Abstract
Although multiple sequence alignments (MSAs) are essential for a wide range of applications from structure modeling to prediction of functional sites, construction of accurate MSAs for distantly related proteins remains a largely unsolved problem. The rapidly increasing database of spatial structures is a valuable source to improve alignment quality. We explore the use of 3D structural information to guide sequence alignments constructed by our MSA program PROMALS. The resulting tool, PROMALS3D, automatically identifies homologs with known 3D structures for the input sequences, derives structural constraints through structure-based alignments and combines them with sequence constraints to construct consistency-based multiple sequence alignments. The output is a consensus alignment that brings together sequence and structural information about input proteins and their homologs. PROMALS3D can also align sequences of multiple input structures, with the output representing a multiple structure-based alignment refined in combination with sequence constraints. The advantage of PROMALS3D is that it gives researchers an easy way to produce high-quality alignments consistent with both sequences and structures of proteins. PROMALS3D outperforms a number of existing methods for constructing multiple sequence or structural alignments using both reference-dependent and reference-independent evaluation methods.
INTRODUCTION
Multiple sequence alignments (MSAs) have a wide range of applications in protein science, such as profile-based similarity searches, structure modeling, functional prediction and phylogenetic analysis. Accurate and fast construction of MSAs has been under extensive research for many years (1,2). Recently emerged consistency-based scoring functions (3), especially those with a probabilistic interpretation (4,5), are superior to general amino acid substitution matrices for MSA construction. Additional evolutionary information from database homologs has also been explored to enhance alignment quality (6). As protein structures generally evolve slower than sequences, structural information, either from available 3D experimental structures (7) or from predicted secondary structures (8,9), can lead to further improvement of MSAs. Our method PROMALS (10) improves alignment quality of distantly related sequences by combining several advanced techniques such as database searching for additional homologs, secondary structure prediction and probabilistic consistency of profile-to-profile comparisons.
Further improvements to PROMALS alignment quality could arise from using constraints on the regions that should be aligned. Such constraints can be defined by structure superposition or other additional expertise knowledge. Structure-based alignments are regarded as high-quality alignments and are routinely used as a gold standard for assessing alignment quality. Ongoing structural genomics initiatives have made great progress toward solving structures for representatives that cover the protein universe. Some alignment algorithms are making use of this information. The program 3DCoffee with web server implementation Expresso (11) automatically combines SAP structural alignments (12) with sequence alignments by using constraints based on structural alignments to derive consistency-based scoring functions. MAFFT (13) server offers an option for the input of alignment constraints, which can be structure-based alignments.
In this article, we explore information from available protein 3D structures by PROMALS. The resulting program, PROMALS3D, brings together sequence and structure-based alignments to generate high-quality multiple alignments consistent with both sequence and structural information. For input sequences, we use similarity searches to retrieve homologs with available 3D structures. The structure-based alignments among these homologs help define high-quality constraints that are combined with sequence-based profile-to-profile alignments enhanced by predicted secondary structures. PROMALS3D can also be used to construct multiple structural alignments for a set of proteins with known 3D structure. PROMALS3D outperforms a number of existing methods for constructing multiple sequence or structural alignments using both reference-dependent and reference-independent evaluation criteria.
MATERIALS AND METHODS
Structural domain database and structural alignment databases
To identify homologs with known structures for target sequences, we used the ASTRAL SCOP40 structural domain database (14,15) (version 1.69, 7290 domains, with <40% sequence identity to each other). Structure-based sequence alignments were made between each pair of domains by three structural comparison programs: DaliLite (16), FAST (17) and TM-align (18). These alignment databases facilitate the use of structural information by allowing lookup of the structure-based sequence alignments for any domain pair, without running the structural comparison programs for them during the multiple sequence alignment process. For each structural domain, we also made PSI-BLAST (19) searches to retrieve homologs that can be used in profile–profile alignments with target sequences.
An overview of PROMALS algorithm
PROMALS (10) is a progressive method that clusters similar sequences and aligns them by a simple and fast algorithm, and applies more elaborate techniques to align the relatively divergent clusters to each other. In the first alignment stage, PROMALS aligns similar sequences using a scoring function of weighted sum-of-pairs of BLOSUM62 (20) scores. The first stage is fast and results in a number of pre-aligned groups (clusters) that are relatively distant from each other. In the second alignment stage, one representative sequence is selected for each pre-aligned group (subsequently referred to as ‘target sequence’). Target sequences are subject to PSI-BLAST searches for additional homologs from UNIREF90 (21) database and to PSIPRED (22) secondary structure prediction. Then a hidden Markov model (HMM) of profile–profile alignment with predicted secondary structures is applied to pairs of representatives to obtain posterior probabilities of residue matches. These probabilities serve as sequence-based constraints that are used to derive a probabilistic consistency scoring function. The representative target sequences are progressively aligned using such a consistency scoring function, and the pre-aligned groups obtained in the first stage are merged into the alignment of representatives to form the final multiple alignment of all sequences.
Incorporating 3D structural information in PROMALS
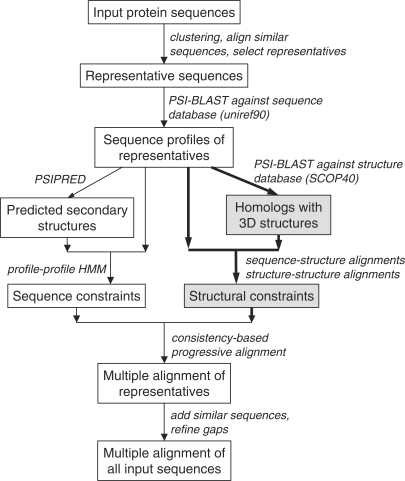
In PROMALS3D, structural constraints are derived for representative sequences with known structures and are combined with sequence-based constraints (Figure 1). First, the program identifies homologs with 3D structures (homolog3D) for target sequences resulting from the first fast alignment stage. For each target sequence, the profile of PSI-BLAST search against the UNIREF90 database are used to initiate a new PSI-BLAST search (one iteration) against the SCOP40 domain database that contains protein domain sequences with known structures. Only structural domains that pass certain similarity criteria (default: e-value <0.001 and sequence identity no <20%) are kept. These structural domains are further filtered to remove redundancy in the following way: if two structural domains pass the criteria and their non-overlapping region is less than 30 residues, only the one with a better e-value is kept. Multiple homolog3Ds could be identified and used for one target sequence if it contains several distinct domains with known structures.
Figure 1.
Flowchart of PROMALS3D method.
Pairwise residue match constraints for two target sequences are derived from sequence-based target-to-homolog3D alignments and structure-based homolog3D-to-homolog3D alignments. For example, if residue A in target S1 is aligned to residue B in homolog3D T1, residue B in homolog3D T1 is aligned with residue C in homolog3D T2 according to a structure comparison program, and residue C in homolog3D T2 is aligned with residue D in target S2, then we deduce that residue A in sequence S1 is aligned with residue D in sequence S2, and this pair is used as a structure-derived constraint (see Figure S1 in Supplementary Data). In the matrix representation, the matrix of structural constraints of two targets S1 and S2 is the product of the multiplication of three residue match matrices: MS1−T1, MT1−T2 and MT2−S2. The alignment between a target sequence and its homolog3D can be the PSI-BLAST alignment, or they can be re-aligned by the profile–profile comparison routine used in PROMALS. For PSI-BLAST alignment between a target and its homolog3D and the structural alignment between two homolog3D, if two residues are aligned, its entry in the residue match matrix is 1, otherwise it is 0. For profile–profile comparison using HMM (10), the entries of a residue match matrix are the posterior probabilities of two residues being aligned (determined by forward–backward algorithm). The structure constraints among target sequences are combined with those constraints derived from profile–profile comparisons in the original PROMALS to deduce a consistency-based scoring function that integrates database sequence profiles, predicted secondary structures and 3D structural information. We used an empirical weight ratio of 1.5 for structure constraints relative to the sequence constraints of profile–profile comparison in the original PROMALS.
Testing the performance of MSA or multiple structural alignment programs
We compared PROMALS3D to other common multiple sequence alignment programs. PROMALS3D, 3DCoffee and Expresso (a web server of 3DCoffee that automatically include 3D information) are multiple alignment programs that use both sequence and structural information. We implemented 3DCoffee by inputting structural alignment constraints to T-Coffee program. We could not obtain the stand-alone version of Expresso and thus manually submitted the 209 ‘twilight zone’ tests in SABmark database to the Expresso server with default options. PROMALS, SPEM (9), MUMMALS (5), ProbCons (4), MAFFT (13), MUSCLE (23), T-Coffee (3) and ClustalW (24) use only sequence information (PROMALS and SPEM also incorporate secondary structure information that is predicted from sequences). The availability of a known structure for every sequence in SABmark database allows us to benchmark MUSTANG, a multiple structural alignment program (25) that uses only 3D structural information. We also tested PROMALS3D performance on only structural information by making consistency measures solely from structural constraints of DaliLite alignments or reference alignments. For reference-dependent evaluation of alignment quality, the alignment quality score (Q-score) is defined as the number of correctly aligned residue pairs in a test alignment divided by the total number of aligned residue pairs in a reference alignment (its value is between 0 and 1). For reference-independent evaluation of alignment quality, we used a number of structure-based scores such as GDT-TS (26) to reflect the similarities of two structures aligned according to a sequence alignment. The details of reference-independent evaluation are described in a previous article (5).
RESULTS AND DISCUSSION
We tested the performance of PROMALS3D and other multiple alignment programs on two alignment benchmark databases, SABmark (27) and PREFAB (23), using reference-dependent and reference-independent evaluation methods (see Materials and methods section). PROMALS3D, 3DCoffee and Expresso use both sequence and 3D structure information. PROMALS3D and MUSTANG can align multiple structures using only 3D structural information. The other programs PROMALS, SPEM, MUMMALS, ProbCons, MAFFT, MUSCLE, T-Coffee and ClustalW do not use 3D structural information.
Tests on SABmark database
SABmark database (version 1.65) has two benchmark sets for testing multiple alignment programs: the ‘twilight zone’ set contains 209 groups of SCOP (version 1.65) fold-level domains with very low similarity, and the ‘superfamilies’ set contains 425 groups of SCOP superfamily-level domains with low to intermediate similarity. For each group, the SABmark database provides a set of pairwise reference alignments for evaluation of alignment quality, instead of a single-reference multiple sequence alignment. Each pairwise reference alignment was derived from the consensus of two structural comparison programs SOFI (28) and CE (29).
Since PROMALS3D uses an ASTRAL domain structural database that is based on a later version of SCOP (1.69) than the one used in SABmark, exact matches or close homologs with structures (homolog3Ds) can be identified for most of the SABmark sequences. Combining structural constraints derived from DaliLite alignments with the profile–profile alignment constraints in original PROMALS, PROMALS3D achieves average Q-scores of 0.603 and 0.805 for the ‘twilight zone’ set and the ‘superfamilies’ set, respectively [Table 1, PROMALS3D (D + S)]. The Q-score improvements over the original PROMALS program without 3D structural information are 0.21 and 0.14, respectively. Such prominent increases of alignment quality are also evident from the reference-independent evaluation using GDT-TS score (Table 1) and other structure-based scores (Table S1 in Supplementary Data). Using DaliLite structural alignments in PROMALS3D gives slightly but significantly better results than using FAST alignments or TM-align alignments (measured by Q-score or GDT-TS), suggesting that DaliLite produces more accurate structural alignments on average. Combining structural alignments made by DaliLite, FAST and TM-align did not yield much improvement over using only DaliLite structural alignments.
Table 1.
Tests on SABmark database
| Method | SABmark-twi (209/10667) | SABmark-sup (425/19092) | ||
|---|---|---|---|---|
| Q-score | GDT-TS | Q-score | GDT-TS | |
| PROMALS3D (D + S) | 0.602 | 0.264 | 0.805 | 0.417 |
| PROMALS3D (F + S) | 0.555 | 0.220 | 0.779 | 0.390 |
| PROMALS3D (T + S) | 0.540 | 0.249 | 0.766 | 0.412 |
| PROMALS3D (D + F + S) | 0.611 | 0.256 | 0.812 | 0.414 |
| PROMALS3D (D + T + S) | 0.603 | 0.264 | 0.805 | 0.421 |
| PROMALS3D (F + T + S) | 0.595 | 0.251 | 0.800 | 0.413 |
| PROMALS3D (D + F + T + S) | 0.616 | 0.260 | 0.812 | 0.420 |
| 3DCoffee (D + S) | 0.574 | 0.252 | 0.802 | 0.421 |
| 3DCoffee (SAP + S) | 0.553 | 0.222 | 0.786 | 0.390 |
| Expresso webserver | 0.508 | 0.206 | – | – |
| PROMALS3D (D/2 + S) | 0.475 | 0.198 | 0.716 | 0.364 |
| 3DCoffee (D/2 + S) | 0.261 | 0.100 | 0.573 | 0.294 |
| 3DCoffee (D/2 + SAP) | 0.255 | 0.095 | 0.572 | 0.289 |
| PROMALS | 0.393 | 0.154 | 0.665 | 0.336 |
| SPEM | 0.326 | 0.124 | 0.628 | 0.318 |
| MUMMALS | 0.196 | 0.081 | 0.522 | 0.278 |
| ProbCons | 0.166 | 0.058 | 0.485 | 0.246 |
| MAFFT-linsi | 0.184 | 0.070 | 0.510 | 0.264 |
| MUSCLE | 0.136 | 0.056 | 0.433 | 0.233 |
| T-Coffee | 0.134 | 0.048 | 0.429 | 0.223 |
| ClustalW | 0.127 | 0.057 | 0.390 | 0.221 |
| MUSTANG | 0.550 | 0.230 | 0.779 | 0.404 |
| PROMALS3D (D) | 0.594 | 0.252 | 0.802 | 0.415 |
The first 13 methods for MSAs use both sequence and 3D structural information. The last two methods assemble multiple alignments solely from structural constraints. The other methods construct multiple alignments using only sequence information (PROMALS and SPEM also use predicted secondary structures). The letters inside the parenthesis after the method names are: ‘D’, using DaliLite structural constraints; ‘F’, using FAST structural constraints; ‘T’, using TM-align structural constraints; ‘S’, using sequence information; ‘SAP’, using SAP structural alignments; ‘D/2’, using DaliLite alignments for half of the sequences; ‘SAP/2’, using SAP alignments for half of the sequences. Q-score is the alignment quality score defined as the number of correctly aligned residue pairs divided by the total number of residue pairs in a reference alignment. GDT-TS is a reference-independent measure of alignment quality based on structural similarity of two structures superimposed according to a test alignment. The ‘twi’ stands for ‘twilight-zone’ set and ‘sup’ stands for ‘superfamilies’ set. The number of multiple alignment tests and pairwise reference alignments are shown in parentheses. The best scores are in bold letters.
Another program that can incorporate 3D structural information is 3DCoffee (7), which we implemented by feeding structural alignment constraints in the T-COFFEE program. The default 3DCoffee program using SAP structural alignments (12) yields significantly worse results than PROMALS3D using DaliLite structural alignments (Table 1). 3DCoffee with DaliLite structural alignment constraints also give better results than 3DCoffee with SAP alignments. These results validate the high quality of DaliLite alignments. Automatic incorporation of SAP structural alignments by 3DCoffee is also available in the Expresso web server. We manually submitted 209 SABmark twilight-zone set alignments to Expresso web server and obtained worse results than running 3DCoffee on our local computers (Table 1).
PROMALS3D and 3DCoffee capture 3D structural information in a similar way through consistency measure. Results on SABmark benchmarks suggest they perform similarly when every sequence has a close homolog3D and the same structural constraints are given. In real-life alignment cases, however, we might not find close homolog3Ds for every sequence, and sequence constraints play a more important role in aligning sequences without 3D structural information. To test this effect, we force PROMALS3D and 3DCoffee to use 3D structural information for only half of the sequences in each alignment in SABmark database [Table 1, PROMALS3D (D/2 + S), 3DCoffee (D/2 + S) and 3DCoffee (SAP/2 + S)]. In these tests, PROMALS3D performs significantly better than 3DCoffee. The average Q-score differences of PROMALS3D (D/2 + S) and 3DCoffee (D/2 + S) are about 0.21 and 0.14 on ‘twilight zone’ set and ‘superfamilies’ set, respectively. These results reflect the superiority of PROMALS3D sequence constraints, which are based on profile–profile comparisons with predicted secondary structures. On the other hand, the sequence constraints of 3DCoffee are based on pairwise sequence alignments in the T-Coffee program.
PROMALS3D can also be used to construct alignments for multiple structures by using only DaliLite pairwise structural alignments as constraints [Table 1, PROMALS3D (D), last line]. PROMALS3D using only structural constraints yields performance slightly worse than combining constraints of structural alignments and profile–profile alignments with predicted secondary structures. We also tested the performance of MUSTANG, a multiple structural alignment program that is based on the consistency of pairwise structural alignments and does not use sequence information. PROMALS3D is significantly better than MUSTANG according to reference-dependent and reference-independent evaluations. These results suggest that PROMALS3D offers a good solution to the multiple structural alignment problem by combining DaliLite structural constraints and sequence constraints of profile–profile comparisons.
As a positive control, we also used pairwise SABmark reference alignments as the sole structural constraints to assemble multiple alignments by the PROMALS3D consistency strategy. SABmark pairwise reference alignments are noted as not entirely consistent with each other (27). Therefore, any method that assembles multiple sequence alignments cannot achieve a perfect average Q-score (would be 1.0) when tested on these pairwise reference alignments. The average accuracies for the multiple alignments assembled from pairwise reference alignments are about 0.71 and 0.87 for the ‘twilight zone’ set and the ‘superfamilies’ set, respectively; suggesting that inconsistency among pairwise reference alignments are more prominent in the more distant ‘twilight zone’ set. The lack of consistency (transitivity) between structural alignments is an intrinsic and unavoidable feature of structure superposition strategies. Superpositions are based on structural closeness, either in Cartesian space or in contact space, and between three structures with residues A, B and C, closeness of (A, B) and (B, C) does not imply that (A, C) are necessarily very close. Sequence alignments are frequently viewed as evolutionary alignments where transitivity applies. Since we consider each aligned site to correspond to a single ancestral site, alignment of (A, B) and (B, C) implies that A and C have the same ancestral site and should be aligned together. Evolutionary alignments are always hypothetical. Structural alignments are purely geometric but are essential benchmarks for accurate structure modeling. Such a difference is addressed by PROMALS3D, which finds the best compromise, consistent with all available sequences and pairwise structural alignments.
The effect of using distant homolog3Ds
The structural constraints between target sequences are deduced from sequence-based target-to-homolog3D alignments and structure-based homolog3D-to-homolog3D alignments. Distant homolog3Ds could affect the quality of these constraints since the quality of target-to-homolog3D alignments could be poor. For this reason, the Expresso webserver of 3DCoffee restricts the use of homolog3Ds only when they show above 60% sequence identity to the targets.
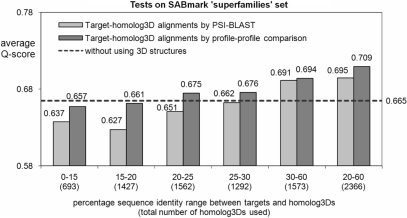
We studied the effect of using distant structural templates by restricting the selected homolog3Ds to certain similarity ranges and examining the alignment quality of using only these homolog3Ds on SABmark alignments. When using only distant homolog3Ds with sequence identity <20% to targets and PSI-BLAST target-to-homolog3D alignments, the average alignment quality score deteriorates as compared to not using 3D structural information (see Figure 2). On the other hand, using structural templates with identity between 20% and 60% results in 3–4% increase in alignment quality scores compared to not using 3D information. These results suggest that homolog3Ds with moderate similarity to targets are still valuable for improving alignment quality.
Figure 2.
The effect of using distant homolog3Ds on SABmark ‘superfamilies’ set.
We reason that increasing the quality of target-to-homolog3D alignments (default are from PSI-BLAST output) can lead to improved quality of structural constraints and the resulting multiple alignments. To test this point, we made alignments between targets and their homolog3Ds using the pairwise profile-to-profile HMMs with predicted secondary structures (the same technique for deriving sequence constraints in PROMALS). These profile–profile target-to-homolog3D alignments indeed yield better quality of multiple sequence alignments than the PSI-BLAST target-to-homolog3D alignments (Figures 2 and S2) when using distant homolog3Ds.
Tests on PREFAB database
PREFAB (23) consists of 1682 alignments (version 4.0), each of which has two sequences with known structures and up to 24 homologous sequences added from database searches for each structure. The reference-dependent evaluation is based on the consensus of FSSP (16) structural alignment and CE alignment of the two sequences with known structures. The average difficulty of PREFAB alignments is less than those of SABmark database, as the original PROMALS has an average Q-score of 0.790 on the PREFAB set of alignments [best among programs not using 3D structural information (10)], as compared to 0.393 and 0.665 on the SABmark ‘twilight-zone’ set and ‘superfamilies’ set, respectively. With the addition of 3D structural constraints from DaliLite alignments, the average Q-score of PROMALS3D on all PREFAB alignments increases to 0.893 (Table 2), which is significantly better than the average Q-score of PROMALS (0.790).
Table 2.
Test on PREFAB database
| Method | Set 1 (0.121/420) | Set 2 (0.185/421) | Set 3 (0.248/420) | Set 4 (0.527/421) | All (0.270/1682) |
|---|---|---|---|---|---|
| PROMALS3D (D + S) | 0.817 | 0.879 | 0.921 | 0.954 | 0.893 |
| PROMALS3D (F + S) | 0.745 | 0.850 | 0.896 | 0.947 | 0.859 |
| PROMALS3D (T + S) | 0.766 | 0.856 | 0.902 | 0.950 | 0.869 |
| PROMALS3D (D + F + S) | 0.818 | 0.886 | 0.919 | 0.952 | 0.894 |
| PROMALS3D (D + T + S) | 0.834 | 0.884 | 0.922 | 0.953 | 0.898 |
| PROMALS3D (F + T + S) | 0.794 | 0.875 | 0.909 | 0.952 | 0.883 |
| PROMALS3D (D + F + T + S) | 0.836 | 0.894 | 0.917 | 0.956 | 0.900 |
| PROMALS | 0.570 | 0.771 | 0.875 | 0.946 | 0.790 |
| SPEM | 0.536 | 0.756 | 0.865 | 0.940 | 0.774 |
| MUMMALS | 0.457 | 0.693 | 0.834 | 0.939 | 0.731 |
| ProbCons | 0.428 | 0.672 | 0.826 | 0.936 | 0.716 |
| MAFFT-linsi | 0.443 | 0.681 | 0.826 | 0.938 | 0.722 |
| MUSCLE | 0.372 | 0.631 | 0.787 | 0.930 | 0.680 |
| ClustalW | 0.299 | 0.536 | 0.726 | 0.906 | 0.617 |
The first seven methods (PROMALS3D) for MSAs use both sequence and 3D structural information. The other methods construct multiple alignments using only sequence information (PROMALS and SPEM also use predicted secondary structures). For the meaning of the letters inside the parenthesis after the method names, refer to Table 1. Average Q-score (see Table 1 for definition) is reported. The total 1682 PREFAB alignments are divided to four semi-equal-sized sets according to sequence identity of the reference alignment. The average sequence identity and the number of alignments are in parentheses beneath the set names. The best scores are in bold letters.
We also sorted the PREFAB alignments according to sequence identity, and divided them into four semi-equal-sized subsets (Table 2). The average sequence identities for the four subsets are 0.121, 0.185, 0.248 and 0.527, respectively. The subset with the lowest average sequence identity is the most difficult, for which we observed the most prominent increase of alignment quality of using structural information (an increase of about 0.25 for average Q-score). For subsets with higher average identity, the improvements of PROMALS3D over PROMALS are less prominent (Table 2). These results suggest that 3D structural information is most valuable for improving alignments of distantly related sequences.
Web server
A web server of PROMALS3D is available at: http://prodata.swmed.edu/promals3d.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Lisa Kinch and James Wrabl for critical reading of the article and helpful comments. We thank Cedric Notredame for help with running 3DCoffee on our local computers. This work was supported in part by NIH grant GM67165 to N.V.G. Funding to pay the Open Access publication charges for this article was provided by Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Edgar RC, Batzoglou S. Multiple sequence alignment. Curr. Opin. Struct. Biol. 2006;16:368–373. doi: 10.1016/j.sbi.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Notredame C. Recent evolutions of multiple sequence alignment algorithms. PLoS Comput. Biol. 2007;3:e123. doi: 10.1371/journal.pcbi.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 4.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pei J, Grishin NV. MUMMALS: multiple sequence alignment improved by using hidden Markov models with local structural information. Nucleic Acids Res. 2006;34:4364–4374. doi: 10.1093/nar/gkl514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson JD, Plewniak F, Thierry J, Poch O. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 2000;28:2919–2926. doi: 10.1093/nar/28.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Sullivan O, Suhre K, Abergel C, Higgins DG, Notredame C. 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J. Mol. Biol. 2004;340:385–395. doi: 10.1016/j.jmb.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 8.Simossis VA, Heringa J. Integrating protein secondary structure prediction and multiple sequence alignment. Curr. Protein Pept. Sci. 2004;5:249–266. doi: 10.2174/1389203043379675. [DOI] [PubMed] [Google Scholar]
- 9.Zhou H, Zhou Y. SPEM: improving multiple sequence alignment with sequence profiles and predicted secondary structures. Bioinformatics. 2005;21:3615–3621. doi: 10.1093/bioinformatics/bti582. [DOI] [PubMed] [Google Scholar]
- 10.Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- 11.Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor WR. Protein structure comparison using iterated double dynamic programming. Protein Sci. 1999;8:654–665. doi: 10.1110/ps.8.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 16.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Weng Z. FAST: a novel protein structure alignment algorithm. Proteins. 2005;58:618–627. doi: 10.1002/prot.20331. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 26.Zemla A, Venclovas C, Moult J, Fidelis K. Processing and analysis of CASP3 protein structure predictions. Proteins. 1999;37(Suppl 3):22–29. doi: 10.1002/(sici)1097-0134(1999)37:3+<22::aid-prot5>3.3.co;2-n. [DOI] [PubMed] [Google Scholar]
- 27.Van Walle I, Lasters I, Wyns L. SABmark–a benchmark for sequence alignment that covers the entire known fold space. Bioinformatics. 2005;21:1267–1268. doi: 10.1093/bioinformatics/bth493. [DOI] [PubMed] [Google Scholar]
- 28.Boutonnet NS, Rooman MJ, Ochagavia ME, Richelle J, Wodak SJ. Optimal protein structure alignments by multiple linkage clustering: application to distantly related proteins. Protein Eng. 1995;8:647–662. doi: 10.1093/protein/8.7.647. [DOI] [PubMed] [Google Scholar]
- 29.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]