Abstract
The interaction between human immunodeficiency virus type 1 (HIV-1) and RNA silencing pathways is complex and multifaceted. Essential for efficient viral transcription and supporting Tat-mediated transactivation of viral gene expression, the trans-activation responsive (TAR) element is a structured RNA located at the 5′ end of all transcripts derived from HIV-1. Here, we report that this element is a source of microRNAs (miRNAs) in cultured HIV-1-infected cell lines and in HIV-1-infected human CD4+ T lymphocytes. Using primer extension and ribonuclease (RNase) protection assays, we delineated both strands of the TAR miRNA duplex deriving from a model HIV-1 transcript, namely miR-TAR-5p and miR-TAR-3p. In vitro RNase assays indicate that the lack of a free 3′ extremity at the base of TAR may contribute to its low processing reactivity in vivo. Both miR-TAR-5p and miR-TAR-3p down-regulated TAR miRNA sensor activity in a process that required an integral miRNA-guided RNA silencing machinery. miR-TAR-3p exerted superior gene downregulatory effects, probably due to its preferential release from HIV-1 TAR RNA by the RNase III Dicer. Our study suggests that the TAR element of HIV-1 transcripts releases functionally competent miRNAs upon asymmetrical processing by Dicer, thereby providing novel insights into viral miRNA biogenesis.
INTRODUCTION
MicroRNAs (miRNAs) are short 21- to 24-nucleotide (nt) endogenous RNA species recognized as key regulators of gene expression that act through the miRNA-guided RNA silencing pathway (for comprehensive reviews, see 1–4). Expressed in almost all eukaryotes examined to date, miRNAs are generated upon processing of miRNA precursors (pre-miRNAs) by the RNase III Dicer (5,6).
Initial evidences for a role of RNA silencing in host defense mechanisms against viruses came from studies in plants. Studying a mechanism known as posttranscriptional gene silencing (PTGS), Hamilton and Baulcombe (7) detected the presence of viral RNA of ∼25-nt in virus-infected plants. These small RNAs were later found to originate from viral double-stranded RNA (dsRNA) processing by DICER-LIKE 1 in Arabidopsis (8). Recent reports suggest that it may play a similar role in human. Indeed, several studies have shown that viral RNAs, such as that of Epstein-Barr virus (EBV) (9), mouse gammaherpesvirus 68, human cytomegalovirus (10), Kaposi's sarcoma-associated herpesvirus (KSHV or HHV8) (10,11) and simian virus 40 (SV40) (12) harbor secondary stem-loop structures that represent a source of miRNAs.
Concerning human immunodeficiency virus type 1 (HIV-1), its interaction with the RNA silencing pathway is complex and multifaceted. Synthetic small interfering RNAs (siRNAs) have been designed against HIV-1 and used successfully to induce sequence-specific degradation of HIV-1 mRNAs and to inhibit viral replication (for a recent review, see 13). This observation implies a certain degree of accessibility of HIV-1 RNA sequences to the RNA silencing machinery in vivo. Two independent observations tend to support that notion: (i) a cluster of cellular miRNAs have been shown to target the 3′ ends of HIV-1 mRNAs and to inhibit virus production in resting CD4+ T cells; (ii) Drosha and Dicer may contribute to inhibit HIV-1 replication (14), suggesting that small RNAs may be released from structured HIV-1 RNAs. Omoto and colleagues (15) identified a miRNA residing in the nef gene (miR-N367) by northern blot, whereas an siRNA embedded in the env gene region (16) was also described.
In this study, we report that HIV-1 trans-activation responsive (TAR) element, a 59-nt stem-bulge-loop RNA structure located at the 5′ end of all HIV-1 mRNA transcripts, represents a source of regulatory miRNAs that are functionally competent in RNA silencing processes.
MATERIALS AND METHODS
DNA constructs
The psiRluc (Renilla luciferase) (psiSTRIKE, Promega), psiNeg vectors and Rluc reporter construct (psiCHECK, Promega), and their use, have been described previously (17). psiTAR was constructed by cloning in psiSTRIKE the HIV-1 TAR sequence. pXP2-LTR-Firefly luciferase (Fluc) and pXP2-LTRΔTAR-Fluc constructs were obtained by cloning either HIV-1 LTR or LTRΔTAR of HXB2 (kindly provided by N.R. Landau, The Salk Institute for Biological Studies, La Jolla, CA, USA) into the BamHI site of pXP2.
The TAR miRNA sensor constructs were prepared by cloning a sequence complementary to either TAR nt 1–35 (left arm) or TAR nt 30–59 (right arm) into the SalI and NotI sites of psiCHECK. pTER+ constructs were prepared by cloning a sequence encoding either a shRNA directed against Dicer mRNA, or a deleted region in Rluc mRNA (used as a negative control), into the pTER plasmid (kindly provided by M. van de Wetering, Hubrecht Laboratory, Utrecht, The Netherlands) (18), as published previously (19).
Cell culture and luciferase activity assays
For TAR miRNA experiments, HEK293 cells were cotransfected with psiTAR or pXP2-LTR-Fluc, the miR-TAR-5p or miR-TAR-3p constructs and pLacZ using calcium phosphate. Mouse embryonic Fmr1 knockout (KO) (STEK TSV-40) (17) and wild-type (WT) (Naïves) (17) fibroblasts were cotransfected with psiTAR and the miR-TAR-5p or miR-TAR-3p sensor constructs using calcium phosphate. Cells were harvested 24–48 h later, and luciferase activities were measured, as described previously (17). β-Gal activity was measured with Galacto-Light Beta-Galactosidase Reporter Gene Assay System (Applied Biosystems).
Cultured 293T-REx + pcDNA6TR cells transfected with either pTER + shDicer or pTER + shNeg were placed under dual blasticidin (10 μg/ml) (InvivoGen) and zeocin (50 μg/ml) (Invitrogen) selection, and clonal cell lines were isolated. Stably transformed cell lines were tested for Dicer expression upon induction with 1 μg/ml of doxycyclin (Dox) (Sigma). Selected cell lines shDICER-6, shDICER-27 and shNEG-13 were pretreated for 12 h with Dox (1 μg/ml) and transfected with plasmids coexpressing the TAR RNA (psiTAR)/miR-TAR-3p sensor or shRluc/Rluc sensor combinations. Cells were harvested 48 h later and luciferase activities were measured, as described previously (17).
H9 cells WT and chronically infected with HIV-1 strain HTLV-IIIMN NIH 1984 (MN cell line) (20) or HTLV-IIIRF NIH 1983 (RF cell line) (21) were obtained from Dr Robert Gallo through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. For TAR miRNA experiments, 5 × 105 cells were transfected with 5 ng of miR-TAR-5p, miR-TAR-3p or an unspecific sensor plasmid, which served as a normalization control, using Lipofectamine 2000 (Invitrogen). Cells were harvested 48 h later and luciferase activities were measured, as described previously (17).
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) analyses were performed as described previously (5,17). The recombinant human Dicer preparation used in the present study was expressed in the Sf9 insect cells used in our baculovirus expression system and purified as described by Provost et al. (5), and was found not to contain Drosophila Dicer-interacting proteins, such as loquacious (22,23) or R2D2 (24). Briefly, WT TAR RNA was transcribed and randomly labeled (α-32P UTP, Perkin Elmer) by in vitro transcription using T7 promoter/polymerase (MEGAshort Script kit, Ambion), purified by denaturing PAGE and incubated in the absence or presence of recombinant human Dicer (5), with or without Tat (obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) (25), for 30 min on ice prior to EMSA. Dicer·TAR RNA complex formation was analyzed by nondenaturing PAGE and autoradiography.
In vitro Dicer RNase assays
32P-labeled WT TAR RNA and various TAR mutants (TAR no loop, TAR G/U loop, TAR 10-nt 3′ overhang, TAR bulge other strand, TAR U5 and TAR U3) were prepared as described in the above section, and incubated in the absence or presence of recombinant human Dicer (5) and/or Tat, in a reaction buffer containing 20 mM Tris·HCl pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM ATP, 5% Superase·In (Ambion), at 37°C for 1 h. The resulting RNA products were analyzed by denaturing PAGE and autoradiography, as described previously (5,17).
Production of virus stocks and infection
Virus particles were produced by calcium phosphate transfection of 293T cells with the virus-encoding vector pNL4-3 (obtained from Dr Malcolm Martin through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) (26). Pseudotyped HIV-1 particles were generated by cotransfection of 293T cells with pNL4-3 and an expression vector coding for the VSV-G full-length envelope protein (27). Virus stocks were normalized for virion content using an in-house sensitive double antibody sandwich ELISA specific for the major core viral p24 protein (28).
Peripheral blood mononuclear cells (PBMCs) were obtained from healthy donors and purified by Ficoll-Hypaque centrifugation. Human T helper cells (i.e. CD4+) were negatively isolated from fresh PBMCs using the CD4+ T cells negative purification kit according to manufacturer's instructions (Miltenyi Biotec Inc., Auburn, CA). CD4+ T cells were activated with PHA-L (1 µg/ml) and IL-2 (30 U/ml) for 48 h and then maintained in RPMI containing 10% fetal calf serum (FCS) (Hyclone Laboratories) and supplemented with penicillin, streptomycin and IL-2 (30 U/ml). Activated cells were infected with a fixed amount of virus (10 ng of p24 per 105 cells) for 4 h, washed, and then maintained in complete RPMI medium. Cells were harvested at 48 h post-infection.
Northern blot analyses
Small RNA (<200 nt) fractions were isolated by using the mirVana miRNA isolation kit (Ambion) from primary human CD4+ lymphocyte T cells infected with mock or VSV-G-pseudotyped NL4-3 viruses and analyzed by northern blotting using 32P-labeled DNA probes recognizing TAR nt 40–59, mature hsa-miR-16-1 and 5S RNA, as described previously (5).
RNase protection assays
Small RNA fractions were isolated from (i) H9 cells WT or chronically infected with HIV-1 strain HTLV-IIIMN or HTLV-IIIRF, or (ii) HEK293 cells transiently transfected using calcium phosphate with pXP2-LTR-Fluc, pXP2-LTRΔTAR-Fluc, psiTAR or psiNeg vectors and harvested 48 h later.
RNase protection assays (RPA) were carried out by using RNA probes complementary to nt 33/64, 40/64, −5/32 and 3/32 of HIV-1 TAR. The RNA probes were synthesized from DNA templates by in vitro transcription using the T7 RNA polymerase (Ambion) and radiolabeled by random incorporation of α-32P UTP (Perkin Elmer), and purified by denaturing PAGE. The radiolabeled probe was hybridized with 1.5 µg of small RNA for 16 h at 57°C. Hybridized RNAs were treated with RNases A/T1 (Ambion) for 30 min at 37°C, heat inactivated and ethanol precipitated. Protected RNAs were resuspended into gel loading buffer II, separated by denaturing PAGE (15%) and visualized by autoradiography.
Primer extension experiments
Small RNA fractions were isolated from HEK293 cells transiently transfected using calcium phosphate with pXP2-LTR-Fluc, pXP2-LTRΔTAR-Fluc, psiTAR or psiNeg vectors, and harvested 48 h later. Briefly, 10 µg of RNA were hybridized with 5′ radiolabeled DNA oligonucleotide complementary to nt 9/23 and 44/61 of TAR RNA for 15 min. After addition of the primer extension mix [20 mM Tris–Cl, 10 mM MgCl2, 1.6 mM dNTP, 50 µg/ml Actinomycin D, 10 mM DTT, 2 U/µl SuperScript II Reverse Transcriptase (Invitrogen), 0.1 U/µl Superase·in (Ambion), pH 8.0], primers were extended for 1 h at 42°C. The reaction was stop by phenol/chloroform extraction and ethanol precipitation. Pellets were resuspended in gel loading buffer II (Ambion) and separated by denaturing PAGE (15%). The extended primers were visualized by autoradiography.
RESULTS
HIV-1 TAR RNA is processed by Dicer in vitro
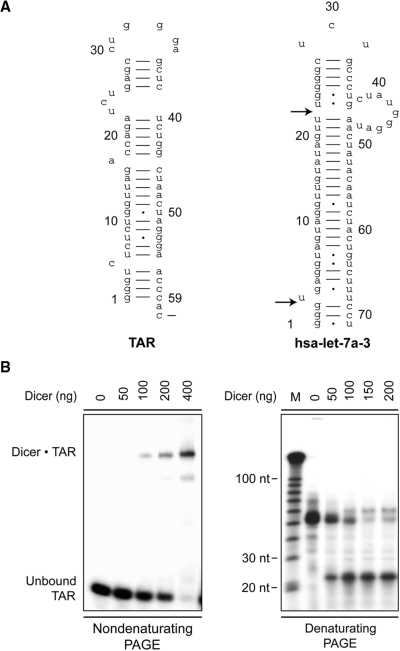
Exploring specific aspects of the interaction between HIV-1 and the host miRNA-guided RNA silencing pathway, we initially asked whether HIV-1 RNA would be susceptible to Dicer cleavage. Essential for efficient viral transcription, HIV-1 TAR element shows important structural similarities with the hsa-let-7a-3 pre-miRNA (Figure 1A), which can be processed into miRNA by recombinant human Dicer in vitro (5). When incubated with 32P-labeled TAR RNA, Dicer was able to bind (see Figure 1B, left panel) and cleave (see Figure 1B, right panel) it into ∼23- to 24-nt miRNA products, making of HIV-1 TAR a potential Dicer substrate.
Figure 1.
HIV-1 TAR RNA is bound and cleaved by recombinant Dicer into ∼23–24-nt miRNA products in vitro. (A) Structural similarities between HIV-1 TAR RNA and the hsa-let-7a-3 pre-miRNA. The arrows indicate the Dicer cleavage sites. (B) Left, EMSA. 32P-labeled TAR RNA was incubated in the absence or presence of Dicer, and the formation of Dicer·TAR complexes was analyzed by nondenaturing PAGE and autoradiography. (B) Right, Dicer RNase assays. 32P-labeled TAR RNA was incubated in the absence or presence of Dicer with MgCl2. The samples were analyzed by denaturing PAGE and autoradiography. M, indicates a 10-nt RNA size marker.
HIV-1 TAR element is the target of the viral transactivating Tat protein, which is expressed during the course of HIV-1 infection and known to act at the RNA level to enhance viral gene expression by >100-fold (29). We thus examined whether Tat could shield TAR RNA and prevent its processing by Dicer. To this end, we determined the experimental conditions under which all TAR RNA is bound by Tat. However, the TAR RNA precomplexed with saturating amounts of Tat remained susceptible to Dicer cleavage (I. Plante and P. Provost, unpublished data).
HIV-1 TAR element is a source of miRNAs in vivo
These in vitro findings were to be interpreted with caution, since the contribution of other proteins and cellular components could not be taken into account. In order to circumvent these issues and to confirm that HIV-1 TAR is a source of miRNAs under more physiological conditions in vivo, we made use of mock- or NL4-3-infected primary human CD4+ T lymphocytes and investigated their small RNA content by northern blot. Previously shown to be expressed in T lymphocytes (30), we were able to detect the mature form of miR-16-1 (see Figure 2A, middle panel, right lane) as well as that of its pre-miR-16-1 and pri-miR-16-1 precursors (I. Plante and P. Provost, unpublished data). Noticeably, the level of miR-16-1 remained unchanged in NL4-3- versus mock-infected primary human CD4+ T lymphocytes (see Figure 2A, middle panel, left lane versus right lane). This is in contrast with a previous study by Bennasser et al. (31), who have reported specific down-regulation of miR-16 levels in HeLa cells overexpressing HIV-1 TAR RNA, a discrepancy that may be accounted for by the difference in the experimental settings (i.e. primary human cells versus an established cell line).
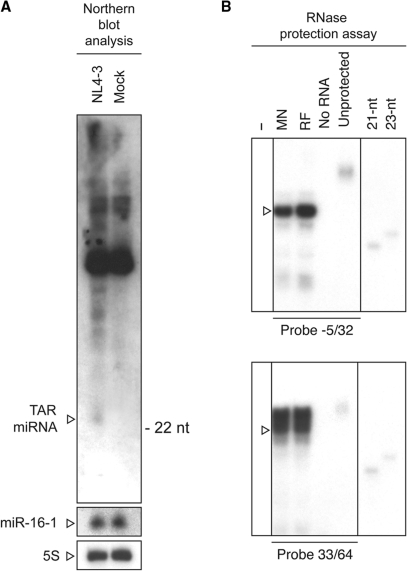
Figure 2.
Detection of miRNAs released from TAR in HIV-1-infected cells. (A) Northern blot analysis of small RNAs (<200 nt) isolated from primary human CD4+ T lymphocytes infected for 48 h with VSV-G-pseudotyped HIV-1 (NL4-3 clone) or mock. A 5′ end labeled probe-recognizing nt 40–59 of HIV-1 TAR (upper panel) was hybridized and detected by autoradiography. The membrane was subsequently probed for the presence of the 22-nt miR-16-1 (middle panel) and of 5S RNA (lower panel). (B) RPA. Small RNAs (<200 nt) isolated from H9 cells wild-type (−) or chronically infected with HTLV-IIIMN NIH 1984 (MN) or HTLV-IIIRF NIH 1983 (RF) HIV-1 strains were analyzed by RPA. Protected RNAs were separated by denaturing PAGE (15%) and visualized by autoradiography. Open arrowheads indicate RNA species expected to be protected by probe −5/32 (upper panel) and 33/64 (lower panel), respectively.
Experiments designed to identify a miRNA derived from HIV-1 TAR RNA in vivo allowed the detection of an RNA product slightly longer than the 22-nt miR-16-1 in NL4-3-, but not in mock-infected, primary human CD4+ T lymphocytes (see Figure 2A, upper panel). Additional bands specific to NL4-3-infected cells that may correspond to cleavage intermediates of TAR were also observed. These results opened the possibility that HIV-1 TAR RNA may represent a source of miRNAs in vivo.
In order to confirm these findings, we used a different experimental approach and analyzed the small RNAs population of two HIV-1-infected cell lines by RPA. When assessing the presence of a miRNA deriving from the left arm of TAR with probe −5/32, a protected RNA species of 27 nt was detected (see Figure 2B, upper panel). Whereas the use of probe 33/64 allowed the detection of ∼28–31 nt RNA species specific to HIV-1-infected cells (see Figure 2B, lower panel). These results suggest that miRNAs may originate from both arms of HIV-1 TAR RNA in vivo.
Delineation of the miRNAs released from HIV-1 TAR
To get a better definition of these viral miRNAs, we performed primer extension in combination with RPA. Using these approaches, we analyzed small RNA preparations derived from HEK293 cells expressing a reporter gene placed under the control of either the HIV-1 5′ LTR WT (LTR-Luc) or lacking the TAR element (LTRΔTAR-Luc).
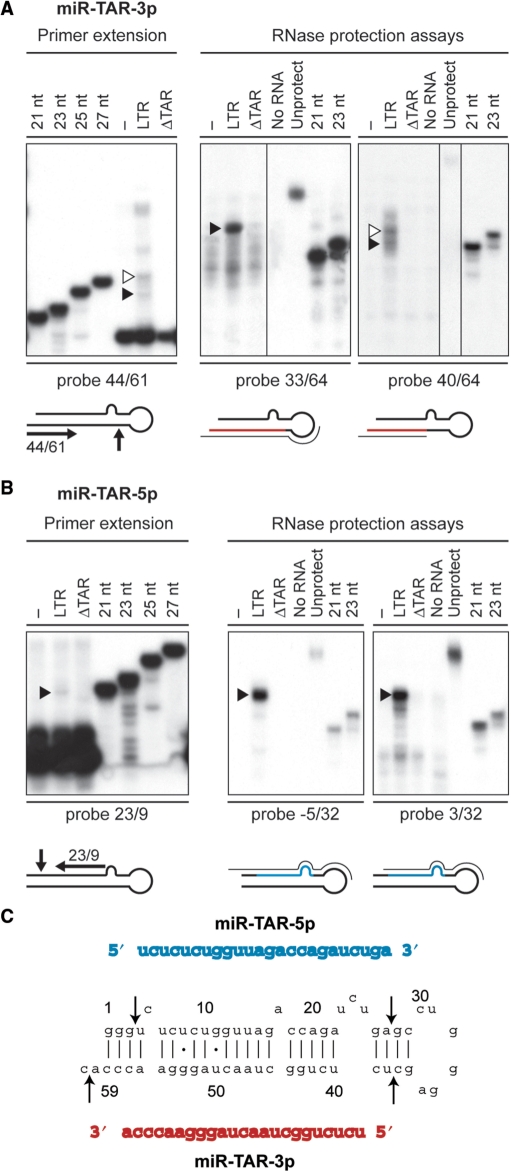
Reactions of primer extension performed with an oligonucleotide complementary to nt 61–44 (probe 44/61) yielded a 24-nt TAR-specific band (see Figure 3A, left panel, filled arrowhead). Elongation of probe 44/61 by 6 nt indicates that nt 38 may represent the 5′ end of the miRNA. Considering the premise that nt 58 of TAR is part of the miRNA, this observation is supported by (i) the 26-nt RNA species obtained by using probe 33/64 (see Figure 3A, center panel) and (ii) a 22-nt band obtained by using probe 29/58 (I. Plante, D.L. Ouellet and P. Provost, unpublished data). A 5′ end formed by nt 34, as suggested by the 28-nt RNA band obtained upon extension of probe 44/61 (see Figure 3A, left panel, open arrowhead), is less likely, since it is not supported by concurrent RPA analysis.
Figure 3.
Delineation of the miRNA duplex derived from the HIV-1 TAR element. (A and B) Small RNAs (<200 nt) isolated from HEK293 cells, transfected with or without pXP2-LTR-Fluc or pXP2-LTRΔTAR-Fluc, were analyzed by primer extension and RPA. (A) miR-TAR-3p. Left, Primer extension analysis using a deoxyribonucleotide complementary to TAR nt 61–44, and run in parallel with DNA size markers. The extended products were analyzed by denaturing PAGE. Right, RPA using RNA probes directed against HIV-1 TAR nt 33/64 and nt 40/64, and run in parallel with RNA size markers. Protected small RNAs were analyzed by denaturing PAGE. Open arrowheads indicate RNA species not supported by concurrent RPA analysis, whereas filled arrowheads indicate the RNA bands considered to establish the consensus TAR miRNA duplex. (B) miR-TAR-5p. Left, Primer extension analysis using a deoxyribonucleotide complementary to nt 23–9. Right, RPA using RNA probes directed against HIV-1 TAR nt −5/32 and nt 3/32. Filled arrowheads indicate the RNA bands considered to establish the consensus TAR miRNA duplex. (C) The consensus miR-TAR-5p:miR-TAR-3p duplex derived from HIV-1 TAR. The arrows indicate the cleavages sites deduced from primer extension and RPA results.
Experimental support for nt 60 as the 3′ end of the miRNA originating from the right arm of TAR comes from (i) a 21-nt RNA obtained by the use of probe 40/64, assuming that nt 40 is part of the miRNA (see Figure 3A, right panel, filled arrowhead), and (ii) an 18-nt fragment observed when using probe 43/71, assuming that nt 43 is part of the miRNA (I. Plante, D.L. Ouellet and P. Provost, unpublished data). Together, the results of our analyses are consistent with a 23-nt RNA being derived from nt 38–60 of the right arm of TAR.
miRNAs are known to be contained within miRNA duplexes that result from Dicer processing of miRNA precursor molecules. Analysis of primer extension reactions performed with an oligonucleotide complementary to nt 23–9 revealed a single band of 20 nt specific to TAR (see Figure 3B, left panel), pointing towards nt 4 as the 5′ end nt. RPA analyses using two different probes covering nt −5 to 32 and nt 3–32 of TAR both revealed protected RNAs as a major band of 25 nt (see Figure 3B, center and right panels). Together, these results are in agreement with the generation of a 24-nt RNA originating from the left arm of TAR and spanning nt 4–27.
The consensus miRNA duplex sequence derived from the HIV-1 TAR element, as deduced from our analyses, is shown in Figure 3C. In compliance with current convention, the miRNA derived from the left arm of the TAR stem has been named miR-TAR-5p, whereas that originating from the right arm of TAR has been defined as miR-TAR-3p.
Structural determinants of HIV-1 TAR RNA processing by Dicer
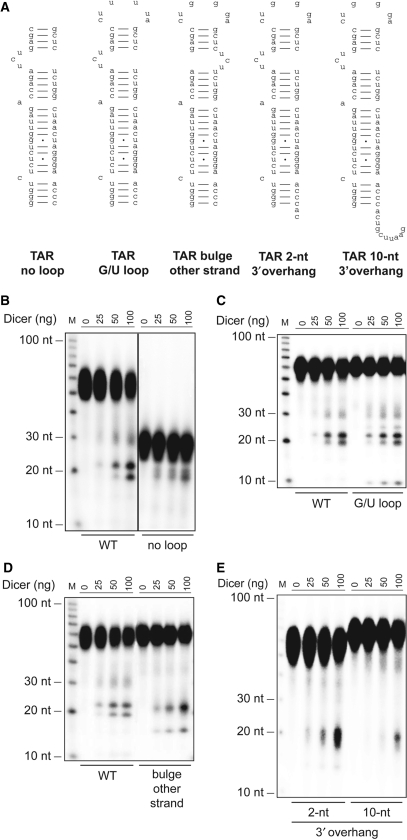
We then wished to identify the structural determinants of TAR RNA processing in Dicer RNase assays in vitro. Stem-bulge-loop RNAs are processed by members of the RNase III family of enzymes, such as Rnt1p (32) and Pac1 (33), through a mode of recognition involving their loop and bulge. To evaluate the importance of the loop, we prepared a TAR RNA substrate lacking the loop sequence (Figure 4A). The resulting TAR RNA duplex was cleaved by Dicer in vitro (Figure 4B), suggesting that the presence of the loop is not a prerequisite for HIV-1 TAR processing.
Figure 4.
Structural determinants of HIV-1 TAR RNA processing by Dicer. (A) Structure of the HIV-1 TAR RNA variants. The structure of wild-type TAR RNA is shown in Figure 1A. (B–E) 32P-labeled TAR RNAs with (wild-type, WT) or (B) without the loop (no loop), (C) with a G/U loop, (D) with the bulge transposed to the right arm (bulge other strand) or (E) with a 2-nt (2-nt 3′ overhang) or 10-nt (10-nt 3′ overhang) extension at the 3′ end were incubated in the absence or presence of recombinant human Dicer with MgCl2. The cleavage products were analyzed by denaturing PAGE and autoradiography.
The RNase III Rnt1p exhibit a certain degree of specificity for RNA substrates bearing AGNN tetraloop, which led us to investigate whether this property is also displayed by the RNase III Dicer. When incubated with Dicer, a G/U loop TAR RNA mutant was cleaved as efficiently as the wild-type sequence (Figure 4C). A difference, however, was the appearance of a ∼10-nt RNA species that may originate from the 32P-labeled, U-rich loop element of the G/U loop mutant (see Figure 4C, last four lanes). These data indicate that TAR RNA processing by Dicer depends neither on the presence nor the sequence of its loop.
We next addressed the contribution of the bulge, a structural element commonly present near the loop of pre-miRNAs, by preparing a substrate in which the 3-nt bulge was transferred to the same position on the other strand, i.e. from the left arm to the right arm of TAR. Analysis of the Dicer RNase reactions by denaturing PAGE revealed a difference in the pattern of small RNAs generated (Figure 4D). This observation suggests that the positioning of the bulge on either arm of the TAR stem may be critical in determining the sequence of the miRNAs released by Dicer.
We then questioned the nature and contribution of the base of the TAR stem. HIV-1 TAR RNA differs from pre-miRNA substrates, since its 3′ extremity is not available, rather extending into the 5′ LTR of HIV-1 mRNAs. Therefore, we mimicked the unavailable 3′ extremity of the HIV-1 TAR RNA by prolonging the 2-nt 3′ overhang to 10 nt. This modification markedly decreased the susceptibility of TAR to cleavage by Dicer in vitro (Figure 4E). The predicted structure of the TAR substrate, as analyzed by MFOLD (34), was preserved (P. Landry and P. Provost, unpublished data), excluding the possibility that global structure rearrangements influenced our data. These results suggest that the extended 3′ extremity of TAR, such as that encountered in the HIV-1 5′ LTR, may hamper its processing by Dicer.
miR-TAR-5p and miR-TAR-3p are functional in gene regulation in vivo
We then asked if the miRNAs derived from TAR are functional and can exert gene regulatory effects in human cells. This aspect was initially addressed by designing a TAR expression construct (psiTAR) based on the psiSTRIKE vector backbone. In preliminary experiments, the RPA patterns of the miRNAs derived from psiTAR, obtained by using probes −5/32 and 33/64, were similar to those obtained upon LTR-Fluc (D.L. Ouellet, I. Plante and P. Provost, unpublished data). These data validated psiTAR as a TAR miRNA expression vector suitable for the assessment of miR-TAR-5p and miR-TAR-3p function in cultured human cells.
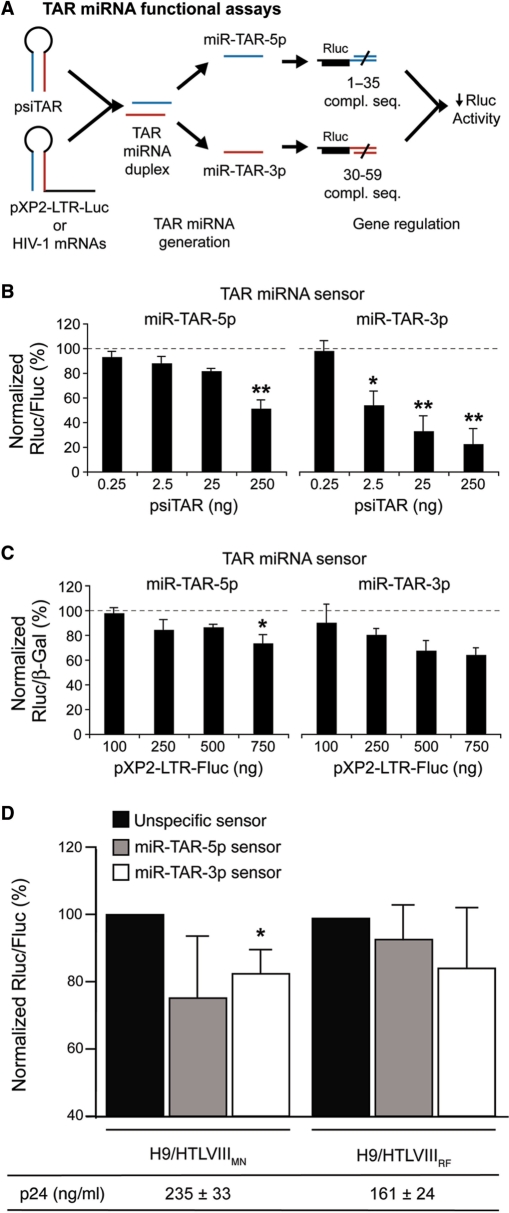
The mammalian expression vectors psiTAR and pXP2-LTR-Fluc were then used in TAR miRNA functional assays established in HEK293 cells, as depicted in Figure 5A. In these assays, HIV-1 TAR was expressed either in the form of an isolated miRNA precursor (via psiTAR) or embedded within its natural 5′ LTR context in a model HIV-1 LTR-Fluc transcript (via pXP2-LTR-Fluc). The gene regulatory effects of the TAR miRNAs was monitored through TAR miRNA sensor constructs specific to either miR-TAR-5p or miR-TAR-3p. As shown in Figure 5B, TAR expression induced a dose-dependent down-regulation of both TAR miRNA sensors, suggesting that the TAR miRNAs entered the miRNA-guided RNA silencing pathway. The degree of inhibition induced by miR-TAR-3p was noticeably greater than that induced by miR-TAR-5p (see Figure 5B, right versus left panels).
Figure 5.
The miRNAs released from the HIV-1 TAR element function in gene regulatory processes in vivo. (A) Experimental scheme of the TAR miRNA functional assays. (B) HEK293 cells were cotransfected with psiTAR and a TAR miRNA sensor construct in which the Rluc reporter gene is coupled with a BS perfectly complementary to the miR-TAR-5p or miR-TAR-3p sequence. Results are expressed as mean ± SEM. (n = 6 experiments, in duplicate). *P < 0.05 or **P < 0.01 versus 0.25 ng psiTAR (one-way ANOVA). (C) HEK293 cells were cotransfected with pXP2-LTR-Fluc, a TAR miRNA sensor construct and pLacZ. Results of Rluc activity were normalized with β-galactosidase activity, and expressed as a percentage of Rluc activity obtained with pXP2-LTRΔTAR-Fluc lacking the TAR element. Results are expressed as mean ± SEM (n = 6–9 experiments, in duplicate). *P < 0.05 versus 100 ng pXP2-LTR-Fluc (one-way ANOVA). (D) H9 cells infected with HIV-1 strain HTLV-IIIMN or HTLV-IIIRF were transfected with miR-TAR-5p or miR-TAR-3p sensor construct and harvested 48 h later. Results of Rluc activity were normalized with those obtained from the unspecific sensor plasmid. Mean ± SEM (n = 2 in duplicate). *P < 0.05 versus the unspecific sensor (Student's t-test). The levels of HIV-1 p24 protein released from either HTLV-IIIMN or HTLV-IIIRF cell lines into the medium were measured by ELISA.
Next, we wished to determine the functionality of the miRNAs derived from TAR when located at the 5′ end of a model HIV-1 mRNA transcript. For that purpose, we measured TAR miRNA sensor activity in the presence of pXP2-LTR-Fluc, and normalized the results with the pXP2-LTRΔTAR-Fluc vector lacking the TAR element so to visualize the changes specifically related to miRNAs derived from TAR. In these experiments, we observed a decrease in reporter gene expression induced by the TAR-containing model HIV-1 transcript (Figure 5C). Again, the effects mediated by miR-TAR-3p were more pronounced than its miR-TAR-5p counterpart (see Figure 5C, right versus left panels). These results attest to the functional competence of the TAR miRNAs derived from the HIV-1 5′ LTR in gene regulatory processes of the host cell.
Whether miR-TAR-5p and miR-TAR-3p are produced and can exert gene regulatory effects in HIV-1-infected cells was examined by assessing the activity of their respective sensor constructs in H9/HTLVIIIMN and H9/HTLVIIIRF cell lines. As shown in Figure 5D, expression of the reporter gene coupled with a sensor specific to miR-TAR-3p was decreased by ∼20% in the MN cell line (P < 0.05), as compared with a reporter gene coupled with an unspecific sensor. When compared with RF cells, the degree of down-regulation was slightly more pronounced in the MN cell line, which also express more p24 proteins. These results indicate the presence in these virus-infected cells of miRNAs derived from HIV-1 TAR element at levels sufficient to be detected in a sensor-based reporter gene system.
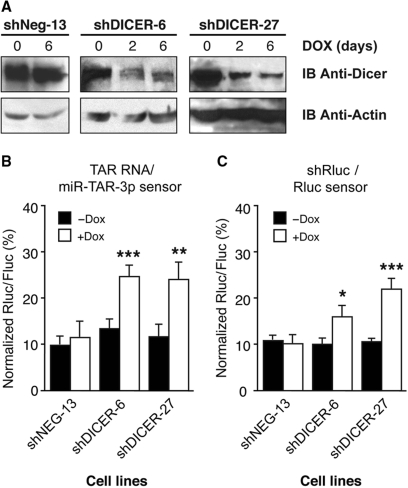
Dicer is required for TAR miRNA function in vivo
Taken together, our results militate for the involvement of Dicer in the gene regulatory roles of HIV-1 TAR miRNAs in vivo. To verify this assertion, we used the shDICER-6 and shDICER-27 cell lines, which have been engineered to conditionally express shRNAs directed against Dicer. A partial, but sustained, down-regulation of Dicer expression was achieved upon Dox treatment, as assessed by western blot (Figure 6A). This coincided with a significant impairment of reporter gene repression mediated by miR-TAR-3p, as compared with untreated cells or shNEG-13 cells expressing basal levels of Dicer (Figure 6B). A comparable defect in RNA silencing was observed when examining Rluc sensor regulation mediated by shRluc expression (Figure 6C). Extending our in vitro data on the ability of recombinant human Dicer to release miRNAs from HIV-1 TAR RNA, these findings confirm a role for Dicer in the regulation of gene expression mediated by TAR miRNAs in vivo.
Figure 6.
Dicer is required for the gene regulatory effects of miR-TAR-3p. (A) Immunoblot (IB) analysis showing Dicer protein down-regulation induced upon Dox treatment (upper panels), in parallel with actin control (lower panels). (B and C) Stable 293T-REx + pcDNA6TR cells conditionally expressing a shRNA directed against Dicer mRNA (shDICER-6 and shDICER-27), or a deleted region in Rluc mRNA (shNeg-13), were induced with 1 µg/ml of Dox for 12 h (+Dox) before transfection with plasmid expressing the (B) TAR RNA/miR-TAR-3p sensor, or (C) shRluc/Rluc sensor combination. Mean ± SEM (n = 4 experiments, in duplicate). *P < 0.05, **P < 0.01 or ***P < 0.005 versus prior induction (−Dox) (Student's t-test).
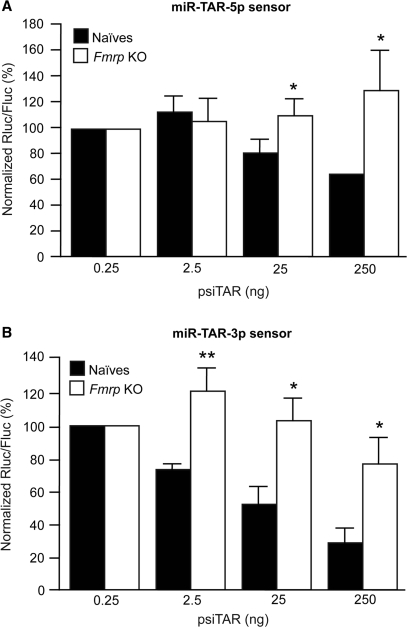
Fragile X mental retardation protein mediates the repressive action of TAR miRNAs
The involvement of the miRNA-guided RNA silencing machinery in these regulatory events was also confirmed in cells lacking fragile X mental retardation protein (FMRP), a component of the effector RNP complex that have been reported to facilitate RNA targeting by Dicer-derived miRNAs and to be required for efficient RNA silencing in mammalian cells (17). To this end, we employed the STEK TSV-40 cell line (17), which we previously validated as a functional representative of four different Fmr1 KO cell lines (17), in parallel with an isogenic cell line established from wild-type mice (Naïves) (17). In addition to correct for cell-to-cell variability, the use of a second reporter gene (Fluc) in our assays allowed us to examine the function of FMRP pertaining to small RNA-mediated RNA silencing. This was particularly important considering that FMRP could act as a negative regulator of translation (35). Expression of TAR in wild-type murine fibroblasts led to a dose-dependent decrease of both miR-TAR-5p and miR-TAR-3p sensor activities (Figure 7A and B). However, silencing of the TAR miRNA sensors was impaired in cells lacking FMRP, indicating the requirement of an integral effector complex to mediate TAR miRNA function in vivo.
Figure 7.
FMRP is required for optimal miR-TAR-5p and miR-TAR-3p function in vivo. (A and B) Fmr1 KO (TSV-40) and wild-type (Naïves) cells were transfected to coexpress TAR RNA (psiTAR) and (A) miR-TAR-5p sensor, or (B) miR-TAR-3p sensor construct. Results are expressed as mean ± SEM (n = 6 experiments, in duplicate). *P < 0.05 or **P < 0.01 versus Naïves (Student's t-test).
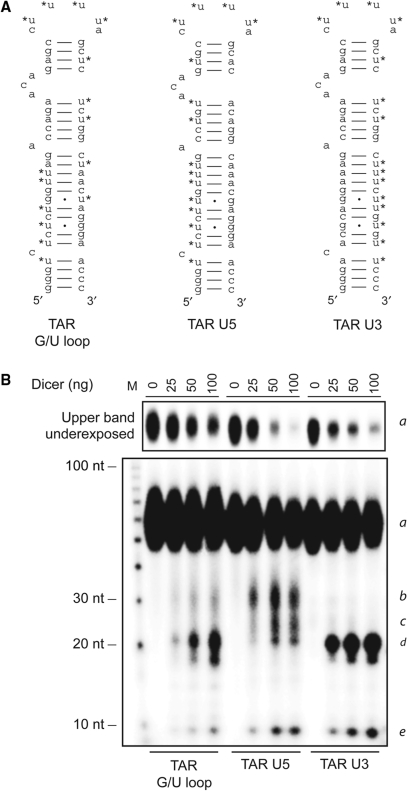
Asymmetric release of miR-TAR-3p upon Dicer processing of HIV-1 TAR RNA
During the course of our studies, we observed that the activity of the miR-TAR-3p miRNA sensor was systematically repressed more than the miR-TAR-5p miRNA sensor upon TAR expression (Figures 5B, C and 7), consistent with a preferential accumulation of the mature miR-TAR-3p in vivo. In order to get further insights into HIV-1 TAR RNA processing into miR-TAR-3p by Dicer, we prepared three substrates designed to monitor the formation of RNA products containing the left arm, the loop and/or the right arm of TAR. This was accomplished by positioning the Us of paired nt combinations on either the left arm (TAR U5) or the right arm (TAR U3) of TAR, while preserving its nt content, base pairing and predicted secondary structure (MFOLD analysis, P. Landry and P. Provost, unpublished data), as shown in Figure 8A. Analysis of the Dicer RNase reactions showed that processing of the TAR U5 and TAR U3 mutants was comparable, but more efficient than that of the TAR G/U loop mutant, as suggested by the decreased intensity of the bands corresponding to the substrate (denoted a, using a single-letter code) (see Figure 8B, upper panel) and the associated increase in the 10-nt, loop-containing RNA species (denoted e) (see Figure 8B, lower panel). It is thus possible that the interversion of the U paired nt combinations may have slightly influenced its susceptibility to Dicer cleavage.
Figure 8.
Asymmetric release of miR-TAR-3p upon HIV-1 TAR processing by Dicer. (A) Structure of the TAR G/U loop RNA or modified G/U loop RNA mutants, in which the A-U and G·U base pairs of the TAR stem were preserved, but positioned to have all the 32P-labeled Us either on the left arm (TAR U5) or the right arm (TAR U3). (B) These TAR substrates were incubated in the absence or presence of recombinant human Dicer with MgCl2. The cleavage products were analyzed by denaturing PAGE and autoradiography. The RNA species detected upon in vitro processing by Dicer contain specific TAR RNA structures and are denoted as follows: a, TAR RNA substrate; b, left arm + loop; c, long left arm precursor of miR-TAR-5p; d, right arm; e, loop.
Nevertheless, informative differences in the band patterns and intensities were noted upon Dicer processing of the TAR U5 and TAR U3 RNA mutants, the latter of which was otherwise similar to the TAR G/U loop RNA (see Figure 8B, lower panel). First, enrichment of the TAR miRNA bands (denoted d) upon labeling of the right arm of the TAR stem, conjugated with the impoverishment of band d associated with unlabeling of the right arm of the TAR stem, is suggestive of its presence in RNA species d. Second, following a similar reasoning, appearance of RNA species b upon labeling of the left arm of TAR is indicative of its presence in this intermediate RNA species. Third, the length of RNA species b differs from that of RNA species d by ∼10 nt (i.e. the length of RNA species e), which likely contains the loop sequence (please refer to Figure 4C) released upon Dicer cleavage of both arms in the upper part of the TAR stem (please refer to Figure 3C for the predicted cleavage sites). RNA species b is thus likely composed of the left arm of TAR with the loop. Fourth, in accordance with this inference, the very low intensity of RNA bands b upon TAR U3 RNA processing is compatible with the absence of intermediate RNA species comprised of the loop and the right arm of TAR. Fifth, appearance of RNA species c upon labeling of the left arm of TAR and its size of ∼24 nt suggest that it may be an RNA intermediate from which nt 1–3 of the left arm of TAR are removed to yield miR-TAR-5p. Collectively, our observations support the concept by which miR-TAR-3p would be released from the TAR element in a process yielding the miR-TAR-5p-loop as an intermediate RNA species.
DISCUSSION
Known to alter the expression of thousands of genes (36), HIV-1 has been shown recently to alter the expression profile of cellular miRNAs. Comparing HIV-1-infected versus noninfected cells by microarray analysis, Triboulet et al. (14) noticed that some miRNAs were up-regulated, whereas others were down-regulated by HIV-1. These findings are not compatible with a general shutdown or stimulation of the miRNA-guided RNA silencing machinery by the virus and rather argue for an alternative mechanism to explain how specific genes and miRNAs are modulated by HIV-1.
Initial studies aimed at investigating the existence of virus-derived miRNAs, through the use of small RNA cloning methods, were unable to provide experimental evidences in the case of HIV-1. In a study by Pfeffer et al. (10), no miRNA of viral origin was found among 1540 small RNA sequences cloned from HeLa cells stably expressing CD4 and CXCR4, and infected with HIV-1, isolate Bru (LAV1). On the other hand, two independent groups have obtained positive signals from the use of northern blotting, compatible with small HIV-1 RNAs (15,16). Here, we characterized and delineated miRNAs originating from the HIV-1 TAR element by making use of northern blotting, primer extension and RPA. As shown for SV40, this latter method may be suitable, better adapted and more sensitive for the detection of viral miRNAs (12).
Why had HIV-1 TAR miRNAs eluded detection for so long? First, low expression levels of some viral miRNAs may render their isolation by cloning methods difficult among the more abundant cellular ribosomal RNAs, transfer RNAs and miRNAs. This is consistent with the facts that miRNA cloning frequency reflects miRNA abundance and that miRNAs cloned in a single copy may be more difficult to detect (37). Second, viral and cellular proteins interacting with the TAR element within the 5′ LTR of HIV-1 mRNAs likely decrease its vulnerability to processing by RNases III. Third, HIV-1 TAR element may partially escape RNases III processing by adopting alternative conformations, such as one called TAR31, which is functionally similar but requires only 31 nt to form (38). Fourth, the lack of a free 2-nt 3′ overhang at the base of TAR in HIV-1 mRNA transcript may hamper its processing, as demonstrated by the marked decrease in TAR RNA processing by Dicer upon extension of its 3′ overhang sequence from 2 to 10 nt. Finally, we cannot exclude the possibility that the TAR miRNAs are modified on the 2′ hydroxyl of the terminal ribose, which would significantly hamper the cloning efficiency of these modified small RNAs, as experienced in plants (39). On the other hand, it is known that the HIV-1 promoter directs, in the absence of Tat, the synthesis of another class of short nonpolyadenylated transcripts with heterogeneous 3′ ends located around position +59 (40). These transcripts may thus represent potential substrates for Dicer and argue for a specific sub-population of TAR as an alternative source of HIV-1 miRNAs.
When examining the TAR miRNA sequences elucidated by primer extension and RPA, we noticed a nt bias at the 5′ end of both miR-TAR-5p and miR-TAR-3p. The presence of a U at this position is in agreement with the preference of the RNases III Drosha and Dicer to cleave on the 5′ side of a U. Likely long enough to be recognized and processed by the RNase III Dicer, the TAR element may be too short for Drosha, whose pri-miRNA substrates typically contain a stem of ∼3 helical turns (∼33 bp) (41). Moreover, TAR miRNAs do not result from cleavage ∼11 bp away from the single-stranded junction at the base of TAR, as would be expected from the Drosha·DGCR8 complex (42). Experimental data rather support the specific involvement of Dicer in HIV-1 TAR RNA processing. First, TAR RNA was efficiently cleaved by recombinant Dicer in vitro. Similar findings were reported recently by Klase et al. (43). Second, the size of the TAR RNA products (∼23–24 nt) detected in vivo correlates with that expected for RNAs issued from Dicer cleavage. Third, TAR RNA was processed into a miRNA duplex, a recognized Dicer signature. The most thermodynamically favorable pairing of the TAR miRNA duplex differs from the sequence structure located in the unprocessed TAR and exhibits two 3-nt 3′ overhangs (D.L. Ouellet, I. Plante and P. Provost, unpublished data). This opens the possibility of a pairing rearrangement within the duplex upon removal of the neighboring base pairs and sequences in the precursor molecule.
The extremities of some viral miRNA duplexes seem to diverge from the general 2-nt 3′ overhang rule established for endogenous miRNAs. Like the TAR miRNA duplex, miRNAs derived from SV40 (12) and KSHV (11) also exhibit atypical 3-nt 3′ overhangs. Computational positioning of mature KSHV miRNA sequences also revealed duplexes bearing 3′ overhangs as short as 1 nt (11). The discrepancies between the viral and endogenous miRNAs may bear the signs of subtle processing irregularities, possibly related to the adaptive capacity of the RNA silencing components to recognize and process internal viral RNA structures like miRNA precursors. Initially reported in Zhang et al. (44), Dicer excision of miRNAs from internal sites is not infrequent, as the long CAG and CUG repeats found within their natural context in transcripts from mutant genes involved in diseases such as myotonic dystrophy type 1, Huntington's disease and spinocerebellar ataxia type 1 are efficiently cleaved by Dicer (45).
Mechanistically, endogenous substrate recognition by Dicer has been proposed to involve anchoring of the pre-miRNA 2-nt 3′ overhang in the pocket formed by its central PAZ domain (6,46). Furthermore, the 2-nt overhang is measured by the alignment of the Dicer RNase III domains rather than by the distance between active residues on one peptide chain, whereas the length of the product (∼21 nt) is determined by the distance between the PAZ domain and the active site (6). Flanked by a single-stranded RNA sequence on its 3′, TAR RNA processing may be suboptimal, perhaps less accurate. In support to this possibility is a study by Vermeulen et al. (47) reporting that shifts in Dicer cleavage sites result from dissimilar processing of substrates bearing overhangs of different lengths. In addition, folding of the TAR element itself may force Dicer dimers to adopt a slightly modified configuration. The 3-nt bulge, in particular, has been shown to introduce a bend of ∼50° into the α helical RNA structure of TAR (48), which otherwise exhibits a certain degree of flexibility. These structural constraints may explain the length of the TAR miRNAs, the nature of the overhangs generated and the relative degree of heterogeneity of the cleavage sites determined for miR-TAR-3p (please refer to the open arrowheads in Figure 3A).
Transactivation by the viral protein Tat is severely compromised upon introduction of G/U mutations in the loop of the TAR element (49). When studied in parallel with the wild-type TAR, however, the G/U loop TAR mutant was processed with similar efficiencies by Dicer in vitro. These observations support the notion that the TAR structure that evolved to support transactivation by Tat remains susceptible to cleavage by Dicer, possibly reflecting the inability of TAR to escape from Dicer surveillance. Evocative of the rapid adaptive evolution of the antiviral pathway in Drosophila (50), this ongoing interaction between HIV-1 and Dicer may be constantly shaping the antiviral functionalities of the host RNA silencing machinery.
When interpreting the results obtained from Dicer RNase assays in vitro, the caveat has to be taken into account that recombinant human Dicer exhibits low processive activity in vitro (5,44), a characteristic that has been attributed to the product remaining bound to the enzyme (44). In more recent studies, Dicer has been shown to operate with TRBP within a pre-miRNA processing complex (51,52) and to interact with Ago2 (53), another component of miRNA-containing ribonucleoprotein (miRNP) complexes (54). Although it has been reported to facilitate pre-miRNA processing in vitro (52), the role of TRBP in modulating Dicer function remains largely undefined. It may be analogous to that played by DGCR8 within the Drosha·DGCR8 complex, in which DGCR8 may interact directly and specifically with pri-miRNAs, and proposed to function as a molecular anchor that determines the Drosha cleavage sites (41). Alternatively, TRBP may contribute to releasing the miRNA product from Dicer and improving biosynthesis of miR-TAR-5p in vivo.
Data obtained from Dicer knockdown and FMRP KO cells show that TAR miRNAs are recognized and channelled through the miRNA-guided RNA silencing pathway. Our TAR miRNA sensor activity assays revealed the miRNAs residing within the TAR element of HIV-1 are functional, as both miR-TAR-5p and miR-TAR-3p exerted downregulatory effects on gene expression, and that these effects were optimal in the presence of Dicer and FMRP. Together, our findings are compatible with TAR miRNAs being extracted from the HIV-1 TAR element by Dicer to be subsequently incorporated into active miRNP complexes in vivo, in which FMRP may act to facilitate their annealing to mRNAs bearing sites of perfect or imperfect complementarity. Whether Dicer mediates action of TAR miRNAs by acting also within miRNP effector complexes in vivo remains to be determined.
Although indicative of the presence of miRNAs derived from HIV-1 TAR element, the response of our sensor constructs in HIV-1-infected cells was relatively modest and less pronounced than that observed in cotransfected 293 cells. This observation may be related to the fact that cells infected with the virus express low physiological levels of TAR RNA, in contrast with vector-based, U6 promoter-driven overexpression of TAR RNA. In addition, expression in these cells of the viral TAR RNA binding Tat protein, which has been proposed as a suppressor of RNA silencing (16), may have hampered Dicer processing of TAR RNA and contributed to attenuate the signal expected. Although a recent study reported that physiological levels of HIV-1 Tat neither inhibited the RNA silencing machinery of infected cells nor induced the accumulation of pre-miRNAs as would be predicted for an inhibitor of Dicer function (55).
As assessed in mRNA target regulation, the silencing properties of miR-TAR-3p were slightly superior than those of miR-TAR-5p, supporting a relative functional asymmetry. This may be related to differences in the thermodynamic stability of the miRNA duplex extremities, as reported in Drosophila where R2D2 has been shown to act as a protein sensor for siRNA thermodynamic asymmetry (56). At that level, the human miRNA-guided RNA silencing machinery may differ from the fly system, as no evidence for a human homolog of R2D2 has been provided yet. Rather, the silencing potency of miR-TAR-3p in vivo paralleled the preferential accumulation of mature miR-TAR-3p upon Dicer processing of TAR in vitro. This latter step may thus represent a novel source of asymmetry between miRNAs comprised within a pre-miRNA. This observation is reminiscent to endogenous miRNA duplexes, where the level of mature miRNA detected is higher than that of its opposite miRNA* strand (3). miR-TAR-3p and miR-TAR-5p may thus qualify as the miRNA and miRNA* of the duplex, respectively.
HIV-1 is known to perturb gene expression programming of infected cells, affecting thousands of genes (36), which likely conceal key determinants of its pathogenesis. As a source of regulatory miRNAs, it is tempting to speculate about a contributory role of the TAR element. Our results indicate that HIV-1 mRNA transcripts are a source of regulatory miRNAs that are functionally competent in RNA silencing processes in vivo. These findings can be reconciled with the recent findings that Dicer may act as natural anti-HIV-1 host defenses in human cells (14). We can thus propose a scenario in which Dicer would interfere with the virus through processing of its structured RNAs, such as TAR, from which miRNA by-products could be released and influence expression of viral and/or host genes, with a potential impact on viral replication.
Biosynthesis and action of TAR miRNAs appear to be guided by the preferential release of miR-TAR-3p along asymmetrical processing events yielding miR-TAR-5p-loop as an intermediate RNA species. The lower efficiency of miR-TAR-5p in regulating gene expression may be inherent to this stepwise maturation process. Considering that miRNAs are usually expressed at low levels and that they act in synergy with other miRNAs, dismissal of a significant role for HIV-1 TAR miRNAs of low abundance would not be prudent. Rather, the possibility that TAR miRNA expression ultimately influences viral replication and/or the efficiency of host antiviral defenses is attractive and warrants further investigations. Our study provides the molecular basis required for elucidating the impact of TAR miRNAs on the gene expression programming of HIV-1-infected cells and for determining their role and importance in viral pathogenesis.
ACKNOWLEDGEMENTS
We are grateful to Maude Baillargeon and Gilles Chabot for graphic illustrations. P.P. is endebt to Edouard W. Khandjian for the kind gift of the Fmr1 KO and Naïves cell lines. L.F. is a Senior Scholar from the Fonds de la Recherche en Santé du Québec (FRSQ). M.J.T. is the recipient of the Canada Research Chair in Human Immuno-Retrovirology (senior level). P.P. is a New Investigator of the Canadian Institutes of Health Research (CIHR) and Junior 2 Scholar from the FRSQ. Funds were provided by Canadian Institutes of Health Research (CIHR) (HOP-83069 to P.P. and M.J.T.). Funding to pay the Open Access publication charges for this article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J. Biomed. Biotechnol. 2006;2006:69616. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 10.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 11.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl Acad. Sci. USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 13.Provost P, Barat C, Plante I, Tremblay MJ. HIV-l and the microRNA-guided silencing pathway: an intricate and multifaceted encounter. Virus Res. 2006;121:107–115. doi: 10.1016/j.virusres.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 15.Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P. Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J. Biomed. Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 21.Popovic M, Sarngadharan MG, Read E, Gallo RC. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 22.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 25.Kashanchi F, Piras G, Radonovich MF, Duvall JF, Fattaey A, Chiang CM, Roeder RG, Brady JN. Direct interaction of human TFIID with the HIV-1 transactivator tat. Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 26.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yee JK, Friedmann T, Burns JC. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43(Pt A):99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 28.Bounou S, Leclerc JE, Tremblay MJ. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4(+)-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J. Virol. 2002;76:1004–1014. doi: 10.1128/JVI.76.3.1004-1014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gatignol A, Jeang KT. Tat as a transcriptional activator and a potential therapeutic target for HIV-1. Adv. Pharmacol. 2000;48:209–227. doi: 10.1016/s1054-3589(00)48007-5. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J. Biol. Chem. 2006;281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- 32.Lamontagne B, Elela SA. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 2004;279:2231–2241. doi: 10.1074/jbc.M309324200. [DOI] [PubMed] [Google Scholar]
- 33.Rotondo G, Huang JY, Frendewey D. Substrate structure requirements of the Pac1 ribonuclease from Schizosaccharmyces pombe. RNA. 1997;3:1182–1193. [PMC free article] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plante I, Provost P. Hypothesis: A role for fragile X mental retardation protein in mediating and relieving microRNA-guided translational repression? J. Biomed. Biotechnol. 2006;2006:16806. doi: 10.1155/JBB/2006/16806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corbeil J, Sheeter D, Genini D, Rought S, Leoni L, Du P, Ferguson M, Masys DR, Welsh JB, Fink JL, et al. Temporal gene regulation during HIV-1 infection of human CD4+ T cells. Genome Res. 2001;11:1198–1204. doi: 10.1101/gr.180201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kloosterman WP, Steiner FA, Berezikov E, de Bruijn E, van de Belt J, Verheul M, Cuppen E, Plasterk RH. Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res. 2006;34:2558–2569. doi: 10.1093/nar/gkl278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter SN, Belanger F, Zheng P, Rana TM. Dynamics of nascent mRNA folding and RNA-protein interactions: an alternative TAR RNA structure is involved in the control of HIV-1 mRNA transcription. Nucleic Acids Res. 2006;34:4278–4292. doi: 10.1093/nar/gkl499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ebhardt HA, Thi EP, Wang MB, Unrau PJ. Extensive 3' modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl Acad. Sci. USA. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheldon M, Ratnasabapathy R, Hernandez N. Characterization of the inducer of short transcripts, a human immunodeficiency virus type 1 transcriptional element that activates the synthesis of short RNAs. Mol. Cell. Biol. 1993;13:1251–1263. doi: 10.1128/mcb.13.2.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 42.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 2007;8:63. doi: 10.1186/1471-2199-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krol J, Fiszer A, Mykowska A, Sobczak K, de Mezer M, Krzyzosiak WJ. Ribonuclease Dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 46.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zacharias M, Hagerman PJ. The bend in RNA created by the trans-activation response element bulge of human immunodeficiency virus is straightened by arginine and by Tat-derived peptide. Proc. Natl Acad. Sci. USA. 1995;92:6052–6056. doi: 10.1073/pnas.92.13.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990;9:4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 51.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 55.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]