Abstract
DNA methylation changes that are recurrent in cancer have generated great interest as potential biomarkers for the early detection and monitoring of cancer. In such situations, essential information is missed if the methylation detection is purely qualitative. We describe a new probe-free quantitative methylation-specific PCR (MSP) assay that incorporates evaluation of the amplicon by high-resolution melting (HRM) analysis. Depending on amplicon design, different types of information can be obtained from the HRM analysis. Much of this information cannot be obtained by electrophoretic analysis. In particular, identification of false positives due to incomplete bisulphite conversion or false priming is possible. Heterogeneous methylation can also be distinguished from homogeneous methylation. As proof of principle, we have developed assays for the promoter regions of the CDH1, DAPK1, CDKN2A (p16INK4a) and RARB genes. We show that highly accurate quantification is possible in the range from 100% to 0.1% methylated template when 25 ng of bisulphite-modified DNA is used as a template for PCR. We have named this new approach to quantitative methylation detection, Sensitive Melting Analysis after Real Time (SMART)-MSP.
INTRODUCTION
In mammalian cells, DNA methylation occurs almost exclusively at the carbon-5 position of cytosine residues within CpG dinucleotides. The CpG dinucleotide is distributed in a non-random fashion throughout the human genome. CpG-depleted regions are interspersed with CpG-rich sequences referred to as CpG islands (1). These islands are often located at promoter regions of protein encoding genes and tend to be unmethylated (2,3).
Aberrant DNA methylation patterns are one of the hallmarks of cancer. In most cancers, promoter hypermethylation correlates with gene silencing. This has been shown for a wide range of tumour suppressor genes including the genes studied here and reviewed in (2,3): the cell-cycle inhibitor gene CDKN2A (p16INK4a), the pro-apoptotic death-associated protein kinase gene DAPK1, the cell-adhesion gene CDH1 and the retinoic acid receptor gene RARB.
In cancer, methylation of some promoter CpG islands can be an early event, and thus the detection of methylation shows great promise as a biomarker for early detection (4–6). Conventional methods for cancer detection are in general not capable of finding pre-neoplastic and small malignant lesions, and are thus not suitable for early detection. Molecular biomarkers in body liquids such as blood, sputum or urine that allow detection and diagnosis of tumours at an early stage would be ideal. However, in these types of samples, tumour-derived material is hard to detect because of the presence of material from normal cells, and thus highly sensitive methods are needed (7). As one example, methylation of the CDKN2A promoter has been detected in the sputum of smokers up to 3 years before they are diagnosed with cancer (8). Detection of low level methylation also shows great potential in the molecular monitoring of established disease after therapy (4). This has already been shown to be feasible in various cancers using DNA derived from plasma or serum (9,10).
Methylation-specific PCR (MSP) (11) is a highly sensitive method for the detection of low level methylation, and can be sensitive to at least 0.1% methylated template. However, MSP is prone to false-positive results (12–14). MSP primers are normally designed to have one or more cytosines of CpG sites at or near the 3′ end. This makes the primers highly selective for methylated template, but also facilitates amplification of incompletely converted sequences in the bisulphite-treated DNA (13). It is thought that bisulphite treatment, in spite of recent improvements in this area, still remains the main source of variability in the analysis of DNA methylation. Recent results show that incomplete conversion may typically be in the order of 2%, even when a commercial kit is used (15,16). This variability can not only lead to false-positive results, but can also impair quantitative assays in a way that leads to overestimation of methylation levels (17), especially when looking at low level methylation.
Different methods for the detection of incompletely converted products co-amplified during the PCR have been proposed (14,16,18). These methods are relatively labour intensive and require removal of the PCR product from the tube for further analysis creating the potential for PCR contamination, or the use of additional probes as in the ConLight-MSP methodology (14).
MSP has been made quantitative by the use of fluorescent TaqMan probes enabling real time detection of MSP products (such as in the MethyLight technique) (19–21). This also eliminates any signal from non-specific amplification. However, the introduction of a probe complicates assay design, and can result in heterogeneously methylated sequences that would otherwise be detected by MSP being missed, because of the need for the probe to hybridize correctly before a signal is observed.
Here we present a new method: Sensitive Melting Analysis after Real Time (SMART)-MSP for sensitive DNA methylation detection based on probe-free real-time PCR. With the use of a new generation of fluorescent dyes, which do not inhibit PCR when they intercalate into double-stranded DNA at saturating levels (22), highly accurate quantification and further analysis of the amplicon by high-resolution melting (HRM), has become possible. As the temperature increases during the HRM step, and the DNA ‘melts’, the dye is released and the signal of fluorescence decreases rapidly. This change in fluorescence is sequence specific and can be monitored by appropriately designed instrumentation.
SMART-MSP provides information that cannot be obtained by electrophoresis, and thus functions as a quality control to avoid false-positive results caused by incomplete conversion or false priming due to less stringent PCR conditions. Primer dimers and non-specific products can be detected as well.
We developed SMART-MSP assays for several tumour suppressor genes that are currently used as molecular biomarkers for early cancer detection or are likely to have a high potential as such. The quantitative accuracy of the assays was tested using a standard dilution series of methylated DNA into unmethylated DNA. Furthermore, we show that false-positive results due to incomplete bisulphite conversion or false priming can be identified by HRM analysis and we have analysed a panel of cell lines, and breast cancer samples by the SMART-MSP methodology.
MATERIALS AND METHODS
Samples and DNA extraction
The investigations were performed after approval by the Peter MacCallum Cancer Centre Ethics of Human Research committee (Projects 02/70 and 02/26). Purified genomic DNA from cell lines (2008, MCF7, HS578T, MCF10A, MDA-MB-468, MDA-MB-231, MDA-MB-435, PC3, SKBr-3, Colo205, RPMI8226, SW948, HL-60 and T47D) was available in our laboratory. The DNA was extracted using the QIAamp® DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Breast cancer samples were provided by the Peter MacCallum Cancer Centre Tissue Bank. DNA from those samples was extracted using the salting out method (23). Universal Methylated DNA (Chemicon, Millipore, Billerica, MA, USA) was used as a fully methylated positive control. DNA from peripheral blood mononuclear cells from normal individuals was used as unmethylated DNA for dilutions. Standard dilution series of 100, 10, 1, 0.1 and 0.01% methylation levels were prepared by diluting the fully methylated DNA into the unmethylated DNA. DNA was quantified with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Whole-genome amplification
Whole-genome amplification (WGA) was performed as described by Umetani et al. (24) with slight modifications. Briefly, for primary WGA, 1 ng of genomic DNA extracted from peripheral blood mononuclear cells in 1 μl was subjected to amplification (2 h at 30°C) with the Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Little Chalfont, UK) according to the manufacturer's instructions. For secondary WGA, 0.1 μl of the unpurified primary WGA product was diluted in water to a final volume of 1 μl and processed as described above. The secondary WGA product was purified using the QIAquick® PCR Purification Kit (Qiagen) according to the manufacturer's instructions, eluted in 50 μl buffer EB, and quantified with a NanoDrop ND-1000 spectrophotometer. The secondary WGA product was used as fully unmethylated DNA (negative control).
Bisulphite modification
Five hundred nanograms of genomic DNA or WGA product was subjected to bisulphite conversion with the EpiTect® Bisulfite kit (Qiagen) according to the manufacturer's instructions. The purified, bisulphite-modified DNA (WGA product) was eluted two times in 20 μl buffer EB. Incomplete bisulphite-modified DNA was prepared by treating the appropriate samples for 20 or 40 min with the bisulphite mix solution and purified as described earlier.
SMART-MSP primer design
The primers were designed to include at least two CpG sites, and one of the cytosines of a CpG site was always placed at or adjacent to the 3′ end. This will make the primers as selective for methylated templates as possible, and ensure that only these are amplified during PCR if sufficiently stringent conditions are chosen. Non-CpG cytosines were included in the primer sequences as well to select against incompletely converted sequences, and at least one of these was placed as close to the 3′ end as possible. Primer Express 1.5 (Applied Biosystems, Foster City, CA, USA) was used to calculate the estimated annealing temperature of the primers. Amplicons were designed to have only one melting domain according to the POLAND software (http://www.biophys.uni-duesseldorf.de/local/POLAND/poland.html) (25). The program Amplify (http://engels.genetics.wisc.edu/amplify/) was used to check for primer dimers, and no single nucleotide polymorphisms (SNPs) were found in any of the designed amplicons using BLAST searching of the SNP database (dbSNP BUILD 127). This is important since SNPs will interfere with the melting profile (if found in between primers) or possibly affect primer binding. The primer sequences and genomic regions spanned, as well as amplicon size and the annealing temperatures (TA) can be found in Table 1.
Table 1.
Primer sequences, annealing temperatures, and amplicon information for the SMART-MSP assays (UCSC Genome Browser, November 2007)
| Gene | Primer sequences (CpG sites in bold and converted Cs as capital Ts or As) | Annealing temperature (°C) | Amplicon size (bp) | CpGs/non-CpG-Cs between the primers | Spanned region |
|---|---|---|---|---|---|
| CDH1 | F-gtgggcgggTcgtTagTtTc | 64 | 86 | 2/8 | 67328555-67328640 of Chr. 16 |
| R-cgctAattAActAaAAAttcacctAccg | |||||
| DAPK1 | F-aggaTagTcggaTcgagTTaacgTc | 67 | 61 | 0/4 | 89302618-89302678 of Chr. 9 |
| R-ttAccgaAtcccctccgcgA | |||||
| CDKN2A | F-gTaTTtTTtTcgagTaTtcgTtTacggc | 63 | 72 | 0/6 | 21964971-21965042 of Chr. 9 |
| R-caaatcctctAAaAAAaccgcgA | |||||
| RARB | F-TcgagaacgcgagcgatTc | 63 | 146 | 5/18 | 25444864-25445008 of Chr. 3 |
| R-gAccaatccaAccgAAAcg | |||||
| COL2A1 | F-gTaatgTTaggagTaTTTtgtgggTa | 65 | 86 | 0/1 | 46667210-46667295 of Chr. 12 |
| R-ctaccccaAAaAaAcccaAtcctA |
PCR and HRM conditions for the SMART-MSP assays
PCR cycling and HRM analysis were performed on the Rotor-Gene 6000™ (Corbett Research, Mortlake, Australia). SYTO® 9 was used as the intercalating dye (Invitrogen, Carlsbad, CA, USA). The reaction mixtures consisted of 25 ng of bisulphite-modified template (theoretical amount), 1x PCR buffer, 2.5 mmol/l MgCl2 final (3 mmol/l in the CDH1 assay), 200 nmol/l of each primer, 200 µmol/l of each dNTP, 5 µmol/l of SYTO 9, 0.5 U of HotStarTaq (Qiagen) (5 U/µl) in a final volume of 20 µl. The cycling protocol started with one cycle of 95°C for 15 min for enzyme activation, followed by 45 cycles of 95°C for 20 s, annealing at the appropriate temperature (Table 1) for 30 s, 72°C for 30 s and one cycle of 95°C for 1 min. HRM was performed from 60°C to 90°C, with a temperature increase at the rate of 0.2°C/s for all assays. The annealing temperature was experimentally determined for each assay to ensure that only methylated templates were amplified. For each assay, a standard dilution series was run to assess the quantitative properties and sensitivity of these. Fully methylated and fully unmethylated control (WGA product), unmodified control and no template control were also included in every run. All samples were analysed in triplicate and each breast tumour sample in duplicate.
Gel electrophoresis analysis
Five microlitres of the PCR products were resolved on 2.5% agarose gels and stained with ethidium bromide.
The CDH1 and RARB MethyLight assays
The same primers were used for the MethyLight assays as for the SMART-MSP assays. The probe sequences were FAM-5′-ttcgcgttgttgattgg-3′-BHQ for CDH1 and FAM-5′-ttgggtatcgtcggggta-3′-BHQ for RARB. The reaction mixtures consisted of 25 ng of bisulphite-modified DNA (theoretical amount), 1x PCR buffer, 250 nmol/l of probe, 3 mmol/l MgCl2, 200 nmol/l of each primer, 200 µmol/l of each dNTP, and 0.5 U of HotStarTaq (Qiagen) (5 U/µl) in a total volume of 20 µl. The cycling protocol started with one cycle of 95°C for 15 min, followed by 45 cycles of 95°C for 20 s and 63°C (64°C in the CDH1 assay) for 40 s. PCR cycling was performed on the Rotor-Gene 6000. All samples were analysed in triplicate and each breast tumour sample in duplicate.
Real time PCR quantification
The relative 2(−ΔΔCT) quantification approach (26) was used. The cycle threshold (CT) value of the control COL2A1 assay (Table 1) is subtracted from the CT value of the target gene for the calibrator sample, the 100% methylated standard. For each test sample, this value is then subtracted from the value resulting from the CT value of the target gene minus the CT value for the COL2A1 control assay. This gives the ‘ΔΔCT’ value, which is then put in to the equation, 2(−ΔΔCT), and multiplied by 100 to give the percentage of methylation relative to the 100% methylated control. For this approach to be valid, the amplification efficiencies of the gene and the control must be approximately equal (26). The take-off values given by comparative quantification (a feature of the Rotor-Gene 6000 Series Software, version 1.7.61) were used as CT values in the calculations. The take-off point is defined as the cycle at which the second derivative is at 20% of the maximum level, which indicates the end of the background noise and the transition into the exponential phase.
RESULTS
The basis of SMART-MSP
Real-time amplification of bisulphite-converted DNA with MSP primers is performed with a fluorescent dye, which does not inhibit the PCR when used at saturating conditions (22). This allows for highly accurate quantitative results to be obtained without the use of probes and for HRM analysis to be performed. Quantification is based on CT values, and thus it is necessary to run a control assay in parallel to normalize for the amount of input DNA in the PCR. We designed a new COL2A1 control assay to estimate the amount of amplifiable template (Table 1), adjacent to the region used previously as an input control in MethyLight-based assays (27). The region we chose has improved selection against the amplification of unconverted DNA (data not shown).
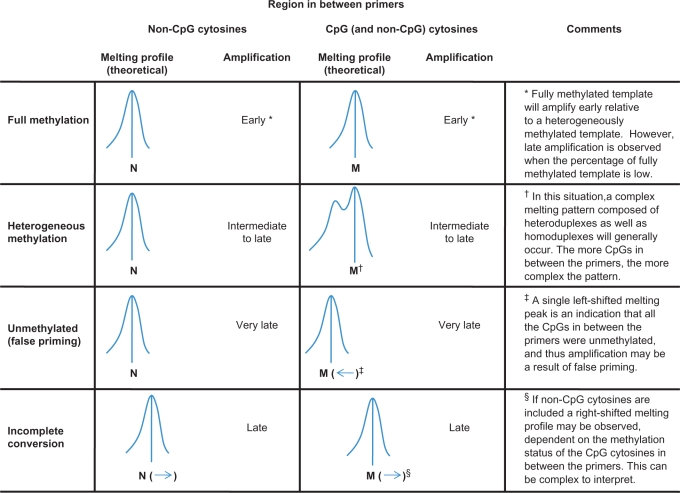
In the SMART-MSP methodology, sensitive melting analysis using HRM is performed immediately after the real-time PCR in a closed-tube system. The kind of information that can be obtained from HRM is dependent on amplicon design, and should be interpreted by considering the amplification data as well (Figure 1). First, when only non-CpG cytosines are included in between the primers, the HRM step can function as a control for amplification of incompletely converted DNA. If incomplete converted sequences are amplified, the amplicon will have a higher GC-content relative to fully converted amplicons, and will therefore melt later. We designed the DAPK1 and CDKN2A assays in this way.
Figure 1.
A schematic overview of SMART-MSP. Bisulphite-modified DNA is amplified in real time using a HRM-compatible intercalating dye to obtain quantitative data. After real-time PCR a HRM step is performed for quality control of the amplicon. The interpretation is made by considering both the real-time PCR and the HRM information. Two different types of SMART-MSP amplicon design are shown here, in combination with the melting profiles and amplification data that can be expected (vertical rows), in different methylation and conversion situations (horizontal rows). Incomplete conversion can be detected most readily when non-CpG cytosines are found in between the primers and no CpG cytosines are found. By including CpG cytosines in between the primers, it can be determined if the region is heterogeneously methylated or unmethylated. If the CpG cytosines in between the primers are unmethylated, the amplification might be a result of false priming. N and M are theoretical temperatures dependent on the amplicon size and sequence.
Second, when CpG sites are included between the primers, the HRM step can be used to assess if these CpG sites are methylated. We designed the CDH1 and RARB assays in this way. Generally, if left-shifted melting is observed, this is an indicator that some or all of the CpG sites are not methylated. A complex melting pattern consisting of heteroduplexes as well as homoduplexes can occur if more than one molecule is amplified during the PCR and the studied region is heterogeneously methylated. Thus, a heterogeneously methylated region can give a melting profile extending to the left, due to the melting of molecules with different CpG positions being methylated and heteroduplex formation between them (Figure 1). Left shifting can also indicate false positives due to false priming. In this case, a single left-shifted peak will be seen. Generally, false priming is associated with very late amplification (Figure 1). False priming can generally be minimized by stringent PCR conditions. Assays should be designed to include as few non-CpG cytosines as possible, preferably none. A conversion control should be performed in parallel when non-CpG cytosines cannot be avoided to be confident that these are converted. Our CDH1 and RARB primers were designed so that they can also be used with a MethyLight probe for validation of the SMART-MSP methodology. Because of this, more non-CpG cytosines are found in between the primers than what would be ideally preferred. Generally, longer products give more intrinsic variation of the melting curves (unpublished results).
Third, small amplicons can be designed that do not include non-CpG cytosines or CpG sites. In this case, only the methylation status of those CpG sites found under the primers are assessed, and the HRM step is used only then to validate that the correct sequence is amplified.
In all the above cases, the presence of primer dimers and non-specific products can be identified on the basis of aberrant melting profiles by HRM analysis (data not shown) without the need for gel electrophoresis.
Melting profiles of the SMART-MSP assays
The melting profiles of a true positive result, obtained by amplifying standards containing methylated template for each assay, were used as references for unknown samples (Figure 2). Since all the amplified dilution standards for a given amplicon have the same melting profile, we could have chosen any one of them as a reference. The melting temperature of each assay was approximately 83.2°C for CDH1, 79.5°C for DAPK1, 75.8°C for CDKN2A and 80.8°C for RARB. Confirmatory gel electrophoresis was performed in the development phase for all assays, and only one band of the expected size was observed (data not shown).
Figure 2.
Melting profiles of a true positive result for each SMART-MSP assay. Universally methylated template was amplified and analysed by HRM analysis. Each assay has a characteristic melting profile. (A) The CDH1 assay. (B) The DAPK1 assay. (C) The CDKN2A assay. (D) The RARB assay.
Sensitivity and quantitative accuracy of the SMART-MSP assays
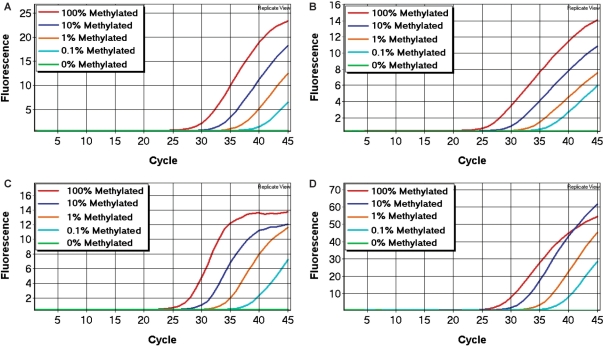
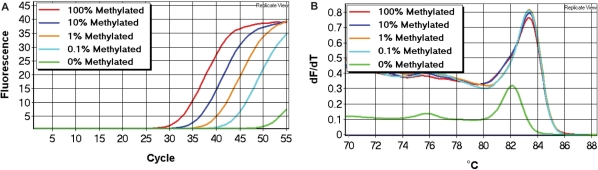
Each SMART-MSP assay was optimized so that amplification only occurred from standards containing methylated template, and no amplification was seen in the fully unmethylated control (WGA product), in the unmodified control or in the no template control. The sensitivity and quantitative accuracy of the assays were determined using the standard dilution series (see Materials and methods section). All our assays were able to reliably detect the 0.1% methylated standards (Figure 3).
Figure 3.
The sensitivity of the SMART-MSP assays. In all assays, the 0.1% methylated standard could be detected with high reproducibility. (A) The CDH1 assay. (B) The DAPK1 assay. (C) The CDKN2A assay. (D) The RARB assay.
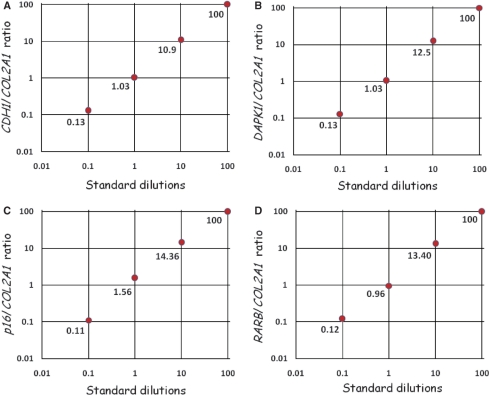
The quantitative accuracy of each assay was determined using the relative 2(−ΔΔCT) quantification approach (see Materials and methods section). Amplification efficiencies of the gene and the control were approximately equal for all assays (data not shown). The calculated values for each standard were plotted versus the dilution factor for each gene (Figure 4).
Figure 4.
The quantitative accuracy of the SMART-MSP assays. The quantitative accuracy of the SMART-MSP technology was assessed using the 2(−ΔΔCT) quantification approach. For each assay the calculated gene/control ratio for each standard is plotted against the dilution factor in a double logarithmic diagram. All assays proved to be quantitatively precise. (A) The CDH1 assay. (B) The DAPK1 assay. (C) The CDKN2A assay. (D) The RARB assay.
All reactions contained approximately equal amounts of template suitable for PCR. This was evident from the similar CT values obtained in the control assays (data not shown). We used the software to obtain a standard curve for the dilution series for calculation of the correlation coefficient (r2) for each assay. The correlation coefficient for each assay (DAPK1: r2 = 0.995, CDKN2A: r2 = 0.998, CDH1: r2 = 0.999, RARB: r2 = 0.995) indicated a strong linear relationship between CT values and given concentrations for all assays.
We also tested the CDH1 and RARB MethyLight assays. These MethyLight assays were quantitatively accurate in the range from 100% down to 0.1% methylated template. The correlation coefficients of the MethyLight assays were: CDH1: r2 = 0.984 and RARB: r2 = 0.983, again indicating a strong linear relationship between CT values and given concentrations for both.
Validation of the DAPK1 and CDKN2A SMART-MSP conversion control assays
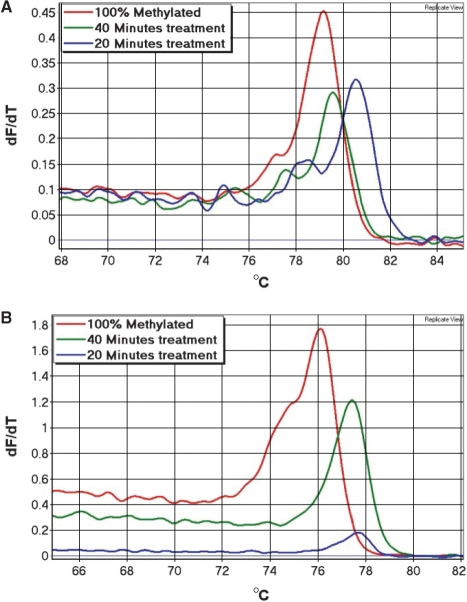
Bisulphite conversion can be assessed by HRM analysis using assays with non-CpG cytosines between the primers. If a right shift of the melting profile is observed, this can only be due to incomplete conversion of some or all of the non-CpG cytosines in between the primers or amplification of non-specific products (Figure 1). Since incompletely converted products are of the same size as true positives, these cannot be distinguished using gel electrophoresis. We generated incompletely converted DNA (see Materials and methods section) to assess whether its amplification showed right-shifted melting profiles in these assays. Amplification was usually seen from these samples and always showed right-shifted melting profiles. As expected, the 100% methylated standard amplified earlier than incompletely converted DNA in both assays, and thus gave higher melting peaks (Figure 5).
Figure 5.
Validation of the conversion control in the DAPK1 and CDKN2A assays. A peripheral blood control sample was bisulphite treated using different times of conversion (20 min, 40 min, normal protocol), and used to test the conversion control of these assays. (A) The DAPK1 assay. A gradual right-shift of the melting peaks was observed as the treatment time decreases. The observed right-shift of the incompletely treated samples indicates that some of the non-CpG cytosines in between the primers were not converted. Thus, these samples could be identified as false positives. (B) The CDKN2A assay. The 40 min treated sample and the 20 min treated sample both showed right-shifted melting peaks. Again, indicating that some of non-CpG cytosines in between the primers were not converted, and thus these samples could also be identified as false positives.
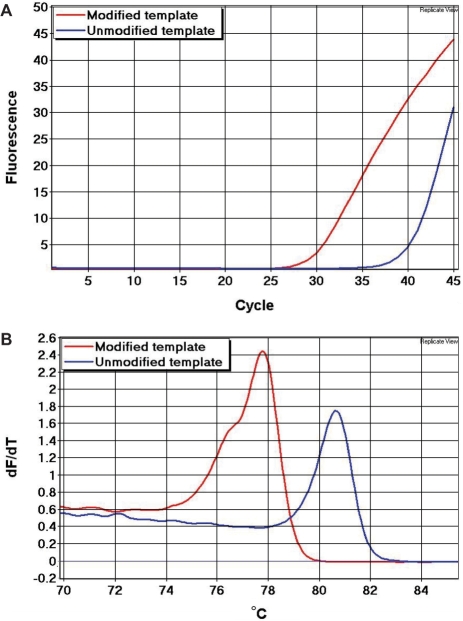
We showed that bisulphite-modified template melts early relative to unmodified template (Figure 6). A primer pair (5′-gggaagatgggatagaagggaataT and 5′-tctAacaAttAtAAActccaaccaccaa) with a limited number of non-CpG cytosines in their sequences (shown in upper case), and thus not particularly discriminatory against unmodified sequences, was used to amplify bisulphite-modified as well as unmodified DNA from the same sample. In this assay, five non-CpG cytosines and no CpG sites are found in between the primers. Since the unmodified amplicon has a higher GC-content, it is more stable and melted later than the bisulphite-modified amplicon.
Figure 6.
Detection of false priming from a whole genome amplified template. We used an assay that selected poorly against unmodified templates. In this assay, five non-CpG cytosines and no CpG sites are found in between the primers. These non-CpG cytosines were converted to uracil in the bisulphite-modified template (red), but not in the unmodified template (blue). Thus, a significant right-shift of the melting profile of the unmodified amplicon is observed. (A) Real-time PCR amplification data. (B) First derivative melting peaks.
Identification of false positives in the CDH1 SMART-MSP assay
By running the CDH1 SMART-MSP assay for an additional 10 cycles, late amplification from the fully unmethylated control (WGA product) occurred. Since this WGA control is not methylated at the two CpG sites in between the primers, we expected to see a readily distinguishable left-shifted melting peak, and thus to be able to identify it as a false-positive result (Figure 1). The melting peak of the unmethylated control (WGA product) was shifted ∼1.2°C to the left compared to the standards containing methylated template (Figure 7).
Figure 7.
Identification of false positives in the CDH1 SMART-MSP assay. The CDH1 SMART-MSP assay was performed with an additional 10 cycles to obtain amplification from the unmethylated control (WGA product) shown in green. (A) Real-time PCR amplification data. (B) The melting peak of the fully unmethylated control was left-shifted by ∼1.2°C relative to melting peaks of the standards containing methylated template, and could thus be identified as a false-positive result.
Screening of cell lines
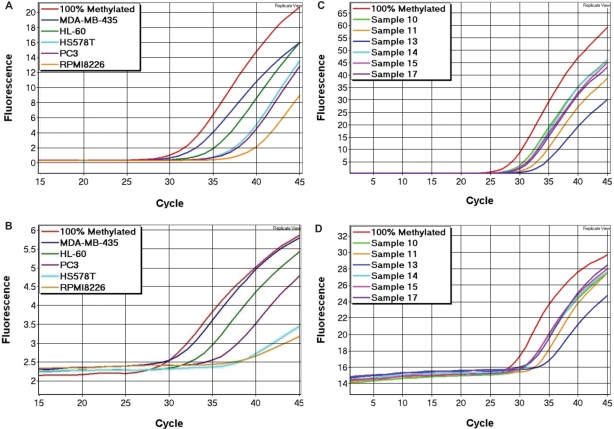
The reliability of our method was tested using a panel of 14 cell lines (2008, MCF7, HS578T, MCF10A, MDA-MB-468, MDA-MB-231, MDA-MB-435, PC3, SKBr-3, Colo205, RPMI8226, SW948, HL-60 and T47D) with the CDH1 and CDKN2A SMART-MSP assays. Five of these cell lines were shown to be methylated at the CDH1 promoter at various levels (HL-60: 100%, MDA-MB-435: 75%, PC3: 10%, HS578T: 6% and RPMI8226: 2%). The amplification data (Figure 8A) were used to calculate the methylation levels as described (see Materials and methods section). These results were validated using the CDH1 MethyLight assay, in which the same five cell lines were shown to be methylated at similar levels (Figure 8B). Our results were consistent with previously published data on these cell lines (28,29).
Figure 8.
Screening of cell lines for CDH1 methylation and breast cancer samples for RARB methylation. (A) CDH1 SMART-MSP amplification data for the positive cell lines. Five out the 14 cell lines screened were shown to be methylated at the CDH1 promoter. (B) CDH1 MethyLight amplification data from the positive cell lines. The data from the MethyLight assay was consistent with the data from the SMART-MSP assay. (C) RARB SMART-MSP amplification data for the positive tumour samples. Six out the 24 samples screened were shown to be methylated at the RARB promoter. (D) RARB MethyLight amplification data from the positive tumour samples. The data from the MethyLight assay was consistent with the data from the SMART-MSP assay.
However, the HS578T cell line was estimated to be methylated at lower levels when using the MethyLight assay relative to the SMART-MSP assay. This may be due to the fact that the probe in the MethyLight assay overlays two CpGs and that these are not consistently methylated in this cell line. The melting profiles obtained by the SMART-MSP assay was left-shifted, indicating that at least one of these two CpGs sites were unmethylated (data not shown).
Five of the 14 cell lines were shown to be methylated by SMART-MSP at various levels at the CDKN2A promoter (T47D: 65%, PC3: 35%, RPMI8226: 30%, Colo205: 1% and SW948: 0.1%). Again, these results were consistent with previously published data on these cell lines (28,30).
Screening of breast cancer samples
The diagnostic applicability of the SMART-MSP methodology was tested using the RARB and CDH1 assays on a panel of 24 breast cancer samples. We found 6 out of the 24 samples to be methylated at the RARB promoter region at levels higher than 5% (76, 57, 44, 33, 29 and 7%). The amplification data (Figure 8C) were used to calculate the methylation levels. Data from the control assay is not shown. These results were validated using the RARB MethyLight assay, in which the same six samples were shown to be methylated at similar levels (Figure 8D). These findings are in agreement with what has recently been reported for this locus in breast cancer (31,32).
None of the samples showed high-level methylation at the CDH1 promoter. We found 11 to be methylated between 0.1% and 0.7% which could be detected with high reproducibility. Furthermore, we found 10 samples to be methylated in the interval from 0.01% to 0.1%, for which the reproducibility between samples was less good. These observed values might only account for a biologically insignificant background methylation level that can be found in normal cells (unpublished results). A recent study has found that a low background of CDH1 methylation is present in normal breast tissue, and that this level did not differ significantly from what was found in adjacent malignant breast tissue (32). Another recent study reports that CDH1 methylation was not found in a cohort of 28 tissue biopsies from 17 breast cancer patients (31). However, the method used in that study was not sensitive enough to detect methylation levels ∼1%. Thus, if CDH1 methylation is detected using a purely qualitative and highly sensitive methodology, positive results might arise due to low background methylation.
We tested the same samples using our CDH1 MethyLight assay with comparable results (data not shown). However, the CDH1 MethyLight assay was not able to detect methylation in four of the samples showing the lowest levels of methylation in the CDH1 SMART-MSP assay.
DISCUSSION
MSP is a widely used method for the detection of DNA methylation. It uses primers specific for methylated (and optionally unmethylated), bisulphite-modified DNA (11). MSP is based on the principle that primers with mismatched 3′-ends will not be capable of extension during the PCR. If a band that corresponds to the amplicon size given by the MSP primers is seen on a gel after PCR, it is concluded that the sample is methylated. MSP is possibly the most sensitive non-quantitative technique available and can detect 0.1% methylation or less.
A major drawback of MSP is its susceptibility to false positives (12–14). This can be due to incomplete conversion of the template, false priming or a sensitivity issue due to the capacity of MSP to amplify very low-level methylation.
Incomplete bisulphite conversion is probably the most problematic cause of false-positive results. When performing MSP experiments, it is thus important to be confident that positive results are not derived from incomplete conversion. MSP primers are normally designed to have two or preferably more cytosines deriving from CpG sites at or near the 3′-end. This makes the primers highly selective for methylated template. However, this also facilitates amplification of incomplete converted sequences in the bisulphite-treated DNA as unconverted sequence resembles methylated sequence. It is possible that only a small subset of the DNA copies suffer a substantially lower conversion rate, which in combination with the high sensitivity of MSP can lead to false positives or overestimation of results. Also, it is possible that the distribution of unconverted sites is non-random, thus making some promoter regions more prone to incomplete conversion than others, and that conversion rates are dependent on DNA quality (16). Thus, in spite of stringent PCR conditions, a false-positive result can arise if bisulphite conversion is incomplete. For these reasons, it is recommended that a control for incomplete conversion is performed. This control should preferably assess the conversion status of non-CpG cytosines in the vicinity of the primer positions. We have shown that having a non-CpG cytosine at the 3′-end is not sufficient to rule out the possible amplification of incomplete converted DNA (Figures 5 and 6).
If the annealing temperature is too low or too many cycles are used, amplification can occur across the 3′ mismatch. This type of false positive can be detected by the use of an appropriate negative control (e.g. WGA product).
Finally, the sensitivity of MSP might lead to false positives because of the amplification of a rare subpopulation of methylated sequences. The tumour sample might be extremely heterogeneous, with only a small proportion of methylated cells. Alternatively, the methylation might derive from normal cells in the tumour biopsy (12,33). In either case, it would be incorrect to call the tumour methylated for that particular gene. This problem arises because MSP is a non-quantitative methodology. Despite these limitations, MSP remains useful, particularly in preliminary screening of a large number of tumour specimens but may not be suitable in a clinical setting (7,34).
Here, we have shown that SMART-MSP can give accurate quantitative data for DNA methylation detection. The combination of MSP with HRM, which is enabled by the use of a HRM-compatible DNA double-stranded intercalating dye, enables the sensitive screening of the region in between the MSP primers. Thus, information is provided that cannot be obtained by electrophoresis. For full utility of the methodology, one or both of two types of primer positioning should be used: (i) only non-CpG cytosines between the primers allowing assessment if (low) levels of amplification are due to incomplete conversion and (ii) CpGs (with as few non-CpG cytosines as possible) between the primers allowing assessment if (low) levels of amplification are due to partial or heterogeneous methylation. Interpretation is made by considering both the quantification and the HRM information (Figure 1). When the melt profile of a true positive is established (Figure 2) there is no need for gel electrophoresis analysis or any further processing, and thus SMART-MSP is a closed-tube method.
HRM analysis can easily detect single base pair changes in the amplified DNA sequence. Thus, by including non-CpG cytosines between the primers, the conversion status of these can be assessed. If cytosines in between the primers are not converted, the amplicons will melt late relative to amplicons derived from fully converted template (Figure 1). By generating incompletely converted DNA sequences, and using these as templates for our CDKN2A and DAPK1 assays, we were able to show that amplification from these usually occurred, and were easily identified as they always corresponded to right-shifted melting peaks (Figure 5). In addition, we amplified unmodified DNA as well as modified DNA derived from the same individual using primers that select poorly between modified and unmodified DNA, and observed highly reproducible right-shifted melting peaks from the unmodified template relative to the modified one (Figure 6).
False-positive results due to false priming can be detected by HRM analysis as well, if CpGs are included in between the primers (Figure 1). By running the CDH1 SMART-MSP assay for an additional 10 cycles, late amplification from the fully unmethylated control (WGA product) was seen. In this case, a left-shifted melting peak was observed due to the two CpGs found in between the primers being unmethylated. This allowed us to readily identify it as a false-positive result (Figure 7). However, sufficiently stringent PCR conditions and adequate primer design are the best insurance against false priming.
A left-shifted peak can also be due to the target sequence being heterogeneously methylated, which can result in heteroduplex formation, and thus the melting profile will often be visually different (Figure 1). The exact number of CpG sites that need to be methylated within MSP primers under given PCR conditions is hard to assess. This is especially important to keep in mind when heterogeneously methylated sequences are studied. Thus, methodologies that utilize MSP primers are only semi-quantitative when heterogeneously methylated DNA is amplified. For this reason, MSP might be less suited in those cases when CpG islands show highly variable methylation, as has been reported for the p15INK4B and CX26 (connexin 26) genes (35–37). However, when including CpG sites in between the primers in the SMART-MSP methodology, it can be assessed whether the studied region is heterogeneously methylated or not, by the HRM analysis.
We were able to reliably detect the 0.1% methylated standard for each assay with high reproducibility using 25 ng of bisulphite-modified DNA as template (Figure 3). Amplification from the 0.01% methylated standard was occasionally observed in all SMART-MSP assays, but generally showed high run to run variations in CT values. This might, in part, be caused by very low methylation levels present in normal blood. However, the quantitative accuracy in the range from 100% methylated template to 0.1% methylated template was not impaired by the fact that we did the dilutions in DNA from normal blood. During WGA of DNA all detectable methylation information is lost, and thus it is possible to create fully unmethylated template. However, results can be biased during normalization for DNA input if the dilutions are done using WGA product, since different genes are not amplified in the exact same proportions. Nevertheless, WGA product is excellent as a negative control, especially when looking at low level methylation.
MSP was originally made quantitative by the use of TaqMan probes (19–21), but quantification without probes using the dye SYBR Green has been reported (38–41). SYBR Green intercalation into double-stranded DNA has been shown to be markedly influenced by salt concentration, by dye/base pair ratios which are not constant during the PCR since more and more double-stranded DNA is generated through each cycle, and to show sequence specific binding (42). For this reason, the number of PCR cycles can influence melting curve analysis (43). Generally, some of the problems associated with SYBR Green can be minimized if high dye/base pair ratios are used. However, SYBR Green cannot be used at saturating concentrations without inhibiting the PCR. These problems have been markedly reduced by the introduction of a new generation of dyes (22).
It has previously been shown that melting analysis can discriminate methylated from unmethylated DNA (44,45). These assays were based on methylation-independent PCR (MIP) primers, and have not become widely used, presumably because of the technical limitations of reagents, instrumentation and data analysis software used at that time. Generally, methods utilizing MIP primers can be compromised by the PCR bias phenomenon (46), but this is not an issue when MSP primers are used.
Melting curve analysis has also been used in combination with the MSP methodology as an alternative to gel electrophoresis (47). This methodology did not provide quantitative data or information that cannot be provided by gel electrophoresis. Melting analysis of MSP products in the presence of SYBR green have also been used to detect primer dimers (38). This can be done by gel electrophoresis as well. This study did provide quantitative data, but these were much less accurate compared to what we have obtained with a dye that does not inhibit PCR.
SMART-MSP is complementary to our previously described methodology using HRM, methylation-sensitive HRM (MS-HRM) (48). SMART-MSP uses MSP primers and quantification is based on CT values instead of melting curve comparisons. Also SMART-MSP can detect the amplification of incompletely converted DNA, and is generally more sensitive. However, the main advantage of SMART-MSP might be that each assay is performed at one annealing temperature; where as in MS-HRM, a range of different temperatures are needed for the sensitive screening of samples showing markedly different methylation levels. In MS-HRM, quantification is based on comparisons with melting profiles of a standard dilution series that needs to be included in every run. This is not necessary when performing SMART-MSP assays which quantify relative to a 100% methylated control and the amplification of a CpG-free control sequence. We are currently using MS-HRM to analyse samples where relatively high levels of methylation are expected whereas SMART-MSP comes into its own to detect low levels of methylation.
Compared to the MethyLight technology, SMART-MSP does not require expensive probes. However, quantification without probes is compromised by primer dimers and non-specific amplification (38). For this reason, there is a need for evaluation of the PCR product, which can be conveniently done with HRM analysis. We observed non-specific amplification from some of our controls when the assays were performed at lower annealing temperatures. These products melted differently, and could be identified as non-specific amplification using gel electrophoresis as well (data not shown). However, when the assays were performed at the optimized annealing temperature, no primer dimers or non-specific products were observed. Without probes, less optimization may be needed and assay design has become easier. The use of HRM can give information about the methylation status of CpGs between the primers, but most importantly, the HRM step can be used as a control to indicate amplification of incompletely modified sequences, false priming or non-specific products. Thus, SMART-MSP is less prone to false-positive results and overestimation of methylation levels. We have shown that the sensitivity of our assays is similar to what has been reported for MethyLight.
In conclusion, SMART-MSP has made quantitative MSP inexpensive, more accurate, and less prone to false positives. It is a closed-tube method based on a high-throughput methodology, and thus it might prove to be the method of choice for the assessment of DNA methylation in clinical samples, particularly when low levels of methylation need to be sensitively and accurately determined.
ACKNOWLEDGEMENTS
We acknowledge Chelsee Hewitt and Angela Tan for critically reading this manuscript, and Stephen Fox and David Westerman for their continued support. This work was funded by grants from the United States Department of Defense Breast Cancer Research Program (BC046207) and the National Health and the Medical Research Council of Australia (350452) to A.D. Funding to pay the Open Access publication charges for this article was provided by Department of Defense (US) Breast Cancer Research Program.
Conflict of interest statement. A provisional patent has been filed for this methodology by the Peter MacCallum Cancer Centre.
REFERENCES
- 1.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 3.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 4.Laird PW. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 5.Shi H, Wang MX, Caldwell CW. CpG islands: their potential as biomarkers for cancer. Expert Rev. Mol. Diagn. 2007;7:519–531. doi: 10.1586/14737159.7.5.519. [DOI] [PubMed] [Google Scholar]
- 6.Toyota M, Issa JP. Epigenetic changes in solid and hematopoietic tumors. Semin. Oncol. 2005;32:521–530. doi: 10.1053/j.seminoncol.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Cottrell SE, Laird PW. Sensitive detection of DNA methylation. Ann. NY Acad. Sci. 2003;983:120–130. doi: 10.1111/j.1749-6632.2003.tb05967.x. [DOI] [PubMed] [Google Scholar]
- 8.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 9.Chan KC, Lo YM. Circulating tumour-derived nucleic acids in cancer patients: potential applications as tumour markers. Br. J. Cancer. 2007;96:681–685. doi: 10.1038/sj.bjc.6603625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taback B, Hoon DS. Circulating nucleic acids and proteomics of plasma/serum: clinical utility. Ann. NY Acad. Sci. 2004;1022:1–8. doi: 10.1196/annals.1318.002. [DOI] [PubMed] [Google Scholar]
- 11.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aggerholm A, Hokland P. DAP-kinase CpG island methylation in acute myeloid leukemia: methodology versus biology? Blood. 2000;95:2997–2999. [PubMed] [Google Scholar]
- 13.Dobrovic A. In: Molecular diagnostics for the clinical laboratorian. 2nd. Coleman WB, Tsongalis GJ, editors. Totowa, NJ: Humana Press; 2005. pp. 149–160. [Google Scholar]
- 14.Rand K, Qu W, Ho T, Clark SJ, Molloy P. Conversion-specific detection of DNA methylation using real-time polymerase chain reaction (ConLight-MSP) to avoid false positives. Methods. 2002;27:114–120. doi: 10.1016/s1046-2023(02)00062-2. [DOI] [PubMed] [Google Scholar]
- 15.Taylor KH, Kramer RS, Davis JW, Guo J, Duff DJ, Xu D, Caldwell CW, Shi H. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 16.Warnecke PM, Stirzaker C, Song J, Grunau C, Melki JR, Clark SJ. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 17.Ehrich M, Zoll S, Sur S, van den Boom D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res. 2007;35:e29. doi: 10.1093/nar/gkl1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki M, Anast J, Bassett W, Kawakami T, Sakuragi N, Dahiya R. Bisulfite conversion-specific and methylation-specific PCR: a sensitive technique for accurate evaluation of CpG methylation. Biochem. Biophys. Res. Commun. 2003;309:305–309. doi: 10.1016/j.bbrc.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo YM, Wong IH, Zhang J, Tein MS, Ng MH, Hjelm NM. Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer Res. 1999;59:3899–3903. [PubMed] [Google Scholar]
- 21.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 22.Gudnason H, Dufva M, Bang DD, Wolff A. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35:e127. doi: 10.1093/nar/gkm671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umetani N, de Maat MF, Mori T, Takeuchi H, Hoon DS. Synthesis of universal unmethylated control DNA by nested whole genome amplification with phi29 DNA polymerase. Biochem. Biophys. Res. Commun. 2005;329:219–223. doi: 10.1016/j.bbrc.2005.01.088. [DOI] [PubMed] [Google Scholar]
- 25.Steger G. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 1994;22:2760–2768. doi: 10.1093/nar/22.14.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J. Mol. Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 29.Reinhold WC, Reimers MA, Maunakea AK, Kim S, Lababidi S, Scherf U, Shankavaram UT, Ziegler MS, Stewart C, Kouros-Mehr H, et al. Detailed DNA methylation profiles of the E-cadherin promoter in the NCI-60 cancer cells. Mol. Cancer Ther. 2007;6:391–403. doi: 10.1158/1535-7163.MCT-06-0609. [DOI] [PubMed] [Google Scholar]
- 30.Wong IH, Ng MH, Lee JC, Lo KW, Chung YF, Huang DP. Transcriptional silencing of the p16 gene in human myeloma-derived cell lines by hypermethylation. Br. J. Haematol. 1998;103:168–175. [PubMed] [Google Scholar]
- 31.Dahl C, Guldberg P. A ligation assay for multiplex analysis of CpG methylation using bisulfite-treated DNA. Nucleic Acids Res. 2007;35:e144. doi: 10.1093/nar/gkm984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng W, Shen L, Wen S, Rosen DG, Jelinek J, Hu X, Huan S, Huang M, Liu J, Sahin AA, et al. Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res. 2007;9:R57. doi: 10.1186/bcr1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggerholm A, Guldberg P, Hokland M, Hokland P. Extensive intra- and interindividual heterogeneity of p15INK4B methylation in acute myeloid leukemia. Cancer Res. 1999;59:436–441. [PubMed] [Google Scholar]
- 34.Mikeska T, Bock C, El-Maarri O, Hubner A, Ehrentraut D, Schramm J, Felsberg J, Kahl P, Buttner R, Pietsch T, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J. Mol. Diagn. 2007;9:368–381. doi: 10.2353/jmoldx.2007.060167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodge JE, List AF, Futscher BW. Selective variegated methylation of the p15 CpG island in acute myeloid leukemia. Int. J. Cancer. 1998;78:561–567. doi: 10.1002/(sici)1097-0215(19981123)78:5<561::aid-ijc6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 36.Tan LW, Bianco T, Dobrovic A. Variable promoter region CpG island methylation of the putative tumor suppressor gene Connexin 26 in breast cancer. Carcinogenesis. 2002;23:231–236. doi: 10.1093/carcin/23.2.231. [DOI] [PubMed] [Google Scholar]
- 37.Cameron EE, Baylin SB, Herman JG. p15(INK4B) CpG island methylation in primary acute leukemia is heterogeneous and suggests density as a critical factor for transcriptional silencing. Blood. 1999;94:2445–2451. [PubMed] [Google Scholar]
- 38.Chan MW, Chu ES, To KF, Leung WK. Quantitative detection of methylated SOCS-1, a tumor suppressor gene, by a modified protocol of quantitative real time methylation-specific PCR using SYBR green and its use in early gastric cancer detection. Biotechnol Lett. 2004;26:1289–1293. doi: 10.1023/B:BILE.0000044922.43572.2d. [DOI] [PubMed] [Google Scholar]
- 39.Chu DC, Chuang CK, Fu JB, Huang HS, Tseng CP, Sun CF. The use of real-time quantitative polymerase chain reaction to detect hypermethylation of the CpG islands in the promoter region flanking the GSTP1 gene to diagnose prostate carcinoma. J. Urol. 2002;167:1854–1858. [PubMed] [Google Scholar]
- 40.Hong YS, Roh MS, Kim NY, Lee HJ, Kim HK, Lee KE, Kwak JY, Kim JY. Hypermethylation of p16INK4a in Korean non-small cell lung cancer patients. J Korean Med. Sci. 2007;22(Suppl):S32–S37. doi: 10.3346/jkms.2007.22.S.S32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomassin H, Kress C, Grange T. MethylQuant: a sensitive method for quantifying methylation of specific cytosines within the genome. Nucleic Acids Res. 2004;32:e168. doi: 10.1093/nar/gnh166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004;32:e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giglio S, Monis PT, Saint CP. Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 2003;31:e136. doi: 10.1093/nar/gng135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guldberg P, Worm J, Gronbaek K. Profiling DNA methylation by melting analysis. Methods. 2002;27:121–127. doi: 10.1016/s1046-2023(02)00063-4. [DOI] [PubMed] [Google Scholar]
- 45.Worm J, Aggerholm A, Guldberg P. In-tube DNA methylation profiling by fluorescence melting curve analysis. Clin. Chem. 2001;47:1183–1189. [PubMed] [Google Scholar]
- 46.Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, Clark SJ. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akey DT, Akey JM, Zhang K, Jin L. Assaying DNA methylation based on high-throughput melting curve approaches. Genomics. 2002;80:376–384. doi: 10.1006/geno.2002.6851. [DOI] [PubMed] [Google Scholar]
- 48.Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35:e41. doi: 10.1093/nar/gkm013. [DOI] [PMC free article] [PubMed] [Google Scholar]