DA neurons. Middle: selection and laser-microdissection of an individual neuron from upper section. Lower: after UV-LMD of individual neuron—section (left) and cap-control (right) demonstrating successful isolation. (C) PCR products after gel electrophoresis of qualitative RT multiplex-nested PCR for individual LMD SN DA neurons and small SN DA pools. Upper and second panel: similar gene-expression profile of dopaminergic marker genes (TH, DAT, Girk2, D2; not Girk2, GAD67, GFAP) in pools of 20 and single SN DA neurons illustrate sensitivity and specificity of the protocol. Third panel: expression profile of an individual calbindin-positive CB (+) SN DA neuron (D and E) Quantitative real-time RT–PCR analysis of Eno2 gene-expression using 10% of cDNA from pools of 15 mouse SN DA neurons as template. Neurons were UV-LMD-collected from either fresh or stored (1-week) and re-used tissue slices. (D) Representative real-time PCR traces testing for Eno2 expression, relative fluorescence levels on a logarithmic scale are plotted against PCR cycles (ΔRN: relative fluorescence, normalized to internal fluorescence marker ROX); threshold-line for determinations of threshold cycles (Ct) indicated by the green line (Δ RN 0.4). (E) No significant difference n.s. between Eno2 detection levels (Ct) in SN DA cell-pools from fresh and frozen tissue sections (fresh: 34.43 ± 0.51, n = 10; re-used: 34.62 ± 0.48 n = 9; p = 0.785).
Cryosectioning of frozen human midbrain tissue blocks
For data shown in Figures 3–5 and Tables 1 and 2, native cryoprotected (−80°C) human midbrain tissue containing the substantia nigra was provided as horizontal tissue-blocks by the German BrainNet (www.brain-net.net; Grant-No. GA 28). Further available details of all human brain samples analyzed in this study are specified in Table 1. Upon arrival, the human midbrain tissue-blocks were cryospotted with mounting medium (Jung) onto UV-C-treated cork discs and long-term stored in boxes at −80°C. For cryosectioning, the tissue was removed from −80°C storage, equilibrated at −35°C for 20 min and afterwards cut into 12 μm sections at −19°C in a cryostat (Leica CM1850). The human midbrain cryosections were mounted on RNAse-free UV-C-treated membrane slides (1 mm PEN-membrane, Microdissect). A fresh, ethanol-cleaned single-use blade (Leica Type 819) was used for each human brain. All experiments were in accordance with the ethic commission of the Philipps-University Marburg (AZ 35/06) and Ulm (277/07 - UBB/se).
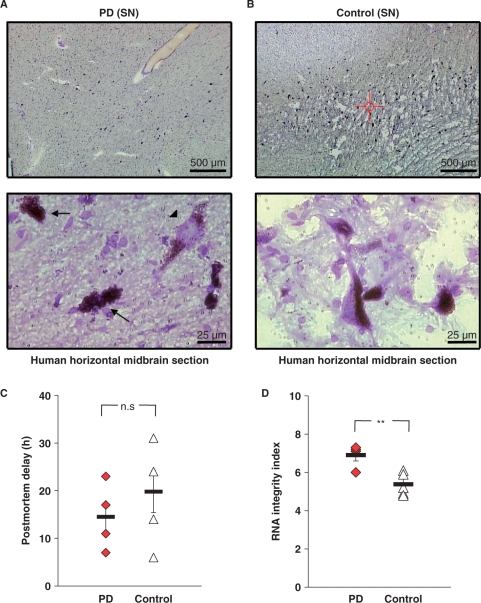
Figure 3.
Assessment of human postmortem midbrain tissue. (A and B) Cresylviolet-stained human postmortem SN nigra tissue showed typical neuromelanin-positive [NM(+)] cells in control and PD brains, whereas in PD tissue the number of NM(+) neurons was reduced. Snap-frozen PD tissue showed better preservation of the macroscopic structure of the midbrain, but cell morphology was not affect by macroscopic differences (A, B, lower panels). In PD sections, dying neurons (arrows) with poor cellular structure and Lewy-body positive neurons (arrowhead) were identified. (C) No significant difference in the respective mean postmortem intervals between PD and control brains (PD: 14 h 30 min ± 3 h 50 min, n = 4; control: 19 h 48 min ± 4 h 23 min, n = 5; P = 0.395). (D) The RNA integrity, as given by RIN-value (Agilent analysis) was significantly higher in the PD brains compared to controls (PD: 6.9 ± 0.3 n = 4; control: 5.4 ± 0.3 n = 5; P = 0.007). Additional data are given in Table 1.
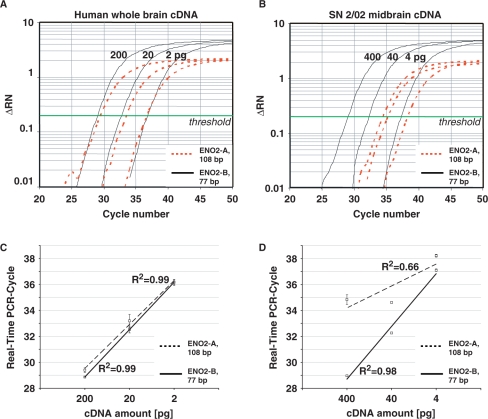
Figure 4.
Effects of RNA integrity and PCR-assay size on real-time quantitative PCR performance. Serial dilutions of high-quality whole brain cDNA (200, 20 and 2 pg; Clontech), and cDNA derived from partly degraded midbrain RNA (400, 40 and 4 pg; brain SN 2/02, RIN = 6.1) were used as templates for ENO2 real-time PCR, employing two different assays for ENO2 with large (ENO2-A, 108 bp) and small (ENO2-B, 77 bp) amplicon size (A and C) With high-quality cDNA as templates, amplification curves (A) and slopes (amplification-efficiencies) of standard curves were similar for both assays (Δ RN: relative fluorescence, normalized to internal fluorescence marker ROX) (C). (B and D) With cDNA from partly degraded RNA as templates, amplification curves (B) and slopes of standard curves (D) were similar as for high-quality cDNA only when using the small amplicon size assay ENO2-B. The larger ENO2-A assay did not allow the generation of a respective reliable standard curve and was not suited for RT–PCR quantification of SN 2/02 RNA sample.
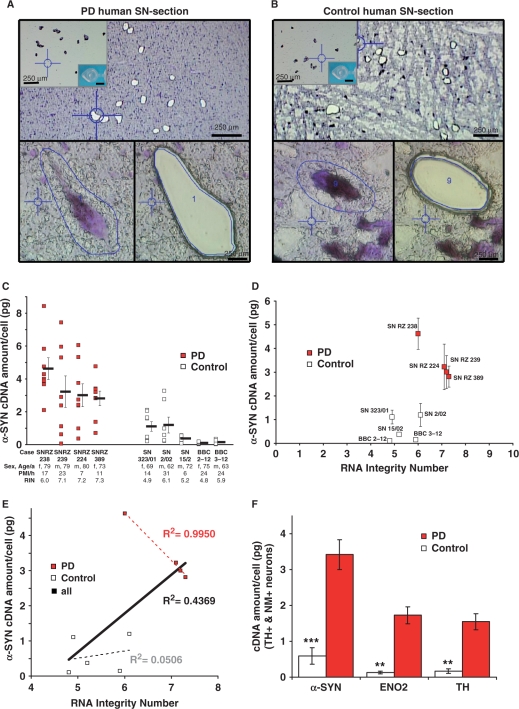
Figure 5.
LMD and mRNA-expression analysis of individual SN DA neurons from human PD and control postmortem brains. (A and B) Pools of neuromelanin-positive [NM(+)] neurons were isolated via LMD of cresylviolet-stained horizontal midbrain cryosections from PD (A) and control brains (B). Upper panel: PD (A) and control (B) cryosections after LMD of small pools of NM+ neurons from SNpc. Lower panels: Representative PD (A) and control (b) SNpc NM(+) neurons before (left) and after dissection (right). Insert: cap control after UV-LMD. Scale bars: 250 μm, 20 μm, respectively. (C) Scatter plot of α-synuclein gene-expression-levels in PD and control brains. α-Synuclein gene-expression of each pool of 15 NM(+) and TH(+) SNpc neurons is given as pg-equivalents of total cDNA derived from human SN-tissue per cell (standard curve quantification), determined via quantitative real-time PCR. Bars represent mean α-synuclein expression for SNpc pools of each brain ± SEM. (D) Plot of the mean α-synuclein cDNA levels (±SEM) against the RNA integrity number for each brain. Brain Bank codes are indicated next to each dot (see Figure 5C and Table 1). (E) Linear regression between mean α-synuclein expression and RNA integrity number for all individual analyzed control and PD brains showed no positive correlation between higher RNA quality of the tissue and detected α-synuclein expression-levels. (controls: black dotted line, R2 = 0.0506; PD brains: red dotted line, R2 = 0.9950; all analyzed brains combined: black line, R2 = 0.4369). Please note that PD brains showed a strong inverse correlation between RNA-integrity and detected α-synuclein expression-levels (red dotted line, R2 = 0.9950). (F) Mean expression levels of α-SYN, TH and ENO2 were significantly higher in individual NM(+) SN DA neurons from PD brains compared to controls (see Tables 1 and 2).
Table 1.
Human postmortem midbrain samples—characteristics and gene-expression
| BBC | Sex, Age/a | CERAD/ Braak | PMI/h RIN | cDNA amount per SN neuron (pg) (n/x) | Disease state | ||||
|---|---|---|---|---|---|---|---|---|---|
| TH | ENO2 | α-SYN | |||||||
| PD | SN RZ 238 | f, 79 | 0, II | 17 | 6.0 | 2.03 ± 0.40 (8/8) | 1.41 ± 0.19 (8/8) | 4.62 ± 0.66 (8/8) | PD |
| SN RZ 239 | m, 79 | 0, 0 | 23 | 7.1 | 1.40 ± 0.40 (8/8) | 2.23 ± 0.55 (8/8) | 3.23 ± 0.96 (8/8) | PD, AD, cardiac infarction, lung cancer | |
| SN RZ 224 | m, 80 | 0, II | 7 | 7.2 | 1.01 ± 0.18 (8/8) | 1.25 ± 0.30 (8/8) | 3.01 ± 0.70 (8/8) | PD, multisystem atrophy | |
| SN RZ 389 | f, 73 | 0, V | 11 | 7.3 | 1.76 ± 0.31 (8/8) | 2.01 ± 0.45 (8/8) | 2.81 ± 0.45 (8/8) | PD, ovary cancer, Sudeck's atrophy | |
| Control | SN 323/01 | f, 69 | A, I | 14 | 4.9 | 0.27 ± 0.08 (8/8) | 0.17 ± 0.08 (8/8) | 1.11 ± 0.29 (8/8) | diabetes, hypertonia, pneumonia, ARDS |
| SN 2/02 | m, 62 | 0, I | 31 | 6.1 | 0.38 ± 0.14 (7/8) | 0.27 ± 0.07 (6/8) | 1.20 ± 0.48 (7/8) | hypertonia, cardiac infarction | |
| SN 15/02 | m, 72 | 0, II | 6 | 5.2 | 0.08 ± 0.02 (8/8) | 0.06 ± 0.02 (8/8) | 0.38 ± 0.06 (8/8) | CMV, diabetes, kidney transplant, nephritis | |
| BBC 2-12 | f, 75 | 0, I | 24 | 4.8 | 0.06 ± 0.02 (8/8) | 0.04 ± 0.01 (5/8) | 0.11 ± 0.02 (8/8) | atherosclerosis | |
| BBC 3-12 | m, 63 | 0, 0 | 24 | 5.9 | 0.07 ± 0.02 (8/8) | 0.12 ± 0.12 (2/8) | 0.15 ± 0.04 (8/8) | liver cancer, hepatorenal syndrome | |
German BrainNet case number (BBC), sex/age, disease states classified according to the Consortium to establish a registry for Alzheimer's disease (CERAD) and according to the classification of Braak and Braak 1991 (Braak), postmortem interval (PMI), RNA integrity number (RIN) and cDNA amount per SN neuron (as pg-equivalents of total cDNA, derived from human SN-tissue) for the genes tyrosine hydroxylase (TH), neuron-specific enolase (ENO2) and α-synuclein (α-SYN) (mean ± SEM) are given for all analyzed PD and control brain tissues. Number of analyzed pools (x) and number of pools which expressed the tested gene (n) are indicated for each brain.
Table 2.
Significantly lower mean mRNA-levels of α-synuclein, tyrosine hydroxylase, and neuron-specific enolase in individual SN DA neurons for Parkinson's brains compared to control brains
| Target gene | Assay size | Parkinson's brains (n = 4) | Control brains (n = 5) | PD/Con | t-test P-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| cDNA amount per neuron (pg) ± SEM | Normalized mean quantity ± SEM | ANOVA P-value | cDNA amount per neuron (pg) ± SEM | Normalized mean quantity ± SEM | ANOVA P-value | ||||
| α-SYN | 62 bp | 3.42 ± 0.41 | 5.81 ± 0.70 | 0.2853 | 0.59 ± 0.24 | 1.00 ± 0.40 | 0.0033 | 5.81 | 0.0004 |
| TH | 73 bp | 1.55 ± 0.22 | 9.05 ± 1.30 | 0.1771 | 0.17 ± 0.07 | 1.00 ± 0.38 | 0.0077 | 9.05 | 0.0060 |
| ENO2 | 77 bp | 1.73 ± 0.24 | 13.22 ± 1.80 | 0.2645 | 0.13 ± 0.04 | 1.00 ± 0.32 | 0.0968 | 13.28 | 0.0056 |
Mean cDNA quantity determined via quantitative real-time RT–PCR of UV-LMD individual neuromelanin-positive SN DA neurons from control and PD brains of α-SYN, TH and ENO2 (compare Table 1). Mean cDNA quantities, normalized to control brains. Amplicon size of the utilized TaqMan real-time PCR assay (assay size) is given as well as the relation of gene-expression between PD and control tissue (PD/Con). ANOVA tests were performed to test for variances of gene-expression within either group (PD and control). P-values of two-tailed t-test indicated significant differences between mean expression levels of all three tested genes between control and PD brains.
Fixation and staining of mouse- and human-brain cryosections prior to UV-LMD
The fixation and staining procedure of cryosections was the same for mouse and human brain sections: immediately after cryosectioning and mounting on PEN-membrane slides, the sections were shortly fixed with RNAse-free ethanol [75% at −20°C, 75%, 95%, 100% (Roth), 100% with molecular sieve 0.3 nm (VWR), at room temperature], stained with sterile-filtered cresylviolet (Sigma, 1% solution in 100% ethanol) and dried in a chamber with Silica Gel (Merck) for at least 40 min at room temperature before use for experiments or long-time storage. For long-term storage, either directly after cutting and fixation/staining, or after UV-laser-microdissection, LMD slides were placed upright in a sterile 50 ml Falcon tube with Silica Gel in the conus, sealed with parafilm and stored at −80°C. Before LMD usage of frozen slides, these sections were allowed to equilibrate (in falcon tube on silicagel) for 15 min at −20°C, followed by 15 min at 4°C and finally for 15 min at room temperature, prior to opening of the falcons and UV-LMD of the sections.
Tissue RNA extraction, RIN measurements and cDNA synthesis
Total RNA of human brain tissue was extracted from spare slices and chippings obtained during the cryosection procedure, without fixation and cresylviolet-staining. RNA from these sections was extracted according to the manufacturer's instructions using suited RNAeasy kits (Qiagen). Eluted total RNA was precipitated with 1/10 vol sodium acetate (Ambion) and 3 vol of 100% ethanol (Sigma) in the presence of 1 μg glycogen (Ambion) and 300 ng poly-inosin (Sigma). The precipitated RNA was washed with 80% ethanol, resuspended in 2 U/μl SUPERase-In (Ambion) and stored at −80°C. For human brains, RNA integrity numbers (RIN) were determined with the Agilent 2100 Bioanalyzer and the RNA 6000 Nano LabChip Kit (46) from the RNA isolated from unfixed, unstained spare cryosections and chippings. Reverse transcription of extracted total RNA was carried out in 1× first-strand buffer (Invitrogen), 10 mM DTT (Invitrogen), 400 U SuperScriptII (Invitrogen), 0.5 mM dNTPs (Pharmacia), 5 μM random hexamer primer (Roche) and 40 U SUPERase-In (Ambion) in a final volume of 10 μl for 2 h at 37°C (Thermomixer, Eppendorf) as described (47).
UV-Laser-microdissection and cDNA synthesis of microdissected cells
A contact-free Leica LMD6000 UV-Laser microdissection system (diode-laser 355 nm; Leica) was used. Cresylviolet-stained neurons were visualized and cut under brightfield (phase contrast, 63×-fold magnification). Individual neurons were harvested by gravity directly into an RNAse-free UV-C-treated cap of a 0.5 ml, nuclease free thin-walled reaction tube (Applied Biosystems). Successful harvesting was controlled by visual inspection of the cap. A mixture for combined cell-lysis and cDNA synthesis was freshly prepared for each experimental day [0.5% NP-40 (Roche Diagnostics), 5 U SUPERaseIn (Ambion) 0.5 mM dNTPs (Pharmacia), 5 μM random hexamer primers (Roche), 500 ng poly-inosin (Sigma), 10 mM Tris–HCl (Sigma), 10 mM DTT (Sigma) in 1× first-strand buffer (Invitrogen)], and was added directly into the cap. The tube (thin-walled PCR, Applied Biosystems) was closed, incubated upside down for 2 min at 72°C (Combibox, Thermostat, Eppendorf), cooled on ice for 1 min, spun down for 60 s full speed (MiniSpin, Eppendorf) and quickly cooled on ice for 10 s, prior to adding 60 U reverse transcriptase (SuperScriptII, Invitrogen). cDNA synthesis was carried out in a final volume of 5 μl at 38°C overnight (Combibox, Eppendorf).
Qualitative and quantitative PCR
Qualitative and quantitative real-time single cell PCR was carried out essentially as described (47,48). For sensitivity experiments, mouse cDNA was split and 80% was used as template for the qualitative marker PCR and 2 × 10% for Eno2 real-time PCRs (duplicate reactions). Mouse-primer-sequences for qualitative marker PCR are as described (48), mouse Eno2 assay: Mm00469062_m1 (FAM, and nonfluorescent quencher-labeled), PCR fragment size 76 bp. For analysis of human LMD samples, 10% aliquots of cDNA (estimating about 1.5 cells) were used to study the expression of each gene. Real-time TaqMan assays were either predesigned by Applied Biosystems or self-designed (Primer Express software 1.5, Applied Biosystems). All predesigned assays were labeled with FAM as reporter and a nonfluorescent quencher. The two self-designed assays (TH and ENO2 assay A = ENO2-A) were labeled with FAM as reporter and TAMRA as quencher. The self-designed TH assay detected all three TH transcript variants. Human TH: Primer: F-1117-GTCCACGCTGTCATGGTTCA; R-1190-CGGCACCATAGGCCTTCA; Probe: 1145-CCCGTTCTGCTTACACAGCCCGAA; fragment size: 73 bp; (ENO2-A): Primer: F-1053-GGCACTCTACCAGGACTTTGTCA; R-1161-GATCCCTACATTGGCTGTGAACTT, Probe-1079-ACTATCCTGTGGTCTCCATTGAGGACCCATT; fragment size: 108 bp. Applied Biosystems identifying assay IDs: human ENO2 assay B = ENO2-B: Hs00157360_m1, fragment size: 77 bp; human α-SYN: HS00240906_m1, fragment size: 62 bp. All assays were tested for performance and reproducibility by generation of relative standard curves using serial dilutions of cDNA generated from brain tissue as templates (over three to four magnitudes, triplicate reactions). Assay performance was assessed on serial dilutions (1:1000, 1:10 000, 1:100 000) of human cDNA derived from SN-tissue [human brain substantia nigra Quick clone cDNA, generated from poly(A)+ RNA; Clontech, 5.6 ng/μl, determined with Picodrop photometer (Biozym)] by analysis of standard deviation for replicates and slopes of standard-curves as described (47,49). cDNA expression levels of small cell pools were calculated in regard to assay performance derived from the respective total cDNA standard curves (slope and Y-intercept of standard curves) as: 10[(Ct−Y-intercept)/slope] and normalized to cell-numbers. Expression levels are given as pg-equivalents of total cDNA derived from SN-tissue per cell (standard curve quantification). Slopes and Y-intercepts for human α-SYN, TH and ENO2-B were −3.41 and 37.26, −3.47 and 37.59 as well as −3.25 and 36.93, respectively. For mouse Eno2, standard curve was generated using cDNA, derived from poly(A)+ RNA from mouse midbrain-tissue; slopes and Y-intercept were −3.54 and 43.2, respectively. Data were not normalized to a reference gene. Reverse transcription conditions were the same for all LMD pools.
Statistics
All data were analyzed with Excel (Microsoft) or Igor (Wavemetrics) software, and given as mean ± SEM. To evaluate statistical significance, we used Student's t-tests, paired t-tests and ANOVA-analysis. Normally distributed, parametric data were compared by a two-tailed, unpaired t-test. A value P < 0.05 was considered to be statistically significant and indicated by asterisk (P < 0.01 and P < 0.001 indicated as ** and ***, respectively).
RESULTS
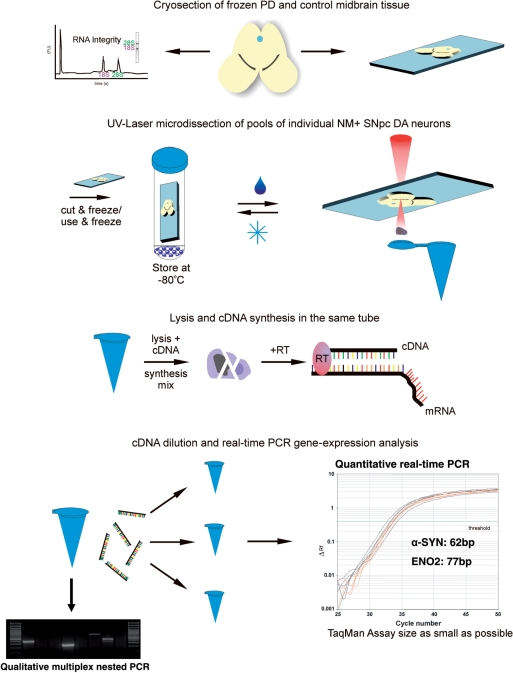
Here, we detail a novel protocol for contact-free UV-Laser-microdissection (LMD) and subsequent RT–PCR-based quantitative gene-expression analysis of single cells and homogeneous small cell pools, optimized for human postmortem tissue. It was developed, optimized and evaluated using a Leica LMD6000 diode-laser (355 nm) setup (50,51). Figure 1 illustrates the general principle of this technique as well as the experimental design of this study for the analysis of α-SYN mRNA levels in individual neuromelanin-positive [NM(+)] DA SN neurons from human iPD brains and controls. To minimize handling steps, we performed cell lysis, cDNA synthesis and qualitative PCR in the same tube in subsequent reactions without a distinct RNA isolation step (see Methods section). An important prerequisite for reliable quantitative gene-expression analysis of LMD-samples is that the RNA quality does not progressively decrease with increasing LMD-harvesting time. In addition, in particular when working with precious human tissues, long-term storage of fixed, stained sections as well as re-use of sections for LMD is highly desired. Thus, as illustrated in Figure 1 and demonstrated in Figure 2, we established and optimized a protocol for sectioning, staining and UV-LMD of brain tissue that allowed (i) long-term storage and re-use of the tissue sections, and (ii) UV-LMD collection of cells over a period of at least 7 h, both without detectable decrease of RNA quality. As shown in Figure 2, we assured that our procedure allowed long-time storage of fixed, stained sections at −80°C after LMD and re-use of sections without detectable loss of RNA quality by harvesting small pools of 15 individual SN DA neurons from coronal mouse-brain sections via UV-LMD, and comparing both qualitative marker-gene expression as well as quantitative real-time PCR results for neuron-specific enolase in parallel. Figure 2A (left panel) illustrates the typical localization of SN DA neurons in coronal mouse brain sections and UV-LMD of the whole substantia nigra pars compacta (SNpc) tissue. Figure 2B shows the UV-LMD harvesting procedure of individual cresylviolet-stained neurons from mouse-brain SNpc. Successful harvesting was verified by optical control of the reaction tube cap after LMD (Figure 2B, lower right panel). After LMD, slides with tissue sections were stored at −80°C in a specially developed storage jar (see Methods section for details). After 1–2 weeks, we thawed the same sections and again collected similar SN DA pools for respective gene-expression analysis. Positive markers for SN DA neurons were: tyrosine hydroxylase (TH), the rate-limiting enzyme for dopamine synthesis, the dopamine transporter (DAT), the dopamine receptors (in particular D2 long and/or short splice variants) and G-protein coupled inwardly rectifying potassium channels (Girk2). Calbindin-d28k (CB), is expressed in small subpopulation of SN DA neurons (as shown in Figure 2C, third panel), l-glutamate decarboxylase 67 (GAD67) is expressed in GABAergic neurons and glial fibrillary acidic protein (GFAP) is a marker for astroglial cells. Expression of all tested genes was detected at tissue level (LMD of a whole substantia nigra containing dopaminergic, GABAergic and astroglial cells (52) (compare Figure 2A, right panel). In contrast, a reproducible expression pattern of only the expected subset of selective DA marker genes (TH, DAT, Girk2, D2) but not those of GABAergic neurons (GAD) or astroglial cells (GFAP) was found in all analyzed TH-positive UV-LMD-pools of SN neurons (n = 20, Figure. 2B and C). These results demonstrated the selectivity of the collected cell pools for DA neurons and thus the specificity of our approach (no accidental LMD collection of GABAergic or glial-cells). Importantly, no significant difference between Eno2 detection levels (i.e. mean Ct-values and variances) in pools of UV-LMD SN DA neurons from fresh slides compared to those from re-used slides was detected (Figure 2D and E; fresh: Ct = 34.43 ± 0.51; n = 10; frozen, re-used: Ct = 34.62 ± 0.48; n = 9; P = 0.78; calculated respective expression per cell (pg equivalents of total cDNA of midbrain-tissue) were: fresh: 29.07 ± 6.58 pg; frozen: 23.15 ± 4.64 pg; P = 0.481; all probes amplified in parallel in the same PCR-run, Eno2, 76-bp PCR-assay).
Figure 1.
Flow-chart of experimental procedure. Flow-chart representing the experimental procedure of the protocol for UV-LMD and quantitative RT–PCR gene-expression analysis of individual SN DA neurons from human postmortem PD brains and controls.
In a similar way, we ensured that gene-expression results were not affected over time during UV-LMD harvesting. We collected comparable pools of 15 SN DA neurons (again identified via expression of the dopaminergic marker-gene set) (i) directly after fixation of tissue-sections and (ii) 3 h after fixation and storage of sections at room temperature (two independent experiments, each LMD-harvesting procedure lasted about 3 h). As desired, we did not detect any significant differences in Eno2 detection thresholds (same PCR-run) between early collected UV-LMD samples [Ct = 34.39 ± 0.24, n = 9; calculated expression (pg equivalents of total cDNA of midbrain-tissue): 23.16 ± 4.72 pg] and samples harvested with a 3–7 h delay [Ct = 34.25 ± 0.32, n = 8; P = 0.553; calculated expression: 26.62 ± 6.62pg; P = 0.671; data not shown].
After having established these crucial prerequisites, we utilized the protocol to harvest neuromelanin-positive [NM(+)] DA neurons of substantia nigra pars compacta (SNpc) from human postmortem brains from iPD patients and matched controls for subsequent quantitative RT–PCR analysis. Human postmortem midbrain samples (four iPD brains and five controls included into this study) were obtained from the German BrainNet (www.brain-net.net). As expected, midbrain-tissue from iPD cases showed remarkable reductions in the numbers of NM(+) SN neurons (Figure 3A and B). We observed that the SN of human control-sections showed reduced tissue integrity with a larger number of ruptures compared to the SN of PD cases. However, the dopaminergic cell morphology was well preserved in both groups (compare Figure 3A and B). Mean age of death was 68.2 ± 2.5 years and 77.8 ± 1.6 years for controls and PD patients, respectively (P = 0.020). The mean postmortem interval was not significantly different (Figure 3C; 19 h 48 min ± 4 h 23 min for controls and 14 h 30 min ± 3 h 30 min for PD patients; P = 0.395). Available clinical details of individual cases are summarized in Table 1.
As different degrees of RNA degradation among the individual postmortem brain samples are to be expected (due to biological and/or methodological issues), comprehensive assessment of respective RNA quality is crucial. We analyzed RNA quality of all human midbrain tissues of this study from spare, unfixed, unstained sections and chippings collected during cryosectioning and before mounting of tissue for LMD using the Agilent Lab-on-a-Chip System, which calculates the RIN. The RIN is a more sophisticated measure of RNA quality, superior to the determination of the 28S/18S ratios (43,46,53). A complex algorithm [detailed in (46)] assigns a RIN score in the range from 1 to 10 to each sample, where a score 10 represents completely intact RNA, and a score 1 fully degraded RNA (46,53). The RNA from the human brain samples in this study was moderately to slightly degraded with RIN numbers ranging between 4.8 and 7.3. The mean RNA integrity in PD brains (RIN = 6.9 ± 0.3) was significantly higher compared to controls (RIN = 5.4 ± 0.3; P = 0.007, Figure 3D). It has been previously reported that the probability of skewed quantitative RT–PCR results dramatically depends on PCR-assay sizes. Assay sizes smaller than 150 bp have been reported to be suited for reliable real-time RT–PCR given a RIN > 5 (53–55). To evaluate this proposed correlation in our study experimentally, we compared two different human ENO2 TaqMan assays (ENO2-A and ENO2-B) with different amplicon sizes of 108 bp and 77 bp, respectively. For real-time PCR, we used two different sets of serial dilutions of human brain cDNA as templates, generated from (i) RNA with very high integrity (whole-brain cDNA purchased from Clontech, RIN-number not determined, but photograph of RNA gel electrophoresis provided, demonstrating the high RNA integrity) and (ii) from the partly degraded RNA isolated from BrainNet sample SN 2/02 (RIN = 6.1). Using the cDNA from nondegraded whole-brain RNA as template, similar over-all PCR efficiencies and respective standard curves were obtained for both, the large and the small ENO2 PCR assay. [Figure 4A and C; standard curve ENO2-A (108 bp): slope: −3.38, R2 = 0.99; ENO2-B (77 bp): slope: −3.65, R2 = 0.99]. However, using the cDNA derived from partly degraded RNA (SN 2/02, RIN = 6.1) as template, we received dramatically different real-time PCR results and respective standard curves depending on assay size. While the 77-bp ENO-assay still allowed quantification over three magnifications of template DNA, similar as for high-quality RNA, a meaningful standard curve for the 108-bp ENO2-assay could not be generated. Thus, reliable gene-expression quantification was not possible with the 108-bp assay using partially degraded RNA (Figure 4B and D, standard curve ENO2-A: slope: −1.69, R2 = 0.66). Given these findings, we decided to use the smallest available α-synuclein real-time TaqMan PCR assay for the quantitative RT–PCR expression analysis of UV-LMD NM(+) SN neurons from the human postmortem brains (see Methods section for details). In addition, we tested the performance of the chosen 62-bp α-synuclein assay on serially diluted cDNAs derived from commercially available SN tissue RNA (Ambion, RIN 6.4) and our cDNA from SN 2/02 (RIN = 6.1). In contrast to the 108-bp ENO2 assay, we observed no significant difference between the slopes of respective standard curves for both cDNA sources as well as a near ideal slope for the α-synuclein assay of −3.38 (R2 = 0.98; data not shown).
Similarly as described earlier for mouse-brain sections, pools of 15 individual NM(+) SN DA neurons with well-defined cell borders were collected via UV-LMD from each human postmortem brain after sectioning, fixation and staining and analyzed (compare Figure 5A and B). Eight individual SN DA pools of each human midbrain were harvested, reverse transcribed and analyzed via quantitative PCR for the RNA-expression-levels of α-SYN (62-bp assay), ENO2 (77-bp assay) and TH (73-bp assay). Individual α-synuclein expression levels for all analyzed cell pools of NM(+) and TH(+) (detected via RT–PCR) SNpc neurons from human PD and control brains are given in the scatter plots of Figure 5C. With our approach, we detected no significant differences between the α-SYN cDNA levels of the different brains within the PD group via ANOVA analysis (Table 2). However, the gene expression of brains from the control group was identified as not homogeneous (ANOVA P-values <0.05 for TH and α-SYN, Table 2), i.e. control brains SN 323/01 and SN 2/02 displayed significantly higher expression levels for TH and α-SYN genes compared to the other control brains. These inhomogeneities were not associated with differences in sex, age, PMI or RIN (Table 1 and Figure 5C–E). Importantly, we detected significantly higher α-synuclein expression levels (about 6-fold) in surviving individual NM(+) and TH(+) SN DA neurons from PD brains compared to unaffected controls (Figure 5C–F; control = 0.59 ± 0.24 pg per cell, n = 5; PD = 3.42 ± 0.41 pg per cell, n = 4, P = 0.0004). As evident from Figure 5D, even for comparable RINs, α-synuclein expression levels were higher in PD brains (compare PD brain SN RZ 238 to control brains SN 2/02 and BBC 3-12). In addition, there was no positive correlation between higher RIN numbers (i.e. higher RNA integrity) and detected higher α-synuclein expression levels for control brains as well as for PD brains (Figure 5E, R2 = 0.0506 for control brains, 0.9950 for PD brains and 0.4369 for all pooled brains). This argues against the possibility that higher α-SYN expression in surviving SN DA neurons from PD brains simply reflected their higher RNA qualities. The strong inverse correlation (R2 = 0.9950) found between RIN number and gene-expression levels for the PD brains further supports this notion, as detected α-SYN gene-expression levels decrease but not increase with higher RIN numbers (reflecting higher RNA integrities).
For ENO2 and TH, we also detected significantly higher gene-expression-levels per cell (about 10-fold) in pools of SN NM- and TH-positive PD brains compared to controls (Figure 5F; ENO2: control: 0.13 ± 0.04 pg, n = 5; PD: 1.73 ± 0.24 pg, n = 4; P = 0.0056; TH: control: 0.17 ± 0.07 pg, n = 5 versus PD: 1.55 ± 0.22 pg, n = 4; P = 0.0060).
To further rule out artificial quantitative RT–PCR results for human TH, ENO2 and α-SYN due to potential differences in the respective rates of progressive RNA degradation in SN DA neurons from human brain sections before and after fixation and staining, we have repeated the complete set of UV-LMD and RT–qPCR experiments for these three genes after one additional year of storage of the unfixed brain tissue-blocks at −80°C. In addition, in contrast to the first set of experiments, where fixed, stained brain sections were directly used for UV-LMD, this time we stored the fixed and stained brain sections on PEN-membrane slides for 1 week at −80°C prior to UV-LMD and subsequent quantitative RT–PCR analysis. Again, we detected significantly higher mRNA levels of α-SYN, TH and ENO2 in SN DA neurons from PD brains compared to controls (see Supplementary Figure). More important, the detected differences in gene-expression levels between PD and control brains for a-SYN, ENO2 and TH were not significantly different compared to those of the previous experimental series (Paired t-tests: a-SYN: PD: P > 0.2; control: P > 0.2; ENO2: PD: P > 0.2; control: P > 0.2; TH: PD: P > 0.2; control P > 0.2; see also Supplementary Figure). This robust replication of the detected quantitative differences in gene-expression between NM(+) and TH(+) SN DA neurons of PD and control brains after extended storage of human brain tissue blocks and fixed stained human brain-tissue sections, demonstrates that these results are likely to reflect genuine biological differences and are rather not significantly affected by progressive RNA degradation due to the experimental procedures.
DISCUSSION
In the present study, we detected significantly higher levels of α-synuclein mRNA in individual neuromelanin (NM)- and TH-positive dopaminergic substantia nigra neurons from postmortem PD brains compared to unaffected controls by combining UV-LMD with quantitative real-time PCR. In contrast to previous tissue-based studies, our approach demonstrates for the first time a cell-type specific transcriptional upregulation of α-synuclein in SN DA neurons (from PD brains), a neuronal key population in PD. In addition, we provide a step-to-step description and extensive experimental evaluation of the utilized single-cell UV-LMD protocol including tissue-section preparation and their long-term storage that allows the reliable application of laser-microdissection with contact-free UV-LMD-systems in combination with quantitative RT–PCR analysis of individual cells, in particular from human postmortem tissue. Based on our well-established RT–PCR protocols for single living SN DA neurons and single fixed SN DA neurons from mouse brain sections (47,49), we optimized, validated and utilized this protocol for fixed postmortem human brain tissue. All data shown here were generated with the Leica LMD6000 system. However, we have also tested our protocol for another contact-free UV-LMD system, the PALM-LMD, again with reliable results (data not shown). In contrast, we have not tested this protocol with the Arcturus IR-LMD system as it utilizes a different technical approach including physical contact, and thus might be less suitable for selective single cell analysis (51). In general, tissue sections for Arcturus IR-LMD exhibit properties distinct from the UV-LMD systems and thus require different fixation and lysis protocols for successful RT–PCR of LMD-samples (56,57).
The quantitative results in this study demonstrated the selectivity, sensitivity and reproducibility of our rapid and easy to perform protocol by analyzing gene-expression of individual dopaminergic neurons from mouse midbrain via qualitative and quantitative RT–PCR (compare Figures 1 and 2). Our detailed protocol allows quantitative RT–PCR based (or after global amplification, microarray based; data not shown) expression profiling of individual cells after UV-LMD, with a similar high sensitivity and reproducibility as described for ‘living’ SN DA cells after cytoplasm harvesting via patch-pipette (47,49,58). We have further successfully applied this protocol for LMD analysis of individual fluorescence labeled mouse dopaminergic midbrain neurons (48) as well as for human SNpc DA neurons from postmortem brain sections (50). We omitted a DNAse digestion step [as described in (58)] for the analysis of human SN DA neurons, as all TaqMan assays used in this study were designed to be intron-spanning, thus amplifying only mRNA-derived cDNA. However, with a respective genomic TaqMan assay we were also able to amplify genomic DNA of single cell samples [compare (58)]. Thus, our protocol is sensitive enough to detect the two genomic DNA copies within a single nucleus.
Here, we employed our protocol to analyze α-synuclein mRNA levels of individual NM(+) and TH(+) DA substantia nigra pars compacta (SNpc) neurons of human postmortem brains from iPD patients and matched controls. As a causal—but mechanistically not well-defined—role of α-SYN for dopaminergic degeneration in PD has been demonstrated, analyzing α-SYN expression in a cell-specific and quantitative fashion is highly relevant. Differences in a-SYN expression levels might reflect different pathophysiologic stages of DA neurons en route either toward degeneration or survival and compensation in PD. As already noted, up- or downregulation of α-SYN expression at the tissue level has been reported in iPD (37–42). Necessarily, none of these tissue-based studies could have been selective for NM(+) SN DA neurons but rather used the highly heterogeneous substantia nigra tissue (and other complex brain tissues), consisting of many different α-SYN-expressing cell-types in addition to DA neurons [but note that Kingsbury et al. (41) carried out in situ hybridization]. The progressive and variable loss of DA SN neurons in iPD, which inevitably alters the relative proportion of DA neurons in the SN, was a further confounding factor for these tissue-based studies. In this context, the described increased ectopic expression of α-SYN in other cell types, e.g. glial cells during the course of the disease (59, 60), might provide further problems for tissue-based studies. Normalization procedures using so-called housekeeping genes as reference genes are of great value when studying gene-expression at the level of tissues (61–63), but they cannot compensate for dwindling content of DA neurons in SN tissue [and are in most cases not homogeneously expressed at the single cell level (49)]. In summary, all these confounding factors are effectively eliminated by cell-specific expression analysis as described here.
With our cell-specific approach with single cell resolution, we compared α-SYN mRNA-levels of small pools (15 neurons each) of individual NM-positive SN DA neurons from controls with the same number of surviving NM-positive SN DA neurons from PD brains via quantitative real-time RT–PCR. We detected about 6-fold higher mRNA-level of α-SYN mRNA in surviving NM(+) and TH(+) SN DA neurons from iPD brains compared to controls. These data do not distinguish between two potential explanations: either an upregulation in α-SYN gene-expression in surviving DA SN neurons in response to the progression of PD, or a selective survival of those SN DA neurons with constitutively elevated α-SYN expression. However, protection of SN DA neurons due to higher α-SYN levels would be at odds with the findings that duplication and triplications of the wild-type SNCA-gene lead to familiar PD (PARK4) (16). On the other hand, a recent study clearly demonstrated that α-SYN protects nerve terminals against injury and neurodegeration (32). In addition to these opposing roles for α-SYN function in vivo, α-SYN expression in neuronal cultures has been described both as beneficial or adverse for neuronal survival (studies summarized in Table 1 of ref. 37,64). As α-SYN appears to be involved in vesicle release (3,31,34), a higher expression of α-SYN might modulate striatal dopamine release in the striatum and compensate for dopamine loss. In this context, it is interesting to note that increased α-SYN expression has also been reported in response to cocaine, amphetamine and ethanol abuse, suggesting an additional pathophysiologic role of α-SYN in the context of drug abuse (65–69). In summary, while we established direct evidence for transcriptional upregulation of α-SYN mRNA in DA SN neurons in iPD, its pathophysiologic consequences—e.g. elevated levels of α-SYN protein but also the potential presence of different α-SYN structural states like unfolded random coiled, soluble oligomers, α-pleaded sheet and insoluble fibrillar structures (20, 21,31,34,64,70)—have still to be elucidated.
An additional level of complexity might be related to changes in alternative splicing of SCNA. Indeed, significantly higher mRNA levels of the α-SYN 112 splice variant have been reported in frontal cortex tissue of DLB brains compared to controls, whereas contrary, the splice variants α-SYN 126 and 140 where expressed at significantly lower levels (42,71). This finding is particularly striking, as the α-SYN 112 splice variant is more prone to form fibrillic conformational structures due to the absence of the dopaminochrome recognition site in exon 5 and the shortened C-terminus of the protein (7). In contrast, α-SYN 126 exhibits diminished aggregation propensity, compared to α-SYN 112 and α-SYN 140 (7). Thus, alternative splicing might also contribute to the heterogeneous findings concerning α-SYN expression, but cell-specific data are missing. The 62-bp TaqMan assay used in our study and the study of Dachsel et al. (39) spans the SNCA-exon 3/4 boundary and therefore detects (but not discriminates) both the α-SYN 140 as well as α-SYN 112 variant, but not the α-SYN 126 splice variant. In contrast, the 104-bp TaqMan assay utilized by Chiba-Falek and Papapetropoulos (37,38) spans the exon4/5 boundary and thus detects both the α-SYN 140 as well as α-SYN 126 variant, but not the α-SYN 112 variant. Although α-SYN 140 has been described as the most relevant and abundantly expressed variant (6), the relative expression ratios of all three splice variants might be crucial in context of iPD (7).
We also detected higher mRNA levels of tyrosine-hydroxylase and neuron-specific enolase (ENO2) in individual surviving SN DA neurons from PD brains, which might indicate a general transcriptional upregulation to counteract the progressive loss within this neuronal population in PD. Respective data for TH-expression in the literature (without single cell resolution) are similarly controversial as those for α-SYN, and splice variants were not addressed systematically (72–78). In general, concerted upregulation of gene-expression has already been described for other related genes at the single-cell level (79,80).
Finally, we focus on specific methodological issues relevant when analyzing human postmortem material, but will not further discuss general methodological issues and caveats of qualitative and quantitative single cell gene-expression profiling that have been extensively reviewed elsewhere (43–45,58,81–83). A prerequisite for the analysis of quantitative gene-expression is that the respective RNA remains stable throughout the fixation/staining and extended LMD-harvesting procedure (performed at room temperature). Currently, the extent of this potential problem for LMD-based gene-expression profiling as well as the use of cresylviolet-stained sections for RNA analysis after LMD is controversially discussed (51,56,82,84–86,87). In our hands, cresylviolet-stained sections gave similar RT–PCR results for individual SN DA neurons after UV-LMD as those collected from unstained fixed sections [where we harvested fluorescence-labeled neurons after retrograde tracing in vivo; (48) Lammel et al., unpublished data]. However, we emphasize, that, in order to avoid RNA degradation, we modified the staining protocol by minimizing incubation times and staining in 100% ethanol, to quickly and completely maintain dehydration of tissue sections. Our protocol for tissue-section preparation allowed the LMD harvesting of samples over several hours at room temperature without significant degradation of RNA. Furthermore, fixed-stained tissue sections of mouse and human brains can be stored after LMD at −80°C for at least 1 week for later re-use without loss of sensitivity (compare Figure 2 and Supplementary Figure). This possibility of re-use is of particular importance, if mRNA of unique postmortem human tissues is to be studied. We also identified the importance of another confounding factor i.e. the variable, mostly suboptimal RNA qualities of the human postmortem tissues, as evident by Agilent RIN analysis. RIN-values allow a very detailed, exact assessment of RNA-integrity, compared to judgment of RNA quality via 28S/18S rRNA analysis (53, 55,88). We would like to emphasize that RIN values of human brain sections were determined in this study after RNA extraction from spare cryosections that were not ethanol-fixed and cresylviolet-stained. Due to the limited amount of RNA from single cells, and as we performed cDNA synthesis of LMD samples without a distinct RNA-isolation step, it was not possible to determine RIN values from UV-LMD samples. However, as the fixation and staining protocol was particularly optimized in order to avoid progressive RNA degradation (as discussed earlier) and procedures and chemicals were exactly the same for all experiments, a potential influence on RNA quality due to fixation and staining is expected to be similar for all samples. In addition, we have demonstrated for mouse brain sections that after fixation and staining, RNA of cryosections was not further degraded—at least for ENO2 mRNA (compare Figure 2D and E). To particularly exclude artificial RT–qPCR results for human TH, ENO2 and α-SYN due to different degrees of progressive RNA degradation in SN DA neurons from human brain sections after fixation and staining, we have repeated UV-LMD and RT–qPCR expression analysis of these genes. We collected SN DA neurons from human midbrain sections which were kept for 1 week at –80°C after cryosectioning, fixation and staining. Sections were cut from the same human brain samples as before (stored for over 1 year at –80°C). The obtained differences in gene-expression levels of SN DA neurons between PD and controls were not significantly different compared to those obtained in the previous experiments on these human brain sections (used for UV-LMD directly after fixation/staining). These results further support that progressive RNA degradation due to the experimental procedure that does not affect the here reported differences in gene-expression (compare Supplementary Figure).
Our comparison of the RIN numbers and the postmortem interval (PMI) of the brains showed, that PMI had no obvious influence on RNA integrity (Figure 3C and D, Table 1), which is in accordance with previous findings (44,45, 89). It has already been demonstrated that expression analysis of degraded mRNA is possible, as long as RIN-values and thus RNA qualities are comparable (53,54,63,88). Furthermore, Fleige and coworkers (53,55) concluded that samples with RIN-differences of +1 are maximally overestimated by factor 3, however most tested genes showed an artificial difference of about 0.7 Ct values per 1 RIN number difference, which would correspond to an artificially 1.6× higher detected cDNA amount. In addition, they showed that this effect was nearly abolished by the use of quantitative PCR amplicon sizes <100 bp (53,55). Here we performed a similar analysis by comparing two different ENO2 assays with (108- and 77-bp amplicon size) and found that quantitative PCR assay-size around 100 bp (in contrast to the 77-bp assay) still did bias the comparison of gene-expression levels if compared RNA had significantly different RNA integrities (compare Figure 4). The α-SYN real-time PCR-assay used in this study generated a very small PCR amplicon of only 62 bp, which was not likely to be affected by RIN differences. Furthermore, samples of control and PD brains with overlapping RIN values also represented the higher expression of α-synuclein in PD (compare Figure 4 and Table 1). The detected differences in ENO2, TH, α-SYN expression-levels were about 13-, 9- and 6-fold, respectively (Table 2). As we have recently shown, the expression of another gene responsible for familiar PD, the p-type ATPase 13A2 (PARK9) was expressed at about 11-fold higher levels in NM(+) SN DA neurons of these PD brains compared to controls (50). Nevertheless, the mean RNA integrity difference of 1.5 between our controls and PD samples provides a factor that might—despite the very small PCR-assay size chosen—contribute to the differences in gene-expression detected here.
Taken together, careful evaluation of RNA integrity is of particular importance when working with—partly degraded—human postmortem material, and absolute differences in gene-expression need to be taken with a degree of caution, even when RNA integrity and PCR amplicon sizes have been taken into account. Unfortunately, in none of the cited tissue-based studies, neither RNA integrities (in particular RIN-values) of individual cases were given, nor the suitability of the utilized PCR assays (all >100 bp) were assessed. In summary, our protocol for tissue-sectioning, fixation, long-term storage, UV-LMD and subsequent one-tube lysis and cDNA-synthesis of individual neurons in combination with smallest possible real-time PCR assay-size and definition of RNA integrity is well suited for cell-specific RT–PCR-based quantitative mRNA expression profiling of partly degraded human postmortem tissue. Compiling carefully controlled cell-specific RNA expression profiles in a quantitative fashion from individual cells will help to sharpen our concepts of cellular identity and diversity and provide important information about the molecular mechanisms of cell-specific adaptations and plasticity—not only in context of PD but in general health and disease states.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank D. Isbrandt for Agilent analysis of human RNA, R. Veh for help with human anatomy, J. Roeper for critical discussion, and Leica Microsystems for providing the LMD6000 and technical support. We are especially grateful to the brain donors. Human brain samples were obtained from BrainNet (GA28). This work was supported by Bundesministerium für Bildung und Forschung, BmbF-NGFN-II (TP: N1NV-S30T10, FKZ: 01GS0471), Gemeinnützige Hertiestiftung (Grant-No: 1.319.204/03/01), The Parkinson's Disease society, UK, the Royal Society. J.G. was supported by a stipend from the Medizinstiftung der Philipps-Universität Marburg and is supported by a PhD Studentship in Neuroscience of the Wellcome Trust, B.L. was a Dorothy Hodgkin Royal Society research fellow, and is supported by the Alfried Krupp prize for young university teachers of the Krupp foundation. Funding to pay the Open Access publication charges for this article was provided by the University of Ulm, Germany.
Conflict of interest statement. None declared.
REFERENCES
- 1.Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson's disease. Annu. Rev. Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 2.Przedborski S. Pathogenesis of nigral cell death in Parkinson's disease. Parkinsonism Related Disord. 2005;11:S3. doi: 10.1016/j.parkreldis.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Farrer MJ. Genetics of Parkinson's disease: paradigm shifts and future prospects. Nat. Rev. Genet. 2006;7:306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 4.Klein C, Lohmann-Hedrich K. Impact of recent genetic findings in Parkinson's disease. Curr. Opin. Neurol. 2007;20:453–464. doi: 10.1097/WCO.0b013e3281e6692b. [DOI] [PubMed] [Google Scholar]
- 5.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 6.Campion D, Martin C, Heilig R, Charbonnier F, Moreau V, Flaman JM, Petit JL, Hannequin D, Brice A, Frebourg T. The NACP/synuclein gene: chromosomal assignment and screening for alterations in Alzheimer disease. Genomics. 1995;26:254–257. doi: 10.1016/0888-7543(95)80208-4. [DOI] [PubMed] [Google Scholar]
- 7.Beyer K. Alpha-synuclein structure, posttranslational modification and alternative splicing as aggregation enhancers. Acta Neuropathol. 2006;112:237–251. doi: 10.1007/s00401-006-0104-6. [DOI] [PubMed] [Google Scholar]
- 8.Halliday GM, McCann H. Exp. Neurol. 2007. Human-based studies on alpha-synuclein deposition and relationship to Parkinson's disease symptoms. doi:10.1016/j.expneurol.2007.1007.1006 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 9.Lee VM, Trojanowski JQ. Mechanisms of Parkinson's disease linked to pathological alpha-synuclein: new targets for drug discovery. Neuron. 2006;52:33–38. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 10.Braak H, Bohl JR, Muller CM, Rub U, de Vos RA, Del Tredici K. Stanley Fahn Lecture 2005: the staging procedure for the inclusion body pathology associated with sporadic Parkinson's disease reconsidered. Mov. Disord. 2006;21:2042–2051. doi: 10.1002/mds.21065. [DOI] [PubMed] [Google Scholar]
- 11.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 12.Norris EH, Giasson BI, Lee VM. Alpha-synuclein: normal function and role in neurodegenerative diseases. Curr. Top. Dev. Biol. 2004;60:17–54. doi: 10.1016/S0070-2153(04)60002-0. [DOI] [PubMed] [Google Scholar]
- 13.Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, Barbour R, Huang J, Kling K, Lee M, et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 2006;281:29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 14.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 15.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 16.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. Alpha-synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 17.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 18.Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 19.Nishioka K, Hayashi S, Farrer MJ, Singleton AB, Yoshino H, Imai H, Kitami T, Sato K, Kuroda R, Tomiyama H, et al. Clinical heterogeneity of alpha-synuclein gene duplication in Parkinson's disease. Ann. Neurol. 2006;59:298–309. doi: 10.1002/ana.20753. [DOI] [PubMed] [Google Scholar]
- 20.Eriksen JL, Przedborski S, Petrucelli L. Gene dosage and pathogenesis of Parkinson's disease. Trends Mol. Med. 2005;11:91–96. doi: 10.1016/j.molmed.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Sulzer D. Multiple hit hypotheses for dopamine neuron loss in Parkinson's disease. Trends Neurosci. 2007;30:244–250. doi: 10.1016/j.tins.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Chiba-Falek O, Kowalak JA, Smulson ME, Nussbaum RL. Regulation of alpha-synuclein expression by poly (ADP ribose) polymerase-1 (PARP-1) binding to the NACP-Rep1 polymorphic site upstream of the SNCA gene. Am. J. Hum. Genet. 2005;76:478–492. doi: 10.1086/428655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, Rocca WA, Schneider NK, Lesnick TG, Lincoln SJ, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson's disease. J. Am. Med. Assoc. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 24.Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: dopamine, vesicles and alpha-synuclein. Nat. Rev. Neurosci. 2002;3:932–942. doi: 10.1038/nrn983. [DOI] [PubMed] [Google Scholar]
- 25.Kirik D, Rosenblad C, Burger C, Lundberg C, Johansen TE, Muzyczka N, Mandel RJ, Bjorklund A. Parkinson like neurodegeneration induced by targeted overexpression of alpha-synuclein in the nigrostriatal system. J. Neurosci. 2002;22:2780–2791. doi: 10.1523/JNEUROSCI.22-07-02780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vila M, Vukosavic S, Jackson-Lewis V, Neystat M, Jakowec M, Przedborski S. Alpha-synuclein up-regulation in substantia nigra dopaminergic neurons following administration of the parkinsonian toxin MPTP. J. Neurochem. 2000;74:721–729. doi: 10.1046/j.1471-4159.2000.740721.x. [DOI] [PubMed] [Google Scholar]
- 27.Kholodilov NG, Neystat M, Oo TF, Lo SE, Larsen KE, Sulzer D, Burke RE. Increased expression of rat synuclein in the substantia nigra pars compacta identified by mRNA differential display in a model of developmental target injury. J. Neurochem. 1999;73:2586–2599. doi: 10.1046/j.1471-4159.1999.0732586.x. [DOI] [PubMed] [Google Scholar]
- 28.Dauer W, Przedborski S. Parkinson's Disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 29.Dauer W, Kholodilov N, Vila M, Trillat AC, Goodchild R, Larsen KE, Staal R, Tieu K, Schmitz Y, Yuan CA, et al. Resistance of alpha -synuclein null mice to the parkinsonian neurotoxin MPTP. Proc. Natl Acad. Sci. USA. 2002;99:14524–14529. doi: 10.1073/pnas.172514599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, et al. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 31.Chandra S, Fornai F, Kwon HB, Yazdani U, Atasoy D, Liu X, Hammer RE, Battaglia G, German DC, Castillo PE, et al. Double-knockout mice for alpha- and beta-synucleins: effect on synaptic functions. Proc. Natl Acad. Sci. USA. 2004;101:14966–14971. doi: 10.1073/pnas.0406283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandra S, Gallardo G, Fernandez-Chacon R, Schluter OM, Sudhof TC. Alpha-synuclein cooperates with CSPalpha in preventing neurodegeneration. Cell. 2005;123:383–396. doi: 10.1016/j.cell.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 33.Chua CE, Tang BL. alpha-synuclein and Parkinson's disease: the first roadblock. J. Cell. Mol. Med. 2006;10:837–846. doi: 10.1111/j.1582-4934.2006.tb00528.x. [DOI] [PubMed] [Google Scholar]
- 34.Beyer K. Mechanistic aspects of Parkinson's disease: alpha-synuclein and the biomembrane. Cell Biochem. Biophys. 2007;47:285–299. doi: 10.1007/s12013-007-0014-9. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Kao SY, Lee FJ, Song W, Jin LW, Yankner BA. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson's disease. Nat. Med. 2002;8:600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- 36.Sidhu A, Wersinger C, Moussa CE, Vernier P. The role of alpha-synuclein in both neuroprotection and neurodegeneration. Ann. NY Acad. Sci. 2004;1035:250–270. doi: 10.1196/annals.1332.016. [DOI] [PubMed] [Google Scholar]
- 37.Papapetropoulos S, Adi N, Mash DC, Shehadeh L, Bishopric N, Shehadeh L. Expression of alpha-synuclein mRNA in Parkinson's disease. Mov. Disord. 2007;22:1057–1059. doi: 10.1002/mds.21466. [DOI] [PubMed] [Google Scholar]
- 38.Chiba-Falek O, Lopez GJ, Nussbaum RL. Levels of alpha-synuclein mRNA in sporadic Parkinson's disease patients. Mov. Disord. 2006;21:1703–1708. doi: 10.1002/mds.21007. [DOI] [PubMed] [Google Scholar]
- 39.Dachsel JC, Lincoln SJ, Gonzalez J, Ross OA, Dickson DW, Farrer MJ. The ups and downs of alpha-synuclein mRNA expression. Mov. Disord. 2007;22:293–295. doi: 10.1002/mds.21223. [DOI] [PubMed] [Google Scholar]
- 40.Rockenstein E, Hansen LA, Mallory M, Trojanowski JQ, Galasko D, Masliah E. Altered expression of the synuclein family mRNA in Lewy body and Alzheimer's disease. Brain Res. 2001;914:48–56. doi: 10.1016/s0006-8993(01)02772-x. [DOI] [PubMed] [Google Scholar]
- 41.Kingsbury AE, Daniel SE, Sangha H, Eisen S, Lees AJ, Foster OJ. Alteration in alpha-synuclein mRNA expression in Parkinson's disease. Mov. Disord. 2004;19:162–170. doi: 10.1002/mds.10683. [DOI] [PubMed] [Google Scholar]
- 42.Beyer K, Lao JI, Carrato C, Mate JL, Lopez D, Ferrer I, Ariza A. Differential expression of alpha-synuclein isoforms in dementia with Lewy bodies. Neuropathol. Appl. Neurobiol. 2004;30:601–607. doi: 10.1111/j.1365-2990.2004.00572.x. [DOI] [PubMed] [Google Scholar]
- 43.Weis S, Llenos IC, Dulay JR, Elashoff M, Martinez-Murillo F, Miller CL. Quality control for microarray analysis of human brain samples: the impact of postmortem factors, RNA characteristics, and histopathology. J. Neurosci. Methods. 2007;165:198–209. doi: 10.1016/j.jneumeth.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Stan AD, Ghose S, Gao XM, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ, Tamminga CA. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11. doi: 10.1016/j.brainres.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webster MJ. Tissue preparation and banking. Prog. Brain Res. 2006;158:3–14. doi: 10.1016/S0079-6123(06)58001-X. [DOI] [PubMed] [Google Scholar]
- 46.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liss B. Improved quantitative real-time RT-PCR for expression profiling of individual cells. Nucleic Acids Res. 2002;30:e89. doi: 10.1093/nar/gnf088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liss B, Haeckel O, Wildmann J, Miki T, Seino S, Roeper J. K-ATP channels promote the differential degeneration of dopaminergic midbrain neurons. Nat. Neurosci. 2005;8:1742–1751. doi: 10.1038/nn1570. [DOI] [PubMed] [Google Scholar]
- 49.Liss B, Franz O, Sewing S, Bruns R, Neuhoff H, Roeper J. Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChip3.1 transcription. EMBO J. 2001;20:5715–5724. doi: 10.1093/emboj/20.20.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramirez A, Heimbach A, Gründemann J, Stiller B, Hampshire D, Cid LP, Goebel I, Mubaidin AF, Wriekat AL, Roeper J, et al. Hereditary Parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 51.Pinzani P, Orlando C, Pazzagli M. Laser-assisted microdissection for real-time PCR sample preparation. Mol. Aspects Med. 2006;27:140–159. doi: 10.1016/j.mam.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Liss B, Bruns R, Roeper J. Alternative sulfonylurea receptor expression defines metabolic sensitivity of K-ATP channels in dopaminergic midbrain neurons. EMBO J. 1999;18:833–846. doi: 10.1093/emboj/18.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Kornacker K. Chipping away at the chip bias: RNA degradation in microarray analysis. Nat. Genet. 2003;35:292–293. doi: 10.1038/ng1203-292. [DOI] [PubMed] [Google Scholar]
- 55.Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol. Lett. 2006:1601–1613. doi: 10.1007/s10529-006-9127-2. [DOI] [PubMed] [Google Scholar]
- 56.Keays KM, Owens GP, Ritchie AM, Gilden DH, Burgoon MP. Laser capture microdissection and single-cell RT-PCR without RNA purification. J. Immunol. Methods. 2005;302:90–98. doi: 10.1016/j.jim.2005.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 58.Liss B, Roeper J. Correlating function and gene expression of individual basal ganglia neurons. Trends Neurosci. 2004;27:475–481. doi: 10.1016/j.tins.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 59.Yazawa I, Giasson BI, Sasaki R, Zhang B, Joyce S, Uryu K, Trojanowski JQ, Lee VM. Mouse model of multiple system atrophy alpha-synuclein expression in oligodendrocytes causes glial and neuronal degeneration. Neuron. 2005;45:847–859. doi: 10.1016/j.neuron.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 60.Braak H, Sastre M, Del Tredici K. Development of alpha-synuclein immunoreactive astrocytes in the forebrain parallels stages of intraneuronal pathology in sporadic Parkinson's disease. Acta Neuropathol. 2007;114:231–241. doi: 10.1007/s00401-007-0244-3. [DOI] [PubMed] [Google Scholar]
- 61.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 62.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 63.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR–a perspective. J. Mol. Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- 64.Dev KK, Hofele K, Barbieri S, Buchman VL, van der Putten H. Part II: alpha-synuclein and its molecular pathophysiological role in neurodegenerative disease. Neuropharmacology. 2003;45:14–44. doi: 10.1016/s0028-3908(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 65.Qin Y, Ouyang Q, Pablo J, Mash DC. Cocaine abuse elevates alpha-synuclein and dopamine transporter levels in the human striatum. Neuroreport. 2005;16:1489–1493. doi: 10.1097/01.wnr.0000175617.39054.ba. [DOI] [PubMed] [Google Scholar]
- 66.Fornai F, Lazzeri G, Bandettini Di Poggio A, Soldani P, De Blasi A, Nicoletti F, Ruggieri S, Paparelli A. Convergent roles of alpha-synuclein, DA metabolism, and the ubiquitin-proteasome system in nigrostriatal toxicity. Ann. NY Acad. Sci. 2006;1074:84–89. doi: 10.1196/annals.1369.007. [DOI] [PubMed] [Google Scholar]
- 67.Fornai F, Lenzi P, Ferrucci M, Lazzeri G, di Poggio AB, Natale G, Busceti CL, Biagioni F, Giusiani M, Ruggieri S, et al. Occurrence of neuronal inclusions combined with increased nigral expression of alpha-synuclein within dopaminergic neurons following treatment with amphetamine derivatives in mice. Brain Res. Bull. 2005;65:405–413. doi: 10.1016/j.brainresbull.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 68.Bonsch D, Greifenberg V, Bayerlein K, Biermann T, Reulbach U, Hillemacher T, Kornhuber J, Bleich S. Alpha-synuclein protein levels are increased in alcoholic patients and are linked to craving. Alcohol Clin. Exp. Res. 2005;29:763–765. doi: 10.1097/01.alc.0000164360.43907.24. [DOI] [PubMed] [Google Scholar]
- 69.Bonsch D, Lenz B, Kornhuber J, Bleich S. DNA hypermethylation of the alpha synuclein promoter in patients with alcoholism. Neuroreport. 2005;16:167–170. doi: 10.1097/00001756-200502080-00020. [DOI] [PubMed] [Google Scholar]
- 70.Cookson MR, van der Brug M. Exp. Neurol. 2007. Cell systems and the toxic mechanism(s) of alpha-synuclein. doi:10.1016/j.expneurol.2007.1005.1022 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beyer K, Humbert J, Ferrer A, Lao JI, Carrato C, Lopez D, Ferrer I, Ariza A. Low alpha-synuclein 126 mRNA levels in dementia with Lewy bodies and Alzheimer disease. Neuroreport. 2006;17:1327–1330. doi: 10.1097/01.wnr.0000224773.66904.e7. [DOI] [PubMed] [Google Scholar]
- 72.Javoy-Agid F, Hirsch EC, Dumas S, Duyckaerts C, Mallet J, Agid Y. Decreased tyrosine hydroxylase messenger RNA in the surviving dopamine neurons of the substantia nigra in Parkinson's disease: an in situ hybridization study. Neuroscience. 1990;38:245–253. doi: 10.1016/0306-4522(90)90389-l. [DOI] [PubMed] [Google Scholar]
- 73.Kastner A, Hirsch EC, Agid Y, Javoy-Agid F. Tyrosine hydroxylase protein and messenger RNA in the dopaminergic nigral neurons of patients with Parkinson's disease. Brain Res. 1993;606:341–345. doi: 10.1016/0006-8993(93)91005-d. [DOI] [PubMed] [Google Scholar]
- 74.Kastner A, Hirsch EC, Herrero MT, Javoy-Agid F, Agid Y. Immunocytochemical quantification of tyrosine hydroxylase at a cellular level in the mesencephalon of control subjects and patients with Parkinson's and Alzheimer's disease. J. Neurochem. 1993;61:1024–1034. doi: 10.1111/j.1471-4159.1993.tb03616.x. [DOI] [PubMed] [Google Scholar]
- 75.Kingsbury AE, Marsden CD, Foster OJ. The vulnerability of nigral neurons to Parkinson's disease is unrelated to their intrinsic capacity for dopamine synthesis: an in situ hybridization study. Mov. Disord. 1999;14:206–218. doi: 10.1002/1531-8257(199903)14:2<206::aid-mds1002>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 76.Gai WP, Vickers JC, Blumbergs PC, Blessing WW. Loss of non-phosphorylated neurofilament immunoreactivity, with preservation of tyrosine hydroxylase, in surviving substantia nigra neurons in Parkinson's disease. J. Neurol. Neurosurg. Psychiatry. 1994;57:1039–1046. doi: 10.1136/jnnp.57.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joyce JN, Smutzer G, Whitty CJ, Myers A, Bannon MJ. Differential modification of dopamine transporter and tyrosine hydroxylase mRNAs in midbrain of subjects with Parkinson's, Alzheimer's with parkinsonism, and Alzheimer's disease. Mov. Disord. 1997;12:885–897. doi: 10.1002/mds.870120609. [DOI] [PubMed] [Google Scholar]
- 78.Tong ZY, Kingsbury AE, Foster OJ. Up-regulation of tyrosine hydroxylase mRNA in a sub-population of A10 dopamine neurons in Parkinson's disease. Brain Res. Mol. Brain Res. 2000;79:45–54. doi: 10.1016/s0169-328x(00)00089-9. [DOI] [PubMed] [Google Scholar]
- 79.Schulz DJ, Baines RA, Hempel CM, Li L, Liss B, Misonou H. Cellular excitability and the regulation of functional neuronal identity: from gene expression to neuromodulation. J. Neurosci. 2006;26:10362–10367. doi: 10.1523/JNEUROSCI.3194-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monyer H, Lambolez B. Molecular biology and physiology at the single-cell level. Curr. Opin. Neurobiol. 1995;5:382–387. doi: 10.1016/0959-4388(95)80052-2. [DOI] [PubMed] [Google Scholar]
- 82.Ginsberg SD, Che S. Combined histochemical staining, RNA amplification, regional, and single cell cDNA analysis within the hippocampus. Lab. Invest. 2004;84:952–962. doi: 10.1038/labinvest.3700110. [DOI] [PubMed] [Google Scholar]
- 83.Kawasaki ES. Microarrays and the gene expression profile of a single cell. Ann. NY Acad. Sci. 2004;1020:92–100. doi: 10.1196/annals.1310.010. [DOI] [PubMed] [Google Scholar]
- 84.Huang J, Qi R, Quackenbush J, Dauway E, Lazaridis E, Yeatman T. Effects of ischemia on gene expression. J. Surg. Res. 2001;99:222–227. doi: 10.1006/jsre.2001.6195. [DOI] [PubMed] [Google Scholar]
- 85.Huang LE, Luzzi V, Ehrig T, Holtschlag V, Watson MA. Optimized tissue processing and staining for laser capture microdissection and nucleic acid retrieval. Methods Enzymol. 2002;356:49–62. doi: 10.1016/s0076-6879(02)56922-6. [DOI] [PubMed] [Google Scholar]
- 86.Kerman IA, Buck BJ, Evans SJ, Akil H, Watson SJ. Combining laser capture microdissection with quantitative real-time PCR: effects of tissue manipulation on RNA quality and gene expression. J. Neurosci. Methods. 2006;153:71–85. doi: 10.1016/j.jneumeth.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 87.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, Strombom L, et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 88.Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jones L, Goldstein DR, Hughes GP, Strand A, Collin F, Dunnett SB, Kooperberg CL, Aragaki A, Olson JM, Augood SJ, et al. Assessment of the relationship between pre-chip and post-chip quality measures for Affymetrix GeneChip expression data. BMC Bioinformatics. 2006;7:211. doi: 10.1186/1471-2105-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]