Abstract
The flowering plant genus Oenothera is uniquely suited for studying molecular mechanisms of speciation. It assembles an intriguing combination of genetic features, including permanent translocation heterozygosity, biparental transmission of plastids, and a general interfertility of well-defined species. This allows an exchange of plastids and nuclei between species often resulting in plastome–genome incompatibility. For evaluation of its molecular determinants we present the complete nucleotide sequences of the five basic, genetically distinguishable plastid chromosomes of subsection Oenothera (=Euoenothera) of the genus, which are associated in distinct combinations with six basic genomes. Sizes of the chromosomes range from 163 365 bp (plastome IV) to 165 728 bp (plastome I), display between 96.3% and 98.6% sequence similarity and encode a total of 113 unique genes. Plastome diversification is caused by an abundance of nucleotide substitutions, small insertions, deletions and repetitions. The five plastomes deviate from the general ancestral design of plastid chromosomes of vascular plants by a subsection-specific 56 kb inversion within the large single-copy segment. This inversion disrupted operon structures and predates the divergence of the subsection presumably 1 My ago. Phylogenetic relationships suggest plastomes I–III in one clade, while plastome IV appears to be closest to the common ancestor.
INTRODUCTION
Plastid chromosomes have received considerable attention to examine phylogenetic relationships among plants as well as to probe into eukaryotic genome evolution. Their endosymbiotic ancestry, limited coding potential, relatively conserved organization and well-defined structure provide a unique source of information to address a wide range of fundamental questions.
At present, sequences of entire plastid chromosomes are available from more than 100, generally distantly related taxa including protist, thallophytic, bryophytic and vascular plants (www.ncbi.nlm.nih.gov). This information has clarified overall evolutionary relationships for most groups of photoautotrophs as well as the outlines of the evolution of the eukaryotic cell with its compartmentalized genome. It emerged that three basic processes shaped that evolution: (i) the restructuring of the genetic potentials of the symbiotic partners (1); this conversion occurred predominantly at the unicellular level (2); (ii) the generation of novel gene sets and genetic programs with the advent of multicellular organisms and (iii) a co-evolution of the cellular genetic compartments that together constitute an integrated genetic system (3,4). The impact of this latter process that operates in relatively short evolutionary time scales becomes apparent after interspecific organelle exchanges, which even between related species can frequently cause serious disturbances in the development of the resulting hybrids or cybrids (5,6). Compartmental co-evolution generates Dobzhansky–Muller incompatibilities responsible for reproduction barriers and consequently represents an important, often neglected, factor in speciation processes (7,8).
Combined with cybrid technology and transplastomic approaches, sequence data of entire plastid chromosomes from closely related species could provide access to molecular mechanisms underlying plant genome evolution (6,9). However, such data are still scarce. Some information is available from the Solanacean family (10–13), from a few related cereal species, notably maize, rice, wheat, barley and sugar cane (14–18), and from a few legumes (19,20). These data have led to refined taxon topologies in some cases, and a recent study of the interspecific plastome–genome cybrids between tobacco and Atropa has allowed identification of a first molecular mechanism that causes plastome–genome incompatibility and could contribute to speciation processes (6,9). Ultimately, such approaches are meaningful only in a context in which the relationships of the plant groups being considered are well understood and other strategies, notably formal nuclear and organelle genetics, can be incorporated to overcome limitations of traditional phylogenetic analyses.
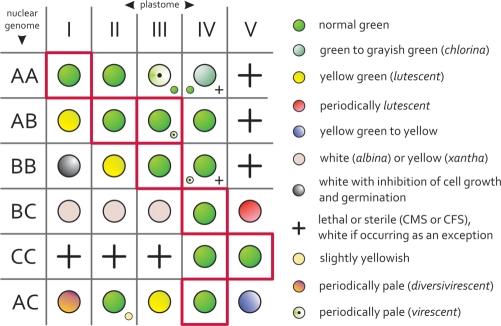
The genus Oenothera is uniquely suited for studying diversification of populations and speciation processes (5,21–25), because it possesses a unique combination of genetic features, and a comprehensive taxonomy, cytogenetics, ecology and classical genetics has been developed over more than a century. The features include the possibility of wide interspecific crossing, biparental transmission of organelles, generation of fertile interspecific plastome–genome cybrids or hybrids, as well as relatively frequent reciprocal translocations of chromosome arms that cause structural heterozygotic genomes. Collectively, these features allow the exchange of plastids and nuclei between species as well as the exchange of individual (or more) chromosome pairs or entire haploid chromosome sets. This often results in fertile but plastome–genome incompatible plants (Figure 1) and/or populations with varying chromosome formulas within species. Subsection Oenothera (=Euoenothera), the most intensely studied of the five subsections in the section Oenothera (5,22–24) is comprised of six basic nuclear genomes and five genetically and molecularly distinguishable plastid types (I–V; 26–29) that are distributed as distinct nuclear genome combinations in approximately a dozen species, predominantly over the North American continent (22,23).
Figure 1.
Plastome–genome compatibility/incompatibility in the subsection Oenothera, redrawn with permission (5,26). A, B and C represent the basic nuclear genotypes, I–V the five genetically distinguishable plastomes. Genotypes boxed in red represent naturally occurring species. Minor symbols indicate variances noted for some nuclear subgenotypes.
To gain causal access to compartmental co-evolution of plant genomes and speciation processes, we have begun to develop cell and molecular biological approaches for the Onagracean genus. In preceding communications we have reported on the complete nucleotide sequence of the plastid chromosome of Oenothera elata subsp. hookeri strain johansen, representing the Oenothera plastome I (30), on an EST library derived from the Oenothera A genome (31), and on cell, tissue culture and transformation protocols of that material (32–34). In this study we present and compare the complete nucleotide sequences of representatives of the five basic Oenothera plastomes and deduce their evolutionary relationship and temporal appearance. In a forthcoming study the phylogenetic relevance of the changes observed and potential plastome-encoded candidates responsible for interspecific plastome–genome incompatibility will be evaluated.
MATERIALS AND METHODS
Plastome sequencing
Fully developed leaves of Oenothera elata subsp. hookeri strain johansen (plastome I; 35), Oe. biennis strain suaveolens Grado (plastome II; 36), Oe. glazioviana strain rr-lamarckiana Sweden (ex genetica: Oe. lamarckiana, platome III; 37), Oe. parviflora strain atrovirens (syn: Oe. cruciata, plastome IV; 38) and Oe. argillicola strain douthat 1 (plastome V; 39) were collected from 5- to 6-week-old greenhouse grown plants [for detailed taxonomy of the subsection Oenothera see (23)]. DNA from plastome I was prepared from intact chloroplasts essentially as described in (40). Molecular cloning, sequencing and sequence analysis were performed as given in (11,30). Total DNA was isolated from plants carrying plastomes I to V, respectively, using the DNeasy™ Plant Mini Kit (Qiagen, Hilden, Germany). Since a number of errors were noted in the original sequence of plastome I (30), especially in the IR as also noted by others (41), its DNA was re-sequenced completely. PCR-derived fragments or fragments subcloned from plastid chromosomes serving as sequencing templates, spanning both DNA strands and overlapping a minimum of 200 bp, were generated over the entire chromosome of these plastomes. Almost the same, approximately 450, oligonucleotide primers (MWG Biotech, Ebersberg, Germany) employed for plastome I were used. Nucleotide sequences were determined by the dideoxy chain termination method (42) using energy-transfer fluorochrome-labeled dideoxynucleotides (43) with an ABI 377 robot (Applied Biosystems, Darmstadt, Germany). Sequence data were first subjected to the BLAST algorithm (44,45) provided by the National Centre for Biotechnology Information (Bethesda, MD, USA). Assembly and evaluation of sequences were performed with the SeqMan 6.1 program (DNASTAR Inc., Madison, WI, USA) using plastome I (accession no. AJ271079) as template for the other plastomes. All plastome sequences were aligned using the MegAlin 6.1 program (DNASTAR Inc., Madison, WI, USA) and the program BioEdit 5.0.9 (North Carolina State University) as alignment editor (46). Gene annotation of plastomes II to V was guided by the annotated plastome I (30).
Verification of inversion breakpoints
Total DNA of Oe. villaricae strain berteriana Schwemmle [syn: berteriana Erlangen (47)] was isolated as described earlier. The primer pair rbcLfor (5′-TGTGGCATATGCCTGCTCTG-3′) and psaI_IVP11rev (5′-GGAGAAATCCATTCTTGTCGTC-3′) deduced from a highly conserved region in the Oenothera plastomes was used to sequence the interval equivalent of the inversion breakpoint between rbcL and accD in the berteriana plastome. PCR was performed with BIO-X-ACT™ Long DNA Polymerase (Bioline GmbH, Luckenwalde, Germany) according to the standard protocol and the product was sequenced. The same strategy was used with the conserved primer pair rps16_IIP3for (5′-GAACAGAAGAAAGGGTGTCGAG-3′) and trnQ_IVP37for (5′-CACTGGAATTGACGAATAACC-3′) for the corresponding region of the inversion between rps16 and trnQ.
Northern analyses
Northern analysis was performed as described (48).
Repeat analysis
Two different types of repeats, palindromes and direct tandems, were analyzed in this study applying the programs palindrome and etandem of the EMBOSS suite (49). For both repeat types, the minimal cut-off identity between two copies was set to 90%. In case of multiple copies for one tandem, each copy was required to have at least one other member matching this constraint. The minimal and maximal copy size investigated were 16 to 100 bp for palindromic and 10 to 100 bp for tandem repeats, respectively. Gap size between palindromes was restricted to a maximal length of 3 kb. Overlapping repeats with sequence similarity were grouped into one repeat motif. For palindromes, both the direct and inverted part of two repeats had to overlap. For each repeat motif, the longest element is provided as its representative. Inversion breakpoints were separately analyzed in the five plastomes of the subsection Oenothera and berteriana Schwemmle.
Computational prediction of sigma-factor and RNA polymerase binding sites
Multiple alignments of intergenic regions delimited either by the 5′ neighboring gene or a maximum size of 600 bp were searched for polymerase binding sites. Due to overlapping binding specificity of different bacterial polymerase-like (PEP) σ-factors in plastids (50), no attempt was made to differentiate between individual factors. The consensus sequence TYRMNN(N)16–20 WANNWT was selected as search pattern which covers a wide range of experimentally reported sites. The regular expression found was similar to but less specific than the consensus suggested by Homann and Link (51) and Kanamaru and Tanaka (52). Binding sites of the phage-type polymerase (NEP promoters) were defined as matches to the consensus ATA0–1N0–1GAA(N)15–23YRT (53,54) representing NEP type Ib promoters. The other two NEP promoter types have been excluded from the analysis as the consensus of NEP type Ia promoters—YRTa—is too low for computational predictions and NEP type II promoters are known from just a single case (50). Candidate binding sites were positioned within the multiple alignments and edited by manual supervision to correct for misaligned regions, e.g. due to small repeats.
Prediction of Shine–Dalgarno sequences
Sequences 50 bp upstream of the start codon were searched for candidate Shine–Dalgarno regions using the program free2bind (55) and the 3′ 16S RNA sequence of Oenothera. A minimum free energy of 4.4 kcal and a maximum distance to the start codon of 23 bp was required for the reported matches.
Evolutionary analyses
Phylogenetic trees were generated from concatenated multiple codon-based alignments of the 47 genes variable in the five Oenothera plastomes. The dataset comprises 44 472 aligned characters present in six species including corresponding sequences of the Lotus japonicus plastome (19) as outgroup for tree rooting. Neighbor-Joining (NJ), Maximum-Likelihood (ML) and Maximum Parsimony (MP) from the PHYLIP package (56) were applied to infer trees. Bootstrap analysis for NJ and ML was performed with 1000 random samples each. In addition, gene-specific phylogenetic trees for all variable genes were determined by NJ and ML. A species tree was then built from individual gene trees using the consense program of PHYLIP. Trees for non-coding sequences were derived from 76 intergenic regions which showed nucleotide substitutions between the five Oenothera plastomes.
Synonymous and non-synonymous substitution rates were estimated applying the yn00 programme of the PAML package (57). F3x4 were selected as substitution matrix and Ka and Ks were determined by the Nei–Gojobori method as implemented in yn00. Rates for protein-coding genes variable among at least two of the five Oenothera plastomes were estimated from pairwise codon-based alignments. For five different plastomes, there are 10 pairwise combinations for each gene, resulting in a total of 780 rates for all and 470 rates for variable genes. Note that the computation of 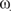 = Ka/Ks is not always applicable (e.g. for Ks = 0). Therefore,
= Ka/Ks is not always applicable (e.g. for Ks = 0). Therefore, 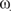 could be determined for only 215 pairwise combinations. To compare average Ka and Ks rates between species, a concatenated alignment of individual protein-coding regions were analyzed. Ycf1, ycf 2 and accD were excluded from the analysis as they contain large portions of repetitive regions and are highly variable between all species.
could be determined for only 215 pairwise combinations. To compare average Ka and Ks rates between species, a concatenated alignment of individual protein-coding regions were analyzed. Ycf1, ycf 2 and accD were excluded from the analysis as they contain large portions of repetitive regions and are highly variable between all species.
RESULTS
The plastid chromosomes of the five Oenothera species that represent the genetically distinguishable basic plastomes of subsection Oenothera were completely sequenced on both strands using a primer-based strategy. The sequences are deposited in GeneBank under the accession nos. AJ271079.3, EU262887, EU262889, EU26890 and EU262891. They include a revised version of plastome I of Oe. elata subsp. hookeri strain johansen (see ‘Materials and Methods’ section) and confirm the corrected ycf2 sequence reported recently (41). A corrigendum of the previous plastome I data will appear (30).
Size, gene content and design of the Oenothera plastid chromosomes
The individual plastome sequences assemble as circular structures of 165 728 bp (plastome I), 164 807 bp (plastome II), 165 225 bp (plastome III), 163 365 bp (plastome IV) and 165 055 bp (plastome V) in size including 56.6% coding regions and 43.4% non-coding regions (spacers and introns). The overall G+C content is 39.1%, about 41.7% in coding and 36.5% in non-coding sequence intervals (Supplementary Table 1), and is thus somewhat higher than found usually (36.8–39.0%) (http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/plastids_tax.html). As expected, the chromosomes display the typical quadripartite anatomy (40,58) characteristic for many plastid chromosomes, i.e. a pair of large inverted repeats that separate two single copy regions, the large single copy (LSC) region and the small single copy (SSC) region. The overall divergence of the chromosomes is expectedly low, between 96.3% and 98.6% similarity and 96.1% to 98.5% identity (Supplementary Table 2), and comparable to that found among various Nicotiana species and Atropa (96.0–98.5 identity; 10,11).
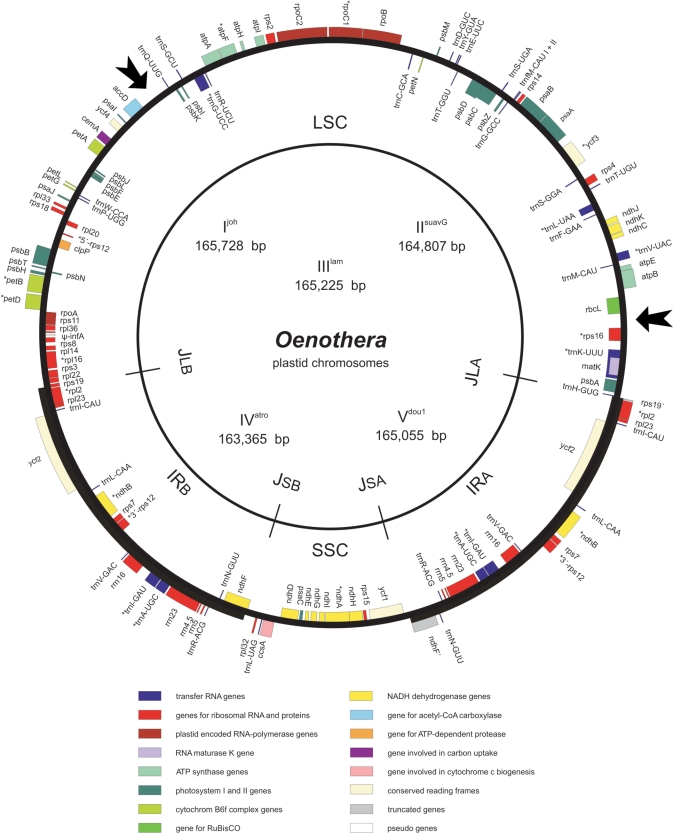
The five Onagracean chromosomes encode an identical set of 113 unique genes with identical gene order and gene clusters, 17 of these are duplicated in the IR (Figure 2). Genes, gene order and gene clusters are essentially colinear with those in the chromosomes from Nicotiana (10), Lotus (19), Atropa (11), spinach (59), Arabidopsis (60) and Eucalyptus (61) except for a large inversion of ∼56 kb in the LSC region (30,40,62,63) that occurred in the intergenic regions between the accD/rbcL and rps16/trnQUUG and reverses the order of genes between rbcL and trnQUUG (Figure 3). Specific also for the Oenothera plastid chromosomes are two copies of the initiator tRNA trnfMCAU which differ by a single nucleotide polymorphism in plastomes I, II, III and IV and are part of a tandem repeat structure (see later).
Figure 2.
Gene map of the Oenothera plastid chromosomes. Arrows mark the inversion breakpoints. Genes drawn on the outside are transcribed clockwise, on the inside counterclockwise.
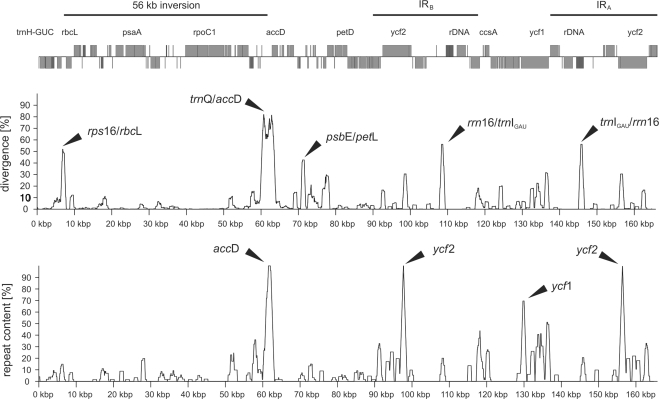
Figure 3.
Overall distribution of sequence divergence and repeat content in the Oenothera plastid chromosomes. Percentage repeat content and divergence on the y-axis were calculated using sliding sequence intervals of 1 kb with a step size of 10 bp along a multiple chromosome alignment. Whole chromosome alignments have been deduced from concatenated multiple alignments of intergenic regions and genes (see ‘Materials and Methods’ section). Repetitive regions frequently overlap with areas of high divergence.
The gene complement of the Oenothera plastid chromosomes is comprised of 4 rRNA genes (16S, 23S, 5S, 4.5S), a total of 31 distinct tRNA genes (the rrn genes and 7 trn genes are duplicated in the IR) and 78 protein-coding loci including ycf1, ycf2, ycf3 and ycf4 (Supplementary Table 3). Sixteen genes contain a single intron, one gene (ycf3) contains two; as in other species, rps12 mRNA is generated by transsplicing. All five species share the same set of introns, one class I intron (trnLUUA) and 17 of class II. Compared to tobacco, clpP lacks both introns, even though the surrounding coding sequences are conserved. In both cases, the deleted sequences coincide precisely with the established intron borders so that the mechanism of intron loss may have involved a processed RNA intermediate. As in various other plastid chromosomes, two pairs of genes overlap, atpB-atpE (4 bp) and psbC-psbD (52 bp); matK is located within the intron of trnKUUU. The 31 tRNA species (including both copies of trnfMCAU) represent 20 amino acid species and are sufficient to satisfy all the requirements for protein synthesis in the organelle. All protein-coding genes use the standard plastid/bacterial code with a predicted methionine/ATG start codon, except of ndhD which starts with an ACG [that is edited as has been shown for plastid chromosome I and IV (64)] and cemA which has been tentatively annotated with an ATA start codon (Supplementary Figure 1). The most common stop codon is TAA (51.3%) as in other plastid chromosomes (65). Seventy-three of the 78 protein-coding genes are present in most angiosperm plastomes. Five, accD, rpl22, rpl23, ycf1 and ycf2, are not generally found, but appear to be functional in Oenothera. Ycf1 and ycf2 with at least 2360 and 2313 codons in length, respectively, are well conserved within all studied dicotyledoneous plants; the former is entirely located within SSC. InfA, present in various species, appears to be a pseudogene in all plastomes, as in tobacco, Arabidopsis and Eucalyptus. SprA, reported from various dicots, e.g. Solanaceae, Arabidopsis or spinach (11,66), is missing. The IRA/LSC, LSC/IRB and IRB/SSC junctions are identical in all five plastomes; the SSC/IRA junction is identical in plastomes I, II and III, but differs by 2 additional bp in plastomes IV and V. The region downstream of ycf1 until ndhF′ is highly polymorphic among all five plastomes. NdhF and rps19 at the border of IRB generate truncated versions at the SSC/IRA and IRA/LSC junctions, respectively. NdhF′ lacks the 5′ end, rps19′ the 3′ end. It is not known whether the truncated versions are functional. Another region with locally conserved homology, noted previously in plastome I and often designated as ycf15, is present in all plastomes. Since it has premature in-frame stop codons generated by frequent insertions of variable sizes and no function or corresponding protein is known (59) we excluded it from the list of genes (Supplementary Table 3).
Sequence divergence and sequence repetition in Oenothera plastomes
The five plastomes are perfectly syntenic. This allowed a comparative analysis of indels and sequence repetition, which are usually difficult to analyze between distant species. Insertions, deletions and repetitions are relatively frequent. Relative to plastome IV, an additional 1456 nt insertions and 3819 nt deletions are found in plastome I. The corresponding nt numbers for the plastomes II, III and V are 1156/2598, 1701/3561, 864/2557, respectively. Indels occur less often in genes and are present in only relatively few polypeptide genes, notably in accD, clpP, ndhD, ndhF, rps18, rpl22, ycf1 and ycf2. These changes in coding potential will be discussed separately (see later and Supplementary Figures 1–4 and Supplementary Tables 4 and 5).
Overall distribution and types of repeats are highly similar between all five plastomes. On average 61 tandem repeats from 55 in plastome IV up to 70 in plastome I were detected, with a mean copy number of 4.5 copies per tandem and an average size of 41 bp per copy. The largest tandem repeat regions which span more than 1 kb and consist of variants of the AAG/TTC trinucleotide sequence are found in all plastomes at the two ycf2 genes in the IR. Expanded tandem repeats are also frequent and overlapping in accD and ycf1 contributing to the substantial sequence divergence of these genes. Rpl32, ndhF and the tandemly repeated trnfMCAUI and trnfMCAUII are associated with small identical tandem repeats in all five plastomes. More interesting are tandem repeats within coding regions, which are specific for particular plastomes, generally of moderate size and low copy number. Such repeats that change coding potential are found in clpP, ndhF and rps18 (Supplementary Figures 2 and 3). The plastome-specific repeat differences associated with ccsA, rpl22, rpl32, rps19 and trnSGCU are located outside the respective coding sequences. On average approximately 70 palindromic repeats with a maximal gap size of 3 kb or less were detected within the five plastome sequences. Palindrome sizes, however, are smaller and far less variable than tandem sizes, ranging from 32 bp (detection limit given by the threshold applied) up to 56 bp. They were detected in accD, ccsA, matK, ndhD, ndhF, ndhJ, petD, psaA, psaB, psbH, rpl32, rpoA, rpoB, rps18, ycf1, ycf2 and ycf4, but no notable changes in coding sequences are caused by palindrome repeats among the plastome types. Exceptions are the highly polymorphic genes accD and ycf2 as well as ndhD in which a palindromic repeat associated with a polyA-stretch shows a frame changing insertion of a single nucleotide (Supplementary Figure 2). Presence and absence of repetitive elements and divergent regions correlate well, but repeat content and high divergence are not strongly linked (Figure 3).
The large inversion
Except for the 56 kb inversion mentioned earlier, no other inversion has been detected in the five plastome sequences. The junctions of the inversion in the intergenic regions between trnQUUG/accD and rps16/rbcL are highly divergent and contain palindromes as well as tandem repeats (Figure 3). Overall the trnQUUG/accD spacer contains more tandem repeats (five to eight, depending on the plastome) than the rps16/rbcL spacer, with a maximum of two tandem repeats in plastome IV and none in plastome V. Three palindromic repeats were detected at the rps16/rbcL junction of plastomes I and II. In the trnQUUG/accD spacer region between two and no palindrome(s) were identified. Of particular note are the elements of clustered palindromes split among both spacer regions which were detected in all five plastomes. Their number varies enormously, between 1 (plastome V) and 17 (plastome III). Also, the same sequence motifs can be present in tandem and palindrome arrangement. The repetition patterns preclude an accurate demarcation of the insertion breakpoints and also affect the highly variable N-terminal region of AccD (Supplementary Figure 1 and see later).
In an attempt to better understand the inversion breakpoints and underlying processes, the corresponding regions in Oe. villaricae strain berteriana Schwemmle, a member of the closely related sister subsection Munzia, were sequenced (rps16/trnQUUG, accession no. EU255777 and rbcL/accD, accession no. EU255778). Subsection Munzia lacks this inversion (62). The berteriana regions corresponding to the Oenothera breakpoints between rps16/trnQUUG and rbcL/accD do not display pronounced divergence. The spacer region between rps16 and trnQ in that plastid chromosome lacks tandem and palindrome repeats. Nevertheless, the entire region is conserved and present in two parts in all five Oenothera plastomes, separated by an interspersed Oenothera-specific sequence interval. The berteriana region between rbcL and accD, in turn, lacks palindromes, but contains two tandem repeats. Approximately 1.5 kb of the berteriana rbcL/accD spacer are unique to the berteriana plastome. Conversely, trnQ/accD spacer sequences between 1.5 and 2.5 kb depending on the Oenothera plastome have no equivalent in the berteriana plastome. The same holds true for the Oenothera rps16/rbcL spacer, in which the number of unique nucleotides differs between approximately 50 and 500. The repeat structure in the rps16/rbcL spacer of the Oenothera plastomes appears to be linked to the inversed arrangement of rbcL and the result of duplication and relocation during the inversion process, as the berteriana equivalent is missing. As mentioned, palindrome repeat copies split among both spacers are not rare in the Oenothera plastomes, but no such cases were detected in berteriana with the selected threshold.
Differences in coding regions among Oenothera plastomes I–V
Three distinct sequence classes, protein-coding loci, tRNA and rRNA genes, have been evaluated for sequence conservation and changes between the five plastomes. Among the 78 protein-coding genes identified (including ycf1 to ycf4), 31 are identical in their nucleotide sequence, while 47 are variable between at least one plastome pair (Supplementary Tables 4 and 5). All carry substitutions of individual nucleotides and eight additionally indels. Sixteen genes, namely atpE, cemA, ndhI, ndhJ, petA, petB, psaB, psaC, psbC, rpl16, rpl33, rpoC1, rps4, rps11, rps14 and rps19 contain only synonymous substitutions which do not change the coding context. Only non-synonymous substitutions causing amino acid changes were detected in 11 genes: ndhB, ndhC, ndhH, psbB, rpl32, rpoA, rpoB, rpoC2, rps2, rps8 and rps15. Rpl22 contains synonymous substitutions and a single bp insertion/deletion, ndhF synonymous substitutions and a multiple base pair indel, rps18 multiple indels and a non-synonymous substitution. Both types of substitutions are found in ndhD, in plastome I together with a single base pair insertion, and a multiple base pair insertion in plastome V. Both, synonymous and non-synonymous substitutions are present in the remaining 16 genes (accD, atpA, atpB, atpF, ccsA, clpP, matK, ndhA, ndhE, petD, psaA, psbA, rps3, ycf1, ycf3 and ycf4). Five of them, clpP, accD, ndhD, ycf1 and ycf2, differ by multiple indels among plastomes. Of the 31 tRNA genes, 30 are identical and 1, trnfMCAUII, is variable in 1 nt. The mutation is not part of the anticodon, should have no or only a negligible effect on the folding of the tRNA, and hence not influence function, last not least since a second, not mutated copy, trnfMCAUI, is present in the Oenothera plastomes. The four rrn genes are identical in all five Oenothera plastomes.
Protein-coding genes with length polymorphisms can be operationally grouped into three categories, those with reading frame shifts (ndhD, rpl22 and rps18), those without reading frame shifts (ycf1, ycf2, accD, clpP and ndhF) and a third class, including atpA and psbB, which are known to differ in electrophoretic mobility of their products in plastome III and plastomes IV and V, respectively, independent of the genotype with which the plastid types are associated with (29). As most of the differences described reside in polypeptide regions known to be highly variable in plastid chromosomes in general, often at the very C-terminus of a polypeptide, it is unlikely that they are of functional and/or evolutionary relevance. Therefore, details of the changes in these 10 loci including verification by SNP, CAPS, Western (cemA) analysis and comparison with corresponding loci of usually 4–8 reference species are presented in Supplementary Figures 1–4 and Supplementary Table 5.
Differences in intergenic regions among plastomes I–V
Intergenic sequence intervals, which house various functional elements, such as promoters, terminators, processing signals or ribosome binding sites, are less conserved among plastomes (76 variable, 37 identical). On average, for each of the 113 intergenic regions, 24.1 indels (2743 for all regions) and 5.4 substitutions (616) have been detected. This contrasts observations made for coding regions in which only few indels have been found.
Approximately 600 bp upstream of each gene were searched for potential promoters of the bacterial type (−10 and −35 boxes) and phage-type polymerases. Various sites previously reported from other species (51,53,67–69) were found in the Oenothera plastomes. Seventy-five genes displayed a putative sigma-factor binding site and 69 genes contained type Ib-like NEP promoters. In total, for 88 genes at least one polymerase binding site could be deduced and in 56 genes promoter predictions were found for both types. For predicted PEP and NEP sites, 39 and 27 promoters had at least one difference in the binding site between one of the five plastomes, respectively. Differences found comprise a relatively wide spectrum of changes, including additional or lacking predicted binding sites as well as single point mutations and spacing differences. Their functional relevance as that of transcriptional start sites remain to be verified.
Regions coding for ribosomal binding sites were detected in 30 genes within the first 23 bp upstream of the initial start codon by their match to the 3′ end of the 16S rRNA. The sequences show no significant differences among the plastomes.
Evolutionary analysis, plastome pedigree
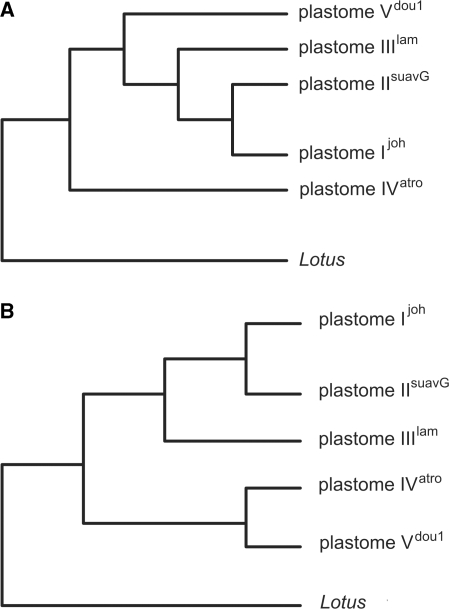
Phylogenetic trees were generated by NJ, ML and MP using the 47 variable genes as well as 76 variable intergenic regions, and rooted by orthologous genes or sequence intervals, respectively, of the Lotus japonicus plastome sequence (19) as outgroup. All three methods grouped plastomes I, II and III in one clade with plastomes I and II being close relatives (bootstrap values 100% for NJ and ML) (Figure 4). ML supported a common ancestor for clade I–III and a clade consisting of IV and V (bootstrap 99%). MP resulted in one maximal parsimonious tree, placing plastome V in one clade with plastomes I–III, while plastome IV is located in a separate branch. This tree is supported both by NJ analysis of the concatenated alignments (bootstrap 96.7% for separation of plastomes V and IV) as well as by a species/consensus tree of the 47 individual NJ gene trees. In the consensus tree, only 28.9% result in a branching pattern identical to the ML topology, while 40% of all gene trees support a separate branch for plastome IV. Hence, we propose that the latter tree topology reflects the phylogenetic relationship between the five Oenothera plastomes (see ‘Discussion’ section).
Figure 4.
Phylogenetic trees of the five Oenothera plastomes. Different tree topologies appear depending on the method. NJ and MP place plastomes I, II, III and V in one clade and plastome IV in a separate branch (A), whereas ML puts I, II and III versus IV and V as two separate clades (B).
Average substitution rates of protein-coding genes between the five plastomes were investigated to derive an estimate of their divergence time. Average Ks values between the five plastomes varied over more than one order of magnitude, from 3 × 10−4 for plastomes I and II up to 3.6 × 10−3 for plastomes I/II and V. Applying a calibration derived for dicotyledoneous chloroplast genes (70), divergence for plastomes ranged generally between 416 000 (II and III) and 830 000 years. The most distant pair (I and V) has diverged approximately 1 million years ago, while divergence of plastomes I and II is estimated to be as recent as 83 000 years ago (Supplementary Table 6).
All data obtained not shown in the manuscript are available upon request.
DISCUSSION
In this study we present the first complete sequences of plastid chromosomes from closely related, morphologically distinct and still interbreeding species, for which an organelle and nuclear genetics including cybrid technology is available. The populations of the North American subsection Oenothera are well characterized taxonomically, geographically and ecologically for more than a century (5,21–24). Interspecific cybrids of the five genetically distinguishable basic plastome types and the six basic genomes of that subsection can exist in all natural or artificial combinations (5,26 and Figure 1). The latter combinations are frequently developmentally impaired, revealing that co-evolution of nuclear and organellar genomes is an important element not only in speciation, but also in pre-speciation processes. In the clade diverging populations become apparent in changing chromosome formulas within species (22) and in intraspecific restriction fragment polymorphism of plastomes (29). The integrated compartmentalized genetic system of the plant cell, as of eukaryotic cells in general, evolves in its entirety generating additional constraints to evolution not found in prokaryotes. A natural or an artificially introduced mutation in any one of the genetic compartments of the cell necessitates a compensatory event or has to fulfil the requirement that the entire system remains genetically balanced, fully functional and phylogenetically fit (28).
The coding potentials of the five plastomes, 113 unique genes, are nearly identical and comparable to those of plastid chromosomes of vascular plants in size, organization, gene clustering and conservation. They deviate from the ‘ancestral form’ of plastid chromosomes by a single kilobase-magnitude inversion in the large single-copy segment. Their sizes are moderately larger than those found generally for higher plants and belong to the largest plastid chromosomes known from vascular plants that usually range between 130 and 160 kb (reviewed in 71). Genes are well-definable, except of the N-terminus of cemA (ycf10) encoding an inner envelope polypeptide involved in CO2 uptake, even after comparison with cemA loci from 50 reference species in which N-termini are not consistent (Supplementary Figure 1). Judged from PCR and Western analysis, the locus does not appear to be a pseudogene; also no evidence was found for a nuclear copy of this gene. This point remains to be settled in general.
Despite their gross conservation and close relationship, Oenothera plastid genomes are remarkably diverged, compared, for instance, to those of the Solanacean genera Atropa belladonna and tobacco (11). All four kinds of mutations that have been reported from plastid chromosomes, point mutations including changes in restriction sites, repetitions, insertions/deletions and inversions occurred. The former are scattered all over the circular chromosome, but found with different frequency in the individual sequence classes (Figure 3). Somewhat less mutations are expectedly found in the IR, probably due to copy-correction of the repeated segments (e.g. 72). Insertions/deletions and repetitions in direct or inverse orientation and restricted to repeat sizes of 16/30 bp or larger with ⩾90% sequence identity are relatively frequent (30,73). Repetitive and divergent regions are correlated suggesting a role of repeats in generating divergent regions, e.g. by illegitime recombination. Slipped mispairing may cause or contribute to changes in a number of instances as well (73–75). Coding sequences are generally well-conserved among the plastomes. Eight genes, almost exclusively non-photosynthetic, differ by indels, nearly a dozen by repeats including plastome-specific changes. Although non-protein and non-RNA coding regions, such as intergenic sequences and introns (together amounting nearly 45% of the chromosomes) evolve faster than genes, they still are quite well-conserved commensurate with the close relationship of plastomes and indicative that functional elements within these sequence intervals are conserved and compactly bundled. The junctions of the IR regions that often cause size differences between spermatophyte plastid chromosomes (e.g. 76) are almost invariant.
Inversions of different sizes in plastid chromosomes have been reported from various taxa including Bryophyta, gymnosperms, grasses, Fabaceae, Ranunculaceae, Caryophyllaceae, Geraniaceae, Asteraceae, Campanulaceae and Onagraceae (http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/plastids_tax.html). Three points of general interest are noteworthy. (i) Features that have been proposed to mediate inversions in plastid chromosomes by homologous or illegitimate, inter- or intramolecular recombination include the presence of dispersed repeated sequences and/or tRNA genes which frequently lie at or adjacent to endpoints (18,77–80). However, none of the elements discussed appears to extend to all evolutionary rearrangements and no satisfactory general model or explanation which would account for their involvement is available. In Oenothera, only one of the junctions lies adjacent to a tRNA gene. The exact breakpoints delimiting the inversion between trnQ and rps16 on the one end and accD and rbcL on the other (63) cannot be assigned by sequence comparison with the non-rearranged berteriana plastome, because of the complex pattern of repetitive sequences, of a lack of readily discernible vestiges of sequences involved bordering inversions, and because causes and mechanisms of the structural alteration that might reveal footprints of the event are not really understood. Single copies of sequences that are repetitive, scrambled and arranged in a relatively complex way in Oenothera are detectable in the Munzia plastome. Whether the structural change is in any way directly related to these elements remains to be shown.
(ii) Mapping of polymorphic restriction fragments in Oenothera plastomes has provided evidence of five preferred sites of mutation (40) which coincide well with sequence data and of which two may be recombinogenically relevant, since they reside at or near the inversion points (Figure 3). An intriguing number of inversions in different materials occurred at nearly identical positions, including these sites e.g. (15,18–20,78,79,81–83). For instance, a 50 kb inversion reported from legumes (19,20,83) resembles, but is not identical, to that found in Oenothera since it includes rps16 besides trnQ at one of the junctions. Two of the three inversions in Adonis and polymorphism in some grasses include the same region (15,79). These mutational ‘hot spots’ may be shared with the non-rearranged plastid chromosomes of tobacco species (84) and restriction analysis indicates that at least some of them contribute to intraplastome variation in Oenothera, which has been noted from all five plastomes (29 and unpublished data). Intraplastome variation is not rare (80,85), genetically virtually neutral, but the relevance of individual events in speciation, remains to be verified. In Oenothera, it would be particularly interesting to see whether the patterns reflect solely mechanistic imprecision with no phylogenetic relevance or correlate in part with geographic trends and genetic drifts of subpopulations in species differing in their chromosome formulas (22,24). Whatever causes and relation to recombinatoric events, it is unlikely that the shared sites reflect random effects. The underlying processes are probably comparable and of general nature, and, at least in some instances, may involve further features, such as transcription, some sequence specificity, elements of secondary structure or sites resembling att-lambda (86,87). Obviously, structural changes in plastid chromosomes are more complex than presently anticipated. An understanding of the requirements and modes of recombination could presumably contribute to clarify functional and evolutionary processes of plastid chromosomes, possibly also some way of transposition of sequence intervals to other organelles, but this can probably not be achieved by mere sequence study and will require more penetrating approaches, such as transformation strategies to determine recombinogenic activity of distinct constructs in vivo before general principles become apparent.
(iii) The Oenothera inversion disrupts known transcriptional linkage. Disruption of operons in plastid DNA of vascular plants by structural rearrangements is considered to be rare; only a few cases have been postulated from legumes and Campanulaceae (79,88–90). However, at present this inference is not fully conclusive due to the recent demonstration of a second plastid located RNA polymerase of nuclear origin (NEP, 67) which has not yet been considered in rearrangement analysis. As the ancestral eubacterial RNA polymerase type PEP, the phage-type NEP polymerase reads the entire plastid chromosome (47) though from different, often multiple and even operon-internal promoters (91). The Oenothera inversion separates accD from rbcL which in non-rearranged plastid chromosomes are members of a larger gene cluster, including rbcL-accD-psaI-ycf4-ycf10-petA. In tobacco, spinach, Arabidopsis and maize, this cluster is co-transcribed from a PEP promoter upstream of rbcL into a giant RNA species of ∼9 kb that is subsequently processed, often rapidly (Figure 5; 92). Removal of rbcL from the operon either required the replacement of the rbcL promoter by that of trnQ, use of the operon-internal NEP promoter in front of accD (93), acquisition of a new PEP promoter 5′ to accD (or trnQ) or use of the NEP promoter upstream of rpoB (c.f. Figure 2). The latter alternative is not likely, because of the general weakness of that promoter. On the other hand, the translocated rbcL promoter may serve the rbcL-rps16-trnKUUU-matK-(psbA-trnHGUG) gene cluster, since an RNA species of corresponding size can be detected in Oenothera (Figure 5). Obviously, the promoter patterns of both regions need to be functionally defined to settle the transcriptional consequences of the inversion.
Figure 5.
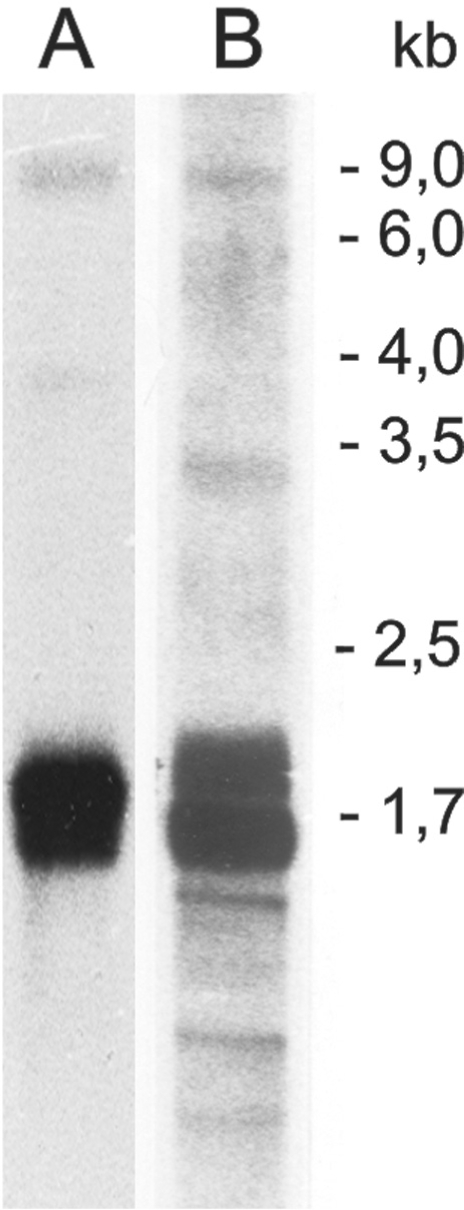
Northern analysis of rbcL operons from rearranged Oe. elata subsp. hookeri (A) and non-rearranged spinach (B) plastid chromosomes.
At the generic level the large inversion shared among all Oenothera plastomes provides a robust phylogenetic marker. The almost identical endpoints and its absence in the closely related South American subsection Munzia, in subsection Raimannia (62) or in the sister group Epilobium (94) suggest that this inversion is not caused by one of the proposed rare parallel inversions (79–81), but has arisen monophyletically within the Oenothera clade and late in the history of the Onagracean complex. It predates the divergence of the subsection, occurred either in the basic plastome IV or in the common ancestor of the subsection, and hence marks a recent split in the history of the genus. Even if the absolute calibration with less than 1 million years based on the molecular clock may require correction, the more than 10-fold difference of divergence times of the five plastomes will remain (Supplementary Table 6). The plastome cladograms determined separately from protein-coding and intergenic regions are nearly congruent and confirm the pedigree deduced from formal genetic approaches on the basis of both compartmental compatibility relations to genomes and differences in multiplication rates of plastids which are independent traits (5,73,95). The genetic determinants for the latter feature reside predominantly in the plastome (96). The pedigree is also consistent with changes in the thylakoid proteome mentioned earlier, notably the psbB variance of plastomes IV and V and that of atpA of plastome III (29 and Supplementary Figure 4), with a 2 bp IR extension shared by plastomes IV and V as well as with various other characters.
Sequences of entire plastid chromosomes have become a powerful research tool, not only to probe into chloroplast biogenesis, function and engineering, but also into eukaryotic genome evolution. The genus Oenothera provides unique opportunities to develop phylogenetic analyses based on mere sequence data into a ‘functional molecular phylogeny’ by applying other kinds of approaches, such as formal genetics combined with those of molecular and cell biology or physiology, to study diversification of populations and speciation processes in wider perspective. The availability of complete Oenothera plastome sequences should facilitate access to unveil molecular mechanisms that contribute to plant genome evolution, specifically to compartmental co-evolution, leading to divergence of populations or new species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by Grants from the Deutsche Forschungsgemeinschaft (SFB-TR1) to R.G.H. and J.M., and by the Hanns-Seidel-Stiftung (Bundesministerium für Bildung und Forschung) to S.G. We highly appreciate the participation of Rainer Maier who passed away in the early phase of the project. We thank Elli Gerick and Ingrid Duschanek for their excellent technical assistance, Peter Poltnigg and Elena Funk for help in sequence evaluation and Holger Grüne for the spinach Northern blot. The CemA antiserum was kindly provided by Jürgen Soll. Funding to pay the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft (SFB-TR1).
Conflict of interest statement. None declared.
REFERENCES
- 1.Herrmann RG. Eukaryotism, towards a new interpretation. In: Schenk HEA, Herrmann RG, Jeon KW, Müller NE, Schwemmler W, editors. Eukaryotism and Symbiosis. Berlin, Heidelberg, New York: Springer; 1997. pp. 73–118. [Google Scholar]
- 2.Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 3.Herrmann RG, Westhoff P. Thylakoid biogenesis: the result of a complex phylogenetic puzzle. In: Aro E-M, Andersson B, editors. Regulation of Photosynthesis. Dordrecht: Kluwer Academic Publishing; 2001. pp. 1–28. [Google Scholar]
- 4.Herrmann RG, Maier RM, Schmitz-Linneweber C. Eukaryotic genome evolution: rearrangement and coevolution of compartmentalized genetic information. Philos. Trans. R. Soc. Lond. Ser. B: Biol. Sci. 2003;358:87–97. doi: 10.1098/rstb.2002.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stubbe W. Oenothera – an ideal system for studying the interaction of genome and plastome. Plant Mol. Biol. Rep. 1989;7:245–257. [Google Scholar]
- 6.Schmitz-Linneweber C, Kushnir S, Babiychuk E, Poltnigg P, Herrmann RG, Maier RM. Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase alpha-subunit mRNA. Plant Cell. 2005;17:1815–1828. doi: 10.1105/tpc.105.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz-Linneweber C, Tillich M, Herrmann RG, Maier RM. Heterologous, splicing-dependent RNA editing in chloroplasts: allotetraploidy provides trans-factors. EMBO J. 2001;20:4874–4883. doi: 10.1093/emboj/20.17.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yukawa M, Tsudzuki T, Sugiura M. The chloroplast genome of Nicotiana sylvestris and Nicotiana tomentosiformis: complete sequencing confirms that the Nicotiana sylvestris progenitor is the maternal genome donor of Nicotiana tabacum. Mol. Genet. Genomics. 2006;275:367–373. doi: 10.1007/s00438-005-0092-6. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz-Linneweber C, Regel R, Du TG, Hupfer H, Herrmann RG, Maier RM. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol. Biol. Evol. 2002;19:1602–1612. doi: 10.1093/oxfordjournals.molbev.a004222. [DOI] [PubMed] [Google Scholar]
- 12.Yukawa M, Tsudzuki T, Sugiura M. The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum) Plant Mol. Biol. Rep. 2005;23:359–365. [Google Scholar]
- 13.Daniell H, Lee SB, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor. Appl. Genet. 2006;112:1503–1518. doi: 10.1007/s00122-006-0254-x. [DOI] [PubMed] [Google Scholar]
- 14.Maier RM, Neckermann K, Igloi GL, Kössel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 15.Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K. Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res. 2004;11:93–99. doi: 10.1093/dnares/11.2.93. [DOI] [PubMed] [Google Scholar]
- 16.Tsudzuki J, Tsudzuki T, Wakasugi T, Kinoshita K, Kondo T, Ito Y, Sugiura M. Comparative analysis of the whole chloroplast genomes from rice, maize and wheat. Endocytobiosis Cell Res. 2004;15:339–344. [Google Scholar]
- 17.Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 2007;115:571–590. doi: 10.1007/s00122-007-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 19.Kato T, Kaneko T, Sato S, Nakamura Y, Tabata S. Complete structure of the chloroplast genome of a legume, Lotus japonicus. DNA Res. 2000;7:323–330. doi: 10.1093/dnares/7.6.323. [DOI] [PubMed] [Google Scholar]
- 20.Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol. Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- 21.Raven PH. Onagraceae as a model of plant evolution. In: Gottlieb LD, Jain SK, editors. Plant Evolutionary Biology: A Symposium Honouring G. Ledyard Stebbins. London: Chapman and Hall; 1988. pp. 85–107. [Google Scholar]
- 22.Cleland RE. Oenothera: cytogenetics and evolution. In: Sutcliffe JF, Mahlberg P, editors. Experimental Botany. Vol. 5. London, New York: Academic Press; 1972. [Google Scholar]
- 23.Dietrich W, Wagner WL, Raven PH. Systematics of Oenothera section Oenothera subsection Oenothera (Onagraceae) In: Anderson C, editor. Systematic Botany Monographs. The American Society of Plant Taxonomists; 1997. Vol. 50. [Google Scholar]
- 24.Harte C. Oenothera: contributions of a plant to biology. In: Frankel R, Grossman M, Linskens HF, Maliga P, Riley R, editors. Monographs on Theoritical and Applied Genetics. Vol. 20. Berlin, Heidelberg, New York: Springer-Verlag; 1994. [Google Scholar]
- 25.Goldschmidt-Clermont M. Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 1998;177:115–180. doi: 10.1016/s0074-7696(08)62232-9. [DOI] [PubMed] [Google Scholar]
- 26.Stubbe W. Genetische Analyse des Zusammenwirkens von Genom und Plastom bei Oenothera. Z. Vererbungsl. 1959;90:288–298. [Google Scholar]
- 27.Herrmann RG. Internatinal Conference on Regulation of Developmental Processes in Plants. Germany: Halle; 1977. Studies on Oenothera plastid DNAs; p. 48. [Google Scholar]
- 28.Herrmann RG, Possingham JV. Plastid DNA - the plastome. In: Reinert J, editor. Results and Problems in Cell Differentiation. Vol. 10. Berlin, Heidelberg, New York: Springer; 1980. “Chloroplasts”, pp. 45–96. [DOI] [PubMed] [Google Scholar]
- 29.Herrmann RG, Seyer P, Schedel R, Gordon K, Bisanz C, Winter P, Hildebrandt JW, Wlaschek M, Alt J, et al. The plastid chromosomes of several dicotyledons. In: Bücher T, Sebald W, Weiß H, editors. Biological Chemistry of Organelle Formation. Berlin, Heidelberg, New York: Springer; 1980. pp. 97–112. [Google Scholar]
- 30.Hupfer H, Swiatek M, Hornung S, Herrmann RG, Maier RM, Chiu WL, Sears B. Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol. Gen. Genet. 2000;263:581–585. doi: 10.1007/pl00008686. [DOI] [PubMed] [Google Scholar]
- 31.Mráček J, Greiner S, Cho WK, Rauwolf U, Braun M, Umate P, Altstatter J, Stoppel R, Mlčochová L, Silber MV, et al. Construction, database integration, and application of an Oenothera EST library. Genomics. 2006;88:372–380. doi: 10.1016/j.ygeno.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Kuchuk N, Herrmann RG, Koop H-U. Plant regeneration from leaf protoplasts of evening primrose (Oenothera hookeri) Plant Cell Rep. 1998;17:601–604. doi: 10.1007/s002990050450. [DOI] [PubMed] [Google Scholar]
- 33.Stubbe W, Herrmann RG. Selection and maintenance of plastome mutants and interspecific genome/plastome hybrids from Oenothera. In: Edelman M, Hallick RB, Chua N-H, editors. Methods in Chloroplast Molecular Biology. Amsterdam: Elesevier Biomedical Press; 1982. pp. 149–165. [Google Scholar]
- 34.Mehra-Palta A, Koop H-U, Goes S, Troidl E-M, Nagy G, Tyagi S, Kofer W, Herrmann RG. Tissue culture of wild-type, interspecific genome/plastome hybrids and plastome mutants of evening primrose (Oenothera): controlled morphogenesis and transformation. Plant Cell Rep. 1998;17:605–611. doi: 10.1007/s002990050451. [DOI] [PubMed] [Google Scholar]
- 35.Cleland RE. Cyto-taxonomic studies on certain Oenotheras from California. Proc. Am. Philos. Soc. 1935;75:339–429. [Google Scholar]
- 36.Stubbe W. Genetische und zytologische Untersuchungen an verschiedenen Sippen von Oenothera suaveolens. Z. indukt. Abstamm- u. Vererbungsl. 1953;85:180–209. [PubMed] [Google Scholar]
- 37.Heribert-Nilsson N. Die Variabilität der Oenothera Lamarckiana und das Problem der Mutation. Z. indukt. Abstamm-u. Vererbungsl. 1912;9:89–231. [Google Scholar]
- 38.de Vries H. Oenothera. Berlin: Gebrüder Borntraeger; 1913. Gruppenweise Artbildung - Unter spezieller Berücksichtigung der Gattung. [Google Scholar]
- 39.Stinson HT. Cytogenetics and Phylogeny of Oenothera argillicola Mackenz. Genetics. 1953;38:389–406. doi: 10.1093/genetics/38.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon KHJ, Crouse EJ, Bohnert HJ, Herrmann RG. Physical mapping of differences in chloroplast DNA of the five wild-type plastomes in Oenothera subsection Euoenothera. Theor. Appl. Genet. 1982;61:373–384. doi: 10.1007/BF00272860. [DOI] [PubMed] [Google Scholar]
- 41.Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenblum BB, Lee LG, Spurgeon SL, Khan SH, Menchen SM, Heiner CR, Chen SM. New dye-labeled terminators for improved DNA sequencing patterns. Nucleic Acids Res. 1997;25:4500–4504. doi: 10.1093/nar/25.22.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 47.Schwemmle J, Haustein E, Sturm A, Binder M. Genetische und zytologische Untersuchungen an Eu-Oenotheren. Teil I bis VI. Z. indukt. Abstamm- u. Vererbungsl. 1938;75:358–800. [Google Scholar]
- 48.Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 2002;31:171–188. doi: 10.1046/j.1365-313x.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 49.Rice P, Longden I, Bleasby A. EMBOSS: The european molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 50.Liere K, Börner T. Transcription of plastid genes. In: Grasser KD, editor. Regulation of Transcription in Plants. Oxford: Blackwell Publishing; 2006. pp. 184–233. [Google Scholar]
- 51.Homann A, Link G. DNA-binding and transcription characteristics of three cloned sigma factors from mustard (Sinapis alba L.) suggest overlapping and distinct roles in plastid gene expression. Eur. J. Biochem. 2003;270:1288–1300. doi: 10.1046/j.1432-1033.2003.03494.x. [DOI] [PubMed] [Google Scholar]
- 52.Kanamaru K, Tanaka K. Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response in higher plants. Biosci. Biotechnol. Biochem. 2004;68:2215–2223. doi: 10.1271/bbb.68.2215. [DOI] [PubMed] [Google Scholar]
- 53.Silhavy D, Maliga P. Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr. Genet. 1998;33:340–344. doi: 10.1007/s002940050345. [DOI] [PubMed] [Google Scholar]
- 54.Kapoor S, Sugiura M. Identification of two essential sequence elements in the nonconsensus type II PatpB-290 plastid promoter by using plastid transcription extracts from cultured tobacco BY-2 cells. Plant Cell. 1999;11:1799–1810. doi: 10.1105/tpc.11.9.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starmer J, Stomp A, Vouk M, Bitzer D. Predicting Shine-Dalgarno sequence locations exposes genome annotation errors. PLoS Comp. Biol. 2006;2:e57. doi: 10.1371/journal.pcbi.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felsenstein J. Seattle: Department of Genetics, University of Washington; 1993. Phylogeny Inference Package (PHYLIP), ver. 3.5. [Google Scholar]
- 57.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 58.Gordon KHJ, Crouse EJ, Bohnert HJ, Herrmann RG. Restriction endonuclease cleavage site map of chloroplast DNA from Oenothera parviflora (Euoenothera plastome IV) Theor. Appl. Genet. 1981;59:281–296. doi: 10.1007/BF00264980. [DOI] [PubMed] [Google Scholar]
- 59.Schmitz-Linneweber C, Maier RM, Alcaraz JP, Cottet A, Herrmann RG, Mache R. The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol. Biol. 2001;45:307–315. doi: 10.1023/a:1006478403810. [DOI] [PubMed] [Google Scholar]
- 60.Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- 61.Steane DA. Complete nucleotide sequence of the chloroplast genome from the Tasmanian blue gum, Eucalyptus globulus (Myrtaceae) DNA Res. 2005;12:215–220. doi: 10.1093/dnares/dsi006. [DOI] [PubMed] [Google Scholar]
- 62.Hachtel W, Neuss A, vom Stein J. A chloroplast DNA inversion marks an evolutionary split in the genus Oenothera. Evolution. 1991;45:1050–1052. doi: 10.1111/j.1558-5646.1991.tb04370.x. [DOI] [PubMed] [Google Scholar]
- 63.Systma KJ, Hahn WJ, Smith JF, Wagner WL. Characterisation and phylogenetic utility of a large inversion in the chloroplast genome of some species in Oenothera (Onagraceae) Am. J. Bot. Suppl. 1993;80:79. [Google Scholar]
- 64.Hupfer H. PhD Thesis. Munich: Ludwig-Maximilians-University; 2002. Vergleichende Sequenzanalyse der fünf Grundplastome der Sektion Oenothera (Gattung Oenothera) – Analyse des Cytochrom-Komplexes. [Google Scholar]
- 65.Meurer J, Lezhneva L, Amann K, Gödel M, Bezhani S, Sherameti I, Oelmüller R. A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell. 2002;14:3255–3269. doi: 10.1105/tpc.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vera A, Sugiura M. A novel RNA gene in the tobacco plastid genome: its possible role in the maturation of 16S rRNA. EMBO J. 1994;13:2211–2217. doi: 10.1002/j.1460-2075.1994.tb06498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hess WR, Börner T. Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- 68.Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 2005;244:1–68. doi: 10.1016/S0074-7696(05)44001-2. [DOI] [PubMed] [Google Scholar]
- 69.Swiatecka-Hagenbruch M, Liere K, Börner T. High diversity of plastidial promoters in Arabidopsis thaliana. Mol. Genet. Genomics. 2007;277:725–734. doi: 10.1007/s00438-007-0222-4. [DOI] [PubMed] [Google Scholar]
- 70.Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 71.Palmer JD. Comparison of chloroplast and mitochondrial genome evolution in plants. In: Herrmann RG, editor. Plant Gene Research. Vol. 6. Berlin, Heidelberg, New York: Springer; 1992. “Cell Organelles”, pp. 99–133. [Google Scholar]
- 72.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl Acad. Sci. USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolfson R, Higgins KG, Sears BB. Evidence for replication slippage in the evolution of Oenothera chloroplast DNA. Mol. Biol. Evol. 1991;8:709–720. doi: 10.1093/oxfordjournals.molbev.a040680. [DOI] [PubMed] [Google Scholar]
- 74.Sears BB, Stoike LL, Chiu WL. Proliferation of direct repeats near the Oenothera chloroplast DNA origin of replication. Mol. Biol. Evol. 1996;13:850–863. doi: 10.1093/oxfordjournals.molbev.a025645. [DOI] [PubMed] [Google Scholar]
- 75.Winter P, Herrmann RG. A five-base-pair-deletion in the gene for the large subunit causes the lesion in the ribulose bisphosphate carboxylase/oxygenase-deficient plastome mutant sigma of Oenothera hookeri. Bot. Acta. 1987;101:42–48. [Google Scholar]
- 76.Goulding SE, Olmstead RG, Morden CW, Wolfe KH. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 77.Howe CJ, Barker RF, Bowman CM, Dyer TA. Common features of three inversions in wheat chloroplast DNA. Curr. Genet. 1988;13:343–349. doi: 10.1007/BF00424430. [DOI] [PubMed] [Google Scholar]
- 78.Ogihara Y, Terachi T, Sasakuma T. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl Acad. Sci. USA. 1988;85:8573–8577. doi: 10.1073/pnas.85.22.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johansson JT. There large inversions in the chloroplast genomes and one loss of the chloroplast gene rps16 suggest an early evolutionary split in the genus Adonis (Ranunculaceae) Plant Syst. Evol. 1999;218:133–143. [Google Scholar]
- 80.Tsumura Y, Suyama Y, Yoshimura K. Chloroplast DNA inversion polymorphism in populations of Abies and Tsuga. Mol. Biol. Evol. 2000;17:1302–1312. doi: 10.1093/oxfordjournals.molbev.a026414. [DOI] [PubMed] [Google Scholar]
- 81.Downie SR, Palmer JD. A chloroplast DNA phylogeny of the Caryophyllales based on structural and inverted repeat restriction site variations. Syst. Bot. 1994;19:236–252. [Google Scholar]
- 82.Hoot SB, Palmer JD. Structural rearrangements, including parallel inversions, within the chloroplast genome of Anemone and related genera. J. Mol. Evol. 1994;38:274–281. doi: 10.1007/BF00176089. [DOI] [PubMed] [Google Scholar]
- 83.Doyle JJ, Doyle JL, Ballenger JA, Palmer JD. The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol. Phylogen. Evol. 1996;5:429–438. doi: 10.1006/mpev.1996.0038. [DOI] [PubMed] [Google Scholar]
- 84.Salts Y, Herrmann RG, Peleg N, Lavi U, Izhar S, Frankel R, Beckmann JS. Physical mapping of plastid DNA variation among eleven Nicotiana species. Theor. Appl. Genet. 1984;69:1–14. doi: 10.1007/BF00262529. [DOI] [PubMed] [Google Scholar]
- 85.Soltis DE, Soltis PS, Milligan BG. Intraspecific chloroplast DNA variation: systematic and phylogenetic implications. In: Soltis PS, Soltis DE, Doyle JJ, editors. Molecular Systematics of Plants. New York: Chapman and Hall; 1992. pp. 117–150. [Google Scholar]
- 86.Howe CJ. The endpoints of an inversion in wheat chloroplast DNA are associated with short repeated sequences containing homology to att-lambda. Curr. Genet. 1985;10:139–145. doi: 10.1007/BF00636479. [DOI] [PubMed] [Google Scholar]
- 87.vom Stein J, Hachtel W. Deletions/insertions, short inverted repeats, sequences resembling att-lambda, and frame shift mutated open reading frames are involved in chloroplast DNA differences in the genus Oenothera subsection Munzia. Mol. Gen. Genet. 1988;213:513–518. doi: 10.1007/BF00339624. [DOI] [PubMed] [Google Scholar]
- 88.Palmer JD, Osorio B, Thompson WF. Evolutionary significance of inversions in legume chloroplast DNAs. Curr. Genet. 1988;14:65–74. [Google Scholar]
- 89.Milligan BG, Hampton JN, Palmer JD. Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol. Biol. Evol. 1989;6:355–368. doi: 10.1093/oxfordjournals.molbev.a040558. [DOI] [PubMed] [Google Scholar]
- 90.Cosner ME, Jansen RK, Palmer JD, Downie SR. The highly rearranged chloroplast genome of Trachelium caeruleum (Campanulaceae): multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr. Genet. 1997;31:419–429. doi: 10.1007/s002940050225. [DOI] [PubMed] [Google Scholar]
- 91.Kapoor S, Wakasugi T, Deno H, Sugiura M. An atpE-specific promoter within the coding region of the atpB gene in tobacco chloroplast DNA. Curr. Genet. 1994;26:263–268. doi: 10.1007/BF00309558. [DOI] [PubMed] [Google Scholar]
- 92.Meurer J, Grevelding C, Westhoff P, Reiss B. The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 1998;258:342–351. doi: 10.1007/s004380050740. [DOI] [PubMed] [Google Scholar]
- 93.Hirata N, Yonekura D, Yanagisawa S, Iba K. Possible involvement of the 5′-flanking region and the 5′ UTR of plastid accD gene in NEP-dependent transcription. Plant Cell Physiol. 2004;45:176–186. doi: 10.1093/pcp/pch021. [DOI] [PubMed] [Google Scholar]
- 94.Schmitz UK, Kowallik KV. Polymorphism and gene arrangement among plastomes of ten Epilobium species. Plant Mol. Biol. 1986;7:115–127. doi: 10.1007/BF00040138. [DOI] [PubMed] [Google Scholar]
- 95.Stubbe W, Steiner E. Inactivation of pollen and other effects of genome-plastome incompatibity in Oenothera. Plant Syst. Evol. 1999;217:259–277. [Google Scholar]
- 96.Chiu W-L, Stubbe W, Sears BB. Plastid inheritance in Oenothera: organelle genome modifies the extent of biparental plastid transmission. Curr. Genet. 1988;13:181–189. [Google Scholar]