Abstract
Regulation of the elongation phase of RNA polymerase II transcription by P-TEFb is a critical control point for gene expression. The activity of P-TEFb is regulated, in part, by reversible association with one of two HEXIMs and the 7SK snRNP. A recent proteomics survey revealed that P-TEFb and the HEXIMs are tightly connected to two previously-uncharacterized proteins, the methyphosphate capping enzyme, MEPCE, and a La-related protein, LARP7. Glycerol gradient sedimentation analysis of lysates from cells treated with P-TEFb inhibitors, suggested that the 7SK snRNP reorganized such that LARP7 and 7SK remained associated after P-TEFb and HEXIM1 were released. Immunodepletion of LARP7 also depleted most of the 7SK regardless of the presence of P-TEFb, HEXIM or hnRNP A1 in the complex. Small interfering RNA knockdown of LARP7 in human cells decreased the steady-state level of 7SK, led to an initial increase in free P-TEFb and increased Tat transactivation of the HIV-1 LTR. Knockdown of LARP7 or 7SK ultimately caused a decrease in total P-TEFb protein levels. Our studies have identified LARP7 as a 7SK-binding protein and suggest that free P-TEFb levels are determined by a balance between release from the large form and reduction of total P-TEFb.
INTRODUCTION
The human positive transcription elongation factor b (P-TEFb), which is composed of Cdk9 and cyclin T1 or cyclin T2 (1–3), stimulates the elongation phase of transcription by reversing the effects of negative elongation factors [for recent reviews see (4,5)]. P-TEFb plays an important role in the transcription of cellular genes (6), and is also a key factor for the expression of the human immunodeficiency virus type 1 (HIV-1) genome (7–9). Previous studies have shown that a complex containing the 7SK small nuclear RNA (snRNA), a 332-nucleotide transcript synthesized by RNAPIII (10,11), and the RNA binding proteins HEXIM1 (12,13) or HEXIM2 (14,15) can interact with P-TEFb and inhibit its kinase activity (16). Signal transduction pathways have been implicated in the general release of P-TEFb from the large form during cardiac hypertrophy (17,18) and upon treatment of cells with the differentiation agent, HMBA (19). Also, transfection of cells with the HIV transactivator, Tat, leads to release of P-TEFb from the large form and the formation of a Tat•P–TEFb complex (20). Recent results from several labs indicate that P-TEFb may play a critical role during development. Poised polymerases have been found on most human genes in embryonic stem cells (21) and on most developmental control genes in Drosophila (22,23).
Recently, soluble human protein complexes containing components of the transcription and RNA processing machineries were analyzed using protein affinity purification coupled to mass spectrometry. Thirty-two tagged polypeptides yielded a network of 805 high-confidence interactions (24). This study revealed that besides its positive (Brd4) (25,26) and negative (HEXIMs and 7SK snRNA) regulators, P-TEFb is tightly connected to many other proteins, including the previously uncharacterized protein BCDIN3 [Bicoid-interacting 3, homolog (Drosophila)]. BCDIN3 is a conserved methyltransferase that has the ability to add a methyl group on the γ-phosphate of 7SK and because of this was renamed the methyl phosphate capping enzyme, MEPCE (24). The addition of this unusual mono-methyl cap structure to RNAPIII-synthesized snRNAs, such as 7SK, was previously shown to occur post-transcriptionally and to be important for protecting the RNA from exonucleolytic degradation (27). Indeed, it has been shown that the cap structure enhances the stability of U6 and 7SK snRNAs and that uncapped U6 snRNA is rapidly degraded (28). In support for a role of capping by MEPCE on 7SK stability, silencing of MEPCE was shown to decrease the steady-state level of cellular 7SK in vivo (24). Here we follow-up on another previously uncharacterized protein, LARP7, which was discovered to be connected with P-TEFb and HEXIM proteins (24).
MATERIALS AND METHODS
Affinity purification of a human LARP7-containing complex
The cDNA encoding human LARP7 (Invitrogen; accession number BC066945) was cloned into the mammalian expression vector pMZI (29) carrying a tandem affinity purification (TAP) tag at its C-terminus and a stable human embryonic kidney (HEK) cell line EcR-293 (derived from HEK 293) carrying this construct was produced (30). The conditions for expression, affinity purification, mass spectrometry identification of proteins and gel filtration chromatography were as previously described (24).
Generation of affinity purified LARP7 antibodies
C-terminally His-tagged recombinant LARP7 was expressed in Escherichia coli BL21 CodonPlus (DE3) RIL cells (Stratagene) from a LARP7 containing pET23a expression vector (Novagene). Cells were induced overnight with 0.1 mM isopropyl 1-thio-β-d-galactopyranoside at 18°C. Purification on Ni-NTA resin and mono S was carried out as previously described for the purification of HEXIM proteins (14). One fraction from the Mono S elution containing predominately a 38 kDa, C-terminal proteolysis product of LARP7 was used as an antigen to generate sheep antibodies (Elmira Biologicals). The LARP7 antibodies were affinity purified from the crude serum by the method previously described for purification of HEXIM2 antibodies (14).
Glycerol gradient analysis
HeLa cells were grown to 90% confluence in two T-150 flasks. Cells were then treated for 1 h with 500 nM Flavopiridol or mock-treated with carrier (0.004% DMSO). Cells were harvested, lysed with a buffer containing 150 mM NaCl, and subjected to glycerol gradient fractionation as previously described (31). Gradient fractions were resolved by 9% SDS–PAGE followed by transfer to 0.45 µm nitrocellulose membranes. The antibodies used for western blotting were: sheep anti-cyclin T1 (ab27963; Abcam), rabbit anti-Cdk9 (sc-8338; Santa Cruz Biotechnology), affinity-purified sheep anti-HEXIM1 (Abcam), rabbit anti-MEPCE (24) and affinity-purified sheep anti-LARP7. Western blots were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (Sigma) and then treated with Super Signal Dura West Extended Duration Substrate (Pierce). Blots were imaged using a cooled charge-coupled camera (UVP).
Immunoprecipitation
Glycerol gradient analyses of Flavopiridol-treated or untreated HeLa cells were performed as described before. fractions 8–11, which contain the majority of LARP7 and the large form of P-TEFb, were pooled. To immunoprecipitate LARP7, affinity purified LARP7 antibodies were covalently attached to Actigel ALD beads (Sterogene) according to the manufacturer's instructions. Glycerol gradient pools were pre-cleared with beads alone at 4°C for 1 h. The flow through from the pre-clearing was then added to beads bound with affinity purified LARP7 or beads alone and rotated for 1 h at 4°C. The beads were washed with IP buffer [10 mM HEPES, 2 mM MgCl2, 10 mM KCl, 0.1% NP-40, 0.5 mM EDTA, 150 mM NaCl, 0.1% PMSF, and 1 unit/ml Complete EDTA Free Protease Inhibitor Cocktail (Roche)]. Trizol reagent (Invitrogen) was then added directly to the beads to extract immunoprecipitated 7SK according to the manufacturer's protocol. Extracted RNA was resolved on a denaturing 6% TBE acrylamide RNA gel, ethidium bromide (EtBr) stained, and visualized using a cooled charge-coupled camera (UVP). RNAs were transferred from the acrylamide gels to 0.2 µm Nytran N nylon membranes (Whatman) and then blocked for 1 h at 65°C in Ultrahybe Hybridization Buffer (Ambion). Radioactively tagged RNA oligos for U2, U6 or 7SK (31) were then added to the hybridization buffer and incubated overnight at 65°C. Blots were visualized using a Packard InstantImager and then exposed to film.
Co-immunoprecipitation of LARP7 associated proteins was similar; however, protein G Sepharose beads (Sigma) were used as the immunoprecipitation substrate to obtain better yields of LARP7 which was bound tightly by the antibodies. After immunoprecipitation, beads were resuspended in SDS loading buffer and boiled for 10 min at 100°C to release the bound protein from the beads. The immunoprecipitated proteins were resolved, blotted and probed with LARP7, Cdk9, MEPCE, and HEXIM1 antibodies as described before.
For co-immunoprecipitation from DRB-treated cells, cells were treated with 100 µM DRB for 1 h, and whole cell extracts were prepared using the same lysis buffer as was used for glycerol gradient analysis. Co-immunoprecipitations were carried out as described before.
Electrophoretic mobility shift assay and 7SK in vitro transcription
The mobility shift assay was carried out as previously described for the binding of HEXIM1 to RNA (31). In short, 0, 10 or 30 ng of recombinant LARP7 was combined in binding buffer (25 mM HEPES pH 7.6, 15% glycerol, 60 mM KCl, 0.1 mM EDTA, 5 mM DTT, 0.01% NP-40, 100 ng/ml BSA, 200 ng/rxn yeast tRNA) with 1 ng α-32P-UTP labeled 7SK for 20 min at room temperature and then resolved on a 4% native Tris polyacrylamide gel. 7SK was transcribed in vitro from a cut pCR2.1 (Invitrogen) template driven by a T7 promoter using T7 polymerase (Stratagene) in the presence of α-32P-UTP.
Small interfering RNA knockdown and luciferase assay
LARP7-specific small interfering RNA (siRNA) (custom synthesized by Integrated DNA Technologies, target sequence: 5′-ACAAGCGAGUAAACAUAUA-3′), MEPCE-specific siRNA (24) and control siRNA (Dharmacon siCONTROL Non-targeting pool), 7SK-specific siRNA (custom synthesized by Integrated DNA Technologies, targeting nucleotides 220-238) were transfected in 50%-confluent HEK 293 cells, TZM-bl cells or HeLa cells using Lipofectamine 2000 (Invitrogen) at a final siRNA concentration of 100 nM. TZM-bl cells, HeLa-cell derivatives that are stably transduced with a LTR-driven firefly luciferase cassette, were obtained from the NIH AIDS Reference and Reagent Program. Cells were transfected with the siRNAs twice at a 24 h interval. At various time intervals post-transfection, cells were lysed and LARP7 expression levels were monitored by western blotting. RNA blot analysis of total RNAs from LARP7-TAP eluate, HEK 293 cells mock-treated or treated with siRNA were carried out as previously described.
For luciferase assays, HEK 293 cells or TZM-bl cells were first mock-transfected or transfected with the control, MEPCE, LARP7 or double (MEPCE and LARP7) siRNAs as indicated before. Following an incubation of 5 h, the media was changed and a second siRNA transfection was performed 24 h later using similar conditions. After an incubation of 3 h, cells were co-transfected with 0.5 µg of the luciferase reporter construct and 0.1 µg of the Tat expressing plasmid or control plasmid. The plasmid encoding Tat (SV CMV Tat), the control plasmid (SV CMVexPA), as well as the LTR-driven firefly luciferase plasmid (pGL2-43-LTR-luc) were described previously (32). Transfected cells were collected 24 h post-final transfection and lysed using the luciferase assay system buffer (Promega). The protein concentration in cell lysates was determined using the Bio-Rad Protein Assay kit (Bio-Rad Laboratories). The luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega).
RESULTS
LARP7 is associated with P-TEFb, HEXIM1/2, MEPCE and 7SK RNA
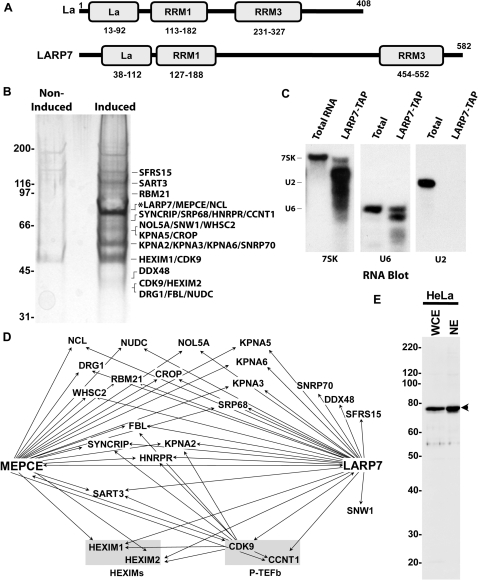
In a recent study (24), a protein interaction network of human transcription and RNA processing machinery was constructed. P-TEFb was found to be associated with its negative regulators (HEXIMs and 7SK snRNA), the 7SK capping enzyme (MEPCE), and among other proteins, a previously uncharacterized protein, LARP7. A bioinformatics analysis revealed that LARP7 is a member of a family of about a dozen La related proteins generated from seven human genes. Of all the family members LARP7 is the most closely related to La. In their amino terminal halves, both La and LARP7 contain the La domain and an associated RNA recognition motif (RRM1) (Figure 1A). A recent structure determination using a co-crystal of La and an RNA oligo (33) strongly suggests that both the La domain and RRM1 are involved in the interaction of La with the 3′ end of newly synthesized RNAPIII transcripts (34,35). Both proteins also contain a second RRM (RRM3) in their carboxyl terminal half (Figure 1A). Previous studies have shown that the La protein is a multifunctional protein that binds to and plays key roles in RNA metabolism (35,36). In the study presented here, we further characterize the association of LARP7 with the 7SK snRNP and determine the functional consequences of knocking down LARP7 in human cells.
Figure 1.
LARP7 is associated with P-TEFb, HEXIM, MEPCE, 7SK snRNA and U6 snRNA. (A) Schematic comparison of La with LARP7 with information obtained by querying NCBI's Conserved Domain Database. La—La domain, RRM1—RNA recognition motif 1, RRM3— RNA recognition motif 3. (B) SDS gel showing the composition of an affinity-purified LARP7-TAP eluate. The tagged protein is indicated by an asterisk. (C) Northern blots were performed on total RNA extracted from HEK 293 cells (T; 900 ng) or RNA extracted from the LARP7-TAP eluate (L; 60 ng) and probed with RNA oligos specific for U2, U6 or 7SK snRNAs. (D) Network highlighting the interactions of LARP7 with various RNA processing factors, P-TEFb (CCNT1/cyclin T1 and Cdk9 subunits), its negative regulatory factors (HEXIM1 and HEXIM2) and the 7SK snRNA capping enzyme MEPCE. (E) Western blot of HeLa whole cell extract (WCE) or HeLa nuclear extract (NE) probed with the affinity purified LARP7 antibody generated in sheep. Arrowhead—LARP7.
To better identify the interaction partners of LARP7, a human HEK 293 cell line was stably transfected with a construct that expressed C-terminally TAP-tagged LARP7 at about the same level as the endogenous LARP7. The proteins found in affinity purified LARP7 complexes were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) after separation of the proteins on an SDS gel. Figure 1B shows the composition of the TAP-tagged LARP7 eluate. Proteins that have been either identified in a control eluate prepared from the non-induced cell extract or represent interactions that had Interaction Reliability scores below a stringent threshold (24) were not listed. RNA extracted from the TAP-tagged LARP7 eluate was subjected to northern analysis. Consistent with the results obtained earlier with MEPCE (24), 7SK and U6 snRNAs, but not U2 snRNA were pulled down with LARP7 (Figure 1C). As we have previously shown for the MEPCE-TAP eluate (24), both 7SK and U6 RNAs were found partially degraded when compared to their respective cellular counterpart isolated from total cell RNA. This is due to extensive incubations of the RNPs in crude extracts. Also relatively less U6 came down with LARP7 compared to that brought down by MEPCE suggesting that LARP7 might preferentially associate with 7SK or that MEPCE might preferentially associate with U6. Combining the data obtained from the previous study in which MEPCE and Cdk9 were tagged (24) with the data obtained here by TAP tagging LARP7 allowed us to construct an interaction network for LARP7, MEPCE and P-TEFb (Figure 1D). Note that arrows point from the tagged protein to the protein discovered to interact. Many of the proteins discovered interact with both MEPCE and LARP7. Not surprisingly, LARP7 is tightly connected to P-TEFb (cyclin T1 and Cdk9 subunits), its negative regulatory factors (HEXIM1 and HEXIM2) and MEPCE (Figure 1D). Together, our results indicate that LARP7 may be a part of the 7SK•HEXIM•MEPCE•P-TEFb snRNP. To facilitate further analysis of the endogenous LARP7 protein, we developed a highly specific LARP7 antibody in sheep that detects a band near the predicted molecular weight (69 kDa) of LARP7 (Figure 1E).
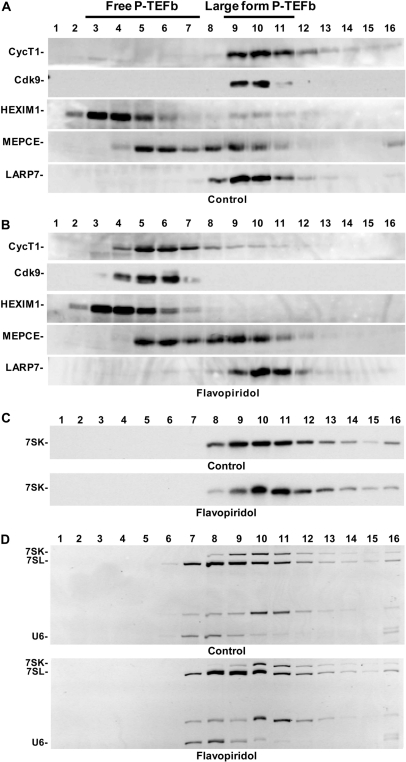
LARP7 co-sediments with large form P-TEFb and 7SK
To analyze the potential association of LARP7 and MEPCE with P-TEFb, HEXIM1 and 7SK, glycerol gradient sedimentation analysis of HeLa cell lysates was carried out. Otherwise identical cultures were either mock-treated or treated with 500 nM Flavopiridol for 1 h. Flavopiridol is a potent P-TEFb inhibitor that leads to inhibition of RNAPII transcription and to the release of P-TEFb and HEXIM1 from the large 7SK containing snRNP (37). As expected, in control cells most of the P-TEFb (cyclin T1 and Cdk9) and about 20% of HEXIM1 were found in fractions 9–11 that contain the 7SK snRNP with inactive P-TEFb (Figure 2A). The 7SK capping enzyme, MEPCE, was previously found in a complex containing P-TEFb, HEXIM1 and 7SK (24). Here slightly less than half of MEPCE was found to co-sediment with P-TEFb in control cells (Figure 2A) and the rest exhibited a slower sedimentation (found toward the top of the gradient). This observation is not surprising since MEPCE was found associated predominantly with U6 snRNA and could potentially be associated with other small RNAs that it targets for capping (24,38). LARP7, on the other hand, completely co-sedimented with P-TEFb and HEXIM1 in the 7SK snRNP (Figure 2A). Flavopiridol treatment led to the expected release of P-TEFb and HEXIM1 from the 7SK snRNP and subsequent shift towards the top of the gradient (Figure 2B). MEPCE sedimentation was unaffected. However, LARP7 shifted about one fraction down the gradient (Figure 2B).
Figure 2.
LARP7 co-sediments with the 7SK snRNP before and after P-TEFb and HEXIM release. (A) Glycerol gradient sedimentation analysis of untreated HeLa cells. Western blotting was performed with the indicated antibodies. (B) Gradient analysis of cells treated with 500 nM Flavopiridol for 1 h. (C) Northern blot of 7SK from gradient samples used in A and B. (D) Ethidium bromide stain of the RNA gels in C prior to northern transfer.
To determine if this shift down the gradient was consistent with LARP7 maintaining an association with 7SK after release of P-TEFb and HEXIM1, we extracted RNA from each of the glycerol gradient fractions and subjected them to northern analysis. In control cells, 7SK appeared in fractions 8–14, peaking in fraction 9; however, after Flavopiridol treatment, 7SK shifted down the gradient and peaked in fraction 10 (Figure 2C). To be sure that the shift in 7SK was not due to differences in the sedimentation or fractionation of the two samples, we visualized other RNAs with EtBr staining of the gel before the transfer. The pattern of sedimentation of other RNAs was identical between the two gradients, but as was found by the northern analysis, the pattern of 7SK changed (Figure 2D). These data strongly suggest that LARP7 remains in a stable complex with the 7SK snRNP even after Flavopiridol treatment and the release of P-TEFb and HEXIM1 from the snRNP. The shift down the gradient is likely due to association of hnRNP proteins (39–41).
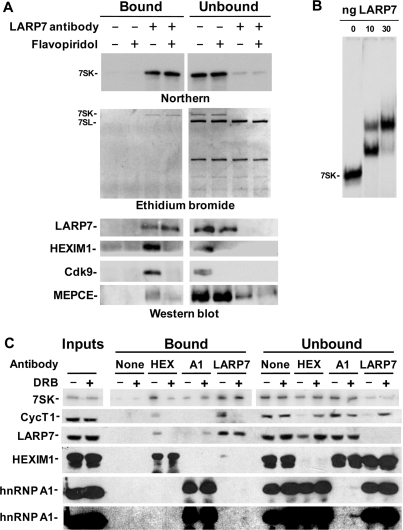
LARP7 is a stable component of the 7SK snRNP
Results presented so far strongly suggest that LARP7 forms a complex with the large form of P-TEFb and remains in complex with 7SK after P-TEFb release. To determine if endogenous LARP7 is actually a component of these complexes, we carried out a series of immunoprecipitation experiments using an affinity purified LARP7 antibody. Glycerol gradient fractions 8–11, which contain ∼80% of LARP7 (Figure 2A and B), were pooled from either control or Flavopiridol-treated cells and passed over beads that lacked antibody or had LARP7 antibody covalently attached. Analysis by either northern blot or EtBr staining of the RNA gel gave the same result. About 90% of 7SK was retained on the beads containing LARP7 antibody, but not on control beads (Figure 3A). Here, because care was taken to avoid ribonucleases, all the 7SK detected was intact. This indicates that LARP7 interacts with full length 7SK and not just the degradation products detected after the TAP purification earlier. Flavopiridol treatment of cells and the subsequent release of P-TEFb and HEXIM did not change the association of 7SK with LARP7 (Figure 3A). Pulldown of 7SK from these samples was highly specific, and did not affect the binding of the other small RNAs found in the gradient fractions (Figure 3A, ethidium bromide). Immunoprecipitation of LARP7 also resulted in the immunodepletion of HEXIM1 and Cdk9 from control gradient samples, and as expected these two proteins were absent from the samples from Flavopiridol-treated cells (Figure 3A, western blot). Some, but not all, of the MEPCE was pulled down with the LARP7 antibody and treatment of cells with Flavopiridol caused less MEPCE to be pulled down (Figure 3A, western blot).
Figure 3.
Immunoprecipitation of LARP7 confirms that it is a stable component of the 7SK snRNP. (A) Glycerol gradient fractions were pooled and subjected to LARP7 immunoprecipitation as described in the text. Nucleic acid and protein bound to the beads were either Trizol extracted or released in SDS loading buffer and analyzed by northern blotting, ethidium bromide staining and western blotting with the indicated antibodies. (B) Autoradiograph from the EMSA of E. coli expressed recombinant LARP7 bound to radioactively labeled 7SK. (C) Co-immunoprecipitation of the 7SK snRNP with HEXIM1 (HEX), hnRNP A1 (A1) and LARP7 antibodies before and after DRB treatment. Samples were then subjected to northern blotting for 7SK and western blotting with the indicated antibodies. Two exposures of the hnRNP A1 western blot are shown.
To determine if LARP7 associates directly with 7SK, we generated recombinant LARP7 in E. coli and used the protein in an electrophoretic mobility shift assay (EMSA) with radioactively labeled 7SK as the probe (Figure 3B). As increasing amounts of LARP7 were added, two 7SK mobility shifts were observed. The two RNPs could be due to the binding of LARP7 to one or two molecules of 7SK or one or two molecules of LARP7 binding to a single molecule of 7SK. From these results we conclude that LARP7 binds directly to 7SK.
To further examine the proteins associated with the 7SK snRNP before and after release of P-TEFb, immunoprecipitations from whole cell extracts of control cells or cells treated with another P-TEFb inhibitor, DRB, were performed using LARP7, HEXIM1 and hnRNP A1 antibodies. Regardless of the presence of the large form of P-TEFb, LARP7 was associated with a significant fraction of 7SK (Figure 3C). Immunoprecipitation of the 7SK snRNP from control cells using a HEXIM1 antibody verified that LARP7 is associated with this complex before but not after release of P-TEFb (Figure 3C). Recent research has shown that HEXIM and P-TEFb are replaced by hnRNP proteins after release from the 7SK snRNP (39,40). Here we confirm that the association of hnRNP A1 with 7SK increases after release, and show that only after release does hnRNP A1 associate with LARP7. Note that the immunoprecipitation with the LARP7 antibody brought down only a small fraction (less than 5%) of the hnRNP A1 protein which is more clearly seen with a longer exposure (Figure 3C, bottom blot). The low fraction bound to 7SK is expected since hnRNP A1 is found in many other RNPs. Overall, these results demonstrate that LARP7 is a stable component of the 7SK snRNP while HEXIM1, P-TEFb and hnRNP A1 are not.
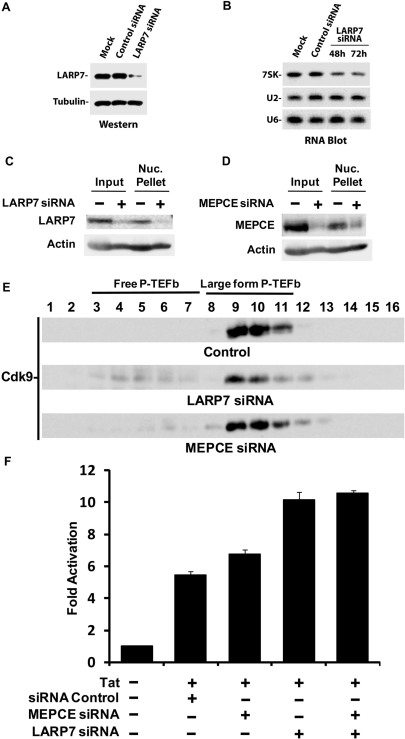
Knockdown of LARP7 decreases the steady-state level of 7SK while increasing free P-TEFb and enhancing Tat-mediated transcription
Given the well-established role of the La protein in the protection of RNAPIII transcripts from 3′ exonucleolytic degradation (34), we asked whether knockdown of LARP7 expression by siRNA would have a negative effect on 7SK snRNA steady-state levels in human cells. Figure 4A shows that reducing the expression of LARP7 by about 70% in vivo (Figure 4A) caused a decrease in the steady-state level of 7SK (Figure 4B). LARP7 knockdown had no effect on U2 or U6 snRNA levels (Figure 4B). This result indicates that LARP7 stabilizes the 7SK snRNA in human cells.
Figure 4.
Knockdown of LARP7 decreases 7SK levels, increases free P-TEFb, and enhances Tat transactivation. (A) LARP7 knockdown (72 h post-transfection) was monitored by western blotting in extracts from HEK 293 cells mock-transfected or transfected with control or LARP7 siRNA (tubulin was used as a loading control). (B) 7SK steady-state levels were assessed by RNA blotting using total RNA extracted from HEK 293 cells mock-transfected or transfected with control or LARP7 siRNA (48 and 72 h post-transfection) and probed with RNA oligonucleotides that specifically detect 7SK, U6 or U2 snRNAs. (C) Knockdown of LARP7 in HeLa cell glycerol gradient input or the leftover nuclear pellet 72 h post-transfection was determined by western blot and comparison with an actin loading control. (D) Same conditions as C, except the siRNA used targeted MEPCE. (E) Glycerol gradient analysis of the large form of P-TEFb after siRNA knockdown of LARP7 and MEPCE using the Cdk9 antibody. (F) HEK 293 cells were mock-transfected or transfected twice with control, MEPCE, LARP7 or double (MEPCE and LARP7) siRNAs and an HIV-1 LTR-luciferase reporter construct. A Tat-expressing plasmid was co-transfected where indicated. Luciferase activity was measured 24 h after transfection. Results are presented as fold activation relative to transfections in the absence of the Tat expression plasmid. N = 3.
Since knockdown of LARP7 resulted in reduced stability of 7SK, we next determined the effect of LARP7 knockdown on the ratio of the forms of P-TEFb and on P-TEFb activity. Glycerol gradient analysis was carried out on control cells or cells treated with either LARP7 or MEPCE siRNA. siRNA treatment resulted in a 70% reduction of both LARP7 and MEPCE (Figure 4C and D). Knockdown of either protein resulted in a small, but significant increase in the amount of free P-TEFb; however, the effect of MEPCE knockdown was less dramatic (Figure 4E). Because the release of P-TEFb is a highly regulated process, we speculated that the increase in free P-TEFb mediated by the loss of LARP7 and MEPCE and subsequent loss of 7SK would affect transcription. LARP7 and MEPCE siRNAs were introduced into human HEK 293 cells, followed by the transient transfection of an HIV-1 LTR-luciferase reporter construct in the presence of the HIV-1 transactivator, Tat. As shown in Figure 4F, knockdown of LARP7 caused a consistent ∼2-fold increase in Tat-mediated transactivation of the HIV-1 LTR. Knockdown of MEPCE had little or no effect either alone or in combination with knockdown of LARP7. Essentially the same results were observed in TZM-bl cells, a stable HeLa-derived cell line containing an integrated luciferase reporter gene driven by the HIV-1 LTR (data not shown). The relatively weak enhancement of Tat-mediated transactivation was most likely due to the 25–50% efficiency of LARP7 knockdown in treated cells (data not shown). Of note, these results compare well with those obtained in an earlier study of the effect HEXIM1 knockdown on transcription from the HIV-1 promoter, which also led to a ∼2-fold increase (12). Together, these data provide evidence that LARP7 plays an important role in stabilizing and maintaining the 7SK snRNP. Knockdown of LARP7 led to both a reduction in 7SK and to an increase in active P-TEFb.
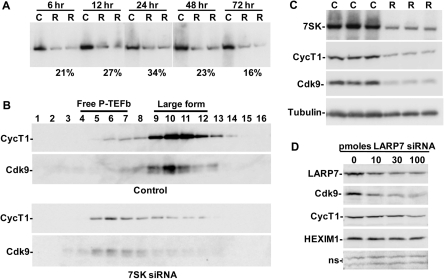
Knockdown of 7SK disrupts the large form of P-TEFb
To determine if the level of 7SK could directly impact the amount of free P-TEFb, siRNA directed against 7SK was used to knock down the snRNA. Previously, 7SK siRNA, but not a non-specific siRNA, was shown to specifically reduce 7SK even though it is a nuclear RNA (42). Total RNA was isolated 6, 12, 24, 48 or 72 h post-transfection from cells treated with siRNA or Lipofectamine 2000 alone. Northern blotting was performed to determine the extent of 7SK knockdown. Knockdown was rapid and persistent, with 7SK levels in siRNA-treated cells reduced to 21% of control cells at 6 h and 16% at 72 h (Figure 5A). The effect of knockdown of 7SK on the forms of P-TEFb was then examined by glycerol gradient sedimentation analysis of cell lysates made 48 h after mock transfection or transfection with 7SK siRNA. In the control cells, the vast majority of P-TEFb was found in the large form, with a small amount in the free form (Figure 5B, Control). The ratio of the free and large forms of P-TEFb was dramatically shifted by 7SK knockdown, with very little P-TEFb remaining in the large form (Figure 5B, 7SK siRNA). We conclude that reduction in the amount of 7SK in cells leads to the release of P-TEFb from the large form and is similar to the effect seen after LARP7 knockdown (Figure 4E) except that when 7SK siRNA is used the reduction in 7SK is much more rapid. However, at the 48 h time point analyzed the absolute signals for cyclin T1 and Cdk9 in 7SK siRNA-treated cells were reduced compared to control cells (Figure 5B).
Figure 5.
Direct knockdown of 7SK disrupts the large form of P-TEFb and decreases total P-TEFb. (A) HeLa cells were mock-treated or treated with 7SK siRNA for the indicated periods of time. One control and two independent siRNA treatments are shown for each time point. C-Control, R-siRNA-treated. (B) HeLa cells were mock-treated or treated with 7SK siRNA for 48 h, subjected to glycerol gradient sedimentation, and analyzed by western blot using the indicated antibodies. (C) HeLa cells were mock-treated or treated with 7SK siRNA for 64 h. Quantitative northern blotting for 7SK and western blotting for cyclin T1 and Cdk9 were performed. C-Control, R-siRNA-treated. (D) HeLa cells were treated with the indicated amount of LARP7 siRNA for 48 h, lysed in SDS loading buffer and western blotted with the indicated antibodies. Non-specific bands (ns) are provided as a loading control.
To explore the possibility that 7SK siRNA treatment affected the overall level of P-TEFb, cells were transfected with siRNA or mock-transfected, lysed with SDS after 64 h, and the lysates were analyzed by northern and western blotting. Northern blotting for 7SK confirmed that it was reduced to ∼20% of control cells (Figure 5C). Knockdown of 7SK caused a reduction in both Cdk9 and cyclin T1 to 30% (Figure 5C), indicating that the absolute amount of P-TEFb is dependent on 7SK levels. Evidently, the prolonged release of active P-TEFb from the disintegrating large form triggers the reduction of P-TEFb. Additionally, reduction of free P-TEFb could be part of the explanation for why only low levels of released P-TEFb were detected 72 h after the LARP7 siRNA treatment of cells (Figure 4E).
Finally, to determine if LARP7 knockdown had a similar effect as 7SK knockdown on total P-TEFb levels, HeLa cells were mock-transfected or transfected with 10, 30 or 100 pmoles of LARP7 siRNA for 48 h. LARP7 knockdown resulted in a significant decrease in both Cdk9 and cyclin T1 levels, paralleling the 7SK results, while HEXIM1 was unchanged (Figure 5D). HEXIM1 levels likely remained the same due to the fact that HEXIM1 in the large form is a small fraction of the total HEXIM1 (Figure 2A). Loss of the 7SK snRNP by either 7SK or LARP7 knockdown underscores the importance of 7SK stability in maintaining proper control over P-TEFb.
DISCUSSION
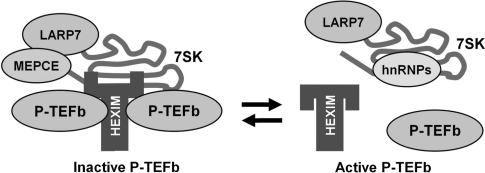
We have examined the association of LARP7 with 7SK, P-TEFb, HEXIM1, MEPCE and hnRNP A1. Although the study began with a TAP-tagged LARP7 pulldown, all other experiments involved analysis of endogenous LARP7. Glycerol gradient sedimentation and immunoprecipitation experiments were used to compare the 7SK snRNP in control cells with the snRNP in cells treated with P-TEFb inhibitors or siRNAs. Our study demonstrated that LARP7 is a stable component of two major 7SK snRNPs that are present before and after the release of P-TEFb (Figure 6). Treatment of cells with P-TEFb inhibitors caused the P-TEFb•HEXIM1•MEPCE•LARP7•7SK complex to be converted into an hnRNPA1•LARP7•7SK complex. Only a small (substoiciometric) amount of MEPCE was found to associate with 7SK and LARP7 after release of P-TEFb. Knockdown of LARP7 led to a reduction of 7SK, the release of P-TEFb from the large form and the activation of transcription from the HIV LTR. Additionally, reduction of 7SK by direct siRNA knockdown or by knockdown of LARP7 caused a time-dependent reduction in the total amount of P-TEFb. This suggests that there is a cellular mechanism that regulates the level of free P-TEFb when it is not inhibited by the 7SK snRNP. Based on bioinformatic analysis LARP7 is the most closely related family member to the founding protein, La. The similarities in the domain structure of La and LARP7 suggest that they may have similar properties. La has been found in association with many nascent RNAPIII transcripts including 7SK (40,41,43). We propose that LARP7 replaces La during 7SK snRNP maturation. This would explain our finding that ∼90% of the 7SK snRNP was immunoprecipitated with LARP7 antibodies (Figure 3). This represents a difference between LARP7 and La in that La is not present in the final mature form of most snRNPs (44–47). The results presented here show that LARP7 protects 7SK from degradation similarly to La's role in protecting other RNPs (34). It is possible that this is accomplished through the binding of the La domain and RRM1 of LARP7 to both the 3′ and 5′ ends of 7SK (33). In addition to its role in RNA protection, La has been implicated in nuclear localization and nucleolar trafficking of RNAs such as U6 and 7SL (36). Binding of LARP7 to 7SK may partially explain why 7SK is predominantly nuclear (48). Interestingly, recent analysis of the crystal structure of La shows that the La domain contains hallmarks of a DNA binding protein and the surfaces that were assumed to be involved in RNA binding are actually free (33). In conjunction with this finding, La has been shown to localize to RNAPIII promoters (49). It would be intriguing if LARP7 had a similar localization to DNA. If so then it could either be used to target inactive P-TEFb to positions on the chromatin for local release to enhance RNAPII transcription, or position 7SK on chromatin for local regulated inhibition of P-TEFb.
Figure 6.
Model of the 7SK snRNP before and after release of P-TEFb. Prior to treatment with P-TEFb inhibitors or cellular stress, the 7SK snRNP contains P-TEFb, a HEXIM dimer, MEPCE, LARP7 and 7SK. Following release, the 7SK snRNP is composed of, LARP7, 7SK and hnRNP proteins.
Binding of La to its RNA targets is a regulated process that is dependent on the 5′ and 3′ ends of the RNA and the phosphorylation state of La. A 5′-monomethyl-guanosine capping of U6 and B2 RNA has been shown to reduce the affinity of La for these RNAs while adenylation of these transcripts completely releases La (50,51). The recent discovery of MEPCE, which monomethylates U6 and 7SK on their 5′ termini may provide a mechanism for 7SK release from La and the subsequent association of LARP7. However, the status of the 5′ end of 7SK bound by LARP7, as well as, the methylation preference of LARP7 has yet to be determined. Alternatively, it is possible that 5′ end capping of 7SK might affect the way that LARP7 and 7SK interact and that could impact the association or dissociation of P-TEFb and HEXIM1. It is also possible that modification of the 3′ end of 7SK could be used for regulation of the protein composition of the 7SK snRNP.
Approximately 70% of 7SK is post-transcriptionally adenylated on the 3′ end (52) and this modification has been shown to affect La binding to other RNAPIII snRNAs. This modification may provide a mechanism for release of 7SK from La or release of P-TEFb from the LARP7•7SK snRNP. Release of P-TEFb from the LARP7•7SK snRNP could be accomplished by altering the association of LARP7 with 7SK in such a way that the secondary structure of the RNA is affected and prevents HEXIM and P-TEFb binding. It is possible that the 25% of 7SK that is associated with P-TEFb (Figure 3A, Northern and 3B, 7SK) might be the fraction of 7SK, ∼30%, that is not adenylated. Phosphorylation of Ser366 of La has been shown to reduce 5′ end binding (53) providing another feasible regulatory mechanism.
Results from the 7SK and LARP7 knockdown experiments performed here provide evidence for the coordination of two cellular mechanisms regulating the level of free P-TEFb. When the level of free P-TEFb was increased by disruption of the large form during siRNA-mediated knockdown of LARP7, an increase in P-TEFb activity was observed. After several days of LARP7 or 7SK siRNA treatment less total P-TEFb was found. This is consistent with an initial increase in P-TEFb activity as the large form is disrupted followed by a slower cellular compensation that leads to reduction in the total amount of P-TEFb protein by increasing turnover or decreasing synthesis of Cdk9 and cyclin T1. The mechanism for the reduction of excess free P-TEFb is likely to be proteosome mediated decay because when P-TEFb subunits were overexpressed most of the P-TEFb was quickly ubiquitylated and degraded by the proteasome (54,55). Degradation of the free P-TEFb would explain why we found only a slight increase in free P-TEFb after LARP7 knockdown. The idea that the amount of free P-TEFb arises from a balance between release from the large form and degradation of the free form is supported by the finding that knockdown of either Cdk9 or cyclin T1 results in a reduction of the total amount of P-TEFb in the cell without grossly affecting transcription of a reporter gene or cell viability (56). In the future, it is important to determine how these mechanisms are used to obtain the selective P-TEFb function required for control of transcription during differentiation and development.
Mechanisms regulating the balance of free versus inactive P-TEFb have been previously characterized. Knockdown of HEXIM1 was shown to have no effect on the absolute levels of P-TEFb or the amount of P-TEFb in the large form because HEXIM1 was functionally replaced by HEXIM2 (14,15). Two studies have shown that long-term treatment of cells with HMBA, caused an initial increase in free active P-TEFb, but ultimately resulted in a shift in the equilibrium back to the large inactive form (19,57). Together these studies provide support for a variety of cellular mechanisms that maintain the P-TEFb equilibrium in cells.
Comparative genomic analysis shows that LARP7 is closely related to the Drosophila melanogaster protein multi sex combs (mxc). The domain structure of mxc is very similar to that of LARP7 and contains a La motif and two RRMs. In flies, mxc is a member of the Polycomb group (PcG) of genes which regulate gene expression during differentiation and development (58). Mxc appears to function as a tumor suppressor because mutations were shown to result in uncontrolled malignant growth (59). Additionally, there is evidence that LARP7, studied under the alias DKFZP564K112, is a marker of gastric cancer as determined by microsatellite instabilotyping (60). Genetic studies in flies show that complete loss of mxc results in cell death after a few divisions while hypomorphic expression results in abnormalities during development (61). Mxc is also implicated in Drosophila hematopoiesis by controlling plasmatocyte proliferation and differentiation (62). It would be interesting to see if the phenotypes observed in flies are related to an interaction between fly P-TEFb and its LARP7 homolog mxc. With these results in mind, it is important that we determine how LARP7 regulates P-TEFb function during cell differentiation, development and cancer progression.
ACKNOWLEDGEMENTS
We are grateful to Guy Poirier and Denis Faubert for mass spectrometry analysis, Mathieu Blanchette and Christian Poitras for help with bioinformatics and Benoit Chabot for assistance with siRNA design. This work was supported by NIH GM35500 (D.H.P.); the Canadian Institutes for Health Research (B.C. and E.A.C.), the Natural Sciences and Engineering Research Council of Canada, Genome Canada and Génome Québec (B.C.); the Association pour la Recherche sur le Cancer, Agence Nationale de Recherche sur le SIDA, Agence Nationale pour la Recherche, Fondation pour la Recherche Médicale (O.B.); the CIHR and the Fonds de la recherche en santé du Québec (C.J.) and the University of Iowa Molecular and Cellular Biology Training Grant (B.J.K.). Funding to pay the Open Access publication charges for this article was provided by NIH GM35500.
Conflict of interest statement. None declared.
REFERENCES
- 1.Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 2.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 3.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 4.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 7.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 8.Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–322. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 12.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 13.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J. Biol. Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- 15.Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J. Biol. Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 16.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sano M, Schneider MD. Cyclin-dependent kinase-9: an RNAPII kinase at the nexus of cardiac growth and death cascades. Circ. Res. 2004;95:867–876. doi: 10.1161/01.RES.0000146675.88354.04. [DOI] [PubMed] [Google Scholar]
- 18.Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 19.Contreras X, Barboric M, Lenasi T, Peterlin BM. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 2007;3:1459–69. doi: 10.1371/journal.ppat.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 27.Shumyatsky GP, Tillib SV, Kramerov DA. B2 RNA and 7SK RNA, RNA polymerase III transcripts, have a cap-like structure at their 5′ end. Nucleic Acids Res. 1990;18:6347–6351. doi: 10.1093/nar/18.21.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shumyatsky G, Wright D, Reddy R. Methylphosphate cap structure increases the stability of 7SK, B2 and U6 small RNAs in Xenopus oocytes. Nucleic Acids Res. 1993;21:4756–4761. doi: 10.1093/nar/21.20.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards D, Beattie B, Emili A, Greenblatt JF. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 2004;3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]
- 30.Jeronimo C, Langelier MF, Zeghouf M, Cojocaru M, Bergeron D, Baali D, Forget D, Mnaimneh S, Davierwala AP, Pootoolal J, et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol. Cell Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q, Cooper JJ, Altwerger GH, Feldkamp MD, Shea MA, Price DH. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 2007;35:2503–2512. doi: 10.1093/nar/gkm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison GP, Lever AM. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ. Structural basis for recognition and sequestration of UUU(OH) 3′ temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol. Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maraia RJ, Bayfield MA. The La protein-RNA complex surfaces. Mol. Cell. 2006;21:149–152. doi: 10.1016/j.molcel.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Curry S, Conte MR. A terminal affair: 3′-end recognition by the human La protein. Trends Biochem. Sci. 2006;31:303–305. doi: 10.1016/j.tibs.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Maraia RJ. La protein and the trafficking of nascent RNA polymerase iii transcripts. J. Cell Biol. 2001;153:F13–18. doi: 10.1083/jcb.153.4.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- 38.Gupta S, Busch RK, Singh R, Reddy R. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of gamma-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J. Biol. Chem. 1990;265:19137–19142. [PubMed] [Google Scholar]
- 39.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 43.Stefano JE. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 44.Guddat U, Bakken AH, Pieler T. Protein-mediated nuclear export of RNA: 5S rRNA containing small RNPs in xenopus oocytes. Cell. 1990;60:619–628. doi: 10.1016/0092-8674(90)90665-2. [DOI] [PubMed] [Google Scholar]
- 45.Boelens WC, Palacios I, Mattaj IW. Nuclear retention of RNA as a mechanism for localization. RNA. 1995;1:273–283. [PMC free article] [PubMed] [Google Scholar]
- 46.Maraia R, Zasloff M, Plotz P, Adeniyi-Jones S. Pathway of B1-Alu expression in microinjected oocytes: Xenopus laevis proteins associated with nuclear precursor and processed cytoplasmic RNAs. Mol. Cell Biol. 1988;8:4433–4440. doi: 10.1128/mcb.8.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steitz JA, Berg C, Hendrick JP, La Branche-Chabot H, Metspalu A, Rinke J, Yario T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J.Cell Biol. 1988;106:545–556. doi: 10.1083/jcb.106.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurney T, Jr, Eliceiri GL. Intracellular distribution of low molecular weight RNA species in HeLa cells. J. Cell Biol. 1980;87:398–403. doi: 10.1083/jcb.87.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fairley JA, Kantidakis T, Kenneth NS, Intine RV, Maraia RJ, White RJ. Human La is found at RNA polymerase III-transcribed genes in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18350–18355. doi: 10.1073/pnas.0506415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Sinha K, Perumal K, Gu J, Reddy R. Accurate 3′ end processing and adenylation of human signal recognition particle RNA and alu RNA in vitro. J. Biol. Chem. 1998;273:35023–35031. doi: 10.1074/jbc.273.52.35023. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Perumal K, Reddy R. Inhibition of translation of mRNAs containing gamma-monomethylphosphate cap structure in frog oocytes and in mammalian cells. Gene Expr. 2000;9:133–143. doi: 10.3727/000000001783992623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinha KM, Gu J, Chen Y, Reddy R. Adenylation of small RNAs in human cells. Development of a cell-free system for accurate adenylation on the 3′-end of human signal recognition particle RNA. J. Biol. Chem. 1998;273:6853–6859. doi: 10.1074/jbc.273.12.6853. [DOI] [PubMed] [Google Scholar]
- 53.Intine RV, Tenenbaum SA, Sakulich AL, Keene JD, Maraia RJ. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol. Cell. 2003;12:1301–1307. doi: 10.1016/s1097-2765(03)00429-5. [DOI] [PubMed] [Google Scholar]
- 54.Garriga J, Bhattacharya S, Calbo J, Marshall RM, Truongcao M, Haines DS, Grana X. CDK9 is constitutively expressed throughout the cell cycle, and its steady-state expression is independent of SKP2. Mol. Cell Biol. 2003;23:5165–5173. doi: 10.1128/MCB.23.15.5165-5173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiernan RE, Emiliani S, Nakayama K, Castro A, Labbe JC, Lorca T, Nakayama Ki K, Benkirane M. Interaction between cyclin T1 and SCF(SKP2) targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell Biol. 2001;21:7956–7970. doi: 10.1128/MCB.21.23.7956-7970.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiu YL, Cao H, Jacque JM, Stevenson M, Rana TM. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1) J. Virol. 2004;78:2517–2529. doi: 10.1128/JVI.78.5.2517-2529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He N, Pezda AC, Zhou Q. Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol. Cell Biol. 2006;26:7068–7076. doi: 10.1128/MCB.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rajasekhar VK, Begemann M. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells. 2007;25:2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- 59.Santamaria P, Randsholt NB. Characterization of a region of the X chromosome of Drosophila including multi sex combs (mxc), a Polycomb group gene which also functions as a tumour suppressor. Mol. Gen. Genet. 1995;246:282–290. doi: 10.1007/BF00288600. [DOI] [PubMed] [Google Scholar]
- 60.Mori Y, Sato F, Selaru FM, Olaru A, Perry K, Kimos MC, Tamura G, Matsubara N, Wang S, Xu Y, et al. Instabilotyping reveals unique mutational spectra in microsatellite-unstable gastric cancers. Cancer Res. 2002;62:3641–3645. [PubMed] [Google Scholar]
- 61.Saget O, Forquignon F, Santamaria P, Randsholt NB. Needs and targets for the multi sex combs gene product in Drosophila melanogaster. Genetics. 1998;149:1823–1838. doi: 10.1093/genetics/149.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Remillieux-Leschelle N, Santamaria P, Randsholt NB. Regulation of larval hematopoiesis in Drosophila melanogaster: a role for the multi sex combs gene. Genetics. 2002;162:1259–1274. doi: 10.1093/genetics/162.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]