Abstract
DNA methylation and chromatin DNaseI sensitivity were analyzed in and adjacent to D4Z4 repeat arrays, which consist of 1 to ∼100 tandem 3.3-kb units at subtelomeric 4q and 10q. D4Z4 displayed hypomethylation in some cancers and hypermethylation in others relative to normal tissues. Surprisingly, in cancers with extensive D4Z4 methylation there was a barrier to hypermethylation spreading to the beginning of this disease-associated array (facioscapulohumeral muscular dystrophy, FSHD) despite sequence conservation in repeat units throughout the array. We infer a different chromatin structure at the proximal end of the array than at interior repeats, consistent with results from chromatin DNaseI sensitivity assays indicating a boundary element near the beginning of the array. The relative chromatin DNaseI sensitivity in FSHD and control myoblasts and lymphoblasts was as follows: a non-genic D4Z4-adjacent sequence (p13E-11, array-proximal)> untranscribed gene standards > D4Z4 arrays> constitutive heterochromatin (satellite 2; P < 10−4 for all comparisons). Cancers displaying D4Z4 hypermethylation also exhibited a hypermethylation-resistant subregion within the 3.3-kb D4Z4 repeat units. This subregion contains runs of G that form G-quadruplexes in vitro. Unusual DNA structures might contribute to topological constraints that link short 4q D4Z4 arrays to FSHD and make long ones phenotypically neutral.
INTRODUCTION
Overall hypomethylation in some cancer specimens and hypermethylation in others was found in NBL2, a tandem array of 1.4 kb repeat units, in a comparison to diverse somatic control tissues. We demonstrated this in ovarian carcinomas and Wilms tumors by hairpin genomic sequencing to analyze methylation at 14 CpG dyads and by southern blotting to assess overall NBL2 methylation at CpG-containing restriction sites (1). NBL2 is located mostly in the short arms of the acrocentric chromosomes. This large tandem array and as well as others, including pericentromeric satellite 2 DNA and subtelomeric D4Z4 repeat arrays, require DNA methyltransferase 3B (DNMT3B) for normal levels of methylation (2). Satellite 2 and other human satellite DNAs are very frequently hypomethylated in diverse cancers compared with a wide variety of control tissues (3). There is clearly much plasticity in the epigenetics of these tandem repeat arrays.
The epigenetics of D4Z4 is of great interest because of the linkage of a short array of these tandem 3.3-kb repeats at 4q35 (Figure 1) to facioscapulohumeral muscular dystrophy (FSHD), a severely debilitating, dominantly inherited disease (4). In the present study, we examined DNA methylation and chromatin structure in D4Z4 and the immediately proximal sequence in various cell populations to gain insights into this array, whose structure must play a central role in FSHD. The very high G+C content of these repeat arrays, 73%, as contrasted with the neighboring 1-kb proximal sequence, which has only 40% G+C, suggests biologically important epigenetic features of the proximal border of the array.
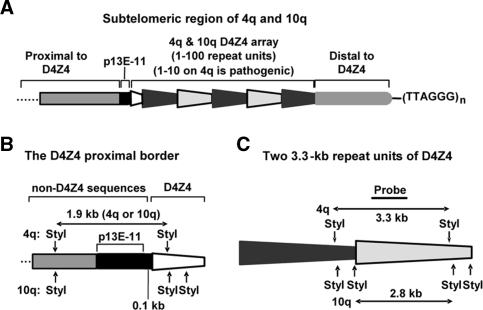
Figure 1.
Maps of the studied D4Z4 and proximal regions. (A) Cartoon (not to scale) showing the size-polymorphic D4Z4 array and p13E-11, a marker sequence. The sequence between the D4Z4 array and the telomeric hexamer repeats is about 15–25 kb. Black or gray trapezoids, essentially identical, tandem D4Z4 repeat units (3.3-kb KpnI fragments). White trapezoid, the 0.9-kb region from the start of the array to the first KpnI site in D4Z4. (B) Maps of StyI sites for chromatin DNaseI sensitivity assays. Left, StyI restriction map of the proximal border of D4Z4 including the StyI fragment spanning the 0.8-kb p13E-11 sequence through 0.3 kb at the beginning of D4Z4. Right, StyI restriction map comparing 4q35 and 10q26 D4Z4 repeat units.
Contraction of the D4Z4 array is tightly linked to the disease in an unelucidated manner. About 95% of FSHD patients have only 1–10 tandem 3.3-kb repeat units in one of the two allelic 4q35 arrays (5,6). Almost all unaffected individuals have 11–100 of these repeat units on both 4q35 arrays. Analysis of the epigenetics of D4Z4 is facilitated by its unusually highly conserved primary structure, compared to many other types of tandem repeats, including satellite DNAs and NBL2. D4Z4 also offers the advantage of having at its proximal border a well-characterized 0.8-kb sequence, p13E-11 (Figure 1A), a hybridization probe for molecular diagnosis of FSHD (7–9).
FSHD is an enigmatic disease. There are subtelomeric D4Z4 arrays at 10q as well as 4q, and both are size polymorphic but only 4q arrays are linked to the disease. Short 10q arrays are phenotypically neutral despite 4q and 10q D4Z4 arrays having 99% homology (GenBank AF117653, AC126281, AL732375). Moreover, there is >95% homology in the 42 kb proximal to D4Z4 at 4q35 and 10q26 and also in the 15–25 kb at 4q35 and 10q26 that is distal to D4Z4 (Figure 1A) all the way to the telomeric TTAGGG repeats (5). FSHD pathogenesis is probably initiated by a short D4Z4 array interacting with a distant 4q35 sequence that is proximal to the array and not present on 10q26 (10). The identity of this sequence is unclear. From three reports of conventional microarray expression analyses on FSHD versus control muscle cell populations (11–13), no candidate FSHD gene present on 4q35, but not 10q26, has emerged. Moreover, 4q35 genes, including a gene-like sequence within the D4Z4 repeat unit, individually tested for involvement in FSHD have produced conflicting results (8,11,14–17).
Because DNA methylation and chromatin structure are often interrelated (18), insights into D4Z4 chromatin might be derived from the study of its methylation patterns. Shortened arrays in FSHD patients are significantly hypomethylated relative to D4Z4 arrays from controls (19), However, they are not as hypomethylated as the normal length D4Z4 arrays of ICF patients (20,21). ICF patients do not show any tendency to muscular disorders. Nonetheless, the FSHD-associated hypomethylation of D4Z4 might be a marker of a less condensed chromatin structure in disease-related short arrays (5).
Several groups reported D4Z4 hypomethylation in cancers, cancer-derived cell lines, and cells treated with demethylating agents or deficient in DNA methyltransferase (22–24). However, interpretation of those studies is complicated by their being PCR-based methylation analyses in which the investigators probably mostly detected highly abundant D4Z4-like sequences rather than specifically D4Z4. In contrast, blot hybridization under stringent conditions provides specificity for D4Z4 arrays (25) and offers the important advantage that a single blot can be serially hybridized with different probes for comparison of DNA methylation or chromatin DNase sensitivity at different sequences. Southern blotting also allows individual assessment of 4q versus 10q D4Z4 arrays by use of one of the rare sequence variations at a restriction site that is 4q- or 10q-specific (26,27).
In this study, we observed that D4Z4 was hypermethylated in some cancers and hypomethylated in others of the same type compared with a diverse set of somatic control tissues. Importantly, the first D4Z4 repeat unit behaved very differently from the rest of the D4Z4 array with respect to changes in DNA methylation. Moreover, the immediately proximal chromatin had a very different accessibility to DNaseI than the array, suggesting a different chromatin structure at the proximal border than in the body of the array. Our results are consistent with a model that we present for the biological importance of the proximal end of the D4Z4 array and a special chromatin structure within the array that could be sensitive to array length in an FSHD-determining manner.
MATERIALS AND METHODS
Tissue samples and DNA isolation
With IRB approval, primary tumor samples were obtained from patients who had not been treated with chemotherapy prior to surgery (28). DNA was purified from quick-frozen samples of tumor tissue or autopsy samples from non-cancer patients or trauma victims by phenol-extraction and ethanol-precipitation.
Cell culture
Lymphoblastoid cell lines (LCLs 149, 724, 112 and 952) were from normal controls (AG14953 and GM18478, Coriell Institute for Medical Research) or FSHD patients (GM17724, Coriell Institute, and WJ952, from Dr Silvere van der Maarel) and propagated in RPMI 1640 (Invitrogen) with 15% fetal bovine serum (FBS, Invitrogen). Normal-control skin fibroblast cell strains 996 and CF702 (GM01996, Coriell Institute, and Tulane Health Science Center, respectively) were derived from skin punch biopsies from non-FSHD patients and grown in MEM (Invitrogen) with 15% FBS and 2 mM l-glutamine. The FSHD myoblast cell strain (FM41j) was derived from a moderately affected, left deltoid biopsy of an FSHD patient. Duly signed patient consent forms were obtained that had been approved by the Institutional Review Boards of Tulane Health Science Center and the University of Mississippi Medical Center in Jackson. The fetal myoblast cell strain (H246) from Dr Stephen Hauschka was derived in 1980 from an anonymous, therapeutically aborted E-79 fetus under a signed patient consent form approved by the University of Washington's Human Subjects Committee. Myoblasts were propagated in F-10 medium (Invitrogen) containing 20% FBS, 1 μM dexamethasone and 10 ng/ml basic fibroblast growth factor (Invitrogen), 100 U/ml penicillin and 100 μg/ml streptomycin in flasks coated with 0.7% gelatin and, for H246, were supplemented with 1.3 mM CaCl2 at 37°C in 5% CO2. They were passaged when no more than about 80% confluent (about 2.5 × 106 cells per T75 flask). Cultures used for experiments were checked by immunocytochemistry to have >90% desmin-positive cells.
DNaseI treatment
Either cells (108 cells) that were permeabilized (LCLs) or nuclei that were gently isolated (LCLs, myoblasts and fibroblasts) were treated with 0–200 U/ml of DNaseI as previously described (29,30) using minor modifications given in the Supplementary Data. Then the DNA was purified by RNase treatment and organic solvent extraction and stored in 10 mM Tris–HCl, pH 7.4, 0.1 mM EDTA, pH 8.0.
Southern blot analysis
DNA was digested with restriction endonucleases and probes were prepared by PCR or directly by excision from recombinant plasmids as described previously (25) or in the Supplementary Data. Complete digestion with restriction endonucleases was verified for each sample with an internal plasmid or phage DNA control (28). When more than one probe was used with a blot, the lower-copy-number probes were used first and the blots were checked for complete stripping before hybridizing to the next probe.
For methylation analysis, tumors were scored for the approximate extent of methylation change relative to the somatic controls by phosphorimager quantitation and assessment of band patterns from X-rays by methods similar to those used previously (28). Scores for cancer DNA methylation (methylation change) relative to diverse somatic control tissues in the same blot were as follows: no appreciable change (0), progressively more hypermethylation (+1, +2 or +3), or more hypomethylation (−1, −2 or −3); −3 was strong hypomethylation, like that of sperm, and +3 was little or no digestion with the CpG methylation-sensitive restriction endonuclease (28). Some blots were hybridized first to the p13E-11 probe (7), followed by stripping and rehybridizing to the D4Z4 probe. Where indicated, the percent methylation was quantitated from the phosphorimager results of the relevant bands, with normalization, when necessary, for the percent overlap with the probe. Methods for statistical analyses of the methylation data are described in Supplementary Data.
For chromatin DNaseI sensitivity assays, the percent remaining DNA (R) in the StyI parent fragment was calculated from phosphorimager data as 100× the signal in the parent fragment after DNaseI treatment divided by that in parent fragment of the analogous sample incubated with buffer instead of DNaseI. Where indicated, R values for D4Z4, p13E-11, B2M, HMBS and IL-2 sequences were normalized to the expected results for a parent band of 1.5 kb, the size of the CST5 and GHRHR parent bands. This was done for each concentration of DNaseI in each blot by using the near-linear slope of the plot of the size of chromosome 1 satellite 2 (Sat2) parent bands (1.3, 1.7, 2.3 and 3.9 kb) versus R for each of those bands. From the slope, R values were extrapolated for hypothetical Sat2 parent bands of 1.5 kb (R1.5-kb Sat2) and the size of the restriction fragment of interest. For B2M, with its 1.2 kb StyI parent fragment, the normalization was RB2M × R1.5-kb Sat2/R1.2-kb Sat2. After quantitation, the blots were stripped and checked for complete removal of signal by phosphorimager analysis before hybridizing to the next probe.
Analysis of guanine quadruplex formation
D4Z4-derived oligonucleotides (5 μM) were heated at 100°C for 10 min, immediately placed on ice, incubated at 37°C for 48 h in 0.1 M KCl, 10 mM Tris-HCl, pH 7.4, and analyzed by circular dichroism spectropolarimetry (CD; Jasco-810, Easton, MD) at 37°C. A 1-mm optical path length, a scanning speed of 50 nm/min, and a response time of 4 s were used for the following oligonucleotides: PQS6, 5′-TGGGGGGGGGGGGTGGGGGGGGA; PQS6-mut, 5′-TGCGAGTGCGAGGTGAGCGTGTA-3′; and PQS5, 5′-AGGGGGAGAGGGGGGAGGGGGGAGGGGGGCGC-3′. The average of three scans was plotted after subtracting the buffer baseline. Non-denaturing polyacrylamide gel electrophoresis (PAGE) was done in a 20% gel at ambient temperature after incubation of PQS 6 or PQS6-mut, as above, subsequent to labeling with [γ-32P]ATP and polynucleotide kinase.
RESULTS
D4Z4 arrays can be hypo- or hypermethylated in cancers in association with NBL2, a heterologous tandem repeat
To examine the nature of epigenetic changes in D4Z4 repeat arrays in cancers relative to various somatic control tissues, DNA samples were digested with the following CpG methylation-sensitive enzymes: EagI (5′-CGGCCG-3′), SmaI (5′-CCCGGG-3′), MluI (5′-ACGCGT-3′), BstUI (5′-CGCG-3′), HpaII (5′-CCGG-3′), HpyCH4IV (5′-ACGT-3′), or BsaAI (5′-YACGTR-3′). Either single digests (EagI, SmaI and MluI) or double digests (BstUI, HpaII, HpyCH4IV and BsaAI plus the CpG methylation-insensitive enzymes KpnI or BglII) were used. After checking all samples for complete digestion, they were blot-hybridized to a 1-kb D4Z4 subfragment under stringent conditions specific for D4Z4 that did not give interference from cross-hybridizing sequences (25). All eight normal postnatal somatic tissues exhibited similar blot-hybridization patterns indicating considerable, but incomplete, methylation of D4Z4. In contrast, the 17 analyzed ovarian epithelial carcinomas and 44 Wilms tumors displayed large differences in D4Z4 methylation (Supplementary Figures 1–3; with summary data from representative samples in Figure 2). As shown in the summary of data from these 61 cancers (Table 1), most specimens from both of these diverse types of cancers were either significantly hypo- or hypermethylated at D4Z4 EagI and SmaI sites relative to somatic control tissues (P = 0.015 and 0.00003 for ovarian carcinomas and P = 0.0006 and 0.00003 for Wilms tumors, respectively). There was a strong association between alterations in D4Z4 methylation at EagI and SmaI sites in both ovarian carcinomas (P < 10−6) and Wilms tumors (P < 10−14). Cancer-linked methylation changes at the other examined CpG-containing restriction sites for a given tumor were also very similar to those at EagI and SmaI sites (Supplementary Figures 2A and 3A and data not shown).
Figure 2.
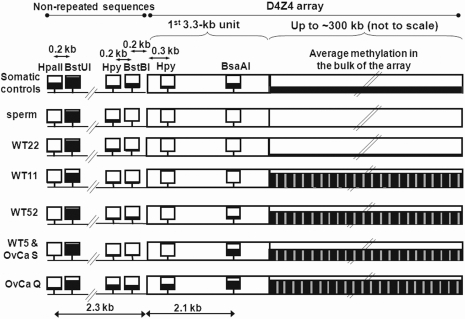
Cancer-linked hyper- or hypomethylation in the bulk of the D4Z4 array, at the very beginning of the array, and immediately proximal to the array in representative samples. The immediately proximal region includes the 0.8-kb p13E-11 sequence, which is 0.1 kb from the beginning of the D4Z4 array. The squares denote analyzed CpG methylation-sensitive restriction sites. The most proximal HpyCH4IV and BsaAI sites in the first repeat unit of the array are shown followed by a long rectangle representing the foreshortened remainder of the array. The height of the black fill-in is approximately proportional to the average extent of methylation. Methylation in the bulk of the D4Z4 array is diagrammed for HpyCH4IV sites because the small number (six) of HpyCH4IV sites per 3.3-kb repeat unit facilitated quantitation. Other CpG-containing restriction sites were more heavily methylated in somatic controls but gave hypo- or hypermethylation in cancers similar to that seen at HpyCH4IV sites. The gray vertical bars in the D4Z4 array in cancer samples represent the subregion of the D4Z4 repeats units that is partially resistant to hypermethylation and located about 1.4 kb distal to the KpnI site (Figure 3A). See Supplementary Figures 1–3 for most of the data used in this figure.
Table 1.
Methylation change scores for ovarian epithelial tumors (carcinomas, LMP tumors, and cystadenomas) and Wilms tumors at EagI or SmaI sites in D4Z4 or HhaI sites in NBL2a
| Sample | D4Z4 EagI | D4Z4 SmaI | NBL2 HhaI | Sample | D4Z4 EagI | NBL2 HhaI | Sample | D4Z4 EagI | D4Z4 SmaI | NBL2 HhaI | Sample | D4Z4 EagI | D4Z4 SmaI | NBL2 HhaI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controlsb | 0 | 0 | 0 | LMP 8 | +1 | nd | WT1 | 0 | 0 | +1 | WT29 | −1 | 0 | +2 |
| OvCa A | 0 | 0 | +1 | LMP 9 | −1 | 0 | WT4 | −2 | −2 | +1 | WT30 | 0 | 0 | +2 |
| OvCa B | 0 | 0 | +1 | LMP 10 | 0 | +1 | WT5 | +3 | +2 | +1 | WT33 | −1 | Y | +2 |
| OvCa C | 0 | 0 | 0 | LMP 11 | 0 | 0 | WT6 | −2 | −3 | +1 | WT34 | Y | 0 | +3 |
| OvCa D | −3 | −3 | −2 | LMP 12 | 0 | +2 | WT7 | −1 | −1 | +1 | WT37 | 0 | Y | +2 |
| OvCa E | 0 | 0 | −1 | LMP13 | −1 | nd | WT8 | −1 | −1 | +3 | WT38 | −1 | −1 | +2 |
| OvCa G | −2 | −2 | −2 | LMP 14 | 0 | 0 | WT9 | 0 | 0 | +1 | WT39 | +2 | +1 | +1 |
| OvCa H | −1 | −1 | +1 | LMP 15 | 0 | +2 | WT11 | +2 | +3 | +3 | WT40 | −1 | −1 | +1 |
| OvCa I | −1 | −2 | +2 | LMP 16 | 0 | +2 | WT13 | −1 | −1 | +1 | WT41 | −1 | −2 | 0 |
| OvCa J | −2 | −1 | +1 | LMP 19 | 0 | +1 | WT15 | +1 | +1 | +3 | WT42 | 0 | 0 | +3 |
| OvCa K | 0 | +1 | +1 | Cystad. 1 | −1 | 0 | WT17 | 0 | 0 | +1 | WT44 | Y | 0 | +1 |
| OvCa L | +1 | +1 | +1 | Cystad. 2 | 0 | 0 | WT18 | −1 | −1 | +2 | WT45 | −1 | −2 | nd |
| OvCa M | 0 | Y | 0 | Cystad. 3 | −1 | +1 | WT19 | 0 | +1 | +1 | WT46 | −1 | 0 | +2 |
| OvCa N | +1 | +1 | +2 | Cystad. 4 | +1 | +2 | WT20 | +1 | +1 | +3 | WT47 | +1 | +1 | 0 |
| OvCa O | +1 | Y | +2 | Cystad. 5 | −1 | 0 | WT21 | +1 | 0 | +3 | WT48 | −2 | −2 | 0 |
| OvCa Q | +3 | +2 | +3 | Cystad. 6 | −1 | 0 | WT22 | −2 | −2 | +1 | WT49 | −1 | −1 | nd |
| OvCa R | 0 | −1 | +1 | Cystad. 7 | −1 | 0 | WT23 | −1 | Y | +1 | WT50 | 0 | 0 | +2 |
| OvCa S | +2 | +3 | +2 | Cystad. 10 | −1 | 0 | WT24 | −2 | −2 | +1 | WT51 | Y | +1 | +1 |
| LMP 1 | −1 | 0 | nd | Cystad. 11 | +1 | nd | WT25 | −1 | −1 | 0 | WT52 | +3 | +2 | +3 |
| LMP 2 | +1 | +2 | nd | Cystad. 17 | 0 | nd | WT26 | 0 | 0 | 0 | WT54 | +2 | +2 | +3 |
| LMP 3 | −1 | nd | nd | Cystad. 20 | −1 | nd | WT27 | +2 | 0 | +1 | WT57 | +3 | +3 | +2 |
| LMP 7 | 0 | +1 | nd | Cystad. 21 | 0 | nd | WT28 | −1 | −1 | +2 | WT67 | −1 | −1 | +1 |
aOvarian carcinomas (OvCa), low malignant potential (LMP) tumors, and cystadenomas (cystad.) as well as Wilms tumors (WT) were scored for methylation change by southern blotting relative to eight diverse somatic control tissues (controls). Scores: −1, −2, and −3, increasing hypomethylation; +1, +2, and +3, increasing hypermethylation; 0, little or no change relative to controls; Y, change in the methylation pattern that is difficult to classify as hypo- or hypermethylation; nd, not determined. Supplementary Figure 1 shows representative blots, which illustrate the hyper- or hypomethylation of most cancers compared to the similar patterns of methylation in the control somatic tissues.
bFour diverse somatic control tissues were present in each cancer DNA blot.
We also found that methylation changes in the subtelomeric 4q and 10q D4Z4 arrays were significantly associated with those in the NBL2 repeats arrays, which are located mostly in the short arms of the acrocentric chromosomes. For this analysis, tumor-linked methylation changes at EagI sites in the D4Z4 arrays were compared with those previously described for HhaI (5′-GCGC-3′) sites in NBL2 (1) using southern-blot-derived methylation scores, that had been validated by hairpin genomic sequencing (1). These sites are present at similar densities (5 and 3 per 3.3- and 1.4-kb repeat unit, respectively) and were analyzed in overlapping sets of tumors (Table 1). This analysis included borderline carcinomas (low malignant potential tumors) and benign tumors (cystadenomas) of epithelial origin (Supplementary Data). Alterations in D4Z4 methylation in the group of all examined tumors were significantly associated with those of NBL2 (P < 0.0001, Kendall's tau correlation). There is probably a common recognition of both dissimilar types of repeat arrays by poorly understood pathways for demethylation and de novo methylation that function during tumorigenesis, just as both tandem repeats are dependent on DNMT3B for normal levels of methylation (1,20).
Spreading of DNA methylation in D4Z4
Several of the cancers had extremely high levels of methylation in consecutive 3.3-kb repeat units of D4Z4 arrays (Supplementary Figures 1–3). For example, the amount of D4Z4 uncleaved by BstUI, HpaII or HpyCH4IV in double digests with the CpG methylation-insensitive KpnI was 52–88% for Wilms tumor 11 (WT11) as compared with 7–14% for control brain DNA, even though BstUI and HpaII sites are respectively about 6 and 12 times as frequent as HpyCH4IV sites in D4Z4 (Supplementary Figure 2A). In single digests of WT11 or WT52 DNA with BstUI or HpyCH4IV, most of the D4Z4 signal was in fragments of more than 10 kb (Figure 2 and data not shown). This indicates complete methylation at these sites for at least three contiguous repeat units. Because BstUI has 37 recognition sites per D4Z4 repeat unit, the long fragments had more than 100 contiguous methylated sites. Other, much smaller, hybridizing fragments were also present indicative of close unmethylated sites in other portions of D4Z4. Not only these two cancer DNA digests but also many other SmaI digests of control and cancer DNAs displayed significantly non-random distributions of D4Z4 fragment sizes consistent with long regions of these arrays being highly methylated and other regions having little methylation in the same sample (Supplementary Figure 1 and Supplementary Data). These observations and the associations between methylation at different restriction sites in D4Z4 suggest that methylation spreads along the arrays during normal development and during tumorigenesis with long regions of complete methylation and interspersed regions of considerably lower methylation.
A methylation-resistant subregion within each 3.3-kb repeat that contains potential G-quadruplexes
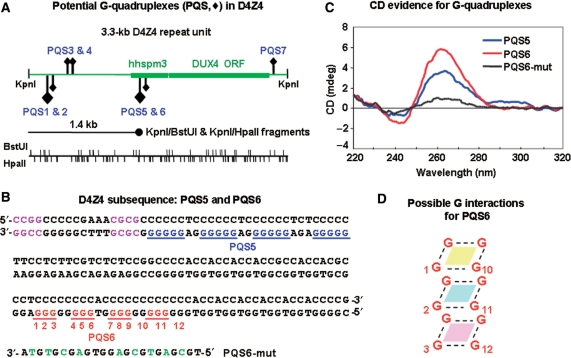
One subregion within the 3.3-kb D4Z4 repeat unit displayed resistance to hypermethylation. An unexpectedly strong D4Z4-hybridizing band at the position of 1.4-kb fragments was seen in BstUI/KpnI and HpaII/KpnI digests from the five examined cancers that had hypermethylation of D4Z4 (WT11, WT52, WT5 and ovarian carcinomas Q and S, Figure 2 and Supplementary Figure 2A and B). This prominent band cannot be explained by trivial reasons, e.g. the distribution of BstUI and HpaII sites in D4Z4 (37 and 71 per 3.3-kb repeat unit, respectively; Figure 3A). It was also observed in BstUI/KpnI and HpaII/KpnI digests of somatic control DNAs, although at a lower intensity, but not in cancer or control samples singly digested with BstUI or HpaII. Therefore, this 1.4-kb fragment should be derived from cleavage at the single KpnI site in each D4Z4 repeat unit and a subregion of the repeat unit located about 1.4 kb distal (Figure 3A). This subregion of the D4Z4 repeat unit is apparently partly resistant to hypermethylation in cancer.
Figure 3.
Potential G-quadruplex sequences (PQS) in D4Z4. (A) Maps of the 3.3-kb D4Z4 repeat unit showing positions of PQS, BstUI and HpaII sites, and the alignment of the unexpectedly strong 1.4-kb band that was observed in BstUI/KpnI and HpaII/KpnI digests (Supplementary Figure 2). Diamonds, PQS; large for G5 and small for G3 runs; upward for those on the forward strand and downward for those on the complementary strand. hhspm3, a sequence that can function as a promoter; DUX4, an open reading frame without a polyadenylation signal (15,16). (B) The D4Z4 subsequence (AF117653) that contains PQS5 and PQS6. Potential unimolecular G-quadruplex structures (blue or red) are shown but there are variations possible in the exact placement of the four runs of G in this subregion that could be involved in unimolecular quadruplexes. In addition, non-adjacent compatible runs of G might participate in quadruplex formation like that seen in bi- and tetramolecular G-quadruplexes. Purple, the closest BstUI and HpaII sites to PQS5; green, mutated residues in PQS6-mut. (C) CD analysis of PQS5, PQS6 and PQS6-mut as single-stranded 23- to 32-nt oligonucleotides. (D) Illustration of one possible orientation for G residues to stack and form a hairpin-type intramolecular G-quadruplex. The numbering corresponds to PQS6 in Panel B, but various other configurations are possible that preserve the planar stacking of four G residues connected by Hoogsteen pairing (dotted lines).
The primary structure of D4Z4 in this subregion is very unusual (Figure 3B). There is an uninterrupted run of 37 pyrimidine residues on the forward strand adjacent to HpaII and BstUI sites located 1395 and 1409 nt, respectively, after the KpnI site of the 3.3-kb D4Z4 repeat unit. Importantly, (G5N2-3)4, a potential guanine quadruplex sequence (PQS5, Figure 3A and B), is complementary to 27 nt of the Y37 sequence. Nearby is a (G3N2)4 sequence (PQS6) that also fits the PQS consensus sequence of four equally long runs of G in the following context: G3-5N1-7G3-5N1-7G3-5. PQS5, which is present 1412 nt after the KpnI site, scores especially high in a PQS prediction program for intramolecular G-quadruplexes (31). Each 3.3-kb D4Z4 repeat unit contains seven PQS that match the above consensus sequence (Figure 3A; GenBank AF117653 and AC126281).
By CD spectroscopy, a single-stranded PQS5 or PQS6 oligonucleotide had a strong positive peak with a maximum at 263 nm and a negative peak centered around 243 nm (Figure 3C). A positive CD peak at about 260 nm and a negative peak at about 240 nm are diagnostic for G-quadruplexes, usually of the parallel type (32) (Figure 3D). PQS6-mut, an oligonucleotide derived from PQS6 but with nine substitutions of C, A or T for G residues, gave a non-specific CD spectrum (Figure 3B and C). Radiolabeled PQS6 and PQS6-mut oligonucleotides were analyzed by non-denaturing PAGE. In addition to a band that migrated as expected for the unstructured oligonucleotide, about half of the signal from PQS6 was in low-mobility bands indicating intermolecular G-quadruplexes (33) (data not shown). For PQS6-mut, only one band with the expected mobility was seen.
Resistance to DNA hypermethylation in the beginning of the D4Z4 array and immediately proximal to it
We were able to compare methylation in the very beginning of the D4Z4 array, in the bulk of this array, and immediately proximal to it by hybridizing blots to a p13E-11 probe, which is a D4Z4-adjacent sequence (Figure 1), and then to the D4Z4 probe. This was done for BstUI/KpnI, HpaII/KpnI, HpyCH4IV/KpnI and BsaAI/BglII digests of cancer and control DNAs (Supplementary Figures 2 and 3). While D4Z4 is extremely rich in CpG's (9.9% CpG), there are only six CpG sites (0.6% CpG) in the 930-bp region immediately proximal to D4Z4. This proximal region encompasses the 0.8-kb p13E-11 sequence (Figure 1). Two of the six proximal CpG's are in HpyCH4IV and BstBI recognition sites and were analyzed by southern blotting (Supplementary Figure 2). Surprisingly, in cancers with overall D4Z4 hypermethylation relative to somatic controls, there was resistance to hypermethylation, and even decreased methylation, in the first approximately 2 kb of the array and immediately proximal to it (Figure 2). A BstUI site located 2.1 kb proximal to the array was the exception (Figure 2). It was tenaciously methylated in all cell populations, including sperm and WT22, which were strongly hypomethylated at D4Z4 compared to somatic controls (Figure 2 and Supplementary Figure 2C and D). Conventional hairpin structures with T-A or T-G base-pairing might form from the T2-5A4-5 sequence and the (TG)8 sequence 10 bp 5′ and 25 bp 3′ to this site, respectively (Supplementary Figure 2E). This might help recruit DNA methyltransferases (34) to keep the site methylated.
Chromatin DNaseI sensitivity of D4Z4 and immediately proximal sequences
Given the unusual DNA methylation changes that we found in and adjacent to this array in cancer and the linkage of D4Z4 arrays to FSHD, we studied the conformation of chromatin in these regions by DNaseI sensitivity assays on cultured cells. Chromatin standards (Supplementary Table 1) and the restriction enzyme for digestion (StyI) of isolated DNA from DNaseI-treated nuclei or permeabilized cells were chosen to allow comparison of test sequences and euchromatin and heterochromatin standards in a single southern blot. The test sequences were 4q35 D4Z4, which is linked to FSHD; 10q26 D4Z4, which is phenotypically neutral; and p13E-11 the sequence immediately proximal to D4Z4 (Figure 1). Because FSHD is mostly a skeletal muscle disease, it was important to test cells from the muscle lineage, namely, FSHD and control myoblast cell strains. These non-transformed cell cultures are generally difficult to obtain, generate and propagate and have limited growth potential before irreversible growth arrest. Moreover, large numbers of cells (108) are required for one chromatin DNaseI sensitivity assay monitored by southern blotting. Therefore, we first tested lymphoblastoid cell lines (LCLs) and control fibroblast cell strains. Lysolecithin efficiently permeabilized LCLs, but not fibroblasts, to DNaseI (Supplementary Figure 4A). We were able to study the chromatin DNaseI sensitivity of fibroblasts and myoblasts by incubating gently isolated nuclei with DNaseI and adjusting DNaseI digestion conditions. A comparison of permeabilized LCL cells and LCL nuclei gave similar results for relative DNaseI sensitivity of the analyzed DNA sequences (Supplementary Table 2).
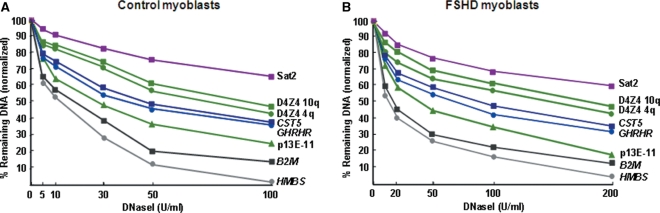
The relative DNaseI sensitivity of standard and test sequences in FSHD and control myoblasts, LCLs and fibroblasts was as follows: constitutively transcribed gene standards (B2M and HMBS) > D4Z4-proximal p13E-11 marker > untranscribed gene standards (CST5, IL-2 and GHRHR) > 4q and 10q D4Z4 > Sat2 juxtacentromeric heterochromatin in chromosome 1 (Figures 4 and 5 and Supplementary Figures 4B and 5). B2M and HMBS are highly transcribed in all studied tissues while CST5, IL-2 and GHRHR are selectively transcribed in only a small number of cell types, not including those in this study (http://www.genecards.org/ and http://expression.gnf.org/cgi-bin/index.cgi). The differences in DNaseI digestion for untranscribed gene standards versus transcribed gene standards and constitutive heterochromatin versus untranscribed gene standards were significant (P < 10−4). It was very surprising that p13E-11, which is only 0.1 kb from D4Z4 (Figure 1) and has no features of a gene (http://exon.gatech.edu/GeneMark/, http://www.softberry.com/berry.phtml, and http://genes.mit.edu/GENSCAN.html), was more sensitive to DNaseI than the untranscribed gene standards in all tested myoblast, fibroblast and lymphoblastoid cell or nuclei populations (P < 10−4).
Figure 4.
The D4Z4-adjacent p13E-11 marker sequence is more accessible to DNaseI than untranscribed gene standards and much more accessible than D4Z4 in myoblast nuclei. The size-normalized (Supplementary Table 2) percentage of DNA remaining in the parent StyI restriction fragment is shown for each examined DNA sequence in DNaseI-treated nuclei from control fetal myoblasts (A) and FSHD myoblasts (B).
Figure 5.
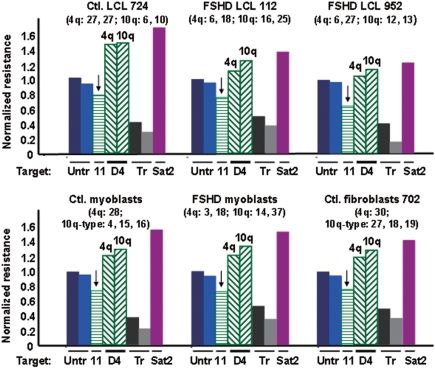
D4Z4 chromatin is quite resistant and the adjacent p13E-11 chromatin is very sensitive to DNaseI in all studied FSHD and control cell populations. Nuclei (FSHD myoblasts, control fetal myoblasts, one control fibroblast cell strain) or permeabilized cells (LCLs) were treated with four or five different concentrations of DNaseI. The results are given for only one concentration of DNaseI (20, 50 or 100 U/ml) but these are representative of the results at each concentration (e.g. Figure 4) and all differences in sensitivity between different types of DNA sequences were highly significant (P < 10−4, see text). The analyzed DNA sequences for each cell type are as follows, from left to right in each bar graph: two untranscribed gene standards (Untr, CST5 and GHRHR), p13E-11 (11; bar denoted by a vertical arrow), D4Z4 (D4, 4q-type and 10q-type arrays), constitutively transcribed gene standards (Tr, B2M and HMBS), and the constitutive heterochromatin standard (Sat2). After size normalization, the percentage of DNA remaining in the parent band for each sequence was divided by that for the average of the untranscribed standards. In parentheses is the number of copies of the D4Z4 repeat unit for the allelic 4q-type arrays (no BlnI site and a single StyI site per 3.3 kb) and the allelic 10q-type arrays (one BlnI site and two StyI sites per 3.3 kb) (51). The control myoblasts and the fibroblasts had one of the 4q D4Z4 arrays replaced by a canonical 10q-type array, as is seen in some individuals, but this has no effect on phenotype (51).
In every studied cell type, both the 3.3- and 2.8-kb D4Z4 parent bands from 4q35 and 10q26, respectively (Figure 1B), were much more resistant to DNaseI than the 1.9-kb region containing the p13E-11 sequence (P < 10−5; Figures 4 and 5). In addition, the D4Z4 arrays were more resistant to DNaseI than untranscribed gene standards and less than the constitutive heterochromatin standard in every FSHD and control cell population (P < 10−4 for both comparisons). Similar results were obtained with or without normalization for the somewhat different sizes of the parent fragments (1.2–3.3 kb; Supplementary Table 2). No DNaseI hypersensitive sites (discrete lower-molecular-weight bands) were seen in the promoter regions or other tested sequences. However, this might be due to optimizing conditions for measuring general DNaseI sensitivity rather than hypersensitive sites.
Among the two studied pairs of similar standards, namely, the untranscribed and the constitutively transcribed gene standards, smaller, but also significant differences, were seen (P < 10−3, Figures 4 and 5). CST5 was slightly more resistant to DNaseI than GHRHR in all samples although both genes should be untranscribed in the examined cells. Similarly, the ubiquitously transcribed B2M was always appreciably more resistant to DNaseI than the other constitutively expressed standard, HMBS. This might reflect gene-specific differences in chromatin structure even when comparing genes with the same expression status. Given the reproducibility of these findings among different cell populations, it is noteworthy that 10q D4Z4 chromatin was generally a little more resistant to DNaseI than 4q D4Z4 chromatin (P = 10−3, Figures 4 and 5).
The length of the D4Z4 arrays needs to be considered in evaluation of their chromatin DNaseI sensitivity because of the frequent correlations between long tandem repeat arrays and heterochromatinization (35,36) and the effect of the size of 4q D4Z4 arrays on disease status. We looked for correlations of DNaseI chromatin resistance and D4Z4 array size as determined by pulsed-field gel electrophoresis (25). Interpretation of the results is complicated by large D4Z4 arrays contributing proportionally more signal than short arrays and the highly polymorphic sizes of these arrays. The most informative of our studied cell populations to look for an effect of D4Z4 array length on the DNaseI resistance was the control LCL 724. It has two short 10q D4Z4 arrays (6- and 10-unit) and two long allelic 4q arrays (both 27 repeat units). The 2.8-kb band from 10q D4Z4 was well separated from the 3.3-kb band from 4q D4Z4. Because 10q and 4q D4Z4 arrays are almost identical in sequence (26), if the chromatin structure of 10q D4Z4 is dependent on the length of the array, this is probably also true for 4q D4Z4. Contrary to a simple correlation between array size and DNaseI resistance, the 10q and 4q D4Z4 arrays of LCL 724 had virtually the same DNaseI resistance (Figure 5 and Supplementary Table 2).
DISCUSSION
A major unanswered question about the molecular genetics of FSHD is how a tandem array of more than ten 3.3-kb D4Z4 units at both 4q35 allelic positions is almost always protective against the disease while one 4q35 D4Z4 array with ten or fewer repeat units usually indicates the presence of the disease by the third decade (37). The in situ ‘ruler’ or biochemical device for measuring D4Z4 array size to establish disease status is likely to involve topological differences between a short array and one containing more than ten units. However, the nature of D4Z4 chromatin is little understood.
We previously reported on histone H4 acetylation of D4Z4 and proximal sequences (8). Histone acetylation is often inversely related to local chromatin compaction (38), although not always (39). D4Z4 chromatin in several human chromosome 4-containing somatic cell hybrids has low levels of H4 acetylation, but not as low as those of a heterochromatin standard. Those PCR-dependent experiments were confined to somatic cell hybrids because of the lack of primers specific for D4Z4 (8). Histone acetylation at p13E-11, the immediately proximal sequence to D4Z4, was at levels similar to those of untranscribed genes rather than to constitutive heterochromatin.
Here we analyzed the general DNaseI sensitivity of D4Z4 and the immediately proximal region under blot-hybridization conditions specific for these sequences (7,25). The DNaseI sensitivity of both 4q and 10q D4Z4 chromatin was intermediate to that of constitutive heterochromatin and inactive genes in FSHD and control myoblasts, fibroblasts and lymphoblastoid cell lines. A caveat is that in comparing short, disease-associated D4Z4 arrays with normal, long D4Z4 arrays, the contribution of the short array to the blot-hybridization signal is obscured by the proportionally greater contribution of the long arrays. However, in a control LCL (724, Figure 5), two short allelic D4Z4 arrays at 10q (6 and 10 repeat units) were not more sensitive to DNaseI than two long 4q D4Z4 arrays (both 27 repeat units) or than untranscribed gene standards. These results suggest a high degree of condensation of D4Z4 chromatin even in short, FSHD-sized repeat arrays, with the caveat that DNaseI sensitivity measures only certain aspects of higher-order chromatin structure.
It has been proposed that heterochromatinization spreads proximally from a normal, long D4Z4 array to a 4q35 gene responsible for initiating the FSHD dysregulation cascade (14,40,41). Contrary to that hypothesis or the idea that long D4Z4 arrays might normally act as insulators (5,41), the region immediately proximal to the D4Z4 array (p13E-11) was more sensitive to DNaseI than the bulk D4Z4 array in all tested cell cultures. Unexpectedly, this D4Z4-adjacent region consistently displayed a DNaseI sensitivity that was even greater than that of untranscribed gene standards. We found no predicted promoter, enhancer, or exon in the 4-kb region proximal to the 4q or 10q D4Z4 arrays that could explain its unexpectedly high sensitivity to DNaseI even in control fibroblasts containing four long D4Z4 arrays (18–30 repeat units per array). The nuclease sensitivity in this region might reflect a local strain in the chromatin conformation.
We could monitor the DNaseI sensitivity of the D4Z4-proximal region and the average sensitivity of the D4Z4 array but not specifically that of the first D4Z4 repeat unit. However, by studying cancer-linked DNA methylation changes, we found evidence for an atypical chromatin structure at the beginning of the D4Z4 array and immediately proximal to it. The apparent spreading of methylation along D4Z4 that we observed seems to have limits to its processivity and to be prone to stop at certain subregions. Specifically, there was a remarkable resistance to cancer-linked hypermethylation at the very beginning of the repeat array and at several CpG sites proximal to the array in cancers that displayed strong hypermethylation in the bulk of the array (Figure 2). The sequence of D4Z4 repeat units is highly conserved throughout the array. Therefore, the differential susceptibility to tumor-linked hypermethylation at the start of the array versus in the bulk of the array is best explained by a special chromatin structure at the junction of D4Z4 and the proximal non-repeated sequence.
The methylation and DNaseI sensitivity findings suggest that there is a boundary element (42) (Figure 6) around the junction of the D4Z4 array and the immediately proximal sequence. Naturally occurring deletions of this D4Z4-proximal region indicate that the exact sequence immediately proximal to D4Z4 cannot be contributing to the phenotype (43). However, there might be a critical interfacing of D4Z4 (first 200 bp, 78% G+C) and a DNA sequence with a more typical base composition for human DNA (immediately proximal 200 bp, 43.5%).
Figure 6.
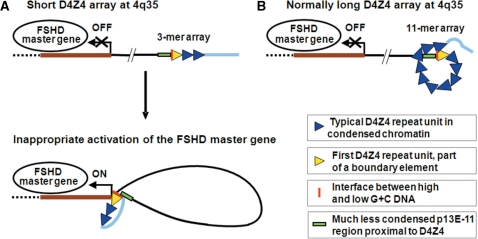
Hypothesized intrachromosomal communication by chromatin looping. (A) A disease-associated long-distance loop is hypothesized to form when the D4Z4 repeat array is contracted to less than 11 repeat units, leading to inappropriate expression in cis of an unelucidated 4q35 FSHD master gene (thick brown line). These abnormal looping interactions might upregulate transcription of the FSHD master gene in cis by direct chromatin-to-chromatin contact and/or alteration of the chromatin protein composition of the promoter of the FSHD master gene. The hypothesized long-distance looping may include interactions with the sequences distal to D4Z4, which are shown as a thick light blue line, ∼15–25 kb ending at the telomeric TTAGGG repeats (5,52). (B) Intra-array looping for arrays with more than 10 repeat units would sequester the proximal end of D4Z4 so that it would not be available for pathogenic long-distance looping. Size-dependent, topological constraints on intra-array looping could explain why the disease severity is often lower when the 4q35 array is in the 8–10 unit range relative to the 1–4 unit range. Looping interactions of D4Z4 sequences might involve G-quadruplexes.
The second region in which we observed resistance to cancer-linked hypermethylation is a subregion of the 3.3-kb D4Z4 repeat unit, about 1.4 kb distal to its single KpnI site. It has runs of G residues that are predicted to form stable G-quadruplexes (Figure 3B). We demonstrated that two of these sequences, PQS5 and PQS6, readily form G-quadruplexes when tested as single-stranded oligonucleotides. Inhomogeneous spreading of DNA methylation along the body of the D4Z4 arrays might be due partly to such unusual DNA secondary structures within the D4Z4 repeat units hindering the spread of DNA methylation in vivo directly or indirectly, by effects on chromatin structure.
Various types of indirect evidence suggest that genomic G-quadruplexes, non-B DNA structures, exist in vivo and mediate transcription control, genomic stability and chromatin packing at telomeres (44,45). In certain functional classes of genes, including those associated with muscle development or contraction, PQS overrepresentation is especially high (46), which is further evidence for their biological relevance. G-quadruplexes can involve Hoogsteen base-pair interactions between runs of G residues within one local sequence on a DNA strand (Figure 3D) or between four equal-length runs of G residues on two or four different strands (44,47). They might also form between two runs of G residues in non-neighboring locations within the same region of a given strand and thereby help establish higher-order packing of chromatin (45).
We propose that G-quadruplex formation in the 3.3-kb D4Z4 repeat unit (Figure 3A) helps organize D4Z4 chromatin to give special intra-array looping in normal, long arrays (Figure 6). We further hypothesize that, within short arrays, abrogation of this intra-array looping allows long-distance looping between the beginning of the array and an FSHD master gene at 4q35 (Figure 6). The identity of this 4q35 gene is still uncertain despite many conventional analyses (8,14–16), including microarray expression studies (11–13). The postulated alternative looping to regulate expression of the FSHD master gene could explain why only short D4Z4 arrays at 4q35 are linked to FSHD by permitting long-distance looping that initiates inappropriate gene expression accounting for the dominant inheritance of this disease.
The hypothesized boundary element at the first D4Z4 repeat unit in normal and FSHD cells (Figure 6) makes it easier to envision pathogenic long-distance chromatin interactions between a short D4Z4 array at 4q35 and another FSHD-related DNA sequence in cis. Given that FSHD is mostly a muscle-specific disease, it is intriguing that the myogenesis-specific homodimeric MyoD protein can specifically recognize G-quadruplexes with a higher affinity than specific duplex sequences (48–50). That this transcription factor preferentially binds to G-quadruplexes between runs of G on different oligonucleotide strands makes the above model a very attractive hypothesis for understanding part of the perplexing near-threshold effect of D4Z4 array size on disease status such that a 4q35 D4Z4 array of <11 repeat units (<36 kb) is diagnostic for FSHD.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the Brain and Tissue Bank for Developmental Disorders at the University of Maryland for generously replacing tissues lost during the Hurricane Katrina flood in New Orleans, LA. Supported in part by NIH grant R01 NS048859 and a grant from the Louisiana Cancer Research Consortium. Funding to pay the Open Access publication charges for this article was provided by the FSH Society.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nishiyama R, Qi L, Lacey M, Ehrlich M. Both hypomethylation and hypermethylation in a 0.2-kb region of a DNA repeat in cancer. Molec. Cancer Res. 2005;3:617–626. doi: 10.1158/1541-7786.MCR-05-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin. Immunol. 2003;109:17–28. doi: 10.1016/s1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 3.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 4.van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 5.van der Maarel SM, Frants RR. The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2005;76:375–386. doi: 10.1086/428361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi M, Ricci E, Colantoni L, Galluzzi G, Frusciante R, Tonali PA, Felicetti L. The facioscapulohumeral muscular dystrophy region on 4qter and the homologous locus on 10qter evolved independently under different evolutionary pressure. BMC Med. Genet. 2007;8:8. doi: 10.1186/1471-2350-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 8.Jiang G, Yang F, van Overveld PG, Vedanarayanan V, van der Maarel S, Ehrlich M. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- 9.Deak KL, Lemmers RJ, Stajich JM, Klooster R, Tawil R, Frants RR, Speer MC, van der Maarel SM, Gilbert JR. Genotype-phenotype study in an FSHD family with a proximal deletion encompassing p13E-11 and D4Z4. Neurology. 2007;68:578–582. doi: 10.1212/01.wnl.0000254991.21818.f3. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich M. In: FSHD Facioscapulohumeral Muscular Dystrophy: Molecular Cell Biology & Clinical Medicine. Cooper DN, Upadhyaya M, editors. New York, NY: BIOS Scientific Pub.; 2004. pp. 253–276. [Google Scholar]
- 11.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 12.Celegato B, Capitanio D, Pescatori M, Romualdi C, Pacchioni B, Cagnin S, Vigano A, Colantoni L, Begum S, Ricci E, et al. Parallel protein and transcript profiles of FSHD patient muscles correlate to the D4Z4 arrangement and reveal a common impairment of slow to fast fibre differentiation and a general deregulation of MyoD-dependent genes. Proteomics. 2006;6:5303–5321. doi: 10.1002/pmic.200600056. [DOI] [PubMed] [Google Scholar]
- 13.Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- 14.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 15.Alexiadis V, Ballestas ME, Sanchez C, Winokur S, Vedanarayanan V, Warren M, Ehrlich M. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochem. Biophys. Acta. 2007;1769:29–40. doi: 10.1016/j.bbaexp.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Matteotti C, Arias C, Corona ED, Nunez NG, Leo O, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 19.van Overveld PG, Enthoven L, Ricci E, Rossi M, Felicetti L, Jeanpierre M, Winokur ST, Frants RR, Padberg GW, van der Maarel SM. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann. Neurol. 2005;58:569–576. doi: 10.1002/ana.20625. [DOI] [PubMed] [Google Scholar]
- 20.Kondo T, Bobek MP, Kuick R, Lamb B, Zhu X, Narayan A, Bourc'his D, Viegas-Pequignot E, Ehrlich M, Hanash SM. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet. 2000;9:597–604. doi: 10.1093/hmg/9.4.597. [DOI] [PubMed] [Google Scholar]
- 21.Tsien F, Sun B, Hopkins NE, Vedanarayanan V, Figlewicz D, Winokur S, Ehrlich M. Hypermethylation of the FSHD syndrome-linked subtelomeric repeat in normal and FSHD cells but not in ICF syndrome cells. Molec. Gen. Metab. 2001;74:322–331. doi: 10.1006/mgme.2001.3219. [DOI] [PubMed] [Google Scholar]
- 22.Cadieux B, Ching TT, Vandenberg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 23.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 24.Cheng JC, Weisenberger DJ, Gonzales FA, Liang G, Xu GL, Hu YG, Marquez VE, Jones PA. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol. Cell. Biol. 2004;24:1270–1278. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehrlich M, Jackson K, Tsumagari K, Camano P, Lemmers RJFL. Improved hybridization analysis of D4Z4 repeat arrays linked to FSHD. Chromosoma. 2007;116:107–116. doi: 10.1007/s00412-006-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cacurri S, Piazzo N, Deidda G, Vigneti E, Galluzzi G, Colantoni L, Merico B, Ricci E, Felicetti L. Sequence homology between 4qter and 10qter loci facilitates the instability of subtelomeric KpnI repeat units implicated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 1998;63:181–190. doi: 10.1086/301906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deidda G, Cacurri S, Grisanti P, Vigneti E, Piazzo N, Felicetti L. Physical mapping evidence for a duplicated region on chromosome 10qter showing high homology with the facioscapulohumeral muscular dystrophy locus on chromosome 4qter. Eur. J. Hum. Genet. 1995;3:155–167. doi: 10.1159/000472291. [DOI] [PubMed] [Google Scholar]
- 28.Nishiyama R, Qi L, Tsumagari K, Dubeau L, Weissbecker K, Champagne M, Sikka S, Nagai H, Ehrlich M. A DNA repeat, NBL2, is hypermethylated in some cancers but hypomethylated in others. Cancer Biol. Ther. 2005;4:440–448. doi: 10.4161/cbt.4.4.1622. [DOI] [PubMed] [Google Scholar]
- 29.Lu Q, Richardson B. Methods for analyzing the role of DNA methylation and chromatin structure in regulating T lymphocyte gene expression. Biol. Proced. Online. 2004;6:189–203. doi: 10.1251/bpo89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward SB, Hernandez-Hoyos G, Chen F, Waterman M, Reeves R, Rothenberg EV. Chromatin remodeling of the interleukin-2 gene: distinct alterations in the proximal versus distal enhancer regions. Nucleic Acids Res. 1998;26:2923–2934. doi: 10.1093/nar/26.12.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kho MR, Baker DJ, Laayoun A, Smith SS. Stalling of human DNA (cytosine-5) methyltransferase at single-strand conformers from a site of dynamic mutation. J. Mol. Biol. 1998;275:67–79. doi: 10.1006/jmbi.1997.1430. [DOI] [PubMed] [Google Scholar]
- 35.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 36.McBurney MW, Mai T, Yang X, Jardine K. Evidence for repeat-induced gene silencing in cultured Mammalian cells: inactivation of tandem repeats of transfected genes. Exp. Cell Res. 2002;274:1–8. doi: 10.1006/excr.2001.5443. [DOI] [PubMed] [Google Scholar]
- 37.Tawil R, van der Maarel SM. Facioscapulohumeral muscular dystrophy. Muscle Nerve. 2006;34:1–15. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- 38.Davie JR, Candido EP. DNase I sensitive chromatin is enriched in the acetylated species of histone H4. FEBS Lett. 1980;110:164–168. doi: 10.1016/0014-5793(80)80063-9. [DOI] [PubMed] [Google Scholar]
- 39.Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, Markovich RP, Murray JC, Wasmuth JJ, Altherr MR, et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- 41.Tam R, Smith KP, Lawrence JB. The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J. Cell Biol. 2004;167:269–279. doi: 10.1083/jcb.200403128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Berger SL. Good fences make good neighbors: barrier elements and genomic regulation. Mol. Cell. 2004;16:500–502. doi: 10.1016/j.molcel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Lemmers RJ, Osborn M, Haaf T, Rogers M, Frants RR, Padberg GW, Cooper DN, van der Maarel S, Upadhyaya M. D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology. 2003;61:178–183. doi: 10.1212/01.wnl.0000078889.51444.81. [DOI] [PubMed] [Google Scholar]
- 44.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 45.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 46.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh K, Gualberto A. MyoD binds to the guanine tetrad nucleic acid structure. J. Biol. Chem. 1992;267:13714–13718. [PubMed] [Google Scholar]
- 49.Yafe A, Etzioni S, Weisman-Shomer P, Fry M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005;33:2887–2900. doi: 10.1093/nar/gki606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Etzioni S, Yafe A, Khateb S, Weisman-Shomer P, Bengal E, Fry M. Homodimeric MyoD preferentially binds tetraplex structures of regulatory sequences of muscle-specific genes. J. Biol. Chem. 2005;280:26805–26812. doi: 10.1074/jbc.M500820200. [DOI] [PubMed] [Google Scholar]
- 51.Lemmers RJL, de Kievit P, van Geel M, van der Wielen MJ, Bakker E, Padberg GW, Frants RR, van der Maarel SM. Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann. Neurol. 2001;50:816–819. doi: 10.1002/ana.10057. [DOI] [PubMed] [Google Scholar]
- 52.Thomas NS, Wiseman K, Spurlock G, MacDonald M, Ustek D, Upadhyaya M. A large patient study confirming that facioscapulohumeral muscular dystrophy (FSHD) disease expression is almost exclusively associated with an FSHD locus located on a 4qA-defined 4qter subtelomere. J. Med. Genet. 2007;44:215–218. doi: 10.1136/jmg.2006.042804. [DOI] [PMC free article] [PubMed] [Google Scholar]