Abstract
Short interfering RNA (siRNA)-based RNA interference (RNAi) is widely used for target gene knockdown in mammalian cells. To clarify the position-dependent functions of ribonucleotides in siRNA, siRNAs with various DNA substitutions were constructed. The following could be simultaneously replaced with DNA without substantial loss of gene-silencing activity: the seed arm, which occupies positions 2–8 from the 5′end of the guide strand; its complementary sequence; the 5′end of the guide strand and the 3′overhang of the passenger strand. However, most part of the 3′ two-thirds of the guide strand could not be replaced with DNA, possibly due to binding of RNA-recognition proteins such as TRBP2 and Ago2. The passenger strand with DNA in the 3′end proximal region was incapable of inducing off-target effect. Owing to lesser stability of DNA–RNA hybrid than RNA duplex, modified siRNAs with DNA substitution in the seed region were, in most cases, incapable to exert unintended gene silencing due to seed sequence homology. Thus, it may be possible to design DNA–RNA chimeras which effectively silence mammalian target genes without silencing unintended genes.
INTRODUCTION
Several nucleic acid-based methods for knockdown of target gene activity have become available over the last several decades, and RNA interference (RNAi) is probably the most prominent. In mammals, short interfering RNA (siRNA), 19-bp long double-stranded (ds) RNA with 2-nt 3′overhangs, is widely used for inducing RNAi (1,2). In Drosophila cells, a heterodimer of Dicer2 and R2D2 is considered to sense the differential stability of siRNA duplex ends in determining the guide strand (3,4). The Dicer2/R2D2 dimer in RLC [RNA-induced silencing complex (RISC) loading complex] is gradually replaced by Argonaute (Ago). Protein-complexed siRNA undergoes unwinding. Recent studies on the structures of archea and eubacteria Ago-like proteins (5–7) and the PAZ domain of human Ago1 (8) have provided a great deal insight into the molecular mechanism of RNAi in mammals and other animals. The 5′ and 3′ends of the guide strand may be anchored in pockets formed in the Mid and PAZ domains of Ago, respectively (4,7,9). The 5′-proximal ‘seed’ nucleotides (10), occupy position 2–8 measured from the 5′end of the guide strand, and are present on the surface of the Mid-PIWI lobe associated with a linker L1 in a quasi-helical form in the RISC, may serve as the entry or nucleation site for mRNA (7).
In active RISCs, target mRNA is cleaved by the RNase H-like slicer activity of Ago (Ago2 in the case of mammals) (11,12). Only siRNA with seed sequence homology to the target may serve as microRNA and induce gene silencing via translational repression rather than mRNA cleavage (13). In addition, recent microarray profiling experiments revealed that unintended reduction in expression of a large number of transcripts can also be observed following the transfection of siRNA without complete sequence complementarity to the transcripts, reminiscent of microRNA-dependent reduction in gene expression (10,14,15).
Certain ribonucleotides in siRNAs undergo substitution with deoxyribonucleotide counterparts without substantial loss of gene-silencing activity. The 3′overhangs are frequently replaced with deoxythymidine to prevent the degradation of siRNA in cells (1,16,17). The passenger strand of siRNA may be substituted totally with the DNA counterpart (18). Chiu and Rana (19) introduced various chemically modified nucleotides into siRNAs and found that the 2′OH of the guide strand near the mRNA cleavage site may not be required for catalytic ribonuclease activity of RISC.
Here, we examined the effects of systematic deoxyribonucleotide substitutions of highly functional siRNAs (20) on gene silencing. The 5′end and the 5′proximal ‘seed’ arm of the guide strand were found capable of being completely replaced with cognate deoxyribonucleotides with little or no loss of gene-silencing activity. In contrast, replacing the 3′ proximal RNA sequence of the guide strand with its DNA counterpart resulted in almost complete loss of gene-silencing activity. Unlike the nonmodified siRNAs, DNA-modified siRNAs, in which the seed region is DNA and the remainder is RNA, could hardly exert reduction of the activity of genes other than the target gene (off-target effect) (21). These findings may indicate most mammalian genes to be effectively knocked down without substantial off-target effect by treating cells with a class of DNA-modified siRNAs possessing a DNA-seed-arm sequence.
MATERIALS AND METHODS
Cell culture and gene-silencing activity assay
Chinese hamster CHO-K1, human HeLa, mouse embryonic stem (ES) cells (E14TG2a) and Drosophila S2 cells were cultured and subjected to gene-silencing assay as described previously (20). Briefly, 1-ml cell suspensions of CHO-K1 (1 × 105 cells/ml), HeLa (1 × 105 cells/ml), E14TG2a (2 × 105 cells/ml) and S2 (1 × 106 cells/ml) were inoculated in a 1.5-cm well 24 h prior to transfection. Cells were transfected with pGL3-Control (1 µg, Promega) or pGL2-Control (1 µg; Promega), both coding for firefly luciferase (luc) gene, and Renilla-luc-gene encoding pRL-SV40 (0.1 µg, Promega) with or without siRNA or the corresponding DNA-modified siRNA. Cells were harvested 24 h after transfection and luc activity was measured using the Dual-Luciferase Reporter Assay System (Promega). RNA, DNA and DNA-modified siRNAs, of which the nucleotide sequences used in this study are listed in Supplementary Table S1, were chemically synthesized (Proligo). Fifty percent inhibitory concentrations (IC50s) of siRNAs and DNA-modified siRNAs were estimated using the Pharmacologic Calculation Program (22).
RT–PCR
Using Lipofectamine 2000 (Invitrogen), E14TG2a or HeLa cells were cotransfected with 50 nM siRNA or DNA-modified siRNA, and pCAGIPuro-EGFP (0.5 µg/ml; 20), which encodes EGFP and puromycin-resistant genes. Puromycin (2 µg/ml; Clontech) was added to the medium 24 h after transfection. RNA was extracted 3 days after transfection using RNeasy (QIAGEN). Possible change in relative fraction of target RNA was examined by RT–PCR using the RNA LA-PCR kit (Takara) and mGapdh mRNA as an internal control. Primers used are listed in Supplementary Table S2.
Electrophoresis mobility shift analysis
Regions encoding a full-length human PACT (23) and human immunodeficiency virus trans-activating response RNA-binding protein 2 (TRBP2) (24) were amplified from a cDNA mixture of a total RNA extracted from HeLa cells by PCR. Used primers are as follows: PACT (5′- TTTTTTTTTTCATATGTCCCAGAGCAGGCACCGCGAGGCC and 5′-TTTTTTTTGCGGCCGCTTACTTTCTTTCTGCTATTATCTTTAAATA) and TRBP2 (5′-TTTTTTTTTTCATATGCTGGCCGCCAACCCAGGCAAGA and 5′-TTTTCCTTTTGCGGCCGCTCACTTGCTGCCTGCCATGATCTTGAGGTA). The amplified fragments were digested with NdeI and NotI and cloned into pET-28a (Novagen). PACT and TRBP2 proteins were produced in Escherichia coli BL21 codon plus and Rosetta (DE3) pLysS, respectively, as amino-terminal hexa-histidine-fusion proteins, and purified with NTA agarose. Binding to siRNA or its DNA counterparts (siDNA) (5 fmol/µl) was carried out in 25 mM Tris–HCl (pH8.0), 150 mM NaCl, 125 mM imidazole, 13% glycerol, 50 ng/µl salmon sperm DNA and 2 U/µl RNasein for 30 min at room temperature. The mobility shift assay was conducted on a 5% polyacrylamide gel and analyzed quantitatively using FLA-2000 image analyzer (Fujifilm).
Target cleavage assay
Luc2-153, the target sequence for the firefly luc siRNA, siLuc2-153, was inserted into the EcoRI/XhoI site of pTREC (25) to generate pTREC-2-153. The construct (0.5 µg) was introduced by transfection into HeLa cells with or without cognate siRNA or DNA-modified siRNA and cells were recovered 24 h after transfection. Total RNA was prepared using QuickPrep Micro mRNA Purification Kit (Amersham). Primer extension was carried out using 32P-labeled primer (5′-CTCGAAGCATTAACCCTCACT-3′) and the Primer Extension System-AMV Reverse Transcription Kit (Promega). Reaction products were size-fractionated by electrophoresis on 6% polyacrylamide gel containing 7-M urea in TBE buffer. Cleavage sites were determined using parallel sequence gel. DNA sequence analysis was carried out by using Thermo Sequence Fluorescent Labeled Primer Cycle Sequencing Kit (Amersham) but fluorescent-labeled dideoxyribonucleotides were replaced with 32P-labeled dideoxyribonucleotides.
Construction of psiCHECK derivatives and the assay of seed-sequence- and passenger-strand-based off-target activity
The plasmids psiCHECK-sm-X1, X2 and X3, respectively, included one, two and three copies of seed-matched (sm) target sequences while psiCHECK-cm-X1-X3, included one to three copies of completely matched (cm) target sequences (see Figure 4, Supplementary Figures S6, S8 and S9). They were constructed as follows. Chemically synthesized oligodeoxynucleotides (29–75 bp) including one to three copies of the same 23-bp sm or cm target sequences with cohesive XhoI/EcoRI ends were inserted into the psiCHECK-1 (Promega) XhoI/EcoRI site, which is situated in the region encoding the 3′UTR of Renilla luciferase mRNA. Human vimentin and mouse Oct4 sequences were used as targets (Supplementary Table S3). The cm sequence is completely matched that of the guide strand of nonmodified or DNA-modified siRNAs, and is expressed as a part of the target mRNA in transfected cells (see Figure 4). In contrast, the sm sequence in the target mRNA consists of two parts. Its 3′-terminal one-third is complementary in sequence to the 8-bp long 5′proximal region of the guide strand of siRNA or DNA-modified siRNA, while the remaining two-thirds, totally nonhomologous (see Figure 4). To construct psiCHECK-sm-X6, which includes six copies of Oct-797 and Luc-36 target sequences (see Supplementary Figure S9), 144-bp of ds insert DNA was synthesized by enzymatic ligation of 71- and 73-bp long chemically synthesized dsDNA fragments with 4-nt long 5′protrusions, and inserted into the XhoI/EcoRI site of psiCHECK-1. psiCHECK-GS and psiCHECK-PS, respectively, were constructed by inserting a PCR-amplified fragment containing target region (nucleotide position 97–856) of HPV16 E6E7 (accession number K02718) into the NotI site of psiCHECK-1 (see Figure 3). Used primers are as follows: 5′-GCGGCCGCAACTGCAATGTTTCAGGACC and 5′-GCGGCCGCTTATGGTTTCTGAGAACAGA. As GS/PS inserts, HPV-16 sequences along with three copies of 23-bp long target sequence of VIM-270 and Oct-797 (Supplementary Table S3) were used. The orientation of the insert was determined by nucleotide sequence analysis and plasmids with different insert orientations were selected. Plasmids expressing the GS and PS targets, respectively, used as psiCHECK-GS and psiCHECK-PS. HeLa cells in a well of 24-well culture plate were transfected simultaneously with one of psiCHECK derivatives (0.1 µg each) and pGL3-Control (Promega, 1 µg), phLuc-Control (see below, 1 µg) and siRNA or DNA-modified siRNA. Cells were harvested 24 h after transfection and luc activity was measured using the Dual-Luciferase Reporter Assay System (Promega). For assay using HVP16 target sequences, HeLa cells in a well of 96-well culture plate were transfected with a psiCHECK derivative (1.6 ng), pZeoSV2-hLuc (0.4 ng) and nonmodified or DNA-modified siRNAs and analyzed 48 h after transfection. pGL3-Control, encoding the firefly luc, served as a control for Renilla luc silencing assay for the siRNAs (siVIM-270, -596, -812 and -1128, siOct-797, and -821, siGRK4-934) along with cognate DNA-modified siRNAs (chiVIM-270, -596, -812 and -1128, chiOct-797, and -821, chiGRK4-934). The gene-silencing assay for siRNAs (siLuc-36, -49, -309, -774 and 2-153) and cognate DNA-modified siRNAs (chiLuc-36, -49, -309, -774 and 2-153) was carried out using as a control phLuc-Control coding for a modified firefly luciferase gene. The assay for five siRNAs (siHPV16-497, 573, -698, -707 and -752) and cognate DNA-modified siRNAs (chiHPV16-497, 573, -698, -707 and -752) was carried out using pZeoSV2-hLuc. phLuc-Control is a derivative of pGL3-Control (Promega) in which the 1.7-kb XbaI/HindIII region encoding firefly luc is replaced with the XbaI/HindIII hLuc fragment of psiCHECK-2 (Promega). pZeoSV2-hLuc is constructed by introducing hLuc fragment into pZeoSV2 (Invitrogen). The siGCY416 is an siRNA for knockdown of GFP and serves as an siRNA control. The nucleotide sequences for the target described above are shown in Supplementary Table S3.
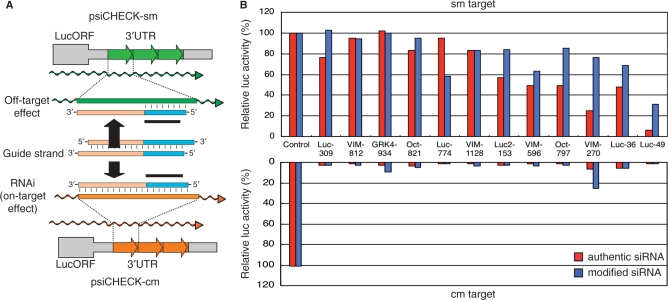
Figure 4.
Comparison of seed-sequence-dependent off-target effects due to RNA and DNA seed arms. Off-target effects of 12 sets of functional siRNA and cognate modified siRNA with 8-bp GS-DNA substitution were examined using psiCHECK-sm and -cm plasmids with various sm and cm targets and the dual luciferase assay. (A) Structures of psiCHECK-sm and -cm plasmids and modified-siRNA-dependent mechanisms of sm and cm target recognition are schematically shown. Three thick green and orange arrows, respectively, indicate sm and cm target sequences inserted. Black thick horizontal bar, seed arm sequence (position 2–8) of the guide strand. DNA sequence in modified siRNA is colored in blue. Only the seed-sequence-including DNA portion of the guide strand possesses a complete complementarity to the sm target sequence which is situated in the 3′UTR of luc mRNA transcribed from psiCHECK-sm. The entire guide strand completely matches the cm sequence in luc mRNA transcribed from psiCHECK-cm. Luc, VIM and Oct, respectively, indicate to contain sequences related to firefly luc, human vimentin and mouse Oct4. (B) HeLa cells were transfected with psiCHECK-1 containing three repeats of sm or cm target sequences (0.1 µg) and either phLuc-Control or pGL3-Control (1 µg) was used as a control. Cells were simultaneously transfected with either nonmodified or DNA-modified siRNA at 50 n M. The lower graph shows silencing effects on cm targets, while the upper graph, those on sm targets. Results shown in the lower graph indicate that not only all nonmodified siRNAs but also modified siRNAs with 8-bp GS-DNA substitution are highly active in silencing cm target sequences, indicating that the guide strand in which an 8-bp region from the 5′ end is DNA is as active in gene silencing as that of cognate authentic siRNA. The upper graph shows that virtually no reduction in sm target expression can be seen in the case of transfection with modified siRNAs with 8-bp GS-DNA substitution. In contrast, occasional reduction in sm target expression occurred in the case of authentic siRNA treatment. These findings may indicate that gene silencing due to transfection of modified siRNAs with 8-bp GS-DNA substitution is associated less frequently, if any, with seed-sequence-dependent off-target effect than classical RNAi.
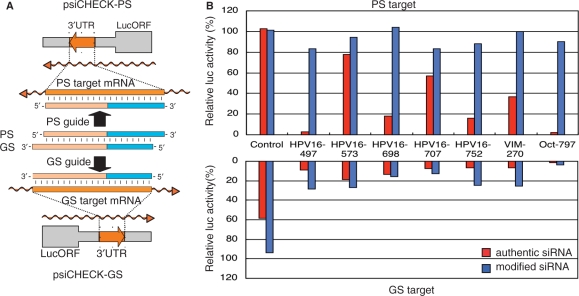
Figure 3.
The absence of passenger-strand-dependent off-target effects from gene silencing due to transfection with modified siRNA with GS-DNA substitution. The action of authentic and modified siRNAs to the guide strand targets (GS targets, lower graph) and passenger strand targets (PS target, upper graph) were examined using five HPV16, one human vimentin and one mouse Oct4 sites. Modified siRNAs with 6-bp GS-DNA substitution were used for five HPV16 targets, while those with 8-bp GS-DNA substitution, for vimentin (VIM) and Oct4 (Oct) targets. (A) Structures of psiCHECK-PS and GS along with PS and GS target recognition mechanisms by modified siRNA with 8-bp GS-DNA substitution are schematically shown. DNA sequence in DNA-modified siRNA is colored in blue. The passenger strand (PS) is completely complementary to the PS target sequence situated in the 3′UTR of luc mRNA transcribed from psiCHECK-PS. The guide strand (GS) completely matches the GS target sequence in luc mRNA transcribed from psiCHECK-GS. (B) HeLa cells were transfected with psiCHECK-PS or -GS and plasmids for luc activity control as shown in ‘Materials and methods’ section. The lower graph shows silencing effects on GS-targets, while the upper graph, those on PS-targets. Results shown in the lower graph indicate that not only all authentic siRNAs but also all seven modified siRNAs with GS-DNA substitution are highly active in silencing GS-targets, indicating that the guide strand (GS) in which a 6- or 8-bp region from the 5′ end is DNA is as active as that of cognate authentic siRNA. In contrast, virtually no reduction in PS-target expression can be seen in the case of transfection with modified siRNAs with GS-DNA substitution, while occasional reduction in PS target expression occurred in the case of non-modified siRNA treatment, indicating that, unlike nonmodified-siRNA-based RNAi, gene silencing due to transfection of modified siRNAs with GS-DNA substitution is associated with no or little PS-dependent off-target effect.
Calculation of melting temperature
Melting temperature (Tm) was calculated based on the nearest-neighbor model (26) and the thermodynamic values for RNA–RNA (27) and RNA–DNA (28). Oligonucleotide and sodium chloride concentrations were assumed to be 100 nM and 100 mM, respectively.
Microarray analysis
HeLa cells (1 × 105 cells/ml) were inoculated in a 1.5-cm well 24 h prior to transfection. Cells were transfected with 50 nM siVIM-270 or chiVIM-270. Total RNA (3 µg) purified using RNeasy Kit (Qiagen) 1 day after transfection was used for hybridization to Human Genome U133 Plus 2.0 GeneChip (Affymetrix) containing ∼47 400 human transcripts according to the manufacturer's protocol. RNA from mock-transfected cells, which were treated with transfection reagent in the absence of nonmodified or DNA-modified siRNAs, was used as a control. To calculate transcript expression values, Microarray Suite 5.0 (MAS5, ref. 29) was used with quantile normalization (30), and transcritps with sufficient hybridization signals to be called present (P) were used in this study. To identify transcripts that were downregulated on the array, we compared the cumulative distribution of expression changes for messages with the site versus those with no canonical site, and calculated the maximum positive cumulative difference between the two distributions. The statistical significance of their dissimilarity was quantified using the Wilcoxon's rank-sum test (31,32) as P-value.
Motif analysis of 3′UTR
The probe sequences were taken from the latest annotation table provided on the Affymetrix Web site (http://www.affymetrix.com). We mapped them to the RefSeq human mRNA sequences (release 24), and then identified the target transcripts. We found 67 220 annotations of the human transcripts, which corresponded to 54 675 of the probesets. Among these, 19 856 transcripts called P by MAS5, of which 16 783 RefSeq entries (25%) with the 3′UTR were considered in this study. sm sequences at positions 2–8, 1–7 and 1–8 from the 5′ end of the guide strand were assigned to the extracted 3′UTR sequences.
Quantitative RT–PCR
Quantitative RT–PCR was carried out using total RNA purified, whose properties were also analyzed using the microarray. RNA was reverse-transcribed using the SuperScript First-Strand Synthesis System for RT–PCR (Invitrogen). The resultant cDNA and SYBR Green PCR Master Mix (Applied Biosystems) were mixed and incubated at 95°C for 10 min before the PCR reaction. The levels of PCR products were monitored with ABI PRISM 7000 sequence detection system and analyzed with ABI PRISM 7000 SDS software (Applied Biosystems). Each reaction was run in triplicate. The expression level of each sample was first normalized to the amount of β-actin and then to the mock-transfection control. The used primer sets are listed in Supplementary Table S4.
RESULTS
Asymmetric requirement of ribonucleotides in siRNA for effective gene silencing
In the present article, the ends of siRNA possessing the 5′ends of the guide and passenger strands, respectively, are referred to as GS and PS ends (Figure 1A). To examine the effects of DNA replacement on gene silencing systematically, six different types of partially DNA-substituted siRNAs, whose structures are illustrated in Figure 1A, were constructed based on the sequences of siLuc-36 (Figure 1) and siLuc2-153 (Supplementary Figure S1). These siRNAs have been shown to be highly functional in firefly luc-gene silencing (20). In three left DNA-modified siRNAs in Figure 1A, ribonucleotides were progressively replaced with cognate deoxyribonucleotides from the PS end, while, in three right ones, deoxyribonucleotide substitutions occurred progressively from the GS end. In replacement with dsDNA, paired ribonucleotides were simultaneously replaced with cognate deoxyribonucleotides. In siRNAs with DNA replacement only in the guide and passenger strand, respectively, guide and passenger strands are DNA–RNA chimeras, with the remaining being RNA. Using these different types of DNA-modified siRNAs, examination was made of possible firefly luc gene activity reduction in three mammalian cells, CHO-K1, HeLa and E14TG2a and Drosophila S2 cells (Figure 1B–G and Supplementary Figure S1A–F).
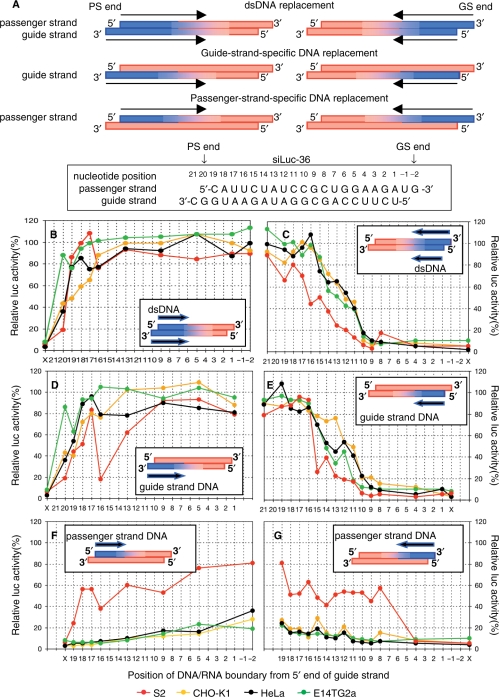
Figure 1.
Effects of the size and position of deoxyribonucleotide substitutions on gene silencing. (A) siRNAs with various DNA substitutions were constructed based on the siLuc-36 sequence, which is shown in the lower margin with nucleotide position measured from the 5′ end of the guide strand, and were subjected to the luc gene assay. Six different types of DNA-modified siRNAs are schematically shown. In three left constructs, RNA sequences were progressively replaced with DNA counterparts from the PS end, which includes 5′ and 3′ ends of passenger and guide strands, respectively. In three right constructs, RNA was replaced with DNA from the GS end, including the 5 ′end of the guide strand and 3′ end of the passenger strand. Both left and right constructs include three types of DNA replacement, that is, double-strand replacement, and guide and passenger-specific replacement. DNA and RNA portions are colored in blue and orange, respectively, and arrows indicate the direction of DNA replacement. (B–G) Ribonucleotides of double (B, C), guide (D, E) and passenger (F, G) strands were progressively replaced with deoxyribonucleotide counterparts from the PS end (B, D, F) or the GS end (C, E, G). Numerals from -2 to 21, which are situated below each graph indicate nucleotide position from the 5′ end (position 1) of the guide strand. (–1, –2) and (20, 21), respectively, correspond to 3′ overhangs at the GS and PS ends. Relative luc activity at position ‘x’ indicates the activity due to authentic siRNA (siLuc-36). S2, CHO-K1, HeLa and E14TG2a cells were transfected with pGL3-Control DNA (1 µg) and pRL-SV40 DNA (0.1 µg) with or without authentic siRNA or the corresponding modified siRNAs with various DNA substitution (50 nM each). Relative luc activity was measured 24 h after transfection. Note that modified siRNA with 10-bp or <10-bp long single or double-stranded DNA substitution from the GS end was capable of inactivating the target luc gene as effectively as authentic siRNA.
DNA replacement of the two siRNA sequences gave results nearly the same with each other in all three mammalian cells. In the case of dsDNA replacement from the PS end, all modified siRNAs except those containing DNA replacement only in the vicinity of the PS end had little, if any, gene-silencing effect on luc target (Figure1B, Supplementary Figure S1A). In contrast, as with nonmodified siRNAs, nearly all luc gene activity was abolished subsequent to transfection with modified siRNAs with dsDNA substitution ≤10 bp (Figure 1C) or 8 bp (Figure S1B) in length from the 5′ end of the guide strand. Furthermore, in mammalian cells, gene-silencing effects due to transfection with modified siRNAs with dsDNA substitution from the GS end were noted strongly correlated with those induced by that with modified siRNAs with DNA substitution only in the guide strand (Figure 1B–E, Supplementary Figures S1A–D and S2A). In contrast, transfection with modified siRNAs with DNA substitution only in the passenger strand gave a small, if any, effect on luc target gene silencing in mammalian cells (Figure 1F and G, and Supplementary Figures S1E and F and S2B). Mammalian gene silencing due to treatment with siRNAs with dsDNA substitution may thus be concluded due primarily to interactions between the guide strand and target mRNA and/or those between the guide strand and protein moieties involved in gene silencing. The contribution of the passenger strand to gene silencing is much greater in Drosophila than mammalian cells, since considerable reduction of luc gene activity was evident in S2 cells transfected with modified siRNAs with DNA substitution only in the passenger strand (Figure 1F and G, and Supplementary Figure S1E and F).
As described below, ribonucleotide residues in the PS-end proximal region are essential to RLC- and/or RISC-protein binding. The 8-bp long GS-end proximal region, which can be replaced with the DNA counterpart without substantial loss of gene-silencing activity, includes the seed region situated from nucleotide position 2 to 8, measured from the 5′end of the guide strand.
DNA-replacement/gene-silencing profiles of the siLuc2-153 sequence were unexpectedly found to shift a few bases to the side of the 5′ end of the guide strand from those of siLuc-36 sequence (Supplementary Figure S3). The apparent profile shift in the vicinity of the PS end may be due in part to different sequence-dependent effects of deoxyribonucleotide replacement in the 3′overhang.
The presence or the absence of 5′terminal phosphate gave no significant difference on gene silencing (Supplementary Figure S4), suggesting that, as with nonmodified siRNA (33), the 5′end of DNA-modified siRNA is phosphorylated within cells. Interferon response due to DNA-modified siRNA was not more profound than that due to nonmodified siRNA (Supplementary Figure S5).
ES-to-differentiated cell fate transition induced by functional DNA-modified siRNA-dependent inactivation of Oct4
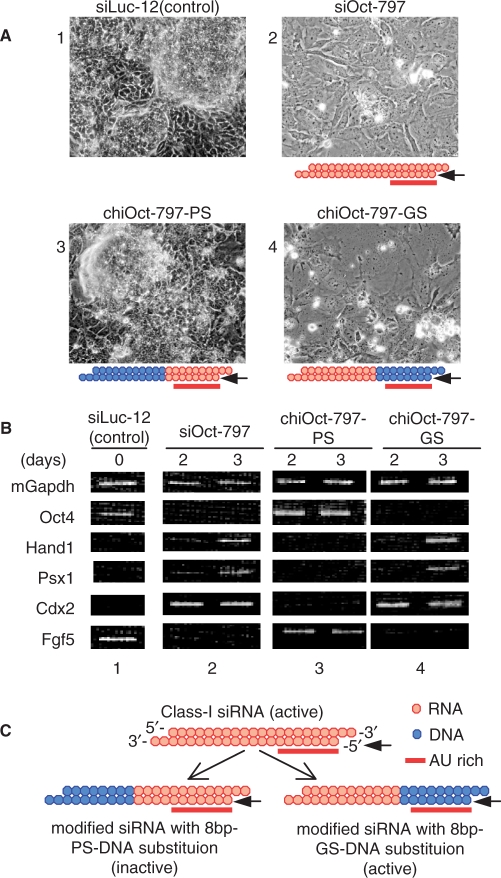
Oct4 is a gene-encoding POU transcription factor that regulates ES cell pluripotency (34). A reduction in Oct4 expression leads to the loss of pluripotency and differentiates ES cells into trophectoderms, which are characterized by flat morphology (34). Indeed, both morphological changes and reduction in Oct4 expression were observed in mouse ES cells transfected with siOct-797, an siRNA specific to Oct4 (Figure 2A1 and 2; B1 and 2). As shown in Figure 2A4 and B4, chiOct-797-GS, a DNA-modified siRNA version of siOct-797, of which the 8-bp long GS-end-proximal region is occupied by DNA, also induced a very similar morphological change and a significant reduction in Oct4 expression. Reduction in Oct4 activity is also known to induce the expression of heart- and neural crest derivatives-expressed protein 1 (Hand1), placenta-specific homeobox 1 (Psx1) and caudal type homeobox 2 (Cdx2) genes and represses Fgf5 expression (34,35). Figure 2B shows that, as with siOct-797, chiOct-797-GS induced Hand1, Psx1 and Cdx2 expression and repressed Fgf5 expression as well. In contrast, changes in neither morphological nor gene expression were observed in ES cells transfected with chiOct-797-PS possessing DNA substitution in the PS-end proximal third (Figure 2A3 and B3).
Figure 2.
Functional assay of modified siRNAs with DNA substitution in mouse E14TG2a cells. The siOct-797 is a highly effective class-I siRNA for Oct4 gene silencing (20). siLuc-12 is an siRNA for knockdown of unrelated luc gene and serves as a control. chiOct-797-PS is a DNA-modified siOct-797, of which the 11-bp long PS-end-proximal region is DNA, while chiOct-797-GS is a DNA-modified siOct-797, of which the 8-bp long GS-end-proximal region is DNA. DNA and RNA are colored in blue and orange, respectively, while the 7-bp long AU- or AT-rich region is indicated by the red horizontal bar. Arrows indicate the guide strands. (A) Morphological change induced by transfection of nonmodified and cognate DNA-modified siRNAs. The structures of nonmodified siRNA or two types of DNA-modified siRNAs are schematically shown in the lower margin of each panel. E14TG2a cells were simultaneously transfected with pCAGIPuro-EGFP (0.5 µg/ml), encoding Enhanced Green Fluorescent Protein (EGFP) and puromycin-resistant genes, and siLuc-12 (negative control), siOct-797 (positive control), chiOct-797-PS or chiOct-797-GS, 50 nM each. Puromycin (2 µg/ml) was added to the medium 24 h after transfection, and possible morphological change was observed under a phase contrast microscope 3 days after transfection. As with the nonmodified functional siRNA (siOct-797), chiOct-797-GS but not chiOct-797-PS induced flat morphology, typical of trophectoderm (34,35). (B) Expression of trophectoderm differentiation marker genes. The mGapdh was used as an internal control. Results of unrelated siLuc-12 transfection is shown in lane 1. As with the nonmodified effective siRNA (siOct-797; lane 2), chiOct-797-GS (lane 4) induced Hand1, Psx1 and Cdx1 expression and repressed Oct4 and Fgf5 expression, indicating that chiOct-797-GS is capable of effectively inactivating Oct4. In contrast, no appreciable gene expression change was induced by chiOct-797-PS (lane 3), indicating chiOct797-PS is an inactive modified siRNA. (C) Structures of functional DNA-modified siRNA. Highly effective siRNAs (class-I siRNAs) can be converted into either active or inactive DNA-modified siRNAs depending on whether DNA arms are situated in the PS or GS terminal halves. Modified siRNA associated with an 8-bp long DNA in the GS-end-proximal region is as active as the parental authentic siRNA. In contrast, modified siRNA with DNA substitution in the PS-end proximal region is inactive.
Taken together, results described above may indicate 8-bp long dsRNA from the 5′guide strand end of functional siRNAs to be replaceable with DNA counterparts without substantial loss of gene silencing activity.
siRNAs in which all ribonucleotides in the 8-bp long ds region from the GS end and those in the 3′overhang of the passenger strand are replaced with cognate deoxyribonucleotides (Figure 2C) are referred to as modified siRNAs with 8-bp GS-DNA substitution from here on and vice versa.
Requirement of 7-bp long GS-end proximal AT-rich DNA sequence for effective gene silencing due to transfection with modified siRNA with 8-bp GS-DNA substitution
In a previous work (20), we classified siRNAs into three groups, class-I, -II and -III and showed class-I siRNAs to be highly functional. Class-I siRNAs satisfy the three following sequence-conditions simultaneously: A/U at the 5′ terminus of the guide strand; G/C at the 5′ terminus of the passenger strand; at least four A/U residues in the 5′ terminal 7 bp of the guide strand. siRNAs opposite in these features give rise to little or no gene silencing in mammalian cells, and are grouped as class-III. In addition, more than 9-bp long G/C stretch is absent from class-I siRNAs. siRNAs other than class-I and III are defined as class-II. This guideline for highly effective or functional siRNAs has been used widely along with those proposed by other investigators (36,37). Modified siRNAs with DNA substitution described above were all designed based on sequences of siRNAs belonging to class-I.
To further examine whether modified siRNAs with 8-bp or less than 8-bp GS-DNA substitution are capable of inducing effective target gene silencing provided that their nonmodified siRNA counterparts belong to class-I, gene-silencing activities of 16 modified siRNAs with 6- or 8-bp GS-DNA substitution were compared with those of cognate authentic siRNAs. Results at 16 target sites are shown in the lower graphs of Figures 3 and 4. Not only all 16 nonmodified siRNAs but also16 DNA-modified siRNAs caused more than 70% of reduction in target gene activity at siRNA concentration of 50 nM.
Target-gene knockdown effect varied depending on siRNA concentration (Supplementary Figure S6). DNA-modified siRNAs exhibited gene-silencing activity virtually similar to that due to the nonmodified cognate siRNAs when the siRNA concentration was 0.5–50 nM (Figures 3 and 4, Supplementary Figure S6), although, at 0.05 nM, gene silencing induced by DNA-modified siRNA was less profound than that induced by nonmodified siRNA counterparts possibly due to weaker seed activity of the DNA seed arm (see below). Calculated IC50 values of the nonmodified siRNAs varied from 1 to 318 pM depending on sequences, while those of DNA-modified siRNA counterparts were from 47 to 5463 pM (Table 1; fold IC50 changes, 1–35).
Table 1.
IC50s of non-modified and DNA-modified siRNAs
| Luc-309 | VIM-812 | GRK4-934 | Oct-821 | Luc-774 | VIM-1128 | Luc2-153 | VIM-596 | Oct-797 | VIM-270 | Luc-36 | Luc-49 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-modified siRNA (pM) | 7 | 25 | 34 | 318 | 13 | 9 | 18 | 17 | 1 | 156 | 167 | 5 |
| DNA-modified siRNA (pM) | 48 | 146 | 744 | 247 | 85 | 75 | 179 | 131 | 47 | 5463 | 270 | 16 |
| IC50 fold change (DNA-modified siRNA/non-modified siRNA) | 7.0 | 5.8 | 21.9 | 0.8 | 6.4 | 8.0 | 10.2 | 7.9 | 33.3 | 35.0 | 1.6 | 3.2 |
It may thus follow that modified siRNAs with 8-bp or <8-bp GS-DNA substitution are capable of inducing effective gene silencing if the concentration is properly chosen and their nonmodified siRNA counterparts belong to class-I.
Involvement of Ago2, TRBP2, PACT and sequence-specific mRNA cleavage in gene silencing due to transfection with modified siRNAs with 8-bp GS-DNA substitution in mammalian cells
TRBP2 (24) and PACT (22) are putative mammalian counterparts of Drosophila RLC protein, R2D2, which senses the differential stability of siRNA duplex ends to determine which strand will be loaded into RISC as the guide strand (3). R2D2 appears to bind more stable RNA end (3). TRBP2, PACT and R2D2 contain dsRNA-binding domains (23,24,38) and, as shown in Figure 5A, are capable of binding to dsRNA but not dsDNA (39,40), suggesting that, in the case of modified siRNAs with 8-bp GS-DNA substitution, these RLC proteins bind only to certain ribonucleotides present in PS terminal two-thirds. Ago2 is a key RNA-binding protein for nonmodified siRNA-based RNAi in mammals (41,42).
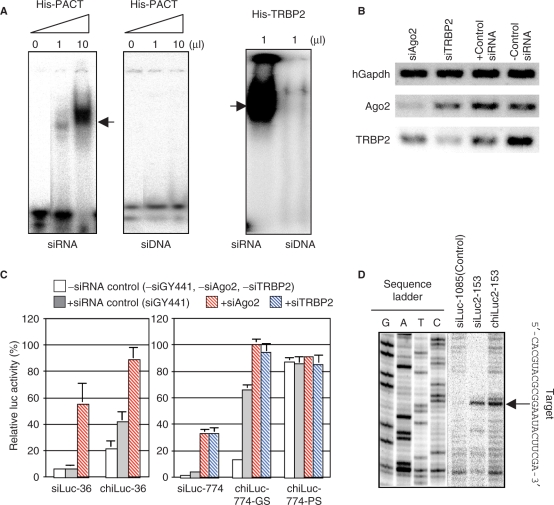
Figure 5.
Involvement of Ago2, TRBP2 and PACT in modified-siRNA-dependent gene silencing. (A) PACT and TRBP2 binding to siRNA (siLuc-36) and cognate siDNA. The 5′end of the guide strand of siRNA or siDNA was phosphorylated with [γ-32P] ATP and the 5′end of the passenger strand was phosphorylated with cold ATP. A mobility shift assay was carried out using PACT and TRBP2 proteins purified from E. coli cells. Note that TRBP2 and PACT are capable of binding to dsRNA but not dsDNA. Arrows indicate the position of protein–nucleic acid complexes. (B) Reduced expression of Ago2 and TRBP2 by RNAi. HeLa cells were transfected with siGY441 (control siRNA), siAgo2 or siTRBP2 (50 nM) along with pCAGIPuro-EGFP (0.5 µg) and subjected to RT–PCR 3 days after transfection. Note that Ago2 and TRBP2 RNA were specifically knocked down by siAgo2 and siTRBP2, respectively. (C) Requirement of Ago2 and TRBP2 for gene silencing due to transfection with modified siRNA with 8-bp GS-DNA substitution. HeLa cells were simultaneously transfected with pGL3-Control DNA (1 µg) and pRL-SV40 DNA (0.1 µg), and a mixture of siRNA and modified siRNA with DNA substitution. Two siRNAs, siLuc-36 (left panel) and siLuc-774 (right panel), and their modified siRNA counterparts, 5 nM each, were used for luc gene silencing. chiLuc-36 and chiLuc-774-GS are functional DNA-modified siRNAs specific to luc knockdown, whereas chiLuc-774-PS is a modified siRNA with DNA replacement in the PS-end-proximal region. Ago2 and TRBP2 were knocked down using 50 nM siAgo2 and siTRBP2, respectively. As an siRNA concentration control, siGY441 (50 nM), an siRNA for EGFP knockdown, was used. Gene-silencing activity was measured 24 h after transfection. Note both nonmodified and DNA-modified siRNA-dependent luc gene-silencing activity to be reduced by 30–50 points by knocking down Ago2 and TRBP2 activity through siAgo2 and siTRBP2 RNAi, respectively. (D) Functional DNA-modified siRNA-dependent target mRNA cleavage. HeLa cells were cotransfected with pTREC-2-153 and siLuc-1085 (a negative control siRNA), siLuc2-153 (target-cleavable siRNA) or its cognate modified siRNA with 8-bp GS-DNA substitution, each 5 nM. RNA was extracted 24 h after transfection, and cleavage sites were determined by primer extension. Sequence ladder was prepared using the same pTREC construct (25). Note that the position of the main cleavage site is precisely identical between RNAi and gene silencing due to transfection with modified siRNA with 8-bp GS-DNA substitution.
We demonstrate that these proteins are essential to functional mammalian gene silencing due to transfection with modified siRNAs with 8-bp GS-DNA substitution in HeLa cells. The activity of TRBP2 and Ago2 were reduced significantly through RNAi with TRBP2 and Ago2-secific siRNAs, respectively (Figure 5B). As shown in Figure 5C, Ago2 and TRBP2 siRNA treatment of HeLa cells not only specifically reduced Ago2 and TRBP2 expression but also prevented siLuc-36 or siLuc-774-based luc gene silencing. By comparison with control siRNA treatment (treatment with siGY441, a siRNA specific for GFP), Ago2 or TRBP2 siRNA treatment was found to increase relative luc activity by 30–50 points in DNA-modified siRNA-dependent gene silencing as in the case of nonmodified-siRNA-dependent RNAi (Figure 5C). A separate experiment showed PACT siRNA treatment prevented DNA-modified-siRNA-based gene silencing to some extent (Supplementary Figure S7). Ago2, PACT and TRBP2 may thus be involved in gene silencing due to modified siRNA with 8-bp GS-DNA substitution as in the case of RNAi with nonmodified siRNAs. However, note that siGY441, a control siRNA, much more strongly prevented DNA-modified-siRNA-dependent gene silencing than RNAi, indicating that certain critical RISC and/or RLC proteins may have less affinity toward modified siRNAs with 8-bp GS DNA substitution than toward nonmodified siRNAs.
In RNAi, target mRNA is cleaved at the point corresponding to the center of the guide strand via RNase H-like activity of the PIWI domain of Ago2 (11,12,16,43). Real-time PCR experiments indicated that the mRNA target is effectively degraded through DNA-modified siRNA treatment (data not shown). Examination was thus made to determine at what point target mRNA is cleaved in DNA-modified-siRNA-dependent gene silencing using chiLuc2-153, a modified siRNA of siLuc2-153 with 8-bp GS-DNA substitution (Figure 5D). The position of the main cleavage site was found to be precisely the same to that of RNAi. In both cases, the main cleavage point was situated between position 10 and 11, as measured from the 5′end of the guide strand. A similar coincidence of target cleavage site was also found when siLuc-36 sequence was examined (data not shown).
These results presented so far may indicate that, as with functional siRNA, functional modified siRNA with 8-bp GS-DNA substitution is loaded into Ago2-including RISC and that the guide strand in its DNA portion serves as guide for target mRNA nucleation and subsequent cleavage. As in the case of nonmodified-siRNA-based RNAi, TRBP2 and PACT are likely to be involved in the formation of RLC including modified siRNA with 8-bp GS-DNA substitution in mammalian cells.
Absence of gene-silencing activity from the passenger strand with DNA substitution in the 3′terminal region
In nonmodified-siRNA-based gene silencing, it may be difficult to evade the possible passenger-strand-dependent silencing of unintended genes (44). As described above (see e.g. Figure 1), if the 3′terminal region of the guide strand of siLuc-36 or siLuc2-153 is substituted with DNA, no or little target gene silencing occurred. The 3′terminal region of the passenger strand of modified siRNA with GS-DNA substitution is always occupied by DNA (see Figure 2C), suggesting that gene silencing due to transfection with functional modified siRNA with GS-DNA substitution is not associated with any activity of inducing passenger-strand-dependent off-target effect.
To further confirm this point, one copy of cm target sequence for guide or passenger strands (GS or PS targets, respectively) were introduced into the 3′UTR of psiCHECK-1 to generate psiCHECK-GS and -PS plasmids, and nonmodified and DNA-modified siRNA-dependent reporter gene silencing effects were examined. All siRNAs used here belong to highly functional class-I. As described above and can be seen in the lower graph of Figure 3B, all 7 DNA-modified siRNAs used here appear to possess a highly functional guide-strand activity. The upper graph of Figure 3B showed that target gene silencing subsequent to transfection with at least four of seven nonmodified siRNAs is associated with apparent passenger-strand-dependent off-target activity. Note that, in all four cases, more than 80% reduction in PS target activity was evident. In contrast, none of seven modified siRNAs with GS-DNA substitution could induce effective silencing of luc gene fused with PS targets. Thus, we conclude that gene silencing due to transfection with functional DNA-modified siRNAs may not be associated with passenger-strand-dependent off-target effects.
Reduction in seed-sequence-based off-target effect in gene silencing due to transfection with functional DNA-modified siRNAs
The 8-bp long DNA of the guide strand in DNA-modified siRNA corresponds in position to the ‘seed’ region of microRNA that is implicated in gene regulation by translational inhibition and/or mRNA degradation (45–49). In nonmodified siRNA, the region of nucleotide position 2–8 measured from the 5′ end of the guide strand may play an essential role in the initial stage of mRNA recognition. The DNA–RNA hybrid is generally less stable thermodynamically than RNA duplex (27,28, 50,51). Thus, the seed activity or capability of inducing inhibition of translation and/or mRNA degradation due to the DNA seed arm in the putative RISC containing the guide strand of modified siRNA with 8-bp GS-DNA substitution is expected to be considerably weaker than that of the nonmodified-siRNA–RISC. In fact, in all cases examined, calculated Tm in the ‘seed’ region of DNA-modified siRNA was considerably lower than that of cognate nonmodified siRNA (Supplementary Table S5).
For further confirmation of this point, three consecutive copies of an identical sm target sequence, which possesses homology only to the seed sequence of nonmodified or DNA-modified siRNAs to be examined, were introduced into the 3′untranslated region of psiCHECK-1 Renilla luc (Figure 4). As a total, 12 sm-targets for highly functional siRNAs were examined as inserts (see Figure 4, Supplementary Figure S8 and Table S3). As described above (see also Figure 3, Supplementary Figure S6), at 50 nM, all DNA-modified siRNAs along with cognate nonmodified siRNAs effectively silenced cm-target genes. A considerable reduction in relative luc activity was observed in sm-target expressing cells transfected especially with certain nonmodified siRNAs possessing a higher G/C content in the seed region (Supplementary Figure S8). However, virtually no sm target knockdown was induced by treatment with DNA-modified siRNAs at least 11 of 12 cases (Figure 4B). The seed activity of authentic siRNA increased with increasing the copy number of sm targets but no such effect was observed in the case of DNA-modified siRNA treatment (Supplementary Figure S9). It may thus follow that most, if not all, functional modified siRNAs with 8-bp GS-DNA substitution may hardly induce seed-sequence-based off-target effects.
Genome-wide analysis of off-target effects due to DNA-modified-siRNA-dependent gene silencing
To further clarify that gene silencing due to modified siRNAs with 8-bp GS-DNA substitution is less frequently associated with off-target effect, genome-wide expression profiles were examined using microarray analysis. siVIM-270 is a functional nonmodified siRNA for human vimentin knockdown (see Figures 3 and 4), while chiVIM-270 is cognate modified siRNA with 8-bp GS-DNA substitution. As a total, 16 783 transcripts were examined by microarray profiling (Figure 6C and D and Supplementary Figure S10).
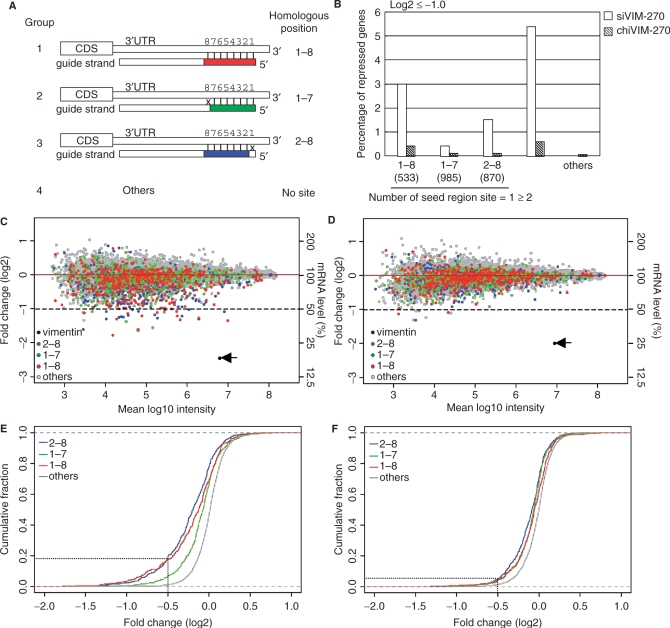
Figure 6.
Microarray-based off-target-effect profiling. HeLa cells were transfected with siVIM-270, a nonmodified siRNA targeting the vimentin transcript, and cognate modified siRNA with 8-bp GS-DNA substitution, and microarray-based expression profiles were examined 24 h after transfection. Change in gene expression is shown by log2 of fold change ratio to the mock transfection. As a total, 16 783 transcripts were examined. (A) Classification of transcripts into four groups based on 3′ UTR homology to the position 1–8 region of the guide strand of siRNA. 3′ UTRs of group-1 transcripts (n = 695) possess homology to the entire position 1–8 region of the guide strand. 3′ UTRs of group-2 transcripts (n = 1195) are completely complementary to the region 1–7 but not position 8. 3′ UTRs of Group-3 transcripts (n = 1039) have homology to the entire seed region (region 2–8) but not position 1. Group-4 transcripts (n = 13 965) are transcripts other than those belonging to group 1–3. They consist of transcripts possessing no homology at nucleotide position 1 and 8, and those possessing 1- or more than 1-bp mismatch in the central seed region (position 2–7). (B) Percentage of transcripts of which activity was reduced to more than 50% are shown. 1–8, 1–7 and 2–8, respectively, group-1, -2 and -3 transcripts with a single site in the 3′ UTR. Greater than two, transcripts of which the 3′ UTR contains two or more than seed region sites (1–7 or 2–8 or 1–8). Seed activity increased with increasing the number of sites possessing seed homology. Note that, in all cases, DNA-modified siRNAs appear much less effective in inducing off-target effect than cognate nonmodified siRNAs. (C, D) Microarray profiling of gene expression subsequent to transfection of siVIM-270 (C) and chiVIM-270 (D). Each point represents the average of 11 observations. Signals of group 1–4 are shown using four different colors. Red, green and blue dots, respectively, represent group-1, -2 and -3 transcripts. Group-4 transcripts or those with no homology site are colored in gray. Group 1–4 signals are separately shown in Supplementary Figure S10. Signals for vimentin belonging to group-4 are colored in black and labeled with arrows. Vertical axis, log2 of fold change ratio to the mock transfection or gene activity levels (%). Horizontal axis, log10 of the multiplied fluorescence intensity of mock transfection. The dotted lines show the level of 50% reduction (log2 ≤ –1) induced by siVIM-270 or chiVIM-270 treatment. (E, F) Cumulative distribution of transcripts downregulated by siVIM-270 (E) and chiVIM-270 (F). Red, green and blue lines, respectively, represent the cumulative fraction of group 1–3 transcripts, respectively. The line for group-4 transcripts is colored in gray.
Homology between the seed region of siRNA and the 3′UTR but not coding sequence of mRNA has been shown to be important for seed-dependent off-target effect (10) and, as described above, the position 1–8 region of the guide strand, consisting of the 5′ very end (position 1) and the seed sequence (position 2–8), is replaceable with DNA without appreciable reduction of gene silencing. The 5′-end-proximal, 7-bp region of class-I siRNA is A/U-rich. Thus, 16 783 transcripts were classified into four groups based on homology between 3′UTR and the guide-strand seed region (Figure 6A). About 5% transcripts (n = 695) possessed homology to the entire position 1–8 region of the guide strand of siVIM-270 (group-1). About 7% transcripts (n = 1195) were completely complementary to the region 1–7 but not position 8 (group-2). Approximately 6% transcripts (n = 1039) had homology to region 2–8 but not position 1 (group-3). All other transcripts (n = 13 965), that is, those with no homology site belong to group-4. Note that a small fraction of transcripts (0.7%; n = 111) with two or more than two homologous regions are doubly counted. In Figure 6C–F, changes in abundance of these four group transcripts monitored with microarrays are shown using four colors.
In the case of siVIM-270 treatment, repression from UTRs of group-3 transcripts with complete seed match was significantly more than those from UTRs of group-1, -2 and -4 (P < 10–3, P < 10–25 and P < 10–154, respectively, Wilcoxon's rank-sum test) (Figure 6E). Similarly, in the case of chiVIM-270 treatment, repression from UTRs of group-3 was significantly more than that those from UTRs of group-1, -2 and -4 (P < 10–5, P < 10–3 and P < 10–54 respectively) (Figure 6F). These results are consistent with previous finding (31) of the importance of the seed homology for microRNA-type targeting specificity.
Results in Figure 6E and F also show that authentic siRNA-dependent repression from UTRs of transcripts with complete seed homology was significantly more than that due to modified siRNA with 8-bp GS-DNA substitution (in the case of group-1 and -3 transcripts, respectively, P < 10–5, and P < 10–24).
The expression of about 20% (n = 184) of group 3 transcripts was reduced to <71% [fold change (log2) ≤ –0.5] subsequent to siVIM-270 treatment (Figure 6C and E), while only 5% (n = 54) of group-3 transcripts were associated with a more than 29% reduction in expression subsequent to chiVIM-270 treatment (Figure 6D and F). The 3′UTRs of group-1 and -3 transcripts are completely homologous to the seed region (positions 2–8) (see Figure 6A). Figure 6C and D shows that the number of group-1/3 transcripts of which the expression levels reduced to more than 50% (fold change of log2 ≤ –1.0) was 47 and 5, respectively, by siVIM-270 and chiVIM-270 treatment. These 47 and 3 transcripts are transcriptional products of 28 and three genes, respectively. As shown in Supplementary Figure S12, 3′UTRs of these gene transcripts did not possess any high homology to the position 9–19 region of the guide strand. In addition, as has been reported previously (31), increased effectiveness of dual sites was evident not only in the case of siVIM-270 treatment but also in the case of chiVIM-270 treatment (Figures 6B, 7A–D and Supplementary Figure S11). The reduction of endogenous signals of vimentin RNA was 79 and 71% by the treatment of siVIM-270 and chiVIM-270, respectively (see the arrows in Figures 6C and D, 7A and B), indicating that chiVIM-270 was slightly less potent compared with siVIM-270. Taken together, these findings may support the notion that gene silencing due to modified siRNA with 8-bp GS-DNA substitution is associated with off-target effect much less frequently, if any, than the nonmodified-siRNA-based RNAi.
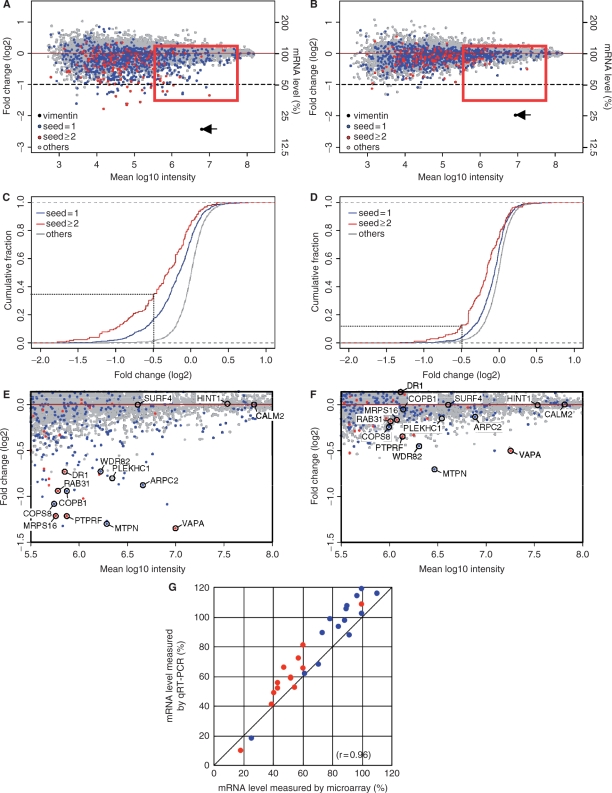
Figure 7.
Microarray analysis of off-target effects due to transfection of transcripts with plural sites for seed-homology within the 3′ UTR. (A, B) Microarray profiles of transcripts extracted from cells treated with siVIM-270 (A) and chi-VIM-270 (B). Vertical axis, log2 of fold change ratio to the mock transfection. Horizontal axis, fluorescence intensity relative to that of mock transfection. Fluorescence intensity is expressed using log10. Blue dots represent transcripts of which the 3′ UTR contains only one site complementary in sequence to the position 2–8 region of the guide strand. Red dots represent transcripts of the 3′ UTR with two or more than two sites completely matching the 2–8 position of the guide strand (see Supplementary Figure S11). The positions of vimentin are indicated as arrows. (C, D) Cumulative fraction of genes or transcripts downregulated by siVIM-270 (C) and chiVIM-270 treatment (D). In the case of siVIM-270 treatment, repression from UTRs of transcripts with two or more than two complete seed matches was significantly more than those from UTRs of group-3 transcripts with a single seed match and transcripts with no homology site (P ≤ 10–8, P ≤ 10–46, respectively, Wilcoxon's rank-sum test). Similarly chiVIM-270-dependent repression from UTRs with two or more than two complete seed matches was significantly more than those from UTRs of group-3 transcripts with a single seed match and transcripts with no homology site (P ≤ 10–8, P ≤ 10–21, respectively). (E) and (F) are enlarged figures of a part of the regions enclosed in (A) and (B), respectively. These two areas shared in common 14 genes (transcripts) labeled with circles and these genes were subjected to quantitative RT–PCR (qRT–PCR) analysis in (G). (G) Comparison of results of microarray analysis with those of qRT–PCR. Fourteen genes selected in (G) and vimentin were subjected to the qRT–PCR and the resultant data were compared with corresponding microarray data. Red and blue dots, respectively, represent data due to siVIM-270 and chiVIM-270 treatment. Note that microarray data is almost linear with those of qRT–PCR, indicating that results of microarray analysis and those of qRT–PCR are essentially identical to each other. The correlation coefficient was estimated at 0.96.
To confirm reliability of microarray data, we performed the quantitative RT–PCR analysis for vimentin mRNA and 14 transcripts (genes) selected from a region enclosed with a rectangle in Figure 7A and B and enlarged in Figure 7E and F. As shown in Figure 7G, transcription levels estimated by quantitative RT–PCR analysis were essentially identical to those obtained by the microarray analysis within a limit of error. The correlation coefficient was estimated at 0.96. Based on these observations, we conclude that gene silencing by modified siRNA with 8-bp GS-DNA substitution is associated with off-target effect much less frequently than that due to nonmodified -siRNA-based RNAi.
DISCUSSION
The present study clearly demonstrated that the GS-end proximal region of siRNA, which includes the seed sequence of the guide strand can be replaced with a DNA counterpart without substantial loss of gene-silencing activity, although most part of the remaining should be RNA. Our results (see Figures 5 and S7) also indicated that gene silencing due to transfection of modified siRNA with 8-bp GS-DNA substitution not only requires the activities of RLC-proteins, TRBP2 and PACT and a RISC-protein, Ago2, but is also associated with target mRNA cleavage. Thus, the molecular mechanism of gene silencing due to transfection of modified siRNA with 8-bp GS-DNA substitution appears very similar, if not identical, to that of nonmodified-siRNA-based RNAi (2,4). In RNAi, siRNA unwinding occurs from the GS end by an unknown unwinding enzyme (20,52–55). Thus, our finding of effective gene silencing induced by DNA-modified siRNAs in mammalian cells may indicate that this enzyme is not specific to dsRNA. Both dsRNA and dsDNA should serve as substrates for the enzyme.
Structural analysis (see Figure 5D) indicated that, as in the case of siRNA-dependent mRNA cleavage, target mRNA is cleaved by modified siRNA with 8-bp GS-DNA substitution mainly at position 10–11, as measured from the 5′ end of the guide strand, which apparently functions as a ruler zero-point in RNAi (43). Although one turn of dsDNA (10.5 bp) is shorter than that of RNA duplex (11–12 bp) by 0.5–1.5 bp (56), no difference in cleavage site could be detected between nonmodified and DNA-modified siRNA-dependent cleavage reactions (Figure 5D). This supports the notion that the helix pitch in the seed region is determined not by polynucleotides but by Ago protein surface properties (7). Results in Figure 4 and Supplementary Figure S8 show that the seed activity of the DNA arm is considerably less than that of the RNA counterpart. Our results indicated that DNA-modified-siRNA-dependent gene silencing is associated with not only little seed-dependent off-target effects but also virtually no passenger-strand-dependent off-target effects, provided that DNA-modified siRNAs possess an 8-bp long DNA guide strand and cognate siRNAs belong to class-I. In contrast, class-I siRNAs possessing a higher Tm of the seed sequence occasionally induced seed-dependent off-target effect (our unpublished data; see also Supplementary Figures S8 and S9).
Since it is possible to design class-I siRNAs for almost all human or mouse genes (20,25), the above finding may indicate that almost all mammalian genes are effectively knocked down with a considerably reduced off-target effect if we use functional modified siRNAs with 8-bp GS-DNA-substitution in place of authentic nonmodified siRNAs. Indeed, a genome-wide analysis using microarray profiling supported the feasibility of this notion experimentally (Figures 6 and 7, and Supplementary Figures S10 and S11).
Our experiments (Figure 5 and Supplementary Figure S7) indicated that DNA-modified-siRNA-dependent gene silencing also requires TRBP2 and PACT as well as Ago2. TRBP2 and PACT are capable of binding to dsRNA but not dsDNA (Figure 5A; 38,40). Ago2 has been shown to interact with ribonucleotide residues of the siRNA guide strand (4–9). Thus, it is quite feasible that, as in the case of nonmodified siRNA, the RNA part of DNA-modified siRNA provides sites for various RLC- and/or RISC–RNA-binding proteins. Figure 5C showed that DNA-modified-siRNA-dependent gene silencing is much more sensitive to unrelated siRNA treatment than nonmodified-siRNA-based RNAi, suggesting that a part of interactions between modified siRNA with 8-bp GS-DNA substitution and RLC/RISC proteins is weaker than those between nonmodified siRNA and RLC/RISC proteins. According to a recent model of RISC (7), the guide arm interacts with Mid-PIWI-L1 regions of Ago2. In addition, our data (see Supplementary Figure S4) suggested that, as with nonmodified siRNA (33), the 5′DNA end of the guide strand of the DNA-modified siRNA is phosphorylated within cells. Thus, the DNA seed arm may possess a weaker affinity to the Mid-PIWI-L1 lobe of Ago2 than the RNA counterpart. In the mRNA cleavage reaction, DNA-modified siRNA is as effective as cognate nonmodified siRNA as far as the oligonucleotide concentration of 50 nM is used. Therefore, RNA in the 3′terminal two-thirds of the guide strand might compensate weak DNA-dependent seed activity with its strong affinity to target mRNA in the case of mRNA cleavage reaction.
As shown previously (20), in highly functional siRNA, the 7-bp long region from the 5′ end of the guide strand is AU rich and the nucleotide pair at position 1 or the 5′ very end of the guide strand is A/U. The position 1 nt may not be involved in the initial mRNA recognition (20). In accordance with the recent reports (31,32), our microarray analysis indicated that the presence of complete homology between 3′UTR and the seed region (position 2–8) of the guide strand is important for recognition of the target sequence by siRNA (see Figure 6E). This may be coincident with the result obtained from crystal structure analysis of archaeal Piwi–RNA complexes (5). Structural analysis indicated that the phosphorylated 5′ end of the guide strand is anchored in a highly conserved deep basic pocket positioned on the surface of the Mid domain of Ago protein, and that the first base pair of the duplex is unwound, separating the 5′ nt of the guide strand from the complementary target mRNA (5). Recently, Jackson et al. (57) also have shown that position-specific, sequence-independent chemical modifications within the seed region reduced silencing of most off-target transcripts complementary to the seed region of the siRNA guide strand. Especially, they showed that the 2′-O-methyl ribosyl modification of nucleotide at the position 2 but not 1 from the 5′GS end decreased sharp position-dependent off-target silencing effect (57). Thus, it is feasible that most of the 5′ terminal nucleotides in the guide strand are essential for establishing conformation of the RLC– and/or RISC–RNA complex and/or executing RNAi. The first 7–8-bp region from the 5′end of the guide strand might have dual functions. That is, nucleotides in the region of position 1–7 might be closely linked to unwinding of siRNA or possible activation of RISC complex and those in the region of position 2–8 appeared involved in target mRNA recognition.
In conclusion, we found that replacement of the seed arm of class-I siRNAs with cognate DNA sequences led to almost complete loss of off-target effects without losing substantial gene-silencing activity in mammalian cells.
ACKNOWLEDGEMENTS
We thank A. Tanaka and N. Fukunishi for technical assistance. This work was supported in part by Special Coordination Fund for promoting Science and Technology to K.S., and grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan to K.U-T. and K.S. Funding to pay the Open Access publication charges for this article was provided by the University of Tokyo Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 3.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 4.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2006;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 5.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI-domain-siRNA-guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 7.Yuan Y-R, Pei Y, Ma J-B, Kuryavyi V, Zhadina M, Meister G, Chen H-Y, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J-B, Ye K, Patel DJ. Structural basis of overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preall JB, Sontheimer EJ. RNAi: RISC gets loaded. Cell. 2005;123:543–545. doi: 10.1016/j.cell.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Birmingham A, Anderson EM, Reynolds A, Ileley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 11.Song JJ, Smith SK, Hannon GJ, Joshua-Tor J. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;205:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 13.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson AL, Burchard J, Schelter J, Chau BM, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbashir S, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohjoh H. RNA interference (RNA(i)) induction with various types of synthetic oligonucleotide duplexes in cultured human cells. FEBS Lett. 2002;19:195–199. doi: 10.1016/s0014-5793(02)02860-0. [DOI] [PubMed] [Google Scholar]
- 19.Chiu Y-L, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNA? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Tallarida RJ, Murray RB. Manual of Pharmacological Calculations with Computer Programs. NY: Springer; 1987. [Google Scholar]
- 23.Lee Y, Hur I, Park S-Y, Kim Y-K, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chendrimada TP, Gregory R, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ui-Tei K, Naito Y, Saigo K. Guidelines for the selection of effective short-interfering RNA sequences for functional genomics. Methods Mol. Biol. 2006;361:201–216. doi: 10.1385/1-59745-208-4:201. [DOI] [PubMed] [Google Scholar]
- 26.Panjkovich A, Melo F. Comparison of different melting temperature calculation methods for short DNA sequences. Bioinformatics. 2005;21:711–722. doi: 10.1093/bioinformatics/bti066. [DOI] [PubMed] [Google Scholar]
- 27.Freier AM, Keirzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 29.Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- 30.Bolstad BM, Irizarry RA, Gautier L, Wu Z. Preprocessing high-density oligonucleotide arrays. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. NY: Springer; 2005. pp. 13–32. [Google Scholar]
- 31.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel KP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen CB, Shomron N, Sndberg R, Hornstein E, Kitzman J, Burge C. Determinants of targeting by endogenous and exogenous microRNAs and siRNA. RNA. 2007;13:1–17. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nykänen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 34.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 35.Chun J-Y, Han Y-J, Ahn K-Y. Psx homeobox gene is X-linked and specifically expressed in trophoblast cells of mouse placenta. Dev. Dyn. 1999;216:257–266. doi: 10.1002/(SICI)1097-0177(199911)216:3<257::AID-DVDY4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 36.Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, Rand TA, Kalidas S, Du F, Kim H-E, Smith DP, Wang Z. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 39.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 40.Patel RC, Sen GC. PACT, a protein activator of the interferon- induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 2003;13:41–46. doi: 10.1016/s0960-9822(02)01394-5. [DOI] [PubMed] [Google Scholar]
- 42.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21 and 22 nt RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotech. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 45.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 46.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 47.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yekta S, Shih I, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 49.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 50.Borer PM, Dengler B, Tinoco I, Jr, Uhlenbeck OC. Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 1974;86:843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- 51.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 53.Schwarz DS, Hutvágner G, Du T, Zu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 54.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 55.Chiu Y-L, Dinesh CU, Chu C-Y, Ali A, Brown KM, Cao H, Rana TM. Dissecting RNA-interference pathway with small molecules. Chem. Biol. 2005;12:643–648. doi: 10.1016/j.chembiol.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 56.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell. Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 57.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2007;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]