Abstract
To better understand DNA recognition and transcription activity by SATB1, the T-lineage-enriched chromatin organizer and transcription factor, we have determined its optimal DNA-binding sequence by random oligonucleotide selection. The consensus SATB1-binding sequence (CSBS) comprises a palindromic sequence in which two identical AT-rich half-sites are arranged as inverted repeats flanking a central cytosine or guanine. Strikingly, the CSBS half-site is identical to the conserved element ‘TAATA’ bound by the known homeodomains (HDs). Furthermore, we show that the high-affinity binding of SATB1 to DNA is dimerization-dependent and the HD also binds in similar fashion. Binding studies using HD-lacking SATB1 and binding target with increased spacer between the two half-sites led us to propose a model for SATB1–DNA complex in which the HDs bind in an antiparallel fashion to the palindromic consensus element via minor groove, bridged by the PDZ-like dimerization domain. CSBS-driven in vivo reporter analysis indicated that SATB1 acts as a repressor upon binding to the CSBS and most of its derivatives and the extent of repression is proportional to SATB1's binding affinity to these sequences. These studies provide mechanistic insights into the mode of DNA binding and its effect on the regulation of transcription by SATB1.
INTRODUCTION
Special AT-rich sequence-binding protein 1 (SATB1) participates in the maintenance of chromatin architecture by organizing it into distinct loops via periodic tethering of matrix attachment regions (MARs) to the nuclear matrix (1–4). In thymocyte nuclei, SATB1 forms a characteristic ‘cage-like’ network that presumably demarcates heterochromatin from euchromatin (2). Furthermore, SATB1 acts as a ‘docking site’ for several chromatin modifiers including ACF, ISWI and HDAC1 (5,6) and these chromatin modifiers were suggested to affect gene expression through histone modifications and nucleosome remodeling at SATB1-bound MARs (5,2). SATB1 also regulates gene expression by recruiting corepressors (HDACs) and coactivators (HATs) directly to promoters (6,7). Post-translational modifications of its N-terminal PDZ-like domain act as molecular switches regulating the transcriptional activity of SATB1 via modulating its association with other proteins (7). The PDZ-like domain is also important for DNA- and chromatin-binding ability of SATB1 through homodimerization (8). In the C-terminal half, amino acids (aa) 346–495 harbor a Cut-like repeat (9) and hence can be referred to as the Cut domain (CD). This region is also referred as the MAR-binding domain (MD) due to its probable role in highly specific recognition of MARs (9). Additionally, SATB1 harbors a homeodomain (HD) spanning aa 641–702 that is believed to act in concert with the MD and direct SATB1 to bind to the core-unwinding element within a MAR with high affinity (10).
Gene profiling studies using RNA from cells overexpressing point mutants of SATB1 defective in phosphorylation or acetylation revealed that SATB1 regulates more than 10% of genes demonstrating the importance of these modifications toward the ability of SATB1 to act as a global regulator of gene expression (7). However, only a limited number of SATB1-binding sites (SBSs) have been characterized so far, most of which were isolated based on their ability to serve as base unpairing regions (BURs) that are hallmark of MARs (1–3). Comparison of these SBSs and various other sequences reported to be bound by SATB1 in vivo did not reveal any specific consensus element, giving rise to the notion that SATB1 binds DNA in a sequence-independent but context-dependent manner. However, such analyses identified an ATC context that has been proposed to be involved in targeting SATB1 (1,2). Due to lack of consensus-binding element the precise mechanism of how SATB1 binds to MARs or non-MAR DNA sequences with high affinity and specificity remains poorly understood. Recently, locus-wide chromatin immunoprecipitation (ChIP) analysis monitoring SATB1 occupancy of the MHC locus showed specific clustering at promoters and MARs suggesting that SATB1 binds to genomic regions in a non-random fashion, and not necessarily dictated by the ATC context (4).
In this study, we set out to understand how SATB1 binds to its target sequences specifically by characterizing its binding targets. We used the approach of systematic evolution of ligands by exponential enrichment (SELEX) (11,12) to isolate a pool of synthetic DNA sequences that were bound with high affinity by SATB1. We found a conserved pattern of 10–12 nucleotide (nt) in all enriched sequences consisting of two inverted AT-rich (4 to 6 nts) repeats resembling the HD-binding site separated by 1–2 non-AT nts. Substitution by cytosine (C) at any position in the conserved HD-binding region ‘TATTAG’ abolishes the DNA-binding activity of SATB1 indicating that it is mediated primarily by the HD. The minor groove-binding agent Distamycin has been shown to abolish the binding of SATB1 to the IgH MAR, indicating that SATB1 binds via the minor groove of the DNA (13). Dimerization mediated by the N-terminal PDZ domain is important for the binding of SATB1 (8). However, SATB1 lacking the PDZ domain, presumably in its monomeric form, has been shown to bind DNA in vitro, albeit at lower affinity via the major groove (9,14,15). Results of our in vitro binding studies in conjunction with the recently solved structure of the N-terminal Cut repeat 1 (CUTr1) (15) led us to propose that SATB1 binds to the inverted consensus palindromic repeats via HD in dimerization-dependent manner via the minor groove, whereas the Cut repeats enhance the binding via hydrogen-bonding interactions in the major groove. As a functional consequence, we demonstrate for the first time that the strength of repression mediated by SATB1 is proportional to its affinity to the target sequence. Collectively, our results provide evidence for sequence specific binding of SATB1 to target DNA and not only to the ATC context as thought before. Furthermore, we demonstrate that high-affinity DNA binding by SATB1 is dimerization-dependent and the binding specificity is mediated by its HD in collaboration with the Cut repeat containing domain (CD).
MATERIALS AND METHODS
SATB1-binding site selection
A synthetic random oligonucleotide library (DDSEL) with two fixed primer regions (5′ DDCI and 3′ DDCII) and a central random 32-base region was synthesized using the reported procedure (16). The oligonucleotide library was desalted and used without any further purification. One hundred nanograms of DDSEL was radiolabeled during synthesis of double-stranded DNA using α-32P dATP (BRIT, India) and unlabeled dGTP, dCTP and dTTP with Escherichia coli Klenow fragment (New England Biolabs, USA). The selection of bound oligonucleotides was performed after incubation with GST:CD + HD (25–100 ng) followed by EMSA as described previously (6). Briefly binding reaction was performed in a 10-μl total volume containing 1× EMSA buffer [10 mM HEPES (pH 7.9), 1 mM dithiothreitol, 2.5 mM MgCl2, 10% glycerol, 1 μg of double-stranded poly(dI–dC), 10 μg of purified bovine serum albumin] and 25–100 ng of pure GST:CD + HD. The bound oligonucleotides were gel extracted by crushing the gel piece and soaking in TE buffer for 12 h followed by phenol–chloroform extraction and ethanol precipitation. The gel-extracted band was radiolabeled during amplification by PCR using DDCI and DDCII primers in the presence of α-32P dATP. This product was used for the next round of selection following the same protocol. The flowchart of the SELEX procedure is shown in Supplementary Figure 1A. Five rounds of iterative selection were carried out by EMSA and the enriched library was cloned into pGEM-T Easy vector (Promega, USA) according to the manufacturer's instructions and transformed into chemically competent E. coli DH5α. More than 200 positive clones were sequenced from which 40 unique sequences were obtained.
Protein expression and purification
Glutathione S-transferase (GST), GST:CD+HD (255–763 aa), GST:CD (346–495 aa), GST:HD (640–763 aa) and GST:PDZ (1–254 aa) were expressed in XL1 blue strain of E. coli (Stratagene) and purified using glutathione-Sepharose affinity columns (GE Healthcare). We also expressed and purified GST:CD+HD (346–763), GST:346–763(ΔHD)-fusion proteins, constructs for which were kindly gifted by Dr T. Kohwi Shigematsu. His-tagged PDZ was expressed in BL21 (DE3) and purified by Ni–NTA affinity resin (Qiagen). GST-fusion proteins were cleaved on column by caspase-6 essentially as described (17).
Electrophoretic mobility shift assay (EMSA) for the determination of dissociation constant (Kd)
Individual oligonucleotides were end labeled with γ-32P ATP using T4 polynucleotide kinase (New England Biolabs, USA). The forward and reverse oligonucleotides for a particular set were mixed together and the NaCl concentration was adjusted to a final concentration to 50 mM in a 50 μl reaction volume. For annealing labeled oligonucleotides, the mixtures were boiled for 10 min and allowed to cool slowly until the samples reached to room temperature. The annealed oligonucleotides were electrophoresed on 12% native polyacrylamide gels and the duplex DNAs were recovered from the excised band of interest and purified as described above. The purified duplexes were used for the EMSA with various proteins to determine the dissociation constants. The concentration (Molar) of protein required to bind 50% of the substrate DNA was considered as a dissociation constant (Kd) of the protein for a particular sequence.
Distamycin competition assay
The radiolabeled CSBS probe was incubated with 50–100 μM of distamycin A (Sigma) at room temperature for 15 min in 1× binding buffer as described before. Proteins were added to the DNA–distamycin complex and incubated for additional 15 min. The positive control was incubated only with the purified protein. The complexes were resolved by 12% native PAGE for 70 min. Gels were dried in vacuum gel drier and exposed to the X-ray film overnight.
Southwestern blot analysis
Southwestern blotting was performed essentially as described previously, with certain modifications (8). Briefly, 0.25, 0.5 and 1.0 μg of various purified proteins were incubated with SDS–PAGE loading buffer at 37°C for 10 min. Proteins were resolved in 10–15% SDS–PAGE depending on the size of the protein used. The resolved proteins from the gels were electrophoretically transferred onto PVDF membrane (Millipore). The proteins on the membrane were renatured by incubating with a blocking and refolding buffer (20 mM Tris pH 7.4, 50 mM NaCl, 1 mM DTT, 0.1% Tween-20 and 5% BSA) for 1 h at room temperature. The membrane was washed four times with binding buffer and then hybridized with the specific probe in hybridization buffer (20 mM Tris pH 7.4, 50 mM NaCl, 1 mM DTT, 0.1% Tween-20 and 0.25% BSA; 4 μg/ml poly(dI–dC), 35 μg/ml salmon sperm DNA supplemented with either 200 ng of 32P-end-filled IgH MAR probe/20 μM of 32P-end-labeled annealed CSBS oligonucleotides) for 30 min at room temperature under gentle agitation. The membrane was subsequently washed four to five times with binding buffer and then exposed to X-ray film (Kodak Biomax) for 12–24 h.
Luciferase reporter assay
Several variants of SATB1-binding consensus sequences were annealed as described above, generating duplex oligonucleotides with staggered ends at both the ends, with XhoI overhang at 5′ end and HindIII overhang at 3′ end. These annealed and phosphorylated oligonucleotides were ligated into XhoI and HindIII-digested pGL3Promoter vector (Promega). Luciferase reporter assays were performed as described (6). Briefly, HEK 293 cells were maintained in DMEM with high glucose and supplemented with 10% fetal calf serum (FCS). Cells were seeded at 0.2 × 106 cells per well in a 12-well plate, 24 h before transfection. A reporter construct was transfected either with control 3X FLAG vector (Sigma) or with 3X FLAG:SATB1 construct. Total DNA used was 2 μg (1 μg of each DNA construct) in every well of a 12-well plate using Lipofectamine 2000 (Invitrogen). The transfected cells were harvested 48 h post-transfection, lyzed by freeze–thaw procedure and the protein content of the lysate was estimated using Bradford reagent (Biorad). Hundred micrograms of lysate was mixed with 100 μl of Luclite substrate (Perkin Elmer) and luciferase activity was measured using Top-Count (Packard). The experiment was performed three times independently and average relative luciferase activity with standard deviation was plotted using Sigma plot. The statistical significance of differences between the samples was calculated using one-way ANOVA (SigmaStat, SPSS Inc.) and the observed P-values were always <0.001.
Mammalian two-hybrid assay
We used the CheckMate mammalian two-hybrid system (Promega Corp. USA) to score for protein–protein interactions. Cloning gene of interest in pBIND vector expresses it as GAL4 DNA-binding domain fusion protein and cloning in pACT fusion construct expresses it as VP-16 activation domain fusion protein. pBIND and pACT fusion constructs were transfected along with a reporter vector, which contains 4× GAL4 responsive element (pG5 luc), and luciferase activity was compared with the control. Specifically, the N-terminal 1–254-aa region of SATB1, which harbors the PDZ domain and C-terminal 255–763 aa region harboring CD+HD were subcloned in pACT and pBIND vectors (Promega) at BamHI and XbaI sites. HEK 293 cells were seeded at 0.5 × 106 cells per well in a 6-well plate (BD Falcon) 24 h before transfection. Cells were transfected with pG5 Luc reporter vector (Promega) using Lipofectamine 2000 reagent (Invitrogen) along with either pBIND fusion construct and pACT empty vector in control and pBIND fusion construct with pACT fusion construct for the experimental set. The DNA was kept constant up to 3 μg (1 μg of each DNA) in every well. The transfected cells were harvested 48 h post-transfection. Harvested cells pellet was washed with PBS and resuspended in 100 μl of lysis buffer. Lysate was prepared by performing three freeze–thaw cycles. Protein in the lysate was estimated by Bradford reagent (Bio Rad). Hundred micrograms of protein was mixed with 100 μl of Luclite (Perkin Elmer) and illumination was measured in Top-Count (Packard). The reading of untransfected wells sample lysate was subtracted from every individual reading and relative luciferase readings were plotted after performing three separate sets of experiments as described above.
RESULTS
In vitro enrichment and cloning of SBSs
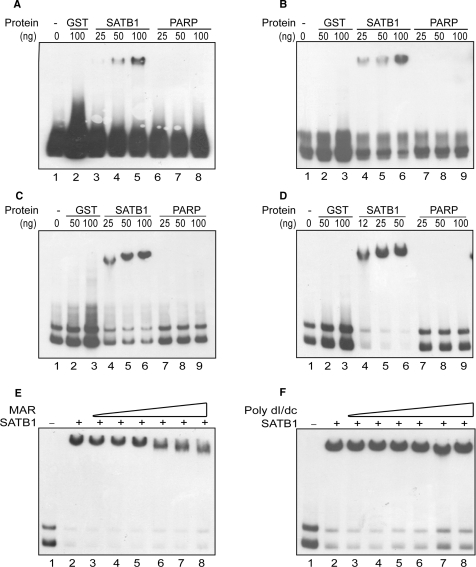
Since SATB1 acts as a transcription factor regulating global gene expression (7), we explored the possibility of consensus-binding element for SATB1. We employed the technique of SELEX for the enrichment of specific SBSs. We used a library of 32-mer random sequences flanked by 24-mers of 5′ and 3′ constant regions that has recently been used to define the DNA-binding consensus motif for HIV-1 transactivator Tat (11). The process of selection using EMSA and amplification was carried out as indicated in Supplementary Figure S1A. Four rounds of selection and amplification resulted in enrichment of highly specific SBSs under conditions that precluded binding with GST or with GST:PARP (Figure 1A–D). The binding of SATB1 to these sequences is specific and strong and could not get competed by 10-fold molar excess of heptameric IgH core MAR (Figure 1E) and up to 6 μg of poly(dI–dC) (Figure 1F). These highly specific SBSs were cloned into pGEM-T Easy vector. Analysis of sequences of more than 200 positive clones yielded about 40 unique sequences after elimination of duplicate sequences (Supplementary Figure S1B). The individual sequences were PCR amplified and radiolabeled for validation of SATB1-binding affinity using EMSA (data not shown). These sequences shared common motifs with many reported SBSs (Supplementary Figure S2) suggesting that variants of SATB1-binding targets were isolated.
Figure 1.
Determination of optimal SATB1-binding sequence. (A–D) The SELEX procedure was performed as described in Materials and Methods section. EMSAs depicting four successive rounds of SATB1-binding site selection following the scheme as depicted in Supplementary Figure S1A. Proteins and their respective amounts (in nanograms) used are indicated above the lanes. The probes used are: (A) SATB1 bound DNA sequences after first round of SATB1 binding. (B) SATB1 bound DNA sequences after second round of SATB1 binding. (C) SATB1 bound DNA sequences after third round of SATB1 binding. (D) SATB1 bound DNA sequences after fourth round of SATB1 binding. (E) Competition of SATB1 binding in EMSA with 1- to 10-fold molar excess of heptameric IgH MAR (lanes 3–8). (F) Competition of SATB1 binding with 1–6 μg (lanes 3–8) of poly(dI–dC) in EMSA with heptameric IgH MAR.
The enriched SBSs possess an AT-rich core that facilitates high-affinity binding and transcription repression mediated by SATB1
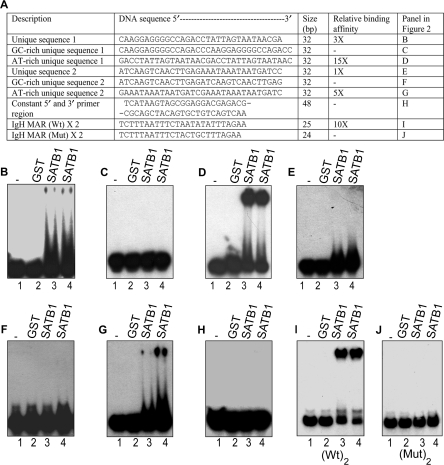
We next determined the DNA-binding affinity of the cloned sequences by EMSAs (Supplementary Figure S3). Two representative cloned sequences were picked to search for the presence of AT-rich region, since SATB1 preferably binds to AT-rich regions (10). We found two distinct regions in the 32-mer variable region (compare first half of 16 nts with the second half in unique sequence 1 and unique sequence 2 in Figure 2A). Strikingly, both these sequences comprised of a region rich in AT and another region containing a mixture of GCA and T but relatively more rich in GC (Figure 2A). Next, we separately synthesized these two sequences only as monomer of 32 nts and without the constant 5′ and 3′ regions. The unique sequence 1 was bound by SATB1 with much higher affinity as compared with unique sequence 2 (Figure 2B and E). Strikingly, one half of these sequences comprising 16 nts was GC-rich whereas another half was AT-rich. A dimeric oligonucleotide of each half was then chemically synthesized. The constant 5′ and 3′ flanking regions were synthesized as a fusion of both sequences. All of these double-stranded oligonucleotides were radiolabeled and used in the EMSA along with wild-type (25 × 2) and mutant IgH MAR dimers (24 × 2) as positive and negative controls for SATB1 binding, respectively (10). Binding to the AT-rich region is stronger (Figure 2D and G) than the parental sequence (Figure 2B and E). The affinities of individual parental 32-mer sequences were considered as one arbitrary unit (1×) and affinities of dimers of the 16-mers were assigned relative to parental sequences. The GC-rich regions were not bound by SATB1 (Figure 2C and F). SATB1 did not bind to the fixed sequences of primers flanking the variable sequence in the SELEX library (Figure 2H). The enhanced binding of SATB1 to dimeric AT-rich regions but not to dimeric GC-rich regions compared with the parental sequences indicates that AT-rich regions are targeted by SATB1. The enhanced binding is also indicative of co-operative binding by SATB1 to the multimeric AT-rich sites. Taken together, these results demonstrate that the AT-rich region forms the core SBS in the SELEX-enriched sequences.
Figure 2.
SELEX-enriched sequences harbor an AT-rich core that is required for DNA binding and transcription regulation by SATB1. (A) Table depicting various sequences studied and their relative binding affinities deduced from EMSAs. Last column denotes the panels depicting the EMSA data for a particular sequence as follows: (B) SELEX-enriched unique sequence 1 of 32-nt length. (C) Dimer of the 16-nt GC-rich region from unique sequence 1. (D) Dimer of the16-nt AT-rich region from unique sequence 1. (E) SELEX-enriched unique sequence 2 of 32-nt length. (F) Dimer of the 16-nt GC-rich region from unique sequence 2. (G) Dimer of the 16-nt AT-rich region from unique sequence 2. (H) The 48-nt fusion of the 5′ and 3′ constant regions from the SELEX library. (I) The dimer of wild-type IgH MAR (wt 25 × 2). (J) A dimer of mutant IgH MAR (mut 24 × 2). For all of the above panels lane 1 denotes free probe; lane 2, with 1 μg GST; lane 3, 10 nM SATB1; lane 4, 20 nM SATB1.
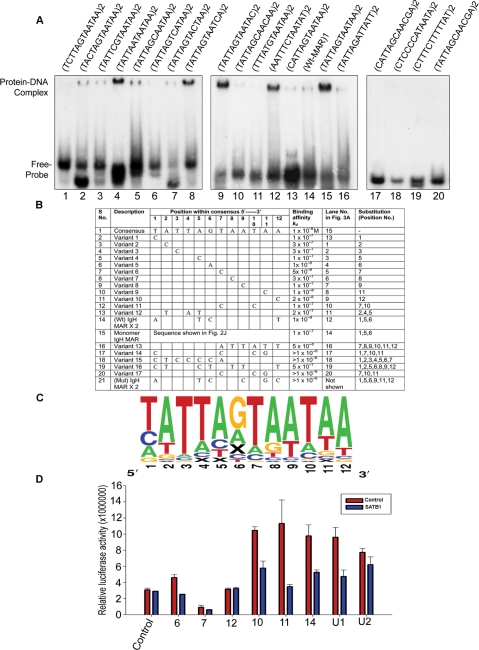
Consensus SATB1-binding sequence (CSBS) harbors a conserved HD-binding element that is essential for transcription regulation by SATB1
We next analyzed the AT-rich regions in 40 unique sequences obtained as described above. We aligned these sequences manually and also using the online software tool MEME (18) and found that the motif TATTAGTAATAA occurs in most of the sequences whereas few sequences harbor its variants (Supplementary Figure S2). Many of the reported SBSs also contained similar motifs (Supplementary Figure S2). We synthesized a dimer of TATTAGTAATAA and also of its variant sequences in which nucleotides were varied at different positions within the 12-mer consensus sequence. We then performed EMSA and determined the SATB1-binding affinity to individual sequences in terms of dissociation constant (Kd). Forty nanomoles of SATB1 was sufficient to bind various enriched sequences (Supplementary Figure S3) and therefore used for the EMSA using synthetic dimeric sequences (Figure 3A). Inclusion of cytosine (C) at positions 1 (lane 13), 2 (lane 1), 3 (lane 2), 5 (lane 3), 8 (lane 6), 9 (lane 7) or at two positions 7 and 10 (lane 10) instead of adenine (A) or thymidine (T) reduced SATB1 binding drastically. Substitution of central guanosine (G) (sixth position) with A (lane 4) or substitution of A or T with C at position 11 (lane 8) or 12 (lane 9) had no significant effect on SATB1 binding. The monomeric wild-type IgH MAR has lower binding activity than the consensus sequence identified here (sequence 15 in Figure 3B). The alignment of the core of the IgH MAR sequence with the CSBS revealed that it varies from CSBS at four places out of 12. At position 1, T is substituted by A and T by A at position 5, central (sixth) position G is replaced by C and at the last position A is replaced by T (Figure 3B, sequence 14). Next, we analyzed SATB1-binding affinity for a synthetic oligonucleotide containing dimer of this sequence and found it to be comparable with the affinity of the CSBS (Figure 3A, lane 12 versus 15), indicating that these substitutions do not have much effect on the binding affinity of SATB1. The relative binding affinities of several variants of CSBS are summarized in Figure 3B. These results indicate that SATB1 binds strongly with the 12-mer CSBS or its variants and a similar sequence is also present in the IgH MAR. Strikingly, the CSBS is divided into two AT-rich regions flanking a central region (1 or 2 nt) that could be either A, C or G. The substitution of A or T residues with a single nucleotide (C) at several positions abrogated the binding affinity drastically, suggesting that SATB1 may exhibit more stringent sequence specificity for binding than the presumed ATC context. The positional mutation analysis along with analysis of the naturally in vitro selected sequences led to propose that a combination of 12-mer palindromic sequence possessing two AT-rich repeats in inverse orientation is essential and sufficient for specific binding by SATB1. The palindromic CSBS is represented in LOGO format in Figure 3C and the percentage frequencies of occurrence of nucleotides at any given position are tabulated in Supplementary Table 1. Analysis of the consensus sequence revealed that it harbors two copies of a conserved HD-binding sequence (TAATA) (Figure 3C) (19) and mutational analysis in this region reduces affinity of SATB1 very significantly. To monitor the effect of positional variations on transcriptional potential of these sequences we cloned several point mutations in the 12-mer SBS as listed in Figure 3B as well as the two unique sequences (Figure 2A) in pGL3P vector. These reporter DNAs were used to transfect 293 cells, either along with 3X FLAG vector or with 3XFLAG-SATB1 construct. Most of these sequences enhanced SV40 promoter-driven transcription except sequence 7, which acted as a silencer of the SV40 promoter-driven transcription (compare red bar for sequence 7 with that of control sequence in Figure 3D) and SATB1 coexpression led to repression of enhancer activity of these sequences (Figure 3D, blue bars corresponding to sequences 6, 7, 10, 11 and 14) whereas there is no effect of SATB1 on sequence 12 which displays weaker affinity to SATB1. The two SELEX-enriched unique sequences enhanced the activity of luciferase reporter driven by SV40 promoter (U1 and U2 in Figure 3D). The stronger SATB1-binding unique sequence (U1) enhanced the luciferase activity to a relatively higher extent compared to the weaker SATB1-binding unique sequence (U2). Furthermore, overexpression of SATB1 resulted in stronger repression with the U1 (>2-fold, Figure 3D) compared to the U2 (∼1.2-fold, Figure 3D). Thus, SBSs generally seem to possess a cis-acting enhancer function, whereas binding of SATB1 to these sequences leads to the repression of enhancer activity that is directly proportional with the binding affinity of SATB1. Taken together, these results suggest that the relative binding affinity of SATB1 to the palindromic HD consensus sequences determines its transcriptional activity.
Figure 3.
Identification of consensus SATB1-binding sites. (A) EMSA using radiolabeled synthetic duplex oligonucleotides. The sequences of various dimeric oligonucleotides are depicted on top of individual lanes. Positions of bound and unbound probes as indicated on left. (B) Table summarizing the various positional variants and their relative binding affinities. (C) Consensus SATB1-binding site (CSBS) derived from the SELEX-enriched sequences is represented in LOGO (http://weblogo.berkeley.edu/logo.cgi) format. The positions of each nucleotide within the 12-mer consensus are indicated below using numbers 1–12. The percentage frequency of occurrence of a specific nucleotide at a position is also shown in the Supplementary Table 1. (D) In vivo reporter assay was performed to monitor the transcriptional activity of dimeric CSBS and its variants. These sequences were separately cloned in the pGL3 promoter vector and their activity toward the modulation of the SV40 promoter derived luciferase reporter and effect of SATB1 toward the regulation of activity of these sequences in a transient transfection experiment are depicted in the histogram. The numbers on X-axis correspond with the serial numbers in the table in (B). Error bars represent standard deviation calculated from triplicates.
N-terminal PDZ-containing domain mediates homodimerization of SATB1 to bind to the specific DNA as a dimer
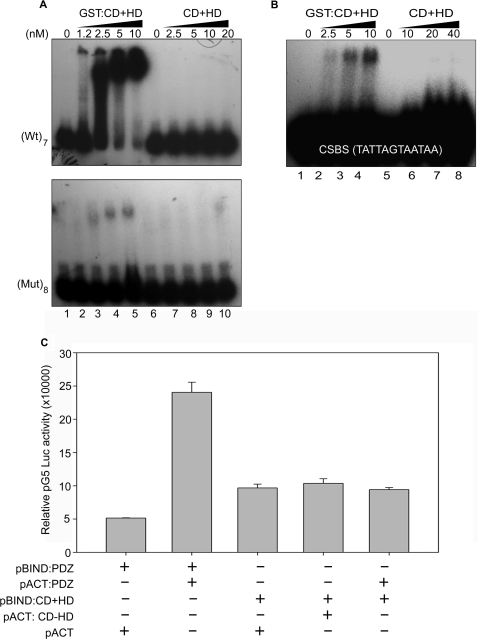
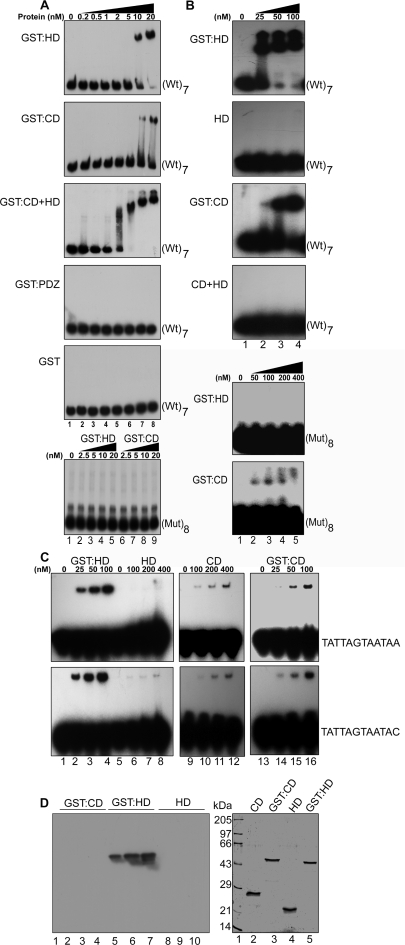
Caspase 6-mediated cleavage of SATB1 was shown to abolish the association of SATB1 with chromatin in vivo during apoptosis and SATB1 lacking N-terminal 96–204 aa region was found to lose its affinity toward MARs in vitro (8). However, few reports also indicated that the first Cut repeat of SATB1 serves as the core DNA-binding domain and contacts DNA as a monomer (9,14,15). To understand the contribution of dimerization of SATB1 toward its high affinity and specific binding to DNA we used a heterologous dimer-forming tag. The GST tag is a widely used fusion tag for affinity purification and is also reported to form a dimer (20). Fusion with GST can restore, at least partially, the oligomerization-dependent function of a protein as shown in case of Bcr-Abl protein (21). Considering this fact we proposed that the GST-fused protein would form a dimer and its GST-free version would form a monomer. We cloned the C-terminal CD + HD region (255–763 aa) into the pC6-2 vector (22,17). The fusion protein was efficiently cleaved on column using caspase-6 and obtained as GST-free CD + HD. The dimeric status of GST:CD+HD and the monomeric status of GST-free CD+HD proteins were confirmed by gel filtration analysis (data not shown). We then compared the binding affinities of GST-tagged and GST-free proteins by EMSA. GST:CD+HD bound the IgH MAR heptamer (Figure 4A, lanes 2–5 in upper panel) with Kd = 2 × 10−9, and the CSBS monomer (Figure 4B, lanes 2–4) with Kd = 1 × 10−8 indicating very high affinity binding. In contrast, the affinity of GST-free CD+HD for the IgH MAR (Figure 4A, upper panel, lanes 7–10) and CSBS (Figure 4B, lanes 6–8) was very low (Kd > 1 × 10−7) indicating that dimerization is essential for the high-affinity binding of SATB1 to CSBS. Next, to monitor whether SATB1 forms dimer in vivo in mammalian cells and to ascertain the dimerization domain, we employed the mammalian two-hybrid system. Mammalian two-hybrid system is similar to yeast two-hybrid system in principle except that the protein–protein interaction is scored as relative luciferase activity in mammalian cells. We confirmed that SATB1 exists as a homodimer in vivo, and homodimerization is mediated by its N-terminal PDZ-containing region (Figure 4C, bar 2 versus bar 1). HDs are also known to bind DNA in a homodimeric form and the dimerization can be mediated via the same domain (23). In case of SATB1, the C-terminal CD+HD region does not show any interaction with another molecule of the same, ruling out HD- or CD-mediated dimerization (Figure 4C, bar 4 versus bar 3). The possibility of hetero-domain interaction in between N-terminal PDZ domain and C-terminal CD+HD was also ruled out since there was no significant increase in the reporter activity (Figure 4C, bar 5 versus bar 3). Thus, dimerization mediated by the N-terminal PDZ-like domain of SATB1 is essential for its high-affinity binding to the CSBS.
Figure 4.
N-terminal PDZ domain mediates dimerization of SATB1 required for DNA-binding activity. (A) Monomeric CD+HD lacks DNA-binding activity. EMSA was performed as described in Materials and Methods section using wild-type IgH MAR (upper panel) and its mutant (lower panel). The concentrations of protein used in the binding reactions are indicated on top of each lane; lanes 1 and 6 represent free probe. (B) EMSA using the consensus SATB1-binding sequence. The proteins and their amounts used are indicated on top of each lane. (C) SATB1 homodimerizes via its N-terminal PDZ-containing domain. Mammalian two-hybrid assay was performed as described in Materials and Methods section to examine homodomain or heterodomain interactions of SATB1. Bar 1 indicates the relative luciferase activity of pBIND:PDZ along with pACT vector, bar 2 indicates the relative luciferase activity of pBIND:PDZ along with pACT:PDZ, bar 3 depicts the relative luciferase activity of pBIND:CD+HD along with pACT vector, bar 4 indicates the relative luciferase activity of pBIND:CD+HD along with pACT:CD+HD, bar 5 depicts the relative luciferase activity of pBIND:CD+HD along with pACT:PDZ. Error bars represent standard deviation calculated from triplicates.
HD and not the CD contributes toward the dimerization-dependent, high affinity and specific binding by SATB1
Studies considering the CD as the principal DNA-binding domain of SATB1 have provided contrasting results such as (i) CD binds as a monomer to DNA (9,14,15) which is surprising since SATB1 is known to exist as homodimer (8, and this study), and (ii) CD shows the major groove recognition (14,15) whereas SATB1 binds predominantly through minor groove as evidenced by competition of its binding by distamycin, a narrow groove binder (13, and Figure 7B, panel 1). To resolve these ambiguities, we expressed and purified GST:CD and GST:HD and compared their binding affinities with that of GST:CD+HD using wild-type IgH MAR heptamer, mutant IgH MAR octamer, and the CSBS. We observed that GST:CD+HD (Kd = 2 × 10−9; Figure 5A, panel 3 and Supplementary Table 2) bound DNA with higher affinity than the GST:HD (Kd = 1 × 10−8; Figure 5A, panel 1 and Supplementary Table 2) or GST:CD (Kd = 5 × 10−8; Figure 5A, panel 2 and Supplementary Table 2). GST:HD binds with 5-fold higher affinity than GST:CD with wild-type IgH MAR (Figure 5A, first two panels) and similar pattern is observed with CSBSs (Figure 5C, compare lanes 2–4 versus 14–16). DNA-binding activity of monomeric HD with that of GST-fused HD with IgH MAR (Figure 5B, panel 2 versus panel 1, Supplementary Table 2) and with two CSBS probes (Figure 5C, compare lanes 6–8 versus 2–4; Supplementary Table 2) followed similar pattern as observed with monomeric CD+HD (Figure 5B, panel 4). Using two variants of the CSBS which bind SATB1 with high affinity also we could not detect the binding of monomeric HD, even at relatively very high concentration of the monomeric HD protein (Figure 5C, lanes 6–8). However, the reduction in the affinity of GST-free CD for binding to the CSBS as compared to dimeric GST:CD is not very significant as compared to the abrupt loss in the binding of monomeric HD (Figure 5C, compare lanes 10–12 versus 6–8). Collectively, these results indicate that in vivo dimerization-dependent binding of SATB1 is presumably mediated via the HD and not by the CD. Next, we compared the relative binding affinities of GST:CD, GST:HD and caspase 6 cleaved monomeric HD proteins by performing Southwestern blot analysis using IgH MAR as a probe (Figure 5D). We found that at concentrations where GST:HD (lanes 5–7) could bind very efficiently there is no binding of either GST:CD (lanes 2–4) or monomeric HD (lanes 8–10) with IgH MAR probe. This result confirms that binding of HD is also dimerization-dependent and that the GST:CD displays very low affinity for the wild-type SBS.
Figure 7.
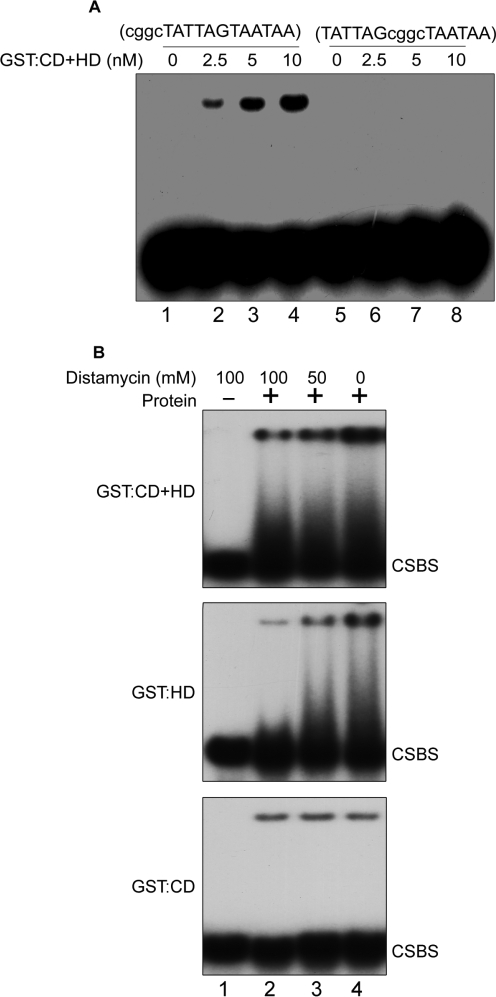
Importance of the distance between the half-sites and minor groove of the palindromic CSBS for binding by SATB1. (A) EMSA was performed using modified CSBS with additional four CG nucleotides at 5′ end (lanes 1–4) or in the center of the inverted palindromic half-sites (lanes 5–8). The concentrations of GST:CD+HD protein used are indicated above each lane. (B) Distamycin-mediated competition of DNA–protein complex. Panel 1, distamycin at indicated concentrations was incubated with the probe DNA prior to addition of protein. Increasing amounts of GST:CD+HD (20 nM), GST:HD (100 nM) and GST:CD (400 nM) were then added (lanes 2–4, respectively) and EMSA was performed as described in Materials and Methods section.
Figure 5.
Homeodomain contributes toward the dimerization-dependent, high affinity and specific binding by SATB1. (A) EMSAs were performed to determine the binding affinity of various domains in SATB1 fused with the dimerization-imparting tag of GST using the heptamer of wild-type IgH MAR (Wt)7 (top five panels). The various proteins used for EMSA are indicated on left-hand side of each panel. The last (sixth) panel shows lack of binding of GST:HD (lanes 2–5) and GST:CD (lanes 6–9) with octameric mutant IgH MAR. (B) Dimer of HD displays more specificity for the IgH MAR than the dimer of CD. Monomeric (GST-free) HD and CD+HD do not bind the same probe even at 100 nM protein concentrations (panel 4). However, GST-fused (dimeric) CD very weakly binds the mutant MAR sequence (panel 6) whereas GST:HD does not bind at all (panel 5). (C) The binding affinities of monomeric and dimeric HD and CD were compared by EMSA using CSBS (top panel) and its variant (lower panel). GST:HD has stronger affinity with the CSBS and its variant (lanes 2–4) as compared to GST:CD (lanes 14–16). Monomeric (GST-free) CD binds with these sequences at relatively higher concentration (lanes 10–12) whereas monomeric HD does not show any affinity at this protein concentration (lanes 6–8). The relative binding affinties (Kd) of various domains with several probes used here are summarized in Supplementary Table 2. (D) Southwestern blot analysis using GST:CD, GST:HD and caspase 6-cleaved monomeric HD proteins and IgH MAR as a probe. Indicated proteins measuring 0.25, 0.5 and 1.0 μg were resolved on 15% SDS–polyacrylamide gels and then transferred onto PVDF membrane and were probed with radiolabeled heptameric IgH MAR probe. Only GST:HD could bind to the probe in a dose-dependent manner (panel 1, lanes 5–7) but there is no binding by GST:CD (panel 1, lanes 2–4) and monomeric HD (panel 1, lanes 8–10). (D, panel 2) Coomassie brilliant blue-stained SDS–polyacrylamide gel for evaluating the quality of proteins used in Southwestern blotting. Proteins (0.5 μg each) were resolved on a 15% SDS–polyacrylamide gel. Lane 1, molecular weight standards; lane 2, caspase 6-cleaved CD; lane 3, GST:CD; lane 4, caspase 6-cleaved HD (640–763); lane 5, GST:HD (640–763).
Dimeric HD is the determinant for the higher affinity and specificity to CSBS variants and IgH MAR, but not the dimeric CD
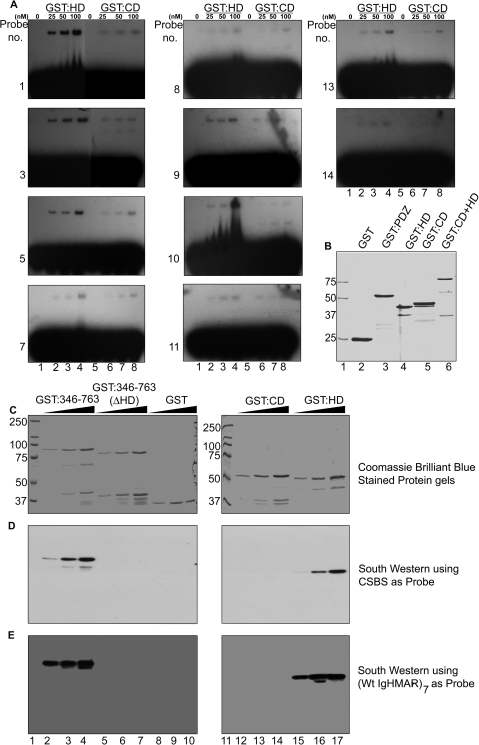
To monitor whether the dimerization-dependent high-affinity binding property of SATB1 is contributed by its HD or CD we compared their relative binding affinities to 10 representative variants (point mutants as listed in Figure 3B) of the CSBS by performing in vitro EMSA using radiolabeled double-stranded oligonucleotides. The relative binding affinities of GST:HD and GST:CD to these CSBS variants are presented as dissociation constants (Kd) in Table 1. The binding affinity of the GST:CD with most of the mutant variants of CSBS is relatively poor compared to that of the GST:HD (Figure 6A, compare lanes 6–8 versus 2–4). However, individual mutations in CSBS bring about drastic changes in the binding exhibited by GST:HD, which correlate well with that observed with the GST:CD+HD (compare the Kds for the GST:CD+HD and GST:HD for individual sequences as indicated in Table 1). The affinity of GST:CD for these sequences is very low and also exhibits very little change with respect to different variants of CSBS (Figure 6A, lanes 6–8 in all panels) indicating nonspecific level of DNA binding by CD of SATB1. Results of this binding analysis suggest that HD provides specific recognition that is similar to what is observed with the complete DNA-binding domain of SATB1 (CD+HD) at least in vitro. This result further strengthens our initial finding through SELEX enrichment that it is the HD-targeted DNA element that is preferentially bound by SATB1. The HD-binding sequence which comprises a palindromic AT-rich sequence is quite flexible and also exhibits better binding with GST:CD indicating that binding of SATB1 would be enhanced at specific sites due to presence of both CD and HD. Thus, the specific and high-affinity binding by SATB1 to CSBS and its variants is presumably mediated by the HD, and CD may play a secondary role by increasing the binding affinity further.
Table 1.
Summary of relative binding affinities of SATB1 domains to various CSBS point mutants
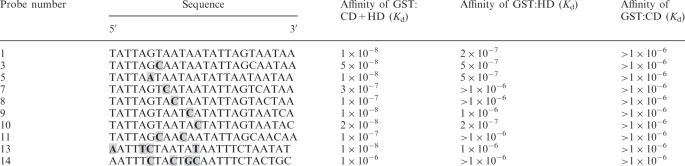 |
The nucleotide sequences of oligonucleotides used for binding by GST:HD and GST:CD as shown in Figure 6 and the dissociation constants for binding of these sequences by the two proteins are listed. Shaded nucleotides depict mutated nucleotides at various positions of the CSBS.
Figure 6.
Dimerization-dependent specific and higher-affinity binding is a property of HD. (A) Comparative EMSA analysis with various CSBS mutants was performed as described in Materials and Methods section. The probe numbers on the left of each panel represent the probes listed in Table 1. These probes were separately incubated with GST:HD (lanes 1–4) or GST:CD (lanes 5–8) at concentrations indicated above each lane. (B) The proteins used in the experiments described above and in Figure 5. Affinity purified recombinant proteins were resolved on a 12% SDS–polyacrylamide gel followed by staining with Coomassie brilliant blue. The positions of molecular weight standards are indicated on the left. (C) Coomassie brilliant blue stained protein gel indicating quality of various proteins used for Southwestern blotting. Lane 1, molecular weight standards; lanes 2–4, GST:346–763; lanes 5–7, GST:346–763 (ΔHD); lanes 8–10, GST; lane 11, molecular weight standards; lanes 12–14, GST:CD and lanes 15–17, GST:HD. For each of the proteins resolved, the three lanes contain 0.25, 0.5 and 1 μg of protein respectively. (D) Southwestern blot using CSBS as probe. (E) Southwestern blot using heptameric IgH MAR as probe.
To directly evaluate and compare the binding affinities of the various proteins we performed Southwestern analysis using GST:346–763 (CD+HD), GST:346–763 ΔHD (lacking 641–702 aa), GST alone, GST:CD and GST:HD. We observed binding of CSBS (Figure 6C) and wild-type heptamer IgH MAR (Figure 6D) with GST:346–763 (lanes 2–5) and with GST:HD (lanes 15–17). Strikingly, no significant binding was observed with GST:CD (Figure 6C and D, lanes 12–14), GST:346–763 ΔHD (Figure 6C and D, lanes 5–7), and GST alone (Figure 6C and D, lanes 8–10). Very faint signal indicating weak binding of GST:346–763 could be detected upon prolonged exposure. These results clearly confirm that dimer of HD makes specific contacts with the DNA at a concentration when there is no binding by GST:CD or GST:346–763 ΔHD. Furthermore, neither HD (Figure 5D, lanes 8–10) nor CD (data not shown for monomer CD) exhibited binding with the probe in their monomeric (GST-free) form. Taken together, these results confirm that dimeric status of HD and not CD is essential for the specific binding of SATB1 with the DNA.
Distance between the half-sites and minor groove of the palindromic CSBS are critical for binding by SATB1
Since the HD consensus is repeated in inverse fashion in the CSBS centered on a single nucleotide, it is imperative that the two half-sites will lie on the opposite sides of the helix considering 10.3 bases per turn of the helix of B-form DNA. We reasoned that if the two half-sites lie on the opposite side of the helix then the binding may be affected by altering the distance between the two if the phasing of the sites is critical for dimerization-dependent binding by SATB1. To monitor this, we designed two sets of oligonucleotides such that we introduced four additional G or C nucleotides either in the center of the two half-sites or on one side maintaining the base composition. Interestingly, binding of SATB1 was abolished when the GC-rich sequence was introduced in the middle as compared to when the GC-rich sequence is introduced on the side (Figure 7A, compare lanes 6–8 with 2–4). This result indicates that altering the distance between the palindromic half-sites of the consensus DNA sequence affects the binding by SATB1. This observation also implies that the dimerization-dependent binding of SATB1 is constrained by the positioning of the AT-rich half sites within the CSBS.
Next, to evaluate the role of minor groove in the binding activity of SATB1, we performed EMSA in presence of distamycin A. In accordance with the earlier report that SATB1 binds to the DNA via minor groove (13), we also found that the minor groove-binding drug distamycin abolishes the DNA binding of GST:CD+HD in a dose-dependent manner (Figure 7B, panel 1). Distamycin also affects binding of GST:HD with CSBS in a similar fashion as observed with SATB1 (Figure 7B, panel 2) whereas there is no effect of distamycin on binding of GST:CD (Figure 7B, panel 3), indicating that the minor groove binding of SATB1 is dictated solely by HD. The minor groove binding of HD in dimeric Pou (one CD) and HD-containing proteins such as Pit 1 (24,25) and HNF1-α (26) and several other similar proteins are well documented.
DISCUSSION
Determination of the optimal recognition site for the SATB family of proteins has important implications for understanding their interaction with DNA and also toward studying the repertoire of genomic targets of SATB1. We speculated that the number of genomic targets of SATB1 could be at least as high as the number of genes it regulates at transcription level (7). Few SBSs have been characterized, however; these belong to various types of genomic sequences and are far too less in number to provide a clear consensus signature that can be used as a universal tool for fishing out the putative SATB1 targets within the genome. We report the delineation of SATB1-binding site preferences from sequence analysis of oligonucleotides selected in vitro from a pool of random sequences. The technology of random oligonucleotide selection has been used for determining the consensus-binding element for a large number of transcription factors and DNA-binding proteins including Pou, CDP, p53 and NFκB (27–30). Interestingly, the consensus sequence derived from our SELEX-enriched sequences shared homologous regions with number of SBSs suggesting that SATB1 may preferentially bind to similar sequences in vivo and may regulate transcription of associated genes. The specific DNA-binding domain has been redefined to be HD and the mode of binding to the CSBS is proposed to be similar to the HNF1 dimer complexed with DNA (26) except that the HD in SATB1 possesses higher affinity and acts as a principal recognition domain.
Delineation of palindromic consensus sequence provides insights into the mechanism of DNA binding by SATB1
Contrasting information is available regarding the mode of DNA binding by SATB1. Full-length SATB1 was shown to bind DNA predominantly through the minor groove (13), whereas its CD (previously referred as MD) was shown to bind via major groove (14,15). The C-terminal HD was shown to be required for providing specificity and enhanced binding to the specific sequence but the mechanism was not understood (10). The 346–495 aa region (CD) of SATB1 harboring a Cut-like repeat was shown to make direct contact with DNA and bind as a monomer. Particularly, mutations at conserved residues Gln402 and Gly403 that make direct contact with the DNA mitigated DNA-binding activity (15). However, in our SELEX study we could not obtain a consensus matching to any of the reported CD-binding sequences harboring the ATCGAT core (28,31). On the contrary, we have found the presence of conserved HD-binding sequences (C/G/ATAATA) [‘TAAT’ as core consensus as described in Ref. (19)] along with a related sequence rich in AT. The consensus sequence we derived reads ‘TATTAGTAATAA’. The underlined sequences highlight the inverse palindromic arrangement of consensus elements that may have important implications for the recognition and high-affinity binding of such sequences by SATB1. A single mutation in the HD consensus region (GTCATA or GTACTA) has detrimental effect toward its binding by SATB1. Furthermore, symmetric positioning of two AT-rich stretches resembling HD-binding consensus in inverse orientation separated with one or two C or G nucleotide(s) is required for the specific binding by SATB1. Thus, the consensus sequence fails to fit the ATC context that was shown to be required for specific binding by SATB1 (1,2,10). Additionally, we found that the spacing between the two AT-rich half-sites is also critical. The dyad symmetry could play a vital role in protein–DNA interaction and regulation of specificity. HDs typically contact DNA by two discrete regions, an N-terminal arm lies in the minor groove and specific DNA contacts are mediated by Arg-3 and Arg-5. The third α-helix or recognition helix fits in the major groove of the recognition site, and Gln-50 and Asn-51 were shown to specifically contact DNA (26,32,33). These residues are conserved in the SATB1 HD and are required for the HD-mediated increase in affinity (10). Mutation studies indicated that the major contribution of the SATB1 HD is mediated by its N-terminal arm, most likely in the minor groove (10). Similarly we also observed the inhibition of SATB1 and HD binding by distamycin. Interestingly, HD was also shown to recognize a short (C/A)TAATA motif that colocalizes with the core unwinding element (10). This motif is identical with the 3′ half of CSBS reported here.
Role of PDZ domain-mediated dimerization in the high-affinity binding by SATB1
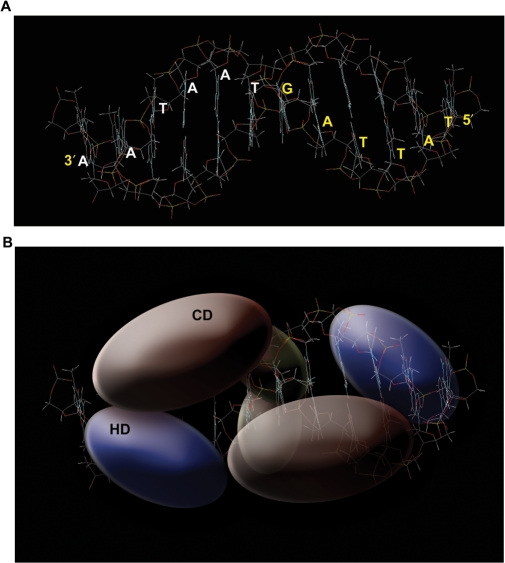
Based on missing nucleoside experiments it was suggested that HD and CD contact the same site simultaneously, possibly from opposite sides of the DNA helix (9). However, this model does not incorporate the fact that SATB1 exists as a homodimer in vivo (8, and this study). It is noteworthy that crystal structure of the even-skipped (eve) HD showed that two HDs are bound by one 10-bp consensus sequence on both faces of the DNA in a tandem fashion (34). This unusual binding mode involving simultaneous occupation of one binding site from both sides of the DNA helix could stabilize the protein–DNA complex. In the transcription factor Oct-1, the bipartite DNA-binding domain is composed of a POU-specific domain (POUs/one CD) and a POU-homeodomain (POUhd) connected by a flexible linker. Solution structure revealed that the left half of the optimal POU-binding site, the octamer ATGCAAAT, is recognized by POUs and the right half by POUhd (35). Interestingly, another POUhd protein LFB1/HNF1 binds as a homodimer to an inverted palindromic consensus-binding element (36). HNF1-α crystal structure indicates that a monomer can occupy more than half site of the DNA when bound to a 21-bp oligonucleotide sequence harboring 13 bp palindrome sequence (26). In light of the findings that SATB1 is a homodimer and that its binding consensus is an inverted palindrome, we propose that the PDZ-like dimerization domain bridges DNA-binding regions of two SATB1 monomeric subunits such that they bind in an antiparallel fashion to the inverse palindromic consensus-binding element (Figure 8). In this model, all three domains have unique contributions toward the high-affinity DNA binding by SATB1. The CD binds DNA through major groove without much specificity and with low affinity whereas the HD binds target DNA specifically through the minor groove and with high affinity. The affinity is increased many folds when both domains are held together in dimeric form by the PDZ domains (or GST). Thus, the dimer of SATB1 may form a clamp-like structure that wraps around the helix via occupying both major and minor grooves (Figure 8). This mode of binding is similar to that of LFB1/HNF1, wherein the DNA-independent dimerization domain is required to increase the DNA-binding affinity, but does not influence the dimer geometry (36). It is not surprising therefore, that the replacement of SATB1's N-terminal PDZ with any other dimerization imparting polypeptide including GST can restore DNA binding to the wild-type levels. Thus, the functional MD of SATB1 is constituted by the CD and HD together and not by the CD alone. The CD may principally occupy the major groove whereas the N-terminal arm of the HD may occupy the minor groove. The recognition helix of the HD may also occupy the major groove but without any significant contribution toward binding, since mutation in the third helix of HD does not affect DNA binding of SATB1 significantly (10). This is yet another unique mode of DNA binding that may provide exceptional stability to the complex, and may therefore explain the remarkable increase in binding specificity and affinity with dimeric CD+HD.
Figure 8.
Model depicting binding of the consensus element by SATB1. (A) The consensus DNA sequence was created using the ‘B’ software (http://www.scripps.edu/mb/case/Biomer/). A pdb file was created from the sequence and rendered in wireframe model assuming B-form of DNA, and then the structure was minimized. This was imported in Photoshop CS2 (Adobe) and labeled with alphabets representing the nucleotides. (B) Three-dimensional representation of SATB1–CSBS complex. The B form of CSBS DNA as obtained using the ‘B’ software was imported to Photoshop CS2 (Adobe) and the cartoon of SATB1–DNA complex was created by adding the colored blobs representing each of the domains of SATB1. The figure represents that one monomer of the dimer occupies one face of DNA and the other monomer occupies the other side where HD recognizes the minor groove whereas CD recognizes major groove and the N-terminal PDZ provides the dimerization function by bridging two monomers together; however, the PDZ domain does not contact with the DNA. Transparent blobs are on the face of the helix away from the reader. For details see text. The sizes and shapes of the blobs depicting the three domains of SATB1 are strictly for schematic presentation purpose only.
Homodimerization of HDs is known to be essential for the DNA binding in several other proteins (23) but in our mammalian two-hybrid study we did not observe any such interaction between C-terminal HD or CD. We observed a very strong interaction mediated via only the N-terminal PDZ-containing domain. Thus, this property of HD in SATB1 is different than the known dimerization of other HDs. Therefore the final proposed model is quite similar with the structure of the dimeric HNF1-α where dimerization domain resides independently and one monomer of the dimer occupies one face of DNA and the other monomer occupies the other side. Additionally, one monomer occupies more than half of the palindrome sequence (26) that could be also true for SATB1 since the distance between the half-sites of CSBS is critical for binding.
Transcription repression by SATB1 is dependent on its affinity toward the binding site
SATB1 is known to act as a repressor at promoters of IL-2 and its receptor (6), and MMTV LTR (37). The AT-rich IgH MAR has been shown to enhance the SV40 promoter in the integrated luciferase system and SATB1 represses this enhancing activity (38). Transcriptional repression by SATB1 occurs via recruitment of HDAC, Sin3A and ACF/ISWI nucleosome remodeling and mobilization complexes to the site of its binding (5,6). SATB1 is also known to act as an activator at the CD8 SBS region (39) and at many genes including β-globin (40), c-Myc (2) and TH2 cytokine genes (3). The major breakpoint region of BCL2 harboring an SBS also activates transcription of BCL2 (41). At any given regulatory sequence, the ability of SATB1 to act as an activator or repressor is governed by a phosphorylation-dependent molecular switch that operates in response to physiological signals (7). However, how SATB1 acts in these two contrasting modes at different genomic sites at the same time is not understood. Analysis of transcriptional activity mediated by the constructs harboring the CSBS and its derivatives revealed their activation potential. Most of these sequences were able to mediate enhanced expression of SV40 promoter-derived expression of luciferase gene. The repression of these by SATB1 was proportional with its in vitro binding affinities to these sequences. This is a unique mode of transcriptional regulation by a factor whose potential as a repressor is dependent on its affinity for the target sequence. Our data clearly argues that there is close relationship between the affinity of SATB1 with its recognition elements and its ability to activate or repress transcription at these sites, thus providing a foundation for understanding the transcriptional signals embedded in the regulatory regions. The precise mechanism of such affinity-based regulation requires further investigation.
Role of HD in regulation of the DNA-binding activity of the full-length SATB1
For a multi-DNA-binding domain containing protein such as SATB1, it is important to find out which domain actually contributes toward DNA-binding activity in vivo. It would be difficult to assign function to a domain based on its DNA-binding sequences in vitro if considered individually. It is plausible that both domains are required for high affinity and specific binding in vivo. Furthermore, binding specificity and affinity could be enhanced by the oligomeric status of the protein. Technically it is difficult to identify and isolate all possible variants of specific binding sites from the complex and heterogeneous chromatin. We therefore exploited the potential of SELEX technique for not only to delineate the specific SATB1-binding consensus sequences, but also to narrow down the region of the protein involved in specific DNA recognition and also to gain insights into the mode of binding. Our study demonstrates that the N-terminal dimerization domain of SATB1 is essential for specific recognition of the DNA substrates, albeit indirectly. Recently, the crystal structure of modified CD has been reported where the interaction between SATB1–CUTr1 and MAR DNA is mediated via a single pair of direct hydrogen bonds, which is atypical of DNA recognition from the major groove side (15). Binding of CD with DNA is very weak and therefore in this study four basic residues were incorporated at the C-terminal to induce DNA interaction. Moreover, it was proposed that the binding by CD with the DNA is poor presumably to enable post-translational modification-mediated regulation by affecting the dimerization status (15). However, it should be noted that the affinity of other human Cut repeats to their cognate binding sites is reported to be very high [Kd < 10−9, Ref. (28)], and that of SATB1 Cutr1 or CD alone is very poor (Kd > 10−7). It is surprising to observe that negatively charged dimeric HD (aa 640–763, pI = 4.81) can bind with very high affinity as compared to the positively charged dimeric CD (aa 346–495, pI = 9.16) at physiological pH, which indicates immense specificity of HD toward the specific target sequence and may therefore serve as the principal determinant for the high-affinity binding by SATB1 to its specific genomic targets. The HD region defined and used in an earlier investigation (10) spans residues 640–702, whereas we have used HD with an extended C-terminal region to match the size of CD (residues 640–763). The C-terminal extension (CTD) by itself does not bind DNA (data not shown) and therefore presumably may act by stabilizing the structure of HD.
It is known that SATB1 binds to the minor groove of the DNA since the minor groove-binder distamycin inhibits the DNA-binding activity of SATB1 (13; this study). Similar pattern of distamycin-mediated inhibition of DNA binding is observed with HD (Figure 7B, panel 2). Distamycin-mediated inhibition of binding to the minor groove of DNA has already been shown for the antennapedia HD (42) and the HD of SATB1 behaves in similar fashion. It has also been recently shown that the monomer CD binds to the DNA via major groove (14). Moreover, since distamycin does not affect binding of CD with DNA, it can be concluded that CD does not make contact via minor groove. The binding by HD of several other cut repeat containing HD proteins have been shown to be mediated via minor as well as major groove HNF1-α (26). Residues R3 and R5 at the N-terminal loop region are conserved and protrude into the minor groove whereas helix 3 makes contact in the major groove where a glutamine residue is very important. Mutation of R3 and R5 have been shown to reduce SATB1 binding by 5-fold whereas mutation in the third helix region reduces DNA binding by just 2-fold, indicating major contribution by the minor groove-binding region and very little contribution of helix 3 of the SATB1 HD (13). Most of the one CD containing POUhd proteins have been shown to have little or half contribution of HD toward the DNA binding (43,44), which does not seem to be true for SATB1 where HD seems to contribute major role in contacting DNA for specific DNA–protein interaction. Therefore, we now provide sufficient evidence to conclude that the ability of SATB1 to bind via the minor groove is contributed by HD and not by the CD. Collectively, our findings establish a distinct and specific role of the HD in defining the binding specificity of SATB1 and provide novel insights into the mode of target recognition and binding by SATB1 for regulation of transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr D. Dandekar for supplying the oligonucleotide library, Dr K. Muniyappa for helpful suggestions and Dr T. Kohwi-Shigematsu for constructs. We also thank Ranveer Jayani, Dimple Notani and Shashikant Gawai for DNA constructs and protein purification. Work in S.G. laboratory was supported by grants from the Department of Biotechnology, Government of India, and the Wellcome Trust, UK. P.K.P., S.S., P.P.K. and S.M. are supported by fellowships from the Council of Scientific and Industrial Research, India. S.G. is an international senior research fellow of the Wellcome Trust, UK. Funding to pay the Open Access publication charges for this article was provided by The Wellcome Trust, UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.deBelle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J. Cell Biol. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- 3.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 4.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 5.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 6.Kumar PP, Purbey PK, Ravi DS, Mitra D, Galande S. Displacement of SATB1-bound histone deacetylase 1 corepressor by the human immunodeficiency virus type 1 transactivator induces expression of interleukin-2 and its receptor in T cells. Mol. Cell. Biol. 2005;25:1620–1633. doi: 10.1128/MCB.25.5.1620-1633.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar PP, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Galande S, Dickinson LA, Mian IS, Sikorska M, Kohwi-Shigematsu T. SATB1 cleavage by caspase 6 disrupts PDZ domain-mediated dimerization, causing detachment from chromatin early in T-cell apoptosis. Mol. Cell. Biol. 2001;21:5591–5604. doi: 10.1128/MCB.21.16.5591-5604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagomi K, Kohwi Y, Dickinson LA, Kohwi-Shigematsu T. A novel DNA-binding motif in the nuclear matrix attachment DNA-binding protein SATB1. Mol. Cell. Biol. 1994;14:1852–1860. doi: 10.1128/mcb.14.3.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickinson LA, Dickinson CD, Kohwi-Shigematsu T. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J. Biol. Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- 11.Dandekar DH, Ganesh KN, Mitra D. HIV-1 Tat directly binds to NFkappaB enhancer sequence: role in viral and cellular gene expression. Nucleic Acids Res. 2004;32:1270–1278. doi: 10.1093/nar/gkh289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manuel M, Rallu M, Loones MT, Zimarino V, Mezger V, Morange M. Determination of the consensus binding sequence for the purified embryonic heat shock factor 2. Eur. J. Biochem. 2002;269:2527–2537. doi: 10.1046/j.1432-1033.2002.02917.x. [DOI] [PubMed] [Google Scholar]
- 13.Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi H, Tateno M, Yamasaki K. Solution structure and DNA-binding mode of the matrix attachment region-binding domain of the transcription factor SATB1 that regulates the T-cell maturation. J. Biol. Chem. 2006;281:5319–5327. doi: 10.1074/jbc.M510933200. [DOI] [PubMed] [Google Scholar]
- 15.Yamasaki K, Akiba T, Yamasaki T, Harata K. Structural basis for recognition of the matrix attachment region of DNA by transcription factor SATB1. Nucleic Acids Res. 2007;35:5073–5084. doi: 10.1093/nar/gkm504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho SP, Britton DH, Bao Y, Scully MS. RNA mapping: selection of potent oligonucleotide sequences for antisense experiments. Methods Enzymol. 2000;314:168–183. doi: 10.1016/s0076-6879(99)14102-8. [DOI] [PubMed] [Google Scholar]
- 17.Purbey PK, Jayakumar PC, Patole MS, Galande S. pC6-2/caspase-6 system to purify glutathione-S-transferase-free recombinant fusion proteins expressed in Escherichia coli. Nat. Protoc. 2006;1:1820–1827. doi: 10.1038/nprot.2006.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 19.Muller M, Affolter M, Leupin W, Otting G, Wuthrich K, Gehring WJ. Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J. 1988;7:4299–4304. doi: 10.1002/j.1460-2075.1988.tb03328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji X, Zhang P, Armstrong RN, Gilliland GL. The three-dimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2-A resolution. Biochemistry. 1992;31:10169–10184. doi: 10.1021/bi00157a004. [DOI] [PubMed] [Google Scholar]
- 21.Maru Y, Afar DE, Witte ON, Shibuya M. The dimerization property of glutathione S-transferase partially reactivates Bcr-Abl lacking the oligomerization domain. J. Biol. Chem. 1996;271:15353–15357. doi: 10.1074/jbc.271.26.15353. [DOI] [PubMed] [Google Scholar]
- 22.Purbey PK, Jayakumar PC, Deepalakshmi PD, Patole MS, Galande S. GST fusion vector with caspase-6 cleavage site for removal of fusion tag during column purification. Biotechniques. 2005;38:360–364. doi: 10.2144/05383BM03. [DOI] [PubMed] [Google Scholar]
- 23.Kasahara H, Usheva A, Ueyama T, Aoki H, Horikoshi N, Izumo S. Characterization of homo- and heterodimerization of cardiac Csx/Nkx2.5 homeoprotein. J. Biol. Chem. 2000;276:4570–4580. doi: 10.1074/jbc.M004995200. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson EM, Li P, Leon-del-Rio A, Rosenfeld MG, Aggarwal AK. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- 25.Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carrière C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 26.Chi YI, Frantz JD, Oh BC, Hansen L, Dhe-Paganon S, Shoelson SE. Diabetes mutations delineate an atypical POU domain in HNF-1 alpha. Mol. Cell. 2002;10:1129–1137. doi: 10.1016/s1097-2765(02)00704-9. [DOI] [PubMed] [Google Scholar]
- 27.Verrijzer CP, Alkema MJ, van Weperen WW, Van Leeuwen HC, Strating MJ, van der Vliet PC. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aufiero B, Neufeld EJ, Orkin SH. Sequence-specific DNA binding of individual cut repeats of the human CCAAT displacement/cut homeodomain protein. Proc. Natl Acad. Sci. USA. 1994;91:7757–7761. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.el Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 30.Kunsch C, Ruben SM, Rosen CA. Selection of optimal kappa B/Rel DNA-binding motifs: interaction of both subunits of NF-kappa B with DNA is required for transcriptional activation. Mol. Cell. Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harada R, Berube G, Tamplin OJ, Denis-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol. Cell. Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 33.Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 34.Hirsch JA, Aggarwal AK. Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity. EMBO J. 1995;14:6280–6291. doi: 10.1002/j.1460-2075.1995.tb00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dekker N, Cox M, Boelens R, Verrijzer CP, van der Vliet PC, Kaptein R. Solution structure of the POU-specific DNA-binding domain of Oct-1. Nature. 1993;362:852–855. doi: 10.1038/362852a0. [DOI] [PubMed] [Google Scholar]
- 36.Tomei L, Cortese R, De Francesco R. A POU-A related region dictates DNA binding specificity of LFB1/HNF1 by orienting the two XL-homeodomains in the dimer. EMBO J. 1992;11:4119–4129. doi: 10.1002/j.1460-2075.1992.tb05505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo J, Lozano MM, Dudley JP. Nuclear matrix binding regulates SATB1-mediated transcriptional repression. J. Biol. Chem. 2005;280:24600–24609. doi: 10.1074/jbc.M414076200. [DOI] [PubMed] [Google Scholar]
- 38.Kohwi-Shigematsu T, Maass K, Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- 39.Banan M, Rojas IC, Lee WH, King HL, Harriss JV, Kobayashi R, Webb CF, Gottlieb PD. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J. Biol. Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- 40.Wen J, Huang S, Rogers H, Dickinson LA, Kohwi-Shigematsu T, Noguchi CT. SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression. Blood. 2005;105:3330–3339. doi: 10.1182/blood-2004-08-2988. [DOI] [PubMed] [Google Scholar]
- 41.Ramakrishnan M, Liu WM, DiCroce PA, Posner A, Zheng J, Kohwi-Shigematsu T, Krontiris TG. Modulated binding of SATB1, a matrix attachment region protein, to the AT-rich sequence flanking the major breakpoint region of BCL2. Mol. Cell. Biol. 2000;20:868–877. doi: 10.1128/mcb.20.3.868-877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorn A, Affolter M, Müller M, Gehring WJ, Leupin W. Distamycin-induced inhibition of homeodomain-DNA complexes. EMBO J. 1992;11:279–286. doi: 10.1002/j.1460-2075.1992.tb05050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenfeld MG. POU-domain transcription factors: pou-er-ful developmental regulators. Genes Dev. 1991;5:897–907. doi: 10.1101/gad.5.6.897. [DOI] [PubMed] [Google Scholar]
- 44.Sturm RA, Herr W. The POU domain is a bipartite DNA-binding structure. Nature. 1988;336:601–604. doi: 10.1038/336601a0. [DOI] [PubMed] [Google Scholar]