Abstract
The DNA structure-selective endonuclease Mus81-Mms4/Eme1 is a context-specific recombination factor that supports DNA replication, but is not essential for DSB repair in Saccharomyces cerevisiae. We overexpressed Mus81-Mms4 in S. cerevisiae, purified the heterodimer to apparent homogeneity, and performed a classical enzymological characterization. Kinetic analysis (kcat, KM) demonstrated that Mus81-Mms4 is catalytically active and identified three substrate classes in vitro. Class I substrates reflect low KM (3–7 nM) and high kcat (∼1 min−1) and include the nicked Holliday junction, 3′-flapped and replication fork-like structures. Class II substrates share low KM (1–6 nM) but low kcat (≤0.3 min−1) relative to Class I substrates and include the D-loop and partial Holliday junction. The splayed Y junction defines a class III substrate having high KM (∼30 nM) and low kcat (0.26 min−1). Holliday junctions assembled from oligonucleotides with or without a branch migratable core were negligibly cut in vitro. We found that Mus81 and Mms4 are phosphorylated constitutively and in the presence of the genotoxin MMS. The endogenous complex purified in either modification state is negligibly active on Holliday junctions. Hence, Holliday junction incision activity in vitro cannot be attributed to the Mus81-Mms4 heterodimer in isolation.
INTRODUCTION
Mus81-Mms4/Eme1 is a eukaryotic endonuclease that supports DNA replication fork recovery and functions in at least one subpathway of meiotic recombination (1–7). Support of DNA replication and the generation of pathological genetic changes by inappropriate applications of recombination during fork recovery may have important consequences to genetic stability (8). As an endonuclease that incises DNA joint molecules associated with replication fork recovery and recombination, Mus81-Mms4/Eme1 is likely a central factor in eukaryotic replication fork support (9–12). Its substrate(s) in vivo, however, are still uncertain. Biochemical studies in vitro demonstrate that the enzyme can cleave a number of DNA joint structures, but a quantitative enzymological analysis determining kinetic parameters for the native enzyme has been lacking.
DNA molecules exchange homologous sequence information during recombinational repair and damage tolerance mechanisms, and undertake structural interactions that require phosphodiester bond hydrolysis for separation. In the traditional double-strand break repair model, the interacting DNA molecules must be separated either by reversal of strand exchange (D-loop disruption), by dissolution (BLM-TOPOIIIα) or by endonucleolytic incision of the structure(s) joining the duplex molecules (4,13).
Prevailing models for DNA double-strand break repair have postulated that a single, symmetric intermediate DNA joint molecule, the Holliday junction, is the target for endonucleolytic separation of the joined duplex molecules (14–17). Support for the existence of this structure comes from physical analyses in bacteria and yeast and from the biochemical description of several endonucleases that deliver paired, symmetric incisions across a four-way branch point (18). Endonucleases that incise Holliday junctions have been characterized from bacteria (RuvC and RusA), archaea (Hjc and Hje) and bacteriophages (T4 endonuclease VII and T7 endonuclease I), but candidate nuclear enzymes have been absent from eukaryotic sources (4). To date, no sequence ortholog of any of the characterized resolvases has been recognized in eukaryotes, apart from the mitochondrial Cce1 that appears to be of bacterial origin (2–4,14).
In association with its interaction partner Mms4 in budding yeast and Eme1 in fission yeast and humans, Mus81 is a facultative associate of the RAD52 epistasis group and has been proposed to be a eukaryotic candidate for Holliday junction processing in vivo (3,4,19–24). Biochemical support for this idea is uneven. Heterodimer preparations from partially purified, eukaryotic sources differ from most highly purified, recombinant sources in their ability to incise synthetic Holliday junctions (2–4,25). Mus81-Eme1 partially purified from Schizosaccharomyces pombe by a tandem affinity purification protocol has shown a capacity to resolve model Holliday junctions to linear duplex products (24). Schizosaccharomyces pombe Mus81-Eme1 recovered in recombinant form from Escherichia coli, however, showed nearly no activity on Holliday junctions in vitro (26,27). Recombinant Saccharomyces cerevisiae Mus81-Mms4 expressed and purified from E. coli was similarly inactive on the same Holliday junction substrates (28). In the case of human Mus81-Eme1, endogenous complex immunoprecipitated or fractionated from HeLa cells (23,29) and recombinant complex immunoprecipitated from a eukaryotic expression source (insect Sf9 cells) (30) has shown an incision capacity on model Holliday junctions, but no Holliday junction incision has been observed for recombinant human complex expressed in E. coli (31,32).
What explains the difference between eukaryotic sources of Mus81-Mms4/Eme1 and recombinant sources in Holliday junction incision activity? One possibility has been the presence of post-translational modifications available in eukaryotic expression systems, but not in E. coli. Another concerns the association of an unidentified junction-targeting factor in partially pure eukaryotic enzyme preparations (3). Still, another possibility relates to the presence of factors in the partially purified preparations that alter junction presentation to Mus81-Mms4/Eme1 by nickase or helicase activity. Most recently, an explanation for the difference between recombinant and endogenous heterodimer abilities to cleave Holliday junctions was proposed by isolation of the first recombinant S. pombe and S. cerevisiae Mus81-Eme1/Mms4 fractions that exhibited Holliday junction cleavage in vitro (20). Gaskell et al. suggest that purification procedures that selected for the endonuclease complex in an oligomeric state greater than the single heterodimer conferred Holliday junction incision ability, in a manner responsive to metal ion concentration.
Because most recombinant preparations incise Holliday junctions poorly, a number of alternative potential physiological targets of Mus81-Mms4/Eme1 have been proposed, including a 3′-flapped structure, variants of a replication fork and the strand exchange intermediate, the displacement loop (D-loop). Whether partially purified from a eukaryotic source or purified from a recombinant source, all preparations of Mus81-Mms4/Eme1 are active on these substrates and it is only the Holliday junction activity that remains unclearly attributable to Mus81 (20,21,23–29,31,32). However, the lack of a quantitative comparison (KM, kcat) does not allow one to distinguish whether substrate recognition or catalysis drives the distinction.
One approach to identify whether eukaryotic-expressed Mus81-Mms4/Eme1 is sufficient for Holliday junction processing in vitro is to isolate the heterodimer to apparent homogeneity from its eukaryotic expression source. This should distinguish whether eukaryotic expression alone is sufficient to confer Holliday junction resolution in vitro, or whether a state of partial purity is necessarily correlated with Holliday junction incision. Furthermore, a strategy to more clearly define Mus81-Mms4 substrate selectivity in vitro entails the design of kinetic assay conditions that supply a direct quantitative comparison among joint molecule substrates. Classically, enzymological assays demand stringency in substrate processing by presenting a limiting quantity of enzyme with an excess of substrate (33). Effective substrate processing is therefore demonstrated as turnover by a catalytic enzyme preparation.
We performed a kinetic analysis of Mus81-Mms4 cleavage, and present a quantitative comparison of its substrate selectivity. Mus81-Mms4 purified from S. cerevisiae exhibits low KM and correspondingly slow turnover on a number of substrates. The incisable joint molecules can be grouped into three classes by differences in KM and kcat; differences among these substrates reveal the need for duplex DNA flanking the branch point for low KM. Furthermore, we find that Holliday junction incision is not an enzymatic property of endogenous S. cerevisiae Mus81 endonuclease in isolation, and not a property likely to be conferred by post-translational modification of the heterodimer in response to genotoxic challenge.
MATERIAL AND METHODS
Cloning of MUS81 and MMS4 into GAL1/10 divergent promoter, 2μ-based overexpression vector
The binary expression vector is based on pJN58, a 2μ shuttle vector bearing the bidirectional yeast promoter GAL1/10 (34). pWDH619 (pJN58 with His10 insertion) was generated by removing the NotI fragment of pWDH423, blunted and cloned into the XbaI site (blunted) of pJN58. MUS81 was PCR-amplified using Elongase® enzyme (Invitrogen™) from plasmid pWDH484 (pGAL-MUS81, a kind gift of S. Brill) using primers olWDH320 and olWDH321 that introduce FLAG and TEV protease recognition sequences, engineered as a cassette flanked by MluI restriction sites. MMS4 was PCR-amplified from WDHY668 yeast genomic DNA using primers olWDH314 and olWDH315 that introduce flanking XhoI and SphI restriction sites. The MUS81 fragment was cloned into the MluI site of pWDH619 (in frame with a His10 sequence) to generate pWDH592, and the MMS4 fragment was cloned into the XhoI/SphI backbone of pWDH597 (in frame with GST and a PreScission™ protease recognition sequence) to generate pWDH620. The EcoNI/SphI fragment of pWDH592 was cloned into the EcoNI/SphI backbone of pWDH620 to generate the double-expression vector, pWDH595. Construction was independently verified by PstI/BamHI and EcoRI/EcoNI double digestions. The GST-MMS4/His10-FLAG-mus81-dd overexpression vector (pWDH596) was generated by mutagenic PCR of pWDH592 using primers olWDH374 and olWDH375, to generate pWDH593. The EcoNI/SphI fragment of pWDH595 was cloned into the corresponding sites of pWDH593 to generate the double-expression vector. Mutation of MUS81 at the XPF-family conserved D414A/D415A sites introduced a diagnostic NheI restriction site, the second such site for the plasmid. Sequencing of all clones confirmed their nucleotide sequences as published in the Saccharomyces Genome Database (SGD) or the engineered MUS81 active site mutations. Primer sequences are available upon request.
Confirmation of fusion protein function by complementation of genotoxin sensitivity of a mus81-Δ mms4-Δ double mutant strain
Complementation analysis was performed by drop dilution assays on solid YPD or YPG agarose with genotoxin where appropriate. WDHY1636 (W303 MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 RAD5) is the wild-type strain and WDHY2129 is the isogenic mms4::KANMX mus81::KANMX double mutant strain. mus81 mms4 cells were transformed with the overexpression vector pWDH619 lacking the MUS81 and MMS4 open reading frames, with pWDH595 expressing N-terminal fusion alleles of MUS81 and MMS4, or with pWDH596 expressing fusion alleles of mus81-dd and MMS4. Cells were spotted at 5-fold dilutions starting at 4 × 104 cells; plates were incubated at 30°C and photographed daily up to 3 days.
Purification of GST-Mms4/His10-FLAG-Mus81 by sequential affinity chromatography
A diploid protease-deficient expression strain, WDHY668 (MATa ura3-52 trp1 leu2-Δ1 his3-Δ200 pep4::HIS3 prb1-Δ1.6R can1 GAL/MATα ura3-52 trp1 leu2-Δ1 his3-Δ200 pep4::HIS3 prb1-Δ1.6R can1 GAL), was transformed with pWDH595 or pWDH596 using a standard lithium acetate transformation protocol and expression was induced for 8 h at 25°C as described in Solinger et al. (35). All purification procedures were performed at 4°C. A total of 120 g of S. cerevisiae cells were thawed in PBS (phosphate-buffered saline: 137 mM NaCl/2.7 mM KCl/10 mM Na2HPO4/1.8 mM KH2PO4) adjusted to 500 mM NaCl, pH 7.5. Cells were mechanically disrupted in a bead chamber with 350 g small glass beads, for 30 s intervals separated by 2 min on ice. Lysate was cleared of cellular debris by centrifugation at 45 000 rpm for 45 min in a Beckman Optima™ LE-80K preparative ultracentrifuge. Solid ammonium sulfate (ICN Biomedicals, Inc.) was slowly added to 25% with constant stirring. Precipitate was removed by centrifugation in a Beckman R2-MC centrifuge, 30 min at 15 000 rpm. The supernatant was then loaded onto an approximately 4 ml-bed volume glutathione-sepharose™ 4B resin (Amersham Biosciences) at 0.3 ml/min. flow rate. After washing the resin with PBS/500 mM NaCl, pH 7.5, GST-tagged material was eluted by competition with 20 mM l-glutathione (Sigma) in PBS/500 mM NaCl, pH 7.5. Fractions were analyzed for protein content by 10% SDS-PAGE and Coomassie Brilliant Blue staining. To remove free glutathione, pooled fractions were dialyzed against 1 l PBS/500 mM NaCl with constant stirring for 1–2 h, then dialyzed against an additional 1 l volume for 1 h. A 2 ml-bed volume Ni2+-NTA resin (Qiagen) was equilibrated with 10 mM imidazole (Fisher Scientific) in PBS/500 mM NaCl, pH 7.5. The dialyzed material was adjusted to 10 mM imidazole and loaded at 0.3 ml/min flow rate. The resin was washed with increasing imidazole stringency to 50 mM imidazole in PBS/500 mM NaCl, pH 7.5. Bound complex was eluted by competition with 250 mM imidazole, pH 7.0. Pooled fractions were dialyzed against two 1 l-volumes of storage buffer: 20 mM Tris–HCl, pH 7.5/500 mM NaCl/10% glycerol/1 mM EDTA/0.2 mM PMSF/0.1 mM DTT, 1–2 h. His10-FLAG-Mus81/GST-Mms4 heterodimer was concentrated by centrifugation at 1500g in a Millipore Ultrafree-15 centrifugal filter device pre-rinsed with 10 ml storage buffer, 10 min at 1500g. Concentrated protein was stored in 10 μl aliquots and flash-frozen in liquid N2 for long-term storage at −80°C. Protein concentration was determined by direct A280 reading using the calculated heterodimer extinction coefficient ɛM at 280 nm = 147 840 cm−1 M−1. Predicted Mr for GST-Mms4 is 106.4 kDa, and for His10-FLAG-Mus81 is 73.6 kDa.
Isolation of heterodimer from MMS- and HU-stressed cells
Heterodimer was purified essentially as described for the wild-type and mutant heterodimer, except with the following modifications. At 6 h during protein overexpression, MMS (Sigma-Aldrich) was added to 0.1% or HU (US Biologicals) was added to 100 mM and cells were grown 2 h before harvest. During extraction and throughout purification, 0.1 mM NaVO3 (Acros Organics) was added to buffers until dialysis prior to the Ni2+-NTA resin [see Bashkirov et al. (36)].
Preparation of DNA oligonucleotide joint molecules for kinetic analysis
Oligonucleotides (Qiagen Operon) were diluted to ∼100 pmol/μl. Non-radiolabeled oligo structures were prepared to define joint molecule concentration in nuclease assays; radiolabeled oligo structures were prepared in parallel to ‘spike’ reactions for nuclease activity detection. Radiolabeling was performed using T4 PNK (New England BioLabs) and 32P-γ-ATP (PerkinElmer). Oligonucleotides were annealed in an MJ Research PTC-200 Peltier thermal cycler under a staged temperature decline program: 95°C/3 min, 65°C/10 min, 37°C/10 min, 25°C/10 min, 0°C/10 min. For non-radiolabeled structures, 600 pmol 50-mers and 1200 pmol 25-mers were annealed in oligo annealing buffer (150 mM NaCl/15 mM Na3C6H5O7) in 60 μl volumes; for radiolabeled structures, one oligonucleotide was first radiolabeled and then 20 pmol radiolabeled oligo and 100 pmol non-radiolabeled 50-mers, 200 pmol non-radiolabeled 25-mers were annealed. The reactions were adjusted to 5% glycerol using DNA loading dye with bromophenol blue and the full volume was loaded onto a 1.5 mm-thick 10 × 10 cm or 10 × 20 cm 10% native PAGE-TBE gel and electrophoresed at 150 V for 65 min. Following electrophoresis, DNA was identified in the PAGE-TBE gel by UV shadowing on Baker-flex® cellulose PEI-F thin layer chromatography paper (J.T. Baker). Bands corresponding to the fully annealed DNA structures were excised and stored at 4°C. For radiolabeled structures, the PAGE-TBE gel was exposed to Kodak Scientific Imaging film for 15 min. Fully annealed joint structure bands were excised using the processed film as a template on a light table. Completely annealed structures were verified by their slow electrophoretic mobility relative to structures annealed with all possible partial oligonucleotide compositions. Heat-denaturation of substrates verified their component strands. DNA joint structures were electroeluted from the polyacrylamide slice in a Millipore Microelutor in TBE buffer at 150 V for 2 h at room temperature or at 4°C. Samples were concentrated in the Millipore Centricon device by centrifugation in a Beckman Avanti J25-I centrifuge at 4000 rpm for 1 h to 1 h 45 min at 4°C. Samples were collected by inverting the Centricon unit with centrifugation at 300g, 2 min, and dialyzed against two volumes of 200 ml TE, pH 7.5 in a Tube-o-dialyzer Medi tube, MWCO 15 000 (GenoTech, Inc.) for 1 h at 4°C. Substrate concentration was determined by spectrophotometry at λ = 260 nm using a NanoDrop® ND-1000 spectrophotometer for non-radiolabeled oligo structures, or by scintillation count to determine c.p.m./μl for radiolabeled oligo structures. When further concentration was needed, DNA samples were transferred to a Microcon YM-10 centrifugal filter device (MWCO 10 000; Millipore/Amicon) and concentrated in a microcentrifuge at 12 000 rpm for 30 min, 4°C. The sequences of oligonucleotides are those reported by Kaliraman et al. and Osman et al. (28,37), other than the oligonucleotides used in preparation of the D-loop structure (DL). These sequences are: [DL-0/olWDH684] 5′-CGTTGGACGCTGCCGAATTCTACCACTGCGTGCCTTGCTAGGACATCTTTGCC CACCTGCAGGTTCACCCATCGC-3′; [DL-1/olWDH685] 5′-GCGATGGGTGAACCTGCAGGTGGGCGGCTGCTCATCGTAGGTTAGTGAATTGGTAGA ATTCGGCAGCGTCCAACG-3′; [DL-2/olWDH686] 5′-GATCGTAAGAGCAAGATGTTCTATAAAAGATGTCCTAGCAAGGCACGCAG-3′; [DL-3/olWDH687] 5′-TATAGAACATCTTGCTCTTACGATC-3′.
Nuclease assays
In salt and pH activity assays, 100 nM 3′-flapped substrate was used (defined by non-radiolabeled substrate, but spiked with radiolabeled 3′-FL as a reporter), and heterodimer was at 5 nM. The radiolabeled spike was confirmed to contribute negligible concentration by DNA DipStick™ analysis (Invitrogen) and NanoDrop® ND-1000 spectrophotometric readings at λ = 260 nm after decay to background. Heterodimer was diluted in standard enzyme diluent (SED: 10 mM Tris–HCl, pH 7.5, 0.5 mg/ml BSA) to 50 nM. Buffers were used at 25 mM reaction concentration, with 10 mM MgCl2/1 mM DTT/0.1 μg/ml BSA. MES, PIPES, MOPS, HEPES, TAPS (all from Sigma) and Tris–HCl (US Biologicals) were titrated as 8× reaction stocks to the appropriate pH at 30°C. Reactions were pre-incubated in a 30°C water bath for 10 min, and were initiated by addition of 1 μl 50 nM heterodimer with gentle mixing. Ionic strength at each pH value was titrated as defined by [NaCl], at 0, 50, 100, 150, 200, 300 and 500 mM. For metal ion characterization, buffer composition was 25 mM HEPES, pH 7.5/100 mM NaCl/1 mM DTT/0.1 μg/ml BSA (New England BioLabs) with the appropriate concentration of the metal acetate salt (magnesium acetate from Fisher; manganese and calcium acetates from Sigma-Aldrich). Aliquots of 0.5 μl were removed from each [substrate] reaction at 3, 6, 10, 15, 20, 30, 45 and 60 min and quenched immediately into pre-aliquoted 0.5 μl volumes of 0.5× nuclease stop buffer (1× is 200 mM EDTA, 2.5% SDS, 10 mg/ml Proteinase K). Nine μl of DNA loading dye (5% glycerol/bromophenol blue) were added to each time point and samples were electrophoresed by 10 × 20 cm native 10% TBE-PAGE at 100 V for 65 min. Gels were equilibrated in 5% glycerol or 3% glycerol/20% methanol for 30 min and vacuum-dried to Whatman® paper at 65°C, then exposed overnight to a phosphorimager screen. For DL analyses, time point volumes were boiled in formamide/bromophenol blue for 2 min at 95°C before electrophoresis on a 7M urea/10% TBE-PAGE denaturing gel at 150 V for 40 min. These gels were equilibrated as described and vacuum-dried to DE81 anion exchanger chromatography paper (Whatman®). The comparison of wild-type heterodimer with the catalytic mutant control was performed at optimized assay conditions, 25 mM HEPES, pH 7.5/3 mM Mg(OAc)2/100 mM NaCl/0.1 mM DTT/0.1 μg/ml BSA.
Kinetic analysis
Substrate concentrations used for initial velocity determinations were chosen to flank a predicted KM on the order of ∼1–10 nM. At least three independent trials were performed for each substrate at 0.5, 1, 2.5, 5, 10, 20, 50 and 100 nM substrate and enzyme fixed at 5 nM, with all substrate concentrations processed in parallel. Nuclease reactions were performed as described for assay optimization, with fixed time points quenched at 0, 3, 6, 10, 15, 20, 30, 45 and 60 min reaction time. Reaction progress was quantified by Storm Phosphorimagery and ImageQuant software. Initial velocities were extrapolated from nonlinear regression curves defined by Graphpad Prism at 30 s reaction time. Michaelis–Menten analysis was performed using GraphPad Prism version 4.03 for Windows, GraphPad Software, San Diego, California USA, www.graphpad.com. Regions of the assay time that followed linear initial velocity were verified and typically lasted between 3 and 5 min or more for each substrate concentration in every reaction. Rates are expressed as nanomolar joint molecule substrate incised/minute, and where calculated for kcat, rates are expressed as number of joint molecule substrates incised per heterodimer molecule per minute.
RESULTS
Purification of GST-Mms4/His10-FLAG-Mus81 heterodimer
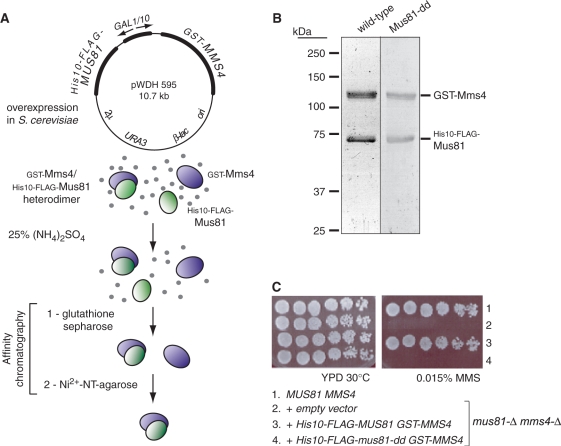
Galactose-induced overexpression of the fusion-tagged heterodimer followed by sequential affinity selection for the N-terminal GST and His10 tags (Figure 1A) resulted in the isolation of GST-Mms4/His10-FLAG-Mus81 heterodimer that is homogeneous by Coomassie staining. Passage of ammonium sulfate-fractionated extract over glutathione-sepharose yielded heterodimer with a slight excess of GST-tagged subunit, as expected if some GST-Mms4 was not associated with Mus81. Passage of the glutathione-sepharose eluants over an immobilized metal chelate resin (Ni2+-NT-agarose) selected for the intact heterodimer with subunits in a 1:1 stoichiometry as determined by Coomassie staining (Figure 1B). The predicted catalytic mutant complex (His10-FLAG-Mus81 D414A, D415A; abbreviated Mus81-dd) behaved like the wild-type heterodimer during its purification.
Figure 1.
Verification of Mus81-Mms4 fusion protein function in vivo and heterodimer purification by sequential affinity chromatography. (A) Overexpression and purification strategy: soluble heterodimer was isolated by sequential affinity selection for the N-terminal GST tag on Mms4 and the His10 tag on Mus81. (B) Coomassie-stained polyacrylamide gel showing the elution products after glutathione-sepharose and Ni2+-NT-agarose selection for subunits in stoichiometric complex. Wild-type and predicted catalytic mutant complexes (Mus81-D414A, D415A; abbreviated Mus81-dd) are shown. Calculated Mr for GST-Mms4 is 106.4 kDa and for His10-FLAG-Mus81/Mus81-dd is 73.6 kDa. (C) Complementation analysis verifies the fusion protein function in vivo. Row 1 is the wild-type strain (WDHY1636); rows 2–4 are the isogenic mus81-Δ mms4-Δ mutant (WDHY2129) transformed with the plasmids listed.
Complementation of mus81-Δ mms4-Δ double mutant genotoxin sensitivity by fusion protein expression
Saccharomyces cerevisiae cells deficient in Mus81-Mms4 are sensitive to alkylation, topoisomerase I inhibition, deoxyribonucleotide depletion, and other perturbations that disturb replication forks by reducing processivity, or by generating template lesions that prohibit replisome progression or provoke its disassembly. We verified that the addition of fusion tags designed for affinity purification of the heterodimer does not interfere with heterodimer function by establishing that the tagged heterodimer (pWDH595) expressed at low level fully complements the genotoxin sensitivity of the double mutant strain (Figure 1C). The heterodimer expression construct is under galactose-inducible control for protein overexpression, but on glucose in the absence of galactose, there is only a low level of transcription from the GAL1/10 promoter. The expression vector bearing instead the mus81-D414A, D415A mutant allele (pWDH596), predicted to encode a catalytically inactive nuclease, does not complement the double mutant strain for sensitivity to 0.015% MMS. This mutant also does not complement sensitivity to the ribonucleotide reductase inhibitor hydroxyurea or the topoisomerase I cleavage complex inhibitor camptothecin (not shown). The fusion complex is therefore active in vivo and competent to perform the DNA joint molecule incision role(s) demanded of native Mus81-Mms4.
Mus81-Mms4 is catalytically active
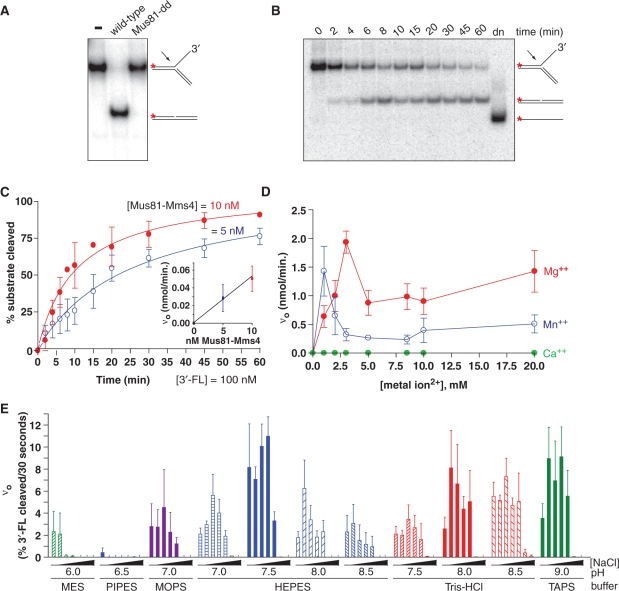
Although evolutionary selection for Mus81-Mms4 has taken place under circumstances expected to generate DNA structures that typically occur with low numerical incidence but regular frequency in cycling cells, a priori it is unclear whether the heterodimer should function stoichiometrically or catalytically with respect to substrate turnover. This has been demonstrated for few structure-selective endonucleases; the 5′-flap endonuclease Fen1 functions catalytically in vitro (38), but work with XPF-Ercc1 and the canonical Holliday junction resolvase RuvC has been typically performed under conditions that do not show catalysis or do not allow substrate turnover (39–43). We therefore assayed whether purified Mus81-Mms4 shows catalytic turnover of DNA joint molecule substrates in vitro. We chose to optimize assay conditions on a 3′-flapped structure because this substrate has consistently shown strong incision by both recombinant and partially purified eukaryotic heterodimer preparations reported to date. We verified that DNA structure incision was dependent on Mus81 catalytic activity; limiting endonuclease (5 nM) was sufficient for turnover of excess 3′-flapped substrate (100 nM) within 30 min at 30°C. No 3′-flapped substrate incision was observed during incubation with an equivalent concentration of the predicted catalytically inactive complex (Figure 2A). The active heterodimer in the presence of 20-fold substrate excess exhibits a time course of 3′-flapped incision that can approach completion within an hour at 30°C (Figure 2B and C). No contaminating helicase, exonuclease or non-specific nicking activities or thermal denaturation of substrate during the assay time course is observed, indicating that the heterodimer preparation and assay design is free of activities that are likely to alter substrate properties from the anticipated annealed structures during incubation. Specifically, there is no loss of label due to phosphatase or 5′-3′ exonuclease, no fragmentation due to contaminating endo- or 3′-5′ exonucleases, and no evidence for helicase or thermal denaturation of substrate during the assay time course (Figures 2 and 3).
Figure 2.
Determination of optimal in vitro assay conditions for Mus81-Mms4 nuclease activity on DNA joint molecules. (A) Phosphodiesterase activity on a 3′-flapped DNA joint structure depends on wild-type Mus81-Mms4 complex in the nuclease assay; the predicted catalytic mutant complex (Mus81-dd) is inactive. Nuclease reactions were performed with 100 nM 3′-flapped structure and 5 nM heterodimer, 30 min at 30°C. (B) Fixed-time point nuclease assays were performed with substrate concentration defined by non-radiolabeled molecules, spiked with a small quantity of 5′-32P radiolabeled molecules to report on the time course of incision by Mus81-Mms4. Denatured substrate (lane 12) demonstrates that time-dependent loss of substrate is due to nucleolytic turnover to duplex with no denaturation of substrate during incubation. (C) Nuclease progress curves with 100 nM 3′-flapped substrate demonstrate catalytic turnover by limiting quantity of endonuclease (10 nM and 5 nM heterodimer shown); Inset, Initial reaction velocity (nmol 3′-flapped substrate converted to product per minute) scales with heterodimer concentration at 5 nM and 10 nM heterodimer. (D) Metal ion response profile. (E) Mus81-Mms4 nuclease activity responds to buffer characteristics and pH, over a range of ionic strengths defined by NaCl concentration. Initial reaction rates on the 3′-flapped substrate were determined in the indicated buffer systems in the presence of 0, 50, 100, 150, 200, 300 or 500 mM NaCl.
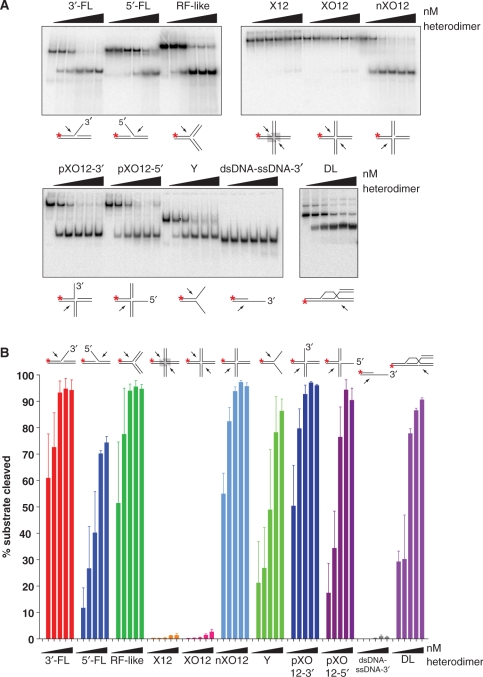
Figure 3.
Mus81-Mms4 from S. cerevisiae is active on a number of DNA joint molecule substrates, but not on intact Holliday junctions in vitro. (A) Heterodimer titration on a number of synthetic DNA joint molecules. Reactions were performed with joint molecule concentration defined by non-radiolabeled substrate at 50 nM and spiked with radiolabeled substrate to report on turnover of the substrate population during 30 min incubation at 30°C. Relative to substrate concentration, Mus81-Mms4 was added to limiting concentration at 5 nM, 10 nM and 20 nM heterodimer, at stoichiometric concentration at 50 nM heterodimer, or at excess concentration at 100 nM heterodimer. (B) Quantitation of A.
We sampled fixed time points during the course of the nuclease assay to gauge product accumulation over time. The progress curves at 5 nM and 10 nM heterodimer show a window of time during which the initial reaction rate remains linear (up to ∼5 min assay time, or 10–20% substrate depletion). The reaction velocity linearity over this extensive window of substrate depletion can probably be explained by a low KM of Mus81-Mms4 for the 3′-flapped substrate; even as substrate concentration drops by turnover to product, the remaining substrate concentration continues to substantially exceed KM (Table 1). When twice the amount of enzyme is added to 100 nM 3′-flapped structure, the initial rate correspondingly responds by a factor of two (Figure 2C).
Table 1.
Mus81-Mms4 kinetic parameters on DNA joint molecules
| Substrate | Vmax (nM/min) | KM (nM) | kcat (min−1) | Catalytic cycle (min) | kcat/KMa (nM−1 min−1) |
|---|---|---|---|---|---|
| 3′-FL | 4.9 ± 0.7 | 5.5 ± 2.6 | 0.97 | 1.03 | 0.19 |
| RF-like | 6.7 ± 0.6 | 7.3 ± 2.0 | 1.35 | 0.74 | 0.19 |
| nXO12 | 6.0 ± 1.7 | 3.1 ± 2.0 | 1.20 | 0.83 | 0.39 |
| pXO12-3′ | 1.6 ± 0.2 | 5.6 ± 1.8 | 0.32 | 3.13 | 0.06 |
| DL | 0.5 ± 0.2 | 1.2 ± 1.6 | 0.09 | 10.75 | 0.08 |
| Y | 1.3 ± 0.04 | 30.4 ± 11.3 | 0.26 | 3.85 | 0.009 |
| XO12 | −b | ||||
| X12 | −b |
akcat/KM traditionally defines a ‘selectivity coefficient’ that can be used to rank substrates for their relative ‘selectability’ by an enzyme. In this case, nXO12 > 3′-FL′ ∼ RF-like > DL > pXO12-3′ > Y.
bThe catalytic parameters could not be determined because of the negligible activity on these substrates.
Mus81-Mms4 is responsive to metal ion character and concentration
Having established that S. cerevisiae fusion-tagged Mus81-Mms4 is catalytically active and shows DNA joint molecule substrate turnover under conditions of limiting enzyme relative to substrate concentration, we defined optimal in vitro assay conditions to profile the enzyme's response to metal ion character and concentration, buffer and pH, and ionic strength as defined by NaCl concentration. We titrated Mg(OAc)2, Mn(OAc)2 and Ca(OAc)2 to define optima by the criterion of initial reaction velocity. Complete 60 min progress curves were collected as in Figure 2C to define the region of initial reaction velocity linearity, with 5 nM heterodimer in the presence of 100 nM 3′-flapped substrate. Like most DNA-acting enzymes, Mus81-Mms4 uses Mg2+ optimally for phosphodiester bond hydrolysis in DNA substrates, with an empirical optimum at approximately 3 mM (Figure 2D). The heterodimer can also use Mn2+ as cofactor, but over a more narrow concentration range with a maximum at less than or equal to 1 mM. Ca2+ is completely ineffective as cofactor, and the enzyme shows absolutely no incision on the 3′-flapped structure at any Ca2+ concentration assayed up to 20 mM.
Mus81-Mms4 is sensitive to buffer, pH and NaCl effects
Although a number of buffers are appropriate and widely applied to biological assays in vitro, some are known to interact non-specifically with certain macromolecules. Although physiological conditions are frequently cited as pH 7.5 with total ionic strength approaching 150 mM, some enzymes are especially active at pH regions outside this standard range. We therefore assayed Mus81-Mms4 initial reaction velocity on the 3′-flapped substrate as a function of pH and ionic strength defined by NaCl concentration (Figure 2E). We chose buffers with pKa values that represented a pH span from 6 to 9, and titrated NaCl at 0, 50, 100, 150, 200, 300 and 500 mM at every pH value represented across the buffers MES, PIPES, MOPS, HEPES, Tris–HCl and TAPS. For the common biological buffers HEPES and Tris–HCl, we assayed initial reaction velocity at several pH values defined at 30°C. First, we observed buffer-specific effects primarily at low pH. PIPES, pH 6.5 is an especially poor buffer for heterodimer activity. Reaction velocity is even poorer than in MES, pH 6.0, suggesting that low pH is not solely responsible for reduced activity. Nuclease activity improves at pH above 7.0, regardless of the buffer identity (Tris–HCl, HEPES or TAPS). Heterodimer nuclease activity is most sensitive to increasing NaCl concentration at low pH, but becomes more resistant to increasing NaCl concentration at higher pH. Mus81-Mms4 shows improved activity at NaCl concentrations approximating physiological ionic strength, but becomes salt-sensitive at concentrations above 200 mM NaCl. Mus81-Mms4 nuclease activity is more robust in the face of increasing ionic strength at pH 8–9. Taking these observations in sum, we chose assay conditions at 25 mM HEPES, pH 7.5/3 mM Mg(OAc)2/100 mM NaCl for nuclease incision assays, being optimal as defined for the 3′-flapped substrate.
Mus81-Mms4 is active on any tested DNA joint molecule that meets minimal criteria of a branch point coincident with a strand discontinuity
Having defined nuclease assay conditions that promote optimal reaction velocity on the 3′-flapped substrate, we examined the heterodimer's substrate selection on a number of DNA joint molecules in vitro. A broad spectrum of DNA joint molecules is readily incised by limiting enzyme (5–10 nM), including the 3′-flapped (3′-FL) and replication fork-like (RF-like) structures, nicked four-way junctions (nXO12), three-way junctions with 3′- or 5′-emanating single-stranded DNA (pXO12-3′ and pXO12-5′), and the displacement loop structure (DL). All these structures share properties previously defined as relevant to Mus81-Mms4/Eme1 recognition and processing, primarily the presence of duplex DNA flanking a phosphodiester backbone discontinuity (nick), which defines a branch point from which emanates duplex or single-stranded DNA. In the case of single-stranded DNA, the polarity must be 5′ → 3′ for optimal cleavage adjacent to the branch point, although this is less important in substrates with three duplex arms (pXO12 structures). Increasing the concentration of enzyme relative to substrate (20–100 nM enzyme) broadens the class of substrates that can be readily incised by S. cerevisiae Mus81-Mms4, including the 5′-flapped (5′-FL) and splayed arm (Y) joints. The apparent selectability of a joint molecule substrate can therefore be modulated by enzyme: substrate stoichiometry (Figure 3A and B). These observations underscore the fact that Mus81-Mms4 is not structure-specific, but rather structure-selective, in vitro.
Mus81-Mms4 in isolation is not active on four-way Holliday junctions
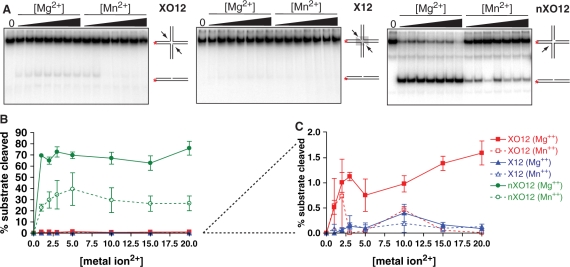
Having shown that Mus81-Mms4 readily incises a number of DNA joint molecules in vitro, and that even structures that are not considered likely targets by structural criteria can be incised when heterodimer approaches or exceeds substrate concentration, we focused on the Holliday junction substrates that have been the most controversial substrate profiled by all reported Mus81-Mms4/Eme1 preparations. S. cerevisiae endogenous heterodimer is nearly inactive on intact four-way oligonucleotide junctions (Figures 3A and B, 4C). This is the case whether the branch point core is fixed at the junction of heterologous arms (XO12) or is free to branch migrate over a span of 12 bp homology shared by the emanating duplex arms (X12). Even at heterodimer: substrate ratios of 2:1, conditions that allow nearly complete incision of a 5′-flapped substrate and splayed arm Y substrate, the Holliday junction structures are nearly untouched by Mus81-Mms4 nuclease. This is true for several preparations of Mus81-Mms4 isolated in our laboratory, ranging in concentration from ∼1 μM to 3 μM heterodimer. We tested whether Mg2+ or Mn2+ concentration influences Mus81-Mms4 capacity to incise intact Holliday junctions (Figure 4). Although the fixed-branch point structure XO12 shows incision up to 20 mM Mg2+, the percent product turnover is nearly negligible (<2% product after 30 min incubation at 30°C). The junction with the branch-migratable core is incised even more poorly, with a small peak in incision near 10 mM Mg2+ and Mn2+.
Figure 4.
Mus81-Mms4 incision on intact Holliday junctions is not enhanced by metal ion cofactor concentration. (A) Four-way junction incision as a function of Mg2+ and Mn2+ concentration. Reactions were performed with 50 nM substrate and 5 nM heterodimer, 30 min at 30°C. (B) Quantitation of assays in A. (C) Magnification of XO12 and X12 response to metal ion concentration.
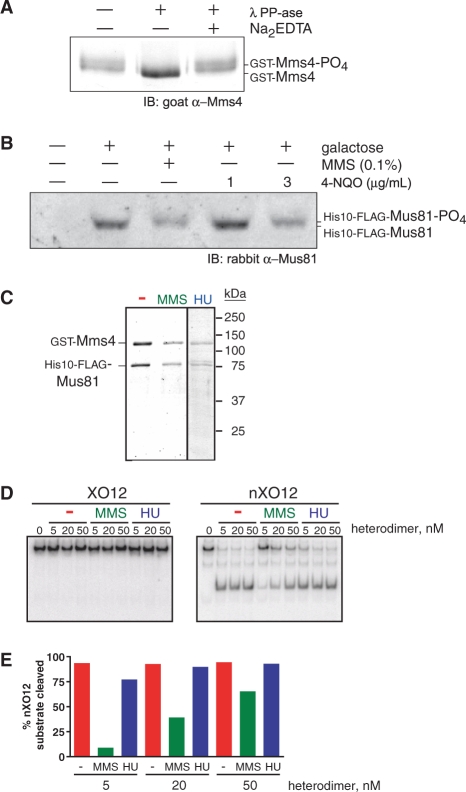
Purified endogenous Mus81-Mms4 is multiply phosphorylated in the absence and presence of genotoxic stress, but these modification states are not sufficient for Holliday junction incision competence in vitro
Because it had been speculated that eukaryotic post-translational modifications could account for the difference in Holliday junction incision activity observed between recombinant and eukaryotic preparations of Mus81-Mms4/Eme1, we examined the phosphorylation status of purified Mus81 and Mms4. Purified Mms4 can be collapsed to a species with faster relative electrophoretic mobility by treatment with phosphatase (Figure 5A). Mus81 does not show a detectable electrophoretic mobility shift under the polyacrylamide gel electrophoresis conditions described. The shift of Mms4 is unlikely due to phosphorylation of GST, as other GST-fusions (Rad54, Rad57) do not show electrophoretic mobility shifts under similar treatment (W.-D.H., data not shown). In addition, mass spectrometric analysis of the purified complex identified a number of constitutive and DNA damage-induced phosphorylated serine and threonine residues on each Mus81 and Mms4 subunit (unpublished data). Mus81 and Mms4 also show electrophoretic mobility shifts in vivo in response to genotoxins including the alkylating agent MMS, the chemical UV mimetic 4-nitroquinoline (4-NQO) and the ribonucleotide reductase inhibitor hydroxyurea (HU) (Figure 5B and C). The heterodimer purified in a modification state induced by genotoxic stress (Figure 5C) remains unable to incise Holliday junctions in vitro (Figure 5D), although the endonuclease is active (Figure 5D and E). It is unclear whether the reduced activity of the heterodimer isolated from MMS-challenged cells reflects a physiologically relevant modification of enzyme behavior; the heterodimer isolated from HU-challenged cells shows nuclease activity similar to the heterodimer isolated from cells in the absence of exogenous genotoxic stress (Figure 5D and E). The reduced specific activity of the preparation from MMS-challenged cells may be associated with chemical modification of the complex by alkylation. In sum, these observations indicate that eukaryotic post-translational modification profiles constitutive to the overexpressed heterodimer complex or modification profiles induced by global genotoxic stress do not confer Holliday junction incision capacity as an enzymatic property latent to the heterodimer. Global dephosphorylation of the complex by PP1 phosphatase treatment also does not alter substrate selection as tested for 3′-flapped and Holliday junction substrates (data not shown).
Figure 5.
Mus81-Mms4 phosphorylation does not contribute to a capacity to incise Holliday junctions in vitro. (A) Purified GST-Mms4 exhibits a phosphatase-sensitive electrophoretic mobility shift. (B) His10-FLAG-Mus81 exhibits a genotoxin-induced electrophoretic mobility shift. (C) Complex purified from cells subjected to genotoxic stress by 0.1% MMS treatment or by 100 mM HU. (D) Heterodimer purified in a modification state induced by genotoxic stress remains incompetent to incise intact Holliday junctions (XO12), although the complex is nucleolytically active (nXO12). The 50 nM substrate was incubated for 30 min in each reaction. (E) Quantitation of data in D.
Determination of KM and kcat on DNA joint molecules
DNA joint molecules exhibiting certain minimal structural features are bound and catalytically processed by Mus81-Mms4/Eme1 (Table 1). Some of these structural features have been defined, including foremost the presence of a hydroxyl or phosphate group at the 5′ deoxyribose position adjacent to a backbone discontinuity at a branch point (44). The discontinuity must occur at the junction of at least three DNA arms, two of which must be duplex DNA for optimal cleavage. We find that nevertheless, the splayed Y structure can be incised, although its initial binding parameter (KM) is highest of all tested substrates. This is consistent with the observation that duplex DNA 3′ to a structural branch point marked by a strand discontinuity is important for optimal enzyme binding to substrate.
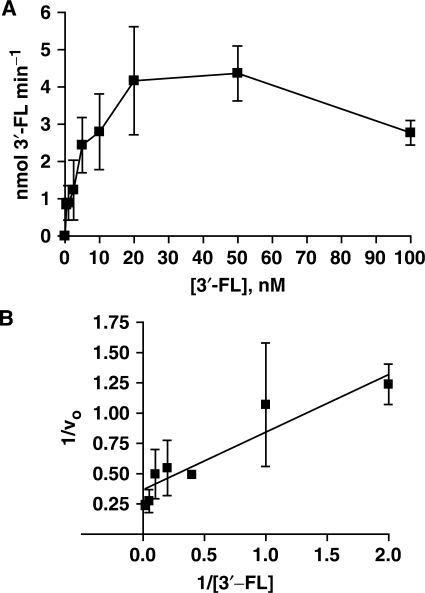
Figure 6 shows an example for a [substrate] versus velocity plot for the 3′-flapped substrate. For the 3′-flapped, RF-like, and pXO12 substrates, expression of the data points at concentrations up to 50 nM in an equation that describes a rectangular hyperbolic function using the KM and Vmax calculated by non-linear regression shows that the data points are appropriately modeled by a rectangular hyperbolic function. Enzyme action on these substrates is therefore accommodated by an Henri–Michaelis–Menten kinetic model. The initial velocity at increasing substrate concentration (100 nM) is reduced relative to the maximal velocity defined by the rectangular hyperbolic region of the [substrate] versus velocity plot, which indicates that at high concentration of these oligonucleotide joint molecules, the endonuclease exhibits substrate inhibition in vitro. On the nicked four-way junction and D-loop structure, substrate inhibition occurs at 10-fold lower substrate concentrations (maximal velocity at 5 nM) relative to that observed for the 3′-flapped, RF-like and pXO12 substrates. The substrate inhibition observed for Mus81-Mms4 may reflect the presence of at least two semi-independent binding sites for at least two discrete substrate regions. These binding sites may be shared between a single heterodimeric Mus81-Mms4 unit, or they may be shared between a homodimeric complex of two Mus81-Mms4 heterodimeric subunits. Analysis and possible implications of substrate inhibition on the mechanism of Mus81-Mms4 are the subject of further analysis to be reported in a subsequent manuscript.
Figure 6.
[Substrate] versus velocity plots for a catalytic preparation of Mus81-Mms4, optimized for pH, ionic strength and metal ion cofactor. (A) 3′-FL structure up to 100 nM substrate; (B) Lineweaver–Burk transformation of data in A.
DISCUSSION
We describe the isolation and characterization of a catalytically active preparation of S. cerevisiae Mus81-Mms4, purified to apparent homogeneity after overexpression in its native eukaryotic source. To our knowledge, we provide the first demonstration of catalytic turnover of DNA joint molecules by a native XPF-family endonuclease. We use this catalytic preparation to supply the first quantitative comparison of DNA joint molecule turnover by a native Mus81 preparation at apparent homogeneity, under conditions of limiting enzyme relative to substrate concentration. We first optimized assay conditions for buffer, pH, ionic strength and metal ion character. Because the anticipated in vivo concentration of a single substrate in a yeast nucleus is low (on the order of 0.5–1 nM), there is no a priori expectation as to whether the enzyme should function catalytically or whether stoichiometric behavior is sufficient (presumably, stoichiometric behavior would be explained by some manner of hysteresis, in which the enzyme adopts an inactive conformation after a catalytic cycle). We show that Mus81-Mms4 performs catalytic turnover on a number of DNA joint molecules that meet minimal structural criteria. We further show that catalysis is not directed to model oligonucleotide Holliday junctions by native Mus81-Mms4 in isolation.
Substrates that are catalytically processed by the enzyme share low KM (on the order of 1–7 nM) and correspondingly slow turnover (estimated on the order of 1–4 min for 3′-flapped, replication fork-like, nicked four-way, partial three-way and splayed Y junctions, and up to 10.8 min for the D-loop). Three substrate classes can be distinguished by their kinetic parameters KM and kcat. Class I substrates (nicked Holliday junction, 3′-flapped junction, and replication fork-like junction) are characterized by low KM (3–7 nM) and highest kcat (∼1 min−1) among the substrates examined. Class II substrates (partial Holliday junction and D-loop) also demonstrate low KM recognition by Mus81-Mms4 (1–6 nM), but lower kcat (≤0.3 min−1) relative to the Class I substrates. The splayed Y junction represents a Class III substrate that is poorly cleaved relative to Class I and II substrates, with both a higher relative KM (∼30 nM) and low kcat (0.26 min−1). Comparing Class I and II substrates to the Class III Y substrate reveals that duplex DNA flanking the branch point is key to a low KM parameter for Mus81-Mms4 substrates, although duplex DNA is not absolutely required 3′ to the branch point for turnover.
Despite the low KM for Class I and II substrates, the average KM is several-fold greater than the anticipated substrate concentration on which Mus81-Mms4 is expected to act in vivo. This may imply that additional factors are needed to target the enzyme to its joint molecule substrate. Mus81-Mms4 was first identified by a physical interaction with the Snf2-like ATPase Rad54, a DNA translocase that promotes Rad51-mediated DNA strand exchange, heteroduplex extension and Rad51 turnover from the DNA strand exchange product (6,45). This physical interaction may suggest a role for Rad54 in placement of Mus81-Mms4 at targeted substrates.
Our kinetic analysis of Mus81-Mms4 isolated from the cognate host demonstrates commonalities and interesting differences to a previous analysis of Mus81-Mms4 isolated from E. coli (25). While the overall substrate selectivity of both preparations appears similar, including exceedingly poor cleavage of intact HJs, the absolute kinetic parameters differ substantially for any given substrate. Moreover, the response to NaCl (improved activity up to 150 mM within pH 7–9 instead of inhibition), a more narrow magnesium optimum (3 mM) and discrimination between Mg2+ and Mn2+ as cofactors distinguish the native enzyme from the bacterial preparation. The reasons for these differences are unclear and may include the presence of different tags, post-translational modifications or differences in assay design and analysis.
Mus81-Mms4 incises a broad class of DNA joint molecules in vitro, but not intact four-way junctions
Coordinated Holliday junction incision in vitro remains an activity that is difficult to attribute with certainty solely to Mus81-Mms4/Eme1. In cases where human HeLa cell Mus81 immunoprecipitates and partially purified S. pombe preparations show activity on Holliday junctions, the duplex products are gapped and flapped (23,24). These products differ from the nicked duplex products associated with symmetric incision by RuvC, RusA, Hjc, Hje and bacteriophage junction-resolving enzymes T4 endonuclease VII and T7 endonuclease I. This suggests that the Holliday junction incision observed for some Mus81-Mms4/Eme1 preparations (i) may occur in a manner distinct from that of other classified four-way junction-specific enzymes, (ii) requires other factors to impose symmetric and coordinated cleavage or (iii) is an off-target outcome of independent incision events that occur at high enzyme: substrate ratios or in the presence of assay components that alter four-way junction presentation.
We show that the apparent selectivity of the enzyme toward substrates can be modulated by protein: substrate concentration ratios. 5′-flapped substrates have been poor substrates for nuclease incision and excluded as potential candidate in vivo targets. At protein: substrate concentration ratios greater than or equal to 1:1, however, even a 5′-flapped substrate can be incised by endogenous S. cerevisiae Mus81-Mms4 (Figure 2). This demonstrates that protein: substrate ratios are in fact critical determinants to apparent selectivity in vitro. The direct comparison of Mus81-Mms4 substrate selectivity under conditions of limiting enzyme is therefore needed to more completely describe Mus81 biochemistry.
Despite the capacity of endogenous Mus81-Mms4 to incise a non-canonical substrate such as the 5′-flap at high protein concentration, S. cerevisiae Mus81-Mms4 is not active on synthetic Holliday junctions, even at excess protein: substrate ratios (2:1). Less than 2% substrate turnover can be demonstrated on 50 nM substrate with 2-fold excess enzyme; this is true for any Mg2+ or Mn2+ concentration tested from 1 to 20 mM. Hence the X12 and XO12 structures are inaccessible for KM and kcat determinations. These observations suggest that S. cerevisiae Mus81-Mms4 does not resolve model Holliday junctions, and by extension to S. pombe and human Mus81-Eme1, may suggest that the heterodimer alone is not responsible for the model Holliday junction processing observed in partially purified eukaryotic preparations. Gaillard et al. (21) proposed that the inability of recombinant S. pombe and human Mus81-Eme1 preparations to cut model Holliday junctions could be explained by an inability of recombinant enzyme to make an initial nick in the junction core. In these studies, S. pombe Mus81-Eme1 has shown greater incision activity on X12 cruciform structures having 12 base-pair homologous cores that allow branch migration, as opposed to XO12 cruciform structures having a fixed branch point. X12 structures can undergo thermal breathing of the interior core with transient single-stranded DNA nature (46). These authors conclude that a coactivator in endogenous preparations may help Mus81-Eme1 open the Holliday junction core at the branch point, as a requirement for initial incision. Although our observations cannot exclude this possibility for S. cerevisiae Mus81-Mms4, our Mus81-Mms4 preparation shows least activity on X12 junctions, and minimal activity on XO12 junctions. This may be explained by an absence of the coactivator proposed by Gaillard et al., or it may alternatively be explained if the preferential incision of X12 substrates in partially purified endogenous preparations relates to a trace contaminating endonuclease that makes an initial nick at the transiently unpaired X12 core. Moreover, enzyme at high concentration excess as performed in these assays may have allowed incision of a partially unpaired (splayed) core, a behavior that is enhanced on the Y substrate in our assays with increasing heterodimer concentration.
Phosphorylation status of endogenous S. cerevisiae Mus81-Mms4 does not confer a latent Holliday junction incision activity to the endonuclease
Given that purified endogenous S. cerevisiae Mus81-Mms4 shows weak activity on model Holliday junctions, we explored alternative global phosphorylation profiles of the native complex. Mus81 complex overexpressed and purified from S. cerevisiae is multiply phosphorylated in a constitutive fashion on both subunits, but this apparently is not sufficient to confer a latent Holliday junction incision activity. Furthermore, enzyme overexpressed and purified from cells challenged by the genotoxin MMS exhibits a different phosphorylation profile, yet remains incompetent to incise Holliday junctions. The damage-modified complex, whether isolated from MMS- or HU-challenged cells, remains competent to incise other structures, however. Eukaryotic post-translational modifications in S. cerevisiae are therefore not sufficient to explain the Holliday junction incision behavior described for partially purified heterodimer preparations from S. pombe and human HeLa cells. If Holliday junction incision is a latent activity of S. cerevisiae Mus81-Mms4, it likely requires an additional factor that functions by a mechanism that remains to be described.
Most challenging to reconcile at this time is the difference in our preparation and the recombinant preparation recently reported by Gaskell et al. (20). These authors propose that recombinant preparations of S. pombe Mus81-Eme1 and S. cerevisiae Mus81-Mms4 can incise Holliday junctions when a purification protocol has selected for oligomeric states greater than the single heterodimer, and that quaternary structural differences account for disparities in Holliday junction incision competence observed to date. Their gel filtration protocol isolated fractions containing Mus81-Mms4/Eme1 with weak Holliday junction incision ability; in these peak fractions, however, strand dissociation of a fraction of the model junction population can also be detected and gel filtration fractions were not normalized for protein concentration. This report further suggests that magnesium ion concentration modulates the oligomeric state of the enzyme, to explain the magnesium-response profile for Holliday junction incision observed in their recombinant preparations. Whereas nicked model Holliday junctions were incised over a broad range of magnesium concentrations (0.5–20 mM), intact Holliday junctions were incised 3- to 4-fold more effectively at magnesium concentrations below 5 mM. Modulation of substrate selectivity by metal ion concentration is without precedent, and a mechanism for oligomerization in response to metal ion concentration remains to be explained. Rather than reflecting different magnesium optima for cleavage of different substrates by Mus81-Mms4/Eme1, the enhanced Holliday junction incision at low Mg2+ concentration may alternatively be interpreted as modulation of a trace activity that responds to metal ion concentration and may alter the substrate presentation to Mus81-Mms4/Eme1. Our results indicate that magnesium or manganese titration does not alter S. cerevisiae Mus81-Mms4 activity on model Holliday junctions. Furthermore, our operative conditions for S. cerevisiae Mus81-Mms4 biochemical assays are not consistent with a requirement for isolation above threshold protein concentrations. Our nuclease assays require dilution of the enzyme 200- to 500-fold for meaningful rate determinations on excess substrate, suggesting that enzyme concentration is not a primary limiting factor for catalytic activity and therefore not likely to bear on active oligomeric state.
The DNA structure selectivity of Mus81-Mms4 is likely to be circumscribed by protein–protein interactions specific to the context in which the joint molecule is generated
Functional specialization of nucleases to targeted substrates has probably been permitted by virtue of an inherent plasticity in substrate selection. This plasticity explains Mus81-Mms4/Eme1 selectivity in vitro. Substrate specificity has been accomplished in vivo by specialization of paralogs in the context of pathways in which their DNA structural targets are generated. A component of the ‘specificity’ of enzymes in vivo stems from their placement relative to potential substrates, and proximity may therefore become central to understanding how the apparent substrate selectivity of Mus81-Mms4/Eme1 in vitro is proscribed to a target or subset of targets in a context-specific manner in vivo.
Consistent with this restriction of substrate specificity in vivo, genetic studies indicate that Mus81-Mms4 has no role in 3′-flap cleavage during single-stranded DNA annealing (SSA) in budding yeast, even though the 3′-flapped structure is a biochemical target in vitro (4,47). Instead, the evolutionarily related endonuclease Rad1-Rad10 is assigned to this pathway, and Mus81-Mms4 is not recruited to the 3′-flap intermediates inherent to this Rad52-promoted context and cannot substitute in the absence of Rad1-Rad10. These observations underscore the probable role of protein–protein interactions in the sanction of substrate specificity by evolutionarily evolved associations within pathways. Substrate assignment to Mus81-Mms4/Eme1 in vivo is most likely to be understood in context of the interacting proteins that generate the DNA substrates the endonuclease targets.
ACKNOWLEDGEMENTS
Thanks are extended to Shamsher Samra, who generated the mus81-dd mutant during a UC Davis Young Scholars Program internship, and to Doug Huseby, who purified the mutant heterodimer. Thanks to Rita Alexeeva for contributions to the expression of heterodimer under HU stress. Michael Rolfsmeier cloned the His10 fragment into plasmid pWDH403 to generate pWDH619. The comments of members of the Heyer laboratory are appreciated, especially those of Xuan Li, Shannon Ceballos, Clare Fasching, Kristina Herzberg, Ryan Janke, Jie Liu, Erin Schwartz, William Wright and Xiao-Ping Zhang. The comments of Neil Hunter, Stephen Kowalczykowski and Jode Plank are also appreciated. We thank Andrei Alexeev and Konstantin Kiianitsa for helpful suggestions during development of the nuclease assay protocol. We thank the Stephen Kowalczykowski, Carol Erickson and John Scholey laboratories for sharing instruments, and Steven Brill for plasmid pGAL-MUS81. K.T.E. specially thanks Toby Ehmsen. An NIH Molecular & Cellular Biology training grant 5 T32 GM007377 partly supported K.T.E. This work was supported by NIH GM58015 to W.-D.H. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM58015.
Conflict of interest statement. None declared.
REFERENCES
- 1.Whitby MC. Making crossovers during meiosis. Biochem. Soc. Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 2.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haber JE, Heyer WD. The fuss about Mus81. Cell. 2001;107:551–554. doi: 10.1016/s0092-8674(01)00593-1. [DOI] [PubMed] [Google Scholar]
- 4.Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem. Sci. 2003;28:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Interthal H, Heyer WD. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- 7.Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 9.Dendouga N, Gao H, Moechars D, Janicot M, Vialard J, McGowan CH. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell Biol. 2005;25:7569–7579. doi: 10.1128/MCB.25.17.7569-7579.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Essers J, Hanada K, Chahwan R, Khaw AK, et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitby MC. Junctions on the road to cancer. Nat. Struct. Mol. Biol. 2004;11:693–695. doi: 10.1038/nsmb0804-693. [DOI] [PubMed] [Google Scholar]
- 12.Osman F, Whitby MC. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair. 2007;6:1004–1117. doi: 10.1016/j.dnarep.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, West SC. Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 2004;5:937–944. doi: 10.1038/nrm1502. [DOI] [PubMed] [Google Scholar]
- 15.Haber JE, Ira G, Malkova A, Sugawara N. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:79–86. doi: 10.1098/rstb.2003.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 17.Holliday R. A mechanism for gene conversion in fungi. Genet.Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 18.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 19.Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaskell LJ, Osman F, Gilbert RJ, Whitby MC. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 22.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 24.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 25.Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair. 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 27.Whitby MC, Osman F, Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 28.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blais V, Gao H, Elwell CA, Boddy MN, Gaillard PH, Russell P, McGowan CH. RNA interference inhibition of Mus81 reduces mitotic recombination in human cells. Mol. Biol. Cell. 2004;15:552–562. doi: 10.1091/mbc.E03-08-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogrunc M, Sancar A. Identification and characterization of human MUS81-MMS4 structure-specific endonuclease. J. Biol. Chem. 2003;278:21715–21720. doi: 10.1074/jbc.M302484200. [DOI] [PubMed] [Google Scholar]
- 32.Ciccia A, Constantinou A, West SC. Identification and characterization of the human Mus81-Eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 33.Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: John Wiley & Sons, Inc.; 1975. [Google Scholar]
- 34.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 35.Solinger JA, Lutz G, Sugiyama T, Kowalczykowski SC, Heyer WD. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
- 36.Bashkirov VI, Bashkirova EV, Haghnazari E, Heyer WD. Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol. Cell Biol. 2003;23:1441–1452. doi: 10.1128/MCB.23.4.1441-1452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 38.Liu R, Qiu J, Finger LD, Zheng L, Shen B. The DNA-protein interaction modes of FEN-1 with gap substrates and their implication in preventing duplication mutations. Nucleic Acids Res. 2006;34:1772–1784. doi: 10.1093/nar/gkl106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsunaga T, Park CH, Bessho T, Mu D, Sancar A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J. Biol. Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- 40.de Laat WL, Appeldoorn E, Jaspers NG, Hoeijmakers JH. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- 41.Gaillard PH, Wood RD. Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 2001;29:872–879. doi: 10.1093/nar/29.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunderdale HJ, Benson FE, Parsons CA, Sharples GJ, Lloyd RG, West SC. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- 43.Bennett RJ, West SC. RuvC protein resolves Holliday junctions via cleavage of the continuous (noncrossover) strands. Proc. Natl Acad. Sci. USA. 1995;92:5635–5639. doi: 10.1073/pnas.92.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West SC. Holliday junctions cleaved by Rad1? Nature. 1995;373:27–28. doi: 10.1038/373027a0. [DOI] [PubMed] [Google Scholar]
- 47.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]