Abstract
Retroviruses replicate by converting their positive sense genomic RNA into double-stranded DNA that is subsequently integrated into the host genome. This conversion is catalyzed by reverse transcriptase (RT) early after virus entry into the target cell and is chaperoned by the nucleocapsid protein (NC). In HIV-1, NC is composed of small basic domains flanking two highly conserved CCHC zinc fingers that specifically interact with the genomic RNA and RT. Through specific interactions with the genomic RNA and RT, and possibly with cellular factors, the NC zinc fingers were found to play critical roles in HIV-1 assembly and budding, and later in proviral DNA synthesis and integration. Therefore, intact NC zinc fingers are needed throughout the virus replication cycle. Here, we report for the first time that deleting either one or the two NC zinc fingers leads to an unexpected premature viral DNA synthesis in virus producer cells and the production of noninfectious particles with a high level of viral DNA. In addition to providing the first example of reverse transcription during the late steps of HIV-1 replication, these findings emphasize the fact that the NC zinc fingers are a major target for new drugs against HIV-1.
INTRODUCTION
Conversion of the positive sense genomic RNA of retroviruses into DNA is a complex multistep process that is initiated from a cellular tRNA annealed to the 5′ end of the genomic RNA and culminates in the synthesis of a double-stranded DNA copy of the genomic RNA flanked by two long terminal repeats (LTR). Reverse transcription is catalyzed by the viral reverse transcriptase (RT) present in a large nucleoprotein complex where viral nucleocapsid protein (NC) molecules coat the genomic RNA (1,2). NC is encoded by Gag and is a small basic protein with nucleic acid-binding and chaperoning properties, found in all retroviruses (3). In lentiviruses, such as HIV-1, NC is formed of small basic domains flanking two highly conserved CCHC zinc fingers (4) (Figure 1).
Figure 1.
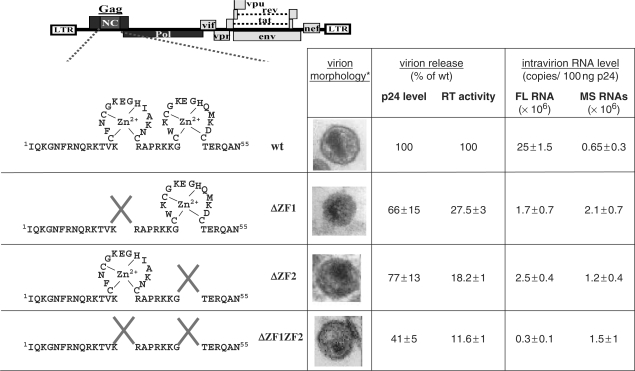
Mutations of the NC zinc finger modify virion morphology, virion release and RNA packaging. Asterisk denotes pictures from electron microscopy analysis that were from (8,13). Amounts of virion released by the wt and mutant viruses were measured by ELISA assay and in vitro RT activity assay. Values were normalized to wt levels (mean of n = 2). Virion full-length (FL) or multispliced (MS) RNA was determined with RT–qPCR (mean of n = 3) (25).
Originally, the intact NC zinc fingers were shown to be required for genomic RNA packaging (5–8) and later to be involved in virus assembly and budding (3,9–11). The selective packaging of the genomic RNA is thought to be directed by specific interactions between NC, notably the zinc fingers, and the packaging Psi signal in the genomic 5′ UTR (12). Recently, we observed that disruption of either one or the two zinc fingers impaired the intracellular Gag trafficking to the budding site, and simultaneously reduced the levels of Gag processing and of virion production (13) probably due to a lost interaction with ALIX (14).
NC also plays key roles in the reverse transcription process that needs its nucleic acid-binding and chaperone activities and its interaction with RT (3,15). Although the exact mechanism by which NC facilitates nucleic acid rearrangements is not completely understood, in vitro assays mimicking the different steps of the reverse transcription reaction clearly showed that NC chaperones primer tRNA annealing to the initiation site (PBS), and the two obligatory strand transfers that are required to generate the complete proviral DNA flanked by two LTR (3,16). However, the precise role of the NC zinc fingers in viral DNA synthesis has been difficult to evaluate in newly infected cells. Current findings show that mutating conserved residues in the first or the second CCHC zinc finger leads to some reverse-transcription defects with reduced DNA synthesis and stability in infected cells, and a drastic reduction of viral DNA integration (8,17,18).
Interestingly, the reverse-transcription reaction appears to be tightly controlled during the late steps of HIV-1 replication since the full-length viral DNA (FL DNA) synthesis is completed only after virions infect target cells (19). How this is regulated is yet poorly understood. Since the NC zinc fingers appear to be required throughout the viral replication cycle, we investigated their role in the temporal control of reverse transcription. We undertook a detailed quantitative analysis of the viral nucleic acid production throughout the replication cycle by qPCR and qRT–PCR. By measuring the effects of NC zinc finger deletions on the conversion of both the genomic and spliced RNA species into DNA, we discovered that viral particles released from the cells expressing HIV-1 NC mutants, contained a high level of DNA. This report shows for the first time that RT can occur in HIV-1-producing cells, converting HIV-1 into a DNA-like viruses similar to hepatitis B virus and spumaviruses. Thus, viral DNA synthesis should be regulated to insure HIV-1 replication in target cells and appears to be a new crucial role for NC.
MATERIALS AND METHODS
Plasmids, cell culture
The HIV-1 pNL4-3 molecular clone was used to generate constructs with deletion of ZF1 (pNL4-3ΔZF1), ZF2 (pNL4-3Δ ZF 2) or both ZF 1 and ZF 2 (pNL4-3Δ ZF 1 ZF 2). These mutant constructs have been described elsewhere (8,13). The human HeLa LTRHIV-1-Luc (20) (kind gift of S. Emiliani), 293T, HeLa cell lines and the stably CD4/CXCR4-coexpressing-293 cells (42CD4) (21) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with glutamine (2 mM), penicillin, streptomycin and 10% (v/v) heat-inactivated fetal calf serum.
Transfection, virus preparation and infection
Transfections of 293T cells were performed with 3 × 106 cells divided 1 day before in 100 mm dishes by calcium phosphate precipitation with 8 μg of HIV-1 plasmid DNA for 48 h. When RT inhibitors were used, 293T cells were pretreated 3 h before transfection with 50 µM AZT or 50 µM Nevirapine and transfection was pursued for 48 h in presence of the drug. In all cases, 6 h after transfection cells were trypsinized, extensively washed with fresh medium and divided into a new plate to eliminate plasmid in excess. The amount of HIV-1 particles in the supernatant was determined using a HIV-1 CA p24 core antigen enzyme-linked immunosorbent assay (ELISA) Kit (Beckman Coulter™) or in vitro standard RT enzyme assay (22). Virions were purified from filtered culture supernatants by centrifugation through a 20% sucrose cushion at 30 000 r.p.m. in an SW32 rotor for 1 h 30 at 4°C.
Infections of the 42CD4 cells were performed in presence of polybrene (2 µg/ml) during 24 h as previously described (23).
For luciferase reporter assay, virions produced from transfected 293T cells were quantified by measuring p24 antigen in culture supernatants (ELISA), purified and treated with DNAse. Virion DNA was extracted and used to transfect HeLa cells stably expressing the HIV-1 promoter/enhancer LTR luciferase construct. HeLa LTRHIV-1-Luc cells were lyzed 72 h posttransfection. Cell protein extracts were standardized by means of the Bradford assay and luciferase activity was monitored using a luminometer.
For the natural endogenous reverse transcription assays (NERT), the virion-containing media of 293T cells were removed 44 h posttransfection, and cell culture was incubated for 4 h in different conditions: standard medium, medium without FBS (minus dNTP), supplemented or not with 100 μM dNTP and with or without 50 µM Nevirapine (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH). To evaluate the effect of time on NERT, the virions produced during the 4 h of culture in standard medium, were collected and incubated for an additional 4 h in absence of cells.
DNA and RNA extraction and analysis
Nucleic acids extraction from virions was performed as previously described (23). Briefly, purified virions were incubated with 8 U of DNAse (RQ1, Promega) at 37°C for 45 min before extraction of the nucleic acids by phenol/chloroform and ethanol-precipitated. DNase treatment reduced contaminations by the transfecting-plasmid DNA (pNL4-3) under the level of the FL DNA. Thus, proportions of pNL4-3 in wt and mutant DNA samples were <20% and 1% of total DNA detected, respectively. Cellular DNA was extracted with DNAzol (MRC) according to the manufacturer's instructions. To avoid any contamination with viral cDNA associated with the particles, cells were trypsinized and extensively washed with PBS before DNA extraction. Cellular RNA was extracted with TriReagent according to manufacturer's instructions. Nucleic acids were quantitated by measuring optical absorption at 260 nm.
To check that intracellular DNA samples were not contaminated by DNA-containing virions, untransfected cells were incubated for 24 h with supernatant containing HIV-1 ΔZF2 virions (issued from 48 h production). After DNA extraction, contaminating multispliced cDNA levels were determined by qPCR.
In vitro reverse transcription for RT–PCR experiments was performed as described (24) with oligo(dT) primer and 1/20 aliquots of virion RNA samples. qPCR was achieved with 2.5% of the RT-d(T) reaction, or with 50 ng of intravirion and cellular nucleic acid samples. Systematically, cellular GAPDH gene level was determined for standardization. qPCR assays were performed with SYBR Green Kit (Roche) with the RotorGene (Labgene) systems. A standard curve was generated from 50 to 500 000 copies of pNL4.3 plasmid. Each assay included a control without RT indicating a DNA contamination level <0.1% of the HIV genomic RNA. Sequences of primers described in Figure 2 and in (23), and detailed PCR conditions will be provided on request. Nucleic acid levels in virion or cell were normalized with respect to p24 (determined by ELISA) and GAPDH gene, respectively.
Figure 2.
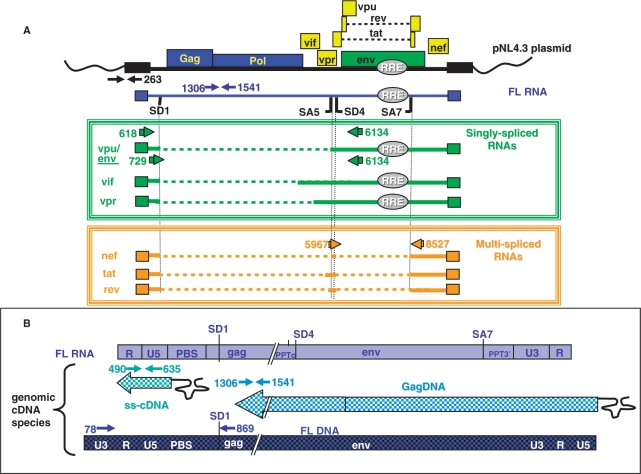
Strategy for RT–PCR and PCR analyses of viral RNA and corresponding cDNA. (A) Schematic representation of templates and primers used for the RT–PCR analyses of HIV-1 RNAs resulting from proviral pNL4-3 plasmid expression. Only the splice sites (SD1/SA5 and SD4/SA7) important for this study are indicated. A color code was used to indicate the specificity of the PCR-primer pairs (arrows) that were used to quantify the pNL4.3 plasmid (black), the full-length RNA (blue), the class of the singly spliced RNAs or DNAs (green) or the multispliced (MS) RNAs (orange). Numbers refer to the position of the elongation start. The same primers were used for the detection of the corresponding cDNAs. (B) Products of the genomic FL RNA reverse transcription. The primer pairs used to detect the intermediate ss-cDNA, GagDNA and the final product FL DNA are presented.
The HIV-1 singly spliced RNAs and corresponding cDNAs were detected in virions (100 ng p24) by standard PCR (25). In addition to the DNAse treatment of the purified virions, an additional DNAse treatment was performed on nucleic acid extracts to allow specific detection of FL RNA by RT–PCR and as a control for the nature of the viral cDNAs.
RESULTS
Deletions of the NC zinc fingers decreased the packaging efficiency of the FL RNA, but not of the viral spliced RNA
To study the control exerted by NC on DNA synthesis by RT, we used HIV-1 mutants with a deletion of either the first (ΔZF1), the second (ΔZF2) or both (ΔZF1ZF2) NC zinc fingers and transfected the corresponding DNA in human 293T cells (Figure 1). As previously reported, the NC mutant particles were noninfectious and were produced in reduced amounts containing less genomic RNA (FL RNA) (Figure 1) (7,8,13). Reduction of genomic RNA packaging was monitored by qRT–PCR (Figure 2A) on purified particles and was found to be from 10% to 1.5% of the wild-type level (Figure 1). In addition, we examined the packaging of the spliced viral RNAs that were shown to be specifically incorporated into HIV-1 particles (25). The NC zinc-finger deletions did not impair their packaging (Figures 1 and S1A) and the mutant particles contained roughly similar levels of genomic and spliced RNAs (Figure 1).
NC zinc-finger deletions resulted in the accumulation of viral DNA in virions
Infection performed with these mutant particles showed that the levels of newly made viral DNA were decreased in target cells (Figure 3, left panel) in agreement with previous results (8,17,18). Indeed, the canonical view of reverse transcription is that it mostly takes place in newly infected cells. However, some reverse transcription can take place prior to cell entry since low levels of viral DNA have been found in HIV-1 virions (23,26–30), and termed NERT for natural endogenous RT. The chaperoning role of NC protein in reverse transcription suggested to us that it could regulate the timing of viral DNA synthesis. Thus, we performed an in-depth analysis of the viral DNA content in HIV-1 NC mutant particles which have been extensively treated with DNase to remove any contaminant plasmid DNA due to cell transfection, and purified. We used qPCR for the quantitative monitoring of ss-cDNA, Gag and FL DNA. The levels of viral DNA resulting from reverse transcription of the spliced RNAs have also been monitored (Figure 2A and B).
Figure 3.
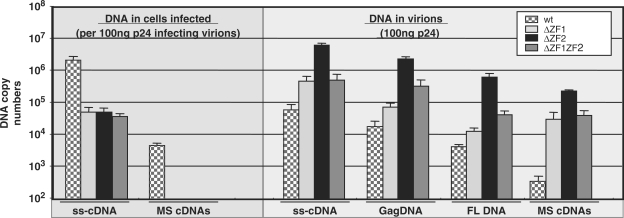
Deletions of the NC zinc fingers change the timing of the reverse transcription. Viral DNA levels were determined by qPCR in wild-type and NC mutant virions (100 ng p24) (right panel). These latter materials were used to infect 42CD4 cells and viral DNA levels were subsequently determined in target cells and expressed for 100 ng p24 particles used to infect cells (n = 3 ± SD) (left panel).
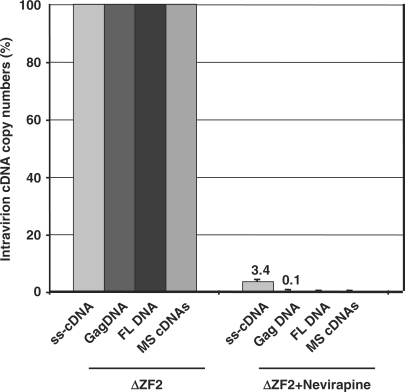
Surprisingly, all NC mutant particles contained a high level of DNA compared with wild-type virions, notably the ΔZF2 virions, leading to DNA-to-RNA ratio increase of up to three orders of magnitude (Figure 3, right panel). Previously, we found that spliced HIV-1 RNAs were reverse-transcribed as efficiently as the genomic RNA (23). Thus, we also monitored the level of spliced DNAs which represent an ideal marker for viral DNA quantitation due to the lack of pNL4.3 plasmid DNA contaminations (Figure 2A). There was also a drastic enhancement of spliced cDNAs in these NC mutant virions, with about a 1000-fold increase in the ΔZF2 particles (Figure 3, right panel and Figure S1B). Similar results were obtained with HIV-1 particles obtained by transfecting HeLa cells (Figure S2).
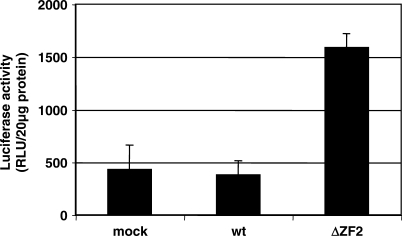
A careful examination of the newly made virion DNA shows that a fraction of it corresponded to the full-length proviral DNA such as in ΔZF2 virions. Interestingly, DNA purified from ΔZF2 particles was active once transfected into the reporter cell-line HeLa LTRHIV-1-Luc since it activated the Tat-mediated luciferase expression (Figure 4).
Figure 4.
Intravirion ΔZF2 DNA expression in cells. Virions were prepared from 293T transfected or not with wild-type or ΔZF2 plasmid and quantified by measuring the p24 antigen in culture supernatants. DNA extracted from wild-type or ΔZF2 purified virions was transfected into the reporter HeLa LTRHIV1-Luc cell-line and luciferase activity was measured in cell lysates and normalized to 20 µg of cell protein extract. (n = 3 ± SD).
Taken together these results suggest that deletion of the zinc fingers did not impair chaperoning of the reverse transcription by NC, in agreement with in vitro data (31). Interestingly, the ΔZF2 virions contained even higher amount of viral DNA products (Figure 3, right panel) than that generated by the wild-type HIV early after cell infection (Figure 3, left panel). Thus, while the NC zinc fingers control at least in part genomic RNA packaging, these conserved NC motifs could also regulate the timing of viral DNA synthesis during the HIV-1 replication.
Viral DNA in zinc finger mutant virions did not result from NERT activity
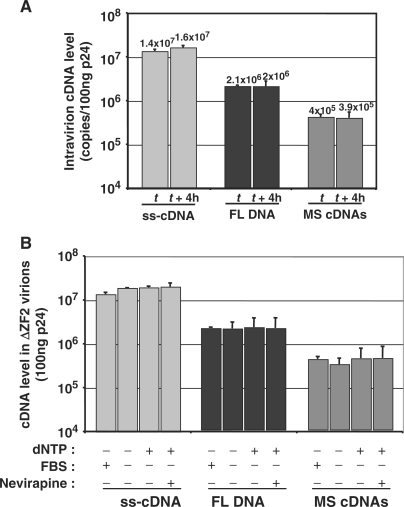
Thus, we asked whether the virion DNA was produced in virus particles by a NERT activity somehow stimulated by the NC zinc-finger deletions. Stimulation of an otherwise marginal cDNA synthesis in virions is usually obtained by adding dNTP to the extracellular milieu together with virion permeabilization agent (32,33). Because the ΔZF1, ΔZF2 and ΔZF1ZF2 particles exhibit an abnormal morphology with a round immature core structure (13) (Figure 1), we hypothesized that an abnormal core structure may confer NERT stimulation through a natural permeability to the dNTP that are possibly present in the fetal bovine serum (FBS) of the cell-culture media.
Since the completion of NERT takes place by 4 h (27), the DNA level in NC mutant particles should increase over time without prior virion permeabilization. However, the DNA level in particles remained the same with or without a 4 h incubation at 37°C (Figure 5A), indicating that viral DNA synthesis was already complete before incubation, probably at the time of particles release from the producer cells. In agreement with this conclusion, addition or deprivation of dNTP or treatment with the HIV-1 RT inhibitor Nevirapine during the 4 h period of virus production did not influence levels of the different viral DNA species found in ΔZF2 particles (Figure 5B).
Figure 5.
No increase of NERT activity over time in ΔZF2 virions. (A) A stock of ΔZF2 virions produced during 4 h (t time) was incubated for 4 h at 37°C (t + 4 h) and viral cDNA levels were determined by qPCR (n = 3 ± SD). (B) The intravirion DNA yield was neither dependent on dNTP concentration nor Nevirapine (50 µM). DNA was analyzed in ΔZF2 virions produced by transfected 293T cells grown during 4 h in the different culture conditions (n = 3 ± SD).
Viral DNA was synthesized in HIV-producer cells
To examine whether reverse transcription did take place in HIV-1 producer cells, we pretreated cells with Nevirapine 3 h prior to DNA transfection, and maintained drug treatment until the viral particles were collected. Under these conditions, virion release and RNA levels in ΔZF2 virions were not affected (Figure S3), but ss-cDNA, Gag and FL DNAs and spliced cDNAs were all found in very low quantities in HIV-1 ΔZF2 virions (Figures 6, S1C and S4A). Similar results were obtained with AZT, except as expected for the initial ss-cDNA product that is known to be poorly responsive to the AZT chain terminator (23,34) (Figures S3 and S4B). Taken together, these findings indicate that viral DNA synthesis can take place in cells producing HIV-1 zinc finger mutant particles.
Figure 6.
Viral DNA synthesis in HIV-transfected cells. Analysis of viral DNA content of ΔZF2 virions released from cells treated or not with RT inhibitor Nevirapine (50 µM) (n = 3 ± SD).
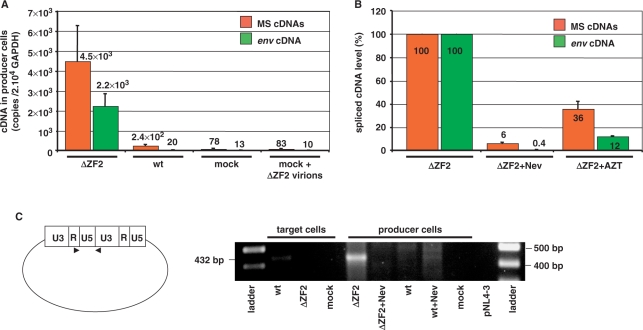
This prompted us to investigate whether newly made ss-cDNA, Gag and FL DNAs were present in HIV-1 producer cells. However, despite extensive DNase treatment of these cells, there was residual plasmid DNA present in the transfected cells. To circumvent this difficulty we analyzed the levels of spliced cDNAs corresponding to the reverse transcription of the spliced viral RNAs. As shown in Figure 7A, spliced viral cDNAs were abundant in cells transfected with the ΔZF2 mutant DNA, but not in those transfected with the wild-type HIV-1 DNA. In support of this, addition of the RT inhibitor Nevirapine or AZT prevented spliced cDNA synthesis in these producer cells (Figures 7B and S5). Upon HIV-1 infection, the double-stranded linear FL DNA is synthesized and the ends can be joined by host ligases generating 2-LTR-circles (35,36). Therefore, detection of circular DNA with an LTR–LTR junction in the ΔZF2 transfected-cell (Figure 7C) indicates that some of the reverse transcripts made in producer cells are double-stranded full-length molecules. Our data clearly showed that NC zinc-finger mutation did not interfere with bona fide reverse transcription process but caused viral DNA synthesis to take place in HIV-1-producing cells, and to generate DNA-containing particles (Figure 8).
Figure 7.
Large amounts of viral DNA in HIV-1 ΔZF2-producing cells. (A) Spliced viral cDNAs were measured by qPCR in 293T cells transfected or not with ΔZF2 or wild-type plasmids (n = 3 ± SD). (B) Effects of Nevirapine (50 μM) and AZT (50 μM) treatments on spliced viral cDNA levels were measured by qPCR in 293T cells producing ΔZF2 virions (n = 3 ± SD). (C) Circular DNA forms were detected by PCR with primers flanking the 2LTR junction in both the infected cells (42CD4) and transfected cells (293T). All DNA samples were normalized to GAPDH gene copy number. A representative experiment is shown.
Figure 8.
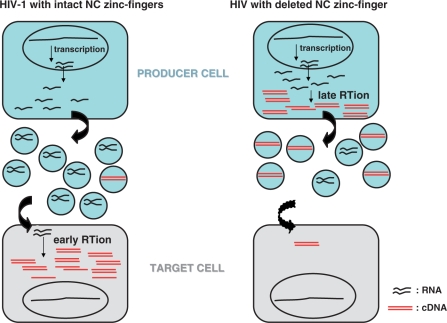
Cartoon depicting conceptual differences between early and late RT activation during HIV-1 replication with intact NC or with a deletion of an NC zinc finger.
DISCUSSION
Soon after infection, viral DNA synthesis is thought to take place within reverse transcription complexes (RTC) containing mature RT, integrase (IN), Vpr, the genomic RNA and NC protein molecules (3,37). The findings reported here favor the notion that the conserved NC zinc fingers exert, either directly or indirectly, a control over the timing of viral DNA synthesis during the late stages of HIV-1 replication. The start of viral DNA synthesis probably follows viral core formation and processing, and necessitates mature RT protein (38–40).
In producer cells, there is an accumulation of Gag and Gag-Pol precursors, containing NC and RT domains, respectively. Nevertheless, mature viral proteins and enzymes are also most probably present as indicated by the detection of mature capsid protein (p24) in HIV-1-producer cells [see for examples (13,14,41,42)]. In addition, both Gag and Gag-Pol precursors can also contribute to the reverse transcription reaction. The RT domain of Pol, which has some enzymatic activity (43), can select the replication tRNALys3 primer (44,45). The Gag–NC was found to chaperone primer tRNALys3 annealing to the genomic primer-binding site (PBS) (46) that is required for the initiation of cDNA synthesis by RT. Such a cellular environment should allow the start of viral reverse transcription and thus the presence of viral DNA in newly formed virions (47) (Figure 3, right panel).
Gag–NC is also implicated in the protease-mediated processing of Gag via its binding to the viral RNA (42,48,49). Although Gag processing was partially impaired when the NC zinc fingers were deleted, mature viral proteins were still easily detected in cells expressing the NC zinc fingers mutants (13,42) and could then endorse the late RT activity observed in cells producing HIV-1 with deleted NC zinc fingers.
Interestingly, the NC zinc fingers are important determinants that are also involved in the control of HIV-1 assembly, trafficking and budding (3,11,50). During these processes, the NC domain mediates important protein–protein interactions with other viral proteins such as Gag-Pol, Vpr and Vif (51), and with cellular proteins, actin (52), ALIX (14), topoisomerase I (53), Staufen (54) and APOBEC3G (3,10). These multiple Gag–NC functions likely contribute to the temporal fine-tuning of the replication steps including the timing of RT (Figure 8).
Then, how could we tentatively explain that viral DNA synthesis can take place in cells producing HIV-1 particles harboring a deletion in either one of the two zinc fingers?
The kinetics of the concerted HIV-1 assembly and budding processes might be slowed down by zinc finger deletions, allowing more time for viral DNA synthesis before particle release. Indeed, mutating or deleting the NC zinc fingers reduced the production of the NC mutant viral particles (Figure 1) with a drastic change in Gag localization (13). More precisely, they caused an accumulation of Gag in the cytoplasm or at the plasma membrane, but not at the level of late endosomes as for wild-type Gag (13), probably due to the fact that Gag–NC interactions with genomic RNA and with the cellular budding factor ALIX were impaired (14). Thus, we favor the notion that late RT is taking place in core structures formed of nonprocessed and processed Gag and Gag-Pol molecules. According to electron microscopy analysis of NC mutant cores (8,13) (Figure 1), it is likely that such intracellular viral core complexes are poorly condensed thus facilitating the reverse transcription reaction.
An alternative mechanism by which zinc finger mutations may stimulate late RT is the alleviation of cofactor(s) activity that negatively regulates RT. Several studies reported a Vif/APOBEC related regulation of the RT (51,55–57). These two proteins interact directly with Gag–NC and genomic RNA (58–60). Interestingly, Vif shares RNA chaperone activity with NC and Gag–NC proteins (51). Thus, in the context of the 293T cells in which APOBEC is poorly expressed, Vif may compete with NC-associated functions in assembly complexes and RTC. Indeed, in vitro Vif inhibits the NC-mediated tRNALys3 annealing and the initiation of RT (51). Then, Vif inhibitory effect could be relieved by the deletion of the NC zinc fingers which prevents Gag–NC/Vif interactions, and consequently, late RT could be stimulated by Gag–NC. It will thus be of interest to investigate viral DNA synthesis in cells expressing HIV-1 NC mutants and APOBEC3G.
The occurrence of late RT inside producer cells is also a property of the hepadnaviruses (e.g. hepatitis B virus) and foamy viruses which lack NC zinc fingers and release viral DNA-containing particles (61). Even though the life cycle of the simian foamy virus is poorly understood (62), the fact that HIV-1 NC zinc finger mutants could engage a similar DNA replication strategy (Figure 8) constitutes a fundamental issue and could bring an alternative explanation for the presence of viral DNA in HIV-1 particles isolated from the peripheral blood and semen of HIV-1-infected patients (29,47).
Last, these new findings on the role of HIV-1 further highlight the fact that the conserved zinc finger motifs should be viewed as a major target for new drugs inhibiting both the late and early steps of HIV-1 replication (3,63).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We wish to thank K.T. Jeang for critical reading of the article. This work was supported by grants from ANRS. L.H. was supported by Sidaction and L.D. by Agence Nationale de Recherche sur le Sida. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Darlix JL, Cristofari G, Rau M, Pechoux C, Berthoux L, Roques B. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. 2000;48:345–372. doi: 10.1016/s1054-3589(00)48011-7. [DOI] [PubMed] [Google Scholar]
- 2.Lener D, Tanchou V, Roques BP, Le Grice SF, Darlix JL. Involvement of HIV-I nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 1998;273:33781–33786. doi: 10.1074/jbc.273.50.33781. [DOI] [PubMed] [Google Scholar]
- 3.Darlix JL, Garrido JL, Morellet N, Mely Y, de Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- 4.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorelick RJ, Nigida SM, Jr, Bess JW, Jr, Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorfman T, Luban J, Goff SP, Haseltine WA, Gottlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorelick RJ, Chabot DJ, Rein A, Henderson LE, Arthur LO. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanchou V, Decimo D, Pechoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix JL. Role of the N-terminal zinc finger of human immunodeficiency virus type 1 nucleocapsid protein in virus structure and replication. J. Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Souza V, Summers MF. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 10.Goff SP. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 11.Muriaux D, Darlix JL, Cimarelli A. Targeting the assembly of the human immunodeficiency virus type I. Curr. Pharm. Des. 2004;10:3725–3739. doi: 10.2174/1381612043382701. [DOI] [PubMed] [Google Scholar]
- 12.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 13.Grigorov B, Decimo D, Smagulova F, Pechoux C, Mougel M, Muriaux D, Darlix JL. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J. Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams MC, Gorelick RJ, Musier-Forsyth K. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein's nucleic acid chaperone function. Proc. Natl Acad. Sci. USA. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 17.Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology. 2006;353:41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varmus H. Retroviruses. Science. 1988;240:1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- 20.du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, et al. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007;26:424–435. doi: 10.1038/sj.emboj.7601517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willey RL, Smith DH, Lasky LA, Theodore TS, Early PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houzet L, Morichaud Z, Mougel M. Fully-spliced HIV-1 RNAs are reverse transcribed with similar efficiencies as the genomic RNA in virions and cells, but more efficiently in AZT-treated cells. Retrovirology. 2007;4:30. doi: 10.1186/1742-4690-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smagulova F, Maurel S, Morichaud Z, Devaux C, Mougel M, Houzet L. The highly structured encapsidation signal of MuLV RNA is involved in the nuclear export of its unspliced RNA. J. Mol. Biol. 2005;354:1118–1128. doi: 10.1016/j.jmb.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, Mougel M. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007;35:2695–2704. doi: 10.1093/nar/gkm153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lori F, di Marzo Veronese F, de Vico AL, Lusso P, Reitz MS, Jr, Gallo RC. Viral DNA carried by human immunodeficiency virus type 1 virions. J. Virol. 1992;66:5067–5074. doi: 10.1128/jvi.66.8.5067-5074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Dornadula G, Alur P, Laughlin MA, Pomerantz RJ. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc. Natl Acad. Sci. USA. 1996;93:12519–12524. doi: 10.1073/pnas.93.22.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trono D. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia viruses. J. Virol. 1992;66:4893–4900. doi: 10.1128/jvi.66.8.4893-4900.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Zhang Y, Spicer TP, Abbott LZ, Abbott M, Poiesz BJ. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res. Hum. Retroviruses. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]
- 30.Warrilow D, Stenzel D, Harrich D. Isolated HIV-1 core is active for reverse transcription. Retrovirology. 2007;4:77. doi: 10.1186/1742-4690-4-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl Acad. Sci. USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 33.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 34.Quan Y, Rong L, Liang C, Wainberg MA. Reverse transcriptase inhibitors can selectively block the synthesis of differently sized viral DNA transcripts in cells acutely infected with human immunodeficiency virus type 1. J. Virol. 1999;73:6700–6707. doi: 10.1128/jvi.73.8.6700-6707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharkey ME, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan JL, Bucy RP, Kostrikis LG, Haase A, Veryard C, et al. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 2000;6:76–81. doi: 10.1038/71569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warrilow D, Harrich D. HIV-1 replication from after cell entry to the nuclear periphery. Curr. HIV Res. 2007;5:293–299. doi: 10.2174/157016207780636579. [DOI] [PubMed] [Google Scholar]
- 38.Katz RA, Skalka AM. The retroviral enzymes. Annu. Rev. Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 39.Stewart L, Vogt VM. Reverse transcriptase and protease activities of avian leukosis virus Gag-Pol fusion proteins expressed in insect cells. J. Virol. 1993;67:7582–7596. doi: 10.1128/jvi.67.12.7582-7596.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogt VM. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 41.Ottmann M, Gabus C, Darlix JL. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J. Virol. 1995;69:1778–1784. doi: 10.1128/jvi.69.3.1778-1784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawson L, Yu XF. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 43.Peng C, Chang NT, Chang TW. Identification and characterization of human immunodeficiency virus type 1 gag-pol fusion protein in transfected mammalian cells. J. Virol. 1991;65:2751–2756. doi: 10.1128/jvi.65.5.2751-2756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barat C, Lullien V, Schatz O, Keith G, Nugeyre MT, Gruninger-Leitch F, Barre-Sinoussi F, LeGrice SF, Darlix JL. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, Khorchid A, Wang J, Parniak MA, Darlix JL, Wainberg MA, Kleiman L. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160(gag-pol) and tRNA(Lys) incorporation into human immunodeficiency virus type 1. J. Virol. 1997;71:4378–4384. doi: 10.1128/jvi.71.6.4378-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Dornadula G, Pomerantz RJ. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J. Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng N, Pettit SC, Tritch RJ, Ozturk DH, Rayner MM, Swanstrom R, Erickson-Viitanen S. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 1997;71:5723–5732. doi: 10.1128/jvi.71.8.5723-5732.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng N, Erickson-Viitanen S. Cleavage of p15 protein in vitro by human immunodeficiency virus type 1 protease is RNA dependent. J. Virol. 1994;68:6207–6214. doi: 10.1128/jvi.68.10.6207-6214.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Resh MD. Intracellular trafficking of HIV-1 Gag: how Gag interacts with cell membranes and makes viral particles. AIDS Rev. 2005;7:84–91. [PubMed] [Google Scholar]
- 51.Henriet S, Sinck L, Bec G, Gorelick RJ, Marquet R, Paillart JC. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 2007;35:5141–5153. doi: 10.1093/nar/gkm542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu B, Dai R, Tian CJ, Dawson L, Gorelick R, Yu XF. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J. Virol. 1999;73:2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi H, Sawa H, Hasegawa H, Nagashima K, Sata T, Kurata T. Topoisomerase I dissociates human immunodeficiency virus type 1 reverse transcriptase from genomic RNAs. Biochem. Biophys. Res. Commun. 2004;313:1073–1078. doi: 10.1016/j.bbrc.2003.12.042. [DOI] [PubMed] [Google Scholar]
- 54.Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dornadula G, Yang S, Pomerantz RJ, Zhang H. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J. Virol. 2000;74:2594–2602. doi: 10.1128/jvi.74.6.2594-2602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dettenhofer M, Cen S, Carlson BA, Kleiman L, Yu XF. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 2000;74:8938–8945. doi: 10.1128/jvi.74.19.8938-8945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo F, Cen S, Niu M, Yang Y, Gorelick RJ, Kleiman L. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J. Virol. 2007;81:11322–11331. doi: 10.1128/JVI.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henriet S, Richer D, Bernacchi S, Decroly E, Vigne R, Ehresmann B, Ehresmann C, Paillart JC, Marquet R. Cooperative and specific binding of Vif to the 5' region of HIV-1 genomic RNA. J. Mol. Biol. 2005;354:55–72. doi: 10.1016/j.jmb.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 59.Bouyac M, Courcoul M, Bertoia G, Baudat Y, Gabuzda D, Blanc D, Chazal N, Boulanger P, Sire J, Vigne R, et al. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J. Virol. 1997;71:9358–9365. doi: 10.1128/jvi.71.12.9358-9365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 61.Yu SF, Baldwin DN, Gwynn SR, Yendapalli S, Linial ML. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 62.Delelis O, Lehmann-Che J, Saib A. Foamy viruses–a world apart. Curr. Opin. Microbiol. 2004;7:400–406. doi: 10.1016/j.mib.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 63.Rice WG, Supko JG, Malspeis L, Buckheit RW, Jr, Clanton D, Bu M, Graham L, Schaeffer CA, Turpin JA, Domagala J, et al. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]