Abstract
In eukaryotic cells, newly synthesized mRNAs acquire a poly(A) tail that plays several fundamental roles in export, translation and mRNA decay. In mammals, PABPN1 controls the processivity of polyadenylation and the length of poly(A) tails during de novo synthesis. This regulation is less well-detailed in yeast. We have recently demonstrated that Nab2p is necessary and sufficient for the regulation of polyadenylation and that the Pab1p/PAN complex may act at a later stage in mRNA metabolism. Here, we show that the presence of both Pab1p and Nab2p in reconstituted pre-mRNA 3′-end processing reactions has no stimulating nor inhibitory effect on poly(A) tail regulation. Importantly, the poly(A)-binding proteins are essential to protect the mature mRNA from being subjected to a second round of processing. We have determined which domains of Nab2p are important to control polyadenylation and found that the RGG-box work in conjunction with the two last essential CCCH-type zinc finger domains. Finally, we have tried to delineate the mechanism by which Nab2p performs its regulation function during polyadenylation: it likely forms a complex with poly(A) tails different from a simple linear deposit of proteins as it has been observed with Pab1p.
INTRODUCTION
Eukaryotic mRNA 3′-end formation is a fundamental step in gene expression that consists in two coupled and successive reactions: a site-specific endonucleolytic cleavage that occurs in the 3′ untranslated region of the RNA, followed by polymerization of adenosine 5′-monophosphate residues initiated from the newly created 3′-hydroxyl end (1–3). These two reactions are performed by a machinery of approximately 1 MDa that is well-conserved from yeast to human. In higher eukaryotes, the cleavage reaction involves cleavage factor Im, cleavage factor IIm [which contains at least hPcf11 and hClp1; (4)], the cleavage stimulation factor (CstF) and the cleavage and polyadenylation specificity factor (CPSF). Recent work brought clear evidence that CPSF comprises the long-sought endonuclease as CPSF-73, which X-ray structure has been recently determined (5). In mammals, the polyadenylation reaction has been extensively studied and most of the requirements and mechanisms involved are now quite well understood. Interactions of mammalian poly(A) polymerase (mPAP) with CPSF subunits CSPF-160 and hFip1p stimulate the enzyme activity by tethering it to the RNA for which it has intrinsically a low affinity (6–10). The poly(A)-binding protein nuclear 1 (PABPN1) interacts with the first polymerized 11 adenosine residues, further increasing the affinity of mPAP for the RNA substrate which switches the enzyme from a distributive to a processive mode (7). Soon after approximately 250 A-long poly(A) tails have been synthesized, mPAP switches back to a distributive mode (11). Although the mechanism responsible for this poly(A) tail length control is not fully understood, it seems that it is achieved through the formation of a 20 nm globular structure, involving bound PABPN1 on the poly(A) that may disrupt the tripartite [CPSF-mPAP-PABPN1] complex (12).
In the yeast Saccharomyces cerevisiae, both cleavage and polyadenylation require cleavage factors IA (CF IA, comprising Rna14p, Rna15p, Pcf11p and Clp1p) and IB (consisting of Nab4p/Hrp1p), and the cleavage and polyadenylation factor CPF. Noticeably, CPF includes Ysh1p, the endonuclease homologue of CPSF-73, and the poly(A) polymerase Pap1p. One major difference between the yeast and mammalian 3′-end processing systems is that in yeast, there is no requirement for a poly(A)-binding protein to induce processive synthesis of the poly(A) tail (13). Instead, the association of Pap1p within CPF enhances its activity (13) and a poly(A)-binding protein is required instead to restrict this action. Interestingly, the protein most similar to PABPN1 in yeast, Rbp29p, has no function in 3′-end formation but rather works in cytoplasmic mRNA metabolism close to translation (14). Despite these discrepancies, the need for a poly(A)-binding protein to control the poly(A) tail length in yeast has been highlighted by several studies. First, the major cytoplasmic poly(A)-binding protein Pab1p has been previously suggested to regulate polyadenylation on its own (15,16) and through the recruitment and stimulation of the Pab1p-dependent poly(A)-nuclease (PAN) (16,17). In addition, it has been shown that the nuclear poly(A)-binding protein Nab2p is involved in mRNA export and in poly(A) tail length regulation both in vivo and in vitro (18). Finally, we have recently obtained evidence that pre-mRNA 3′-end processing can be fully reconstituted in vitro by means of either Pab1p or Nab2p as regulation factors in the absence of PAN (19). These results established that mRNA polyadenylation in yeast is regulated by poly(A)-binding proteins, like in mammals. However, the reason for this requirement remains unclear.
In the present study, we compared Pab1p, Nab2p or the combination of both, in their ability to regulate poly(A) tail length in vitro with yeast cell extracts and in reconstituted systems with purified components. Addition of both proteins did not show any dramatic stimulating or inhibitory effect on polyadenylation. Although Pab1p and Nab2p could both prevent a mature polyadenylated mRNA from being re-cleaved by the yeast 3′-end processing machinery, only Nab2p was able to inhibit its re-adenylation. Finally, we showed that Nab2p uses its RGG-box and zinc-finger domains in conjunction to bind onto the poly(A) tail and regulate its length. The resulting ribonucleoparticule seemed to protect efficiently poly(A) species that surprisingly parallels the physiological poly(A) tails added to mRNAs in yeast.
MATERIALS AND METHODS
Yeast strains
The pab1 deletion is rescued by deletion of the RPL46 gene in strain YAS394 (20). Strains expressing both N-terminal TAP-tagged Rna15p and Fip1p, or TAP-Fip1p alone (YSD39 and YSD10, respectively) have been described in Dheur et al. (19). Because of the severe growth defect of YAS394, TAP-tagging of either Fip1p or Rna15p in a pab1Δ strain has been constructed in the same way in YRP1130 [mtr1-3 pab1Δ; described in (21)] to produce YNV1 and YNV2, respectively.
Constructions of nab2 mutant alleles
Sequencing of the nab2-21 allele [coming from pnab2-21, (18)] cloned in pGEX4T1 showed that the initial deletion was done on a NAB2 variant allele as previously described (22), encoding a Nab2 protein containing only two of the nine QQQP repeats present in the N-terminal region of the protein (referred to as svNab2p for short variant Nab2p). We used this svnab2-21 and the already available NAB2 alleles to rebuild the svNAB2 and nab2-21 alleles encoding the corresponding proteins with two and nine QQQP repeats, respectively. We cloned these alleles in pETM30 which allowed the production of Nab2p with a GST tag. This tag is removable with the TEV protease. Therefore, pGEX4T1-svnab2-21(EcoRI/SalI) and pETM30-NAB2 (NcoI/SalI) were digested with HindIII/SalI and the fragment released from the first one was cloned into the second to produce pETM30-svnab2-21. pETM30-NAB2 and pETM30-svnab2-21 were digested with PstI/SalI. The fragment released from one digestion was ligated to the restricted vector from the other one to produce pETM30-nab2-21 and pETM30-svNAB2, respectively. nab2ΔRGG was amplified by PCR from pGEX2TK-nab2ΔRGG (23) and cloned in pETM30 to produce pETM30-nab2ΔRGG (NcoI/XhoI). pETM30-NAB2 was digested with NcoI/PstI, filled-in and self-ligated to produce pETM30-RGG-(CCCH)7Cter. pETM30-NAB2 was digested with PstI/SalI, filled-in and self ligated to produce pETM30-NterQrich. pETM30-NterQrich was then digested with NcoI/XhoI and the released fragment was cloned in pETM10 (NcoI/XhoI) to produce pETM10-NterQrich. nab2Δ(CCCH)7 and (CCCH)7Cter were amplified by PCR and cloned in pETM11 (NcoI/XhoI) to produce pETM11-nab2Δ(CCCH)7 and pETM11-(CCCH)7Cter, respectively. nab2ΔQrich was amplified by PCR with Accuprime Supermix (Invitrogen) on pETM30-NAB2 using divergent primers containing a BamHI restriction site. The resulting PCR product was digested by BamHI and self-ligated to produce pETM30-nab2ΔQrich.
RNA preparations
Capped and radiolabeled CYC1 and CYC1pre transcripts used in the pre-mRNA 3′-end processing assays were synthesized in vitro as described previously (24). To produce the mature polyadenylated CYC1 RNA (referred to as CYC1pA), 6 pmol of radiolabeled CYC1 RNA were cleaved and polyadenylated with 1.8 mg of wild type BMA64 yeast extract for 75 min at 30°C (the assay was performed as a 12-fold standard 50 µl reaction each containing 0.5 pmol of RNA and 150 µg of cell extract). The processed RNA was then extracted, precipitated and gel-purified on a 6% polyacrylamide-8.3 M urea gel after visualization by autoradiography. Poly(A)20, poly(A)34, poly(A)60 and poly(A)200 were prepared from heterogeneous poly(A) (Amersham) by UV-shadowing as described previously (25). Poly(A)34 contained RNA molecules of 33, 34 and 35 nucleotides (nt), whereas poly(A)200 contained RNA of sizes ranging from 190 to 250 nt. Poly(A)20 initially contained RNA molecules of 19, 20 and 21 nt. Poly(A)20, poly(A)34, poly(A)200 were 5′-end labeled with ATP-[gamma-32P] and T4 polynucleotide kinase, using ‘forward buffer’ conditions as described by the manufacturer (Stratagene). The 5′-radiolabeled poly(A)20 mixture was subsequently separated on a 15% polyacrylamide-8.3 M urea gel and poly(A)20 was specifically excised from it.
In vitro processing assays
Pre-mRNA 3′-end processing competent extracts were prepared following a previously described spheroplast procedure (26). Tandem affinity purification (TAP) of [CF IA+CPF] was performed according to Rigaut et al. (27), with the following modifications that enhanced the amount of complexes and their polyadenylation activity: 2 l of culture of OD600 = 2 were used instead of 1 l at OD600 = 4; the entire purification protocol (extract preparation and complex purification) was conducted in 1 day to avoid multiple freeze/thaw cycles. Purification was analyzed the following day by SDS–PAGE and silver staining, and the relevant fractions were dialyzed against buffer D supplemented with 20% glycerol, divided into 20 µl aliquots, flash-freezed and kept at −80°C.
Pre-mRNA cleavage and polyadenylation were assayed essentially as previously described (24,28). For polyadenylation kinetics, 12-fold reaction mixtures containing 360 µg of pab1Δ 3′-end processing extracts were supplemented with either 6 µg of recombinant Pab1p or 5.28 µg of recombinant Nab2p and preincubated for 15 min at 30°C. Polyadenylation reactions were both initiated at the same time by addition of radiolabeled CYC1pre RNA. Aliquots of 20 µl were withdrawn from each reactions at the time points indicated. The reactions were immediately stopped by transferring them into 42°C prewarmed stop-reaction mixtures containing proteinase K, and further incubated for 30 min at 42°C. RNAs were then precipitated and loaded on a 6% polyacrylamide-8.3 M urea gel and visualized by autoradiography.
For reconstituted reactions, 1 µl of TAP-purified [CF IA+CPF] or 4 µl of TAP-purified CPF alone were incubated with 60 fmol of radiolabeled RNA, 40 ng of recombinant Nab4p/Hrp1p (unless otherwise mentioned) and with the indicated amounts of either Nab2p, Pab1p or nab2 mutant proteins in a 20 µl final volume. Poly(A) polymerase inhibition assays or 3′-end accessibility tests were realized with 24 ng of recombinant Pap1p. This amount of protein equals the amount of Pap1p contained in 4 µl of our TAP-purified CPF (determined by Western blot, our unpublished results). Poly(A) polymerase inhibition assays were done with 4.5 nM of poly(A)34 which is similar to the RNA concentrations used in cleavage/polyadenylation reactions. All reactions were conducted at 30°C for 1 h, stopped and analyzed as described earlier.
Micrococcal nuclease assays
These protection assays were conducted in 20 mM HEPES-KOH (pH 7.9), 50 mM KCl, 2.5 mM β-mercaptoethanol, 50 µl final volume, with 1 pmol of 5′-end radiolabeled poly(A)200 and ∼25 pmol (1.5 µg) of purified recombinant Nab2p, nab2ΔRGGp or Pab1p (an amount sufficient to saturate the RNA, assuming that approximately 20 nt are covered per molecule, resulting in a final protein concentration 50-fold above the Kdapp). The RNA/protein complexes were assembled for 25 min at 30°C. Digestions were then initiated by addition of 1 mM CaCl2 and S7 nuclease (Roche) to the specified final concentrations, and the incubation was continued for 30 min at 30°C (unless otherwise mentioned). Reactions were terminated by addition of 5 mM EGTA and RNA fragments were immediately phenol/chloroform extracted, ethanol precipitated and subjected to electrophoresis on a 12% polyacrylamide-8.3 M urea gel. Visualization was done by autoradiography. Densitometric analysis was performed with the ImageJ program. Densitogram of the molecular weight marker in each gel was used to construct a calibration curve of log(MW) as a function of relative migration distances that was used to determine the size of the poly(A) species observed.
RESULTS
Comparison of Nab2p and Pab1p polyadenylation regulation activities in vitro
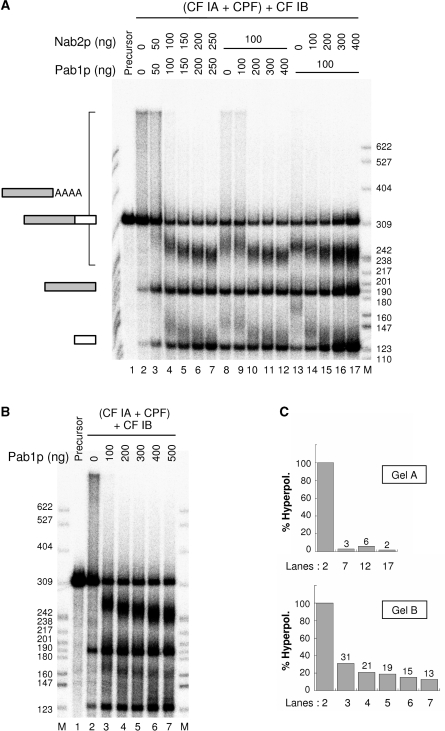
It has been previously established that both Pab1p and Nab2p were capable of regulating polyadenylation in vitro. In vivo, it has been shown that overexpression of the PAB1 gene could suppress the nab2 deletion. Nevertheless, the hyperpolyadenylation defect associated to it could not be corrected (15–19). The requirement for Nab2p and Pab1p to regulate polyadenylation in vivo and in vitro could perhaps partly reflect a collaboration between the two proteins in this process. In agreement with this hypothesis, Dunn et al. (29) have suggested that Nab2p could help the loading of Pab1p onto the poly(A) tail. Since this property may involve a direct interaction between the two proteins, we have addressed this possibility by mixing 2 µM of each of them at room temperature, and loaded the mixture on a Superdex-200 gel-filtration column. This technique has already been employed to show an interaction between Pap1p and Fip1p (30). In the present case, no complex formation between Nab2p and Pab1p could be seen. Each protein behaved on this column as a single monomer of ∼95 kDa in comparison to gel-filtration standard markers (data not shown). Nevertheless, it is still possible that this interaction may only occur during polyadenylation, presumably mediated by the RNA. Therefore, we tested the putative cooperation of a combination of Pab1p and Nab2p in a reconstituted reaction with yeast factors purified by TAP-tagging, and purified recombinant Pab1p, Nab2p and Nab4p/Hrp1p (see Supplementary Figure 1, and Experimental procedures). Two different assays were done: simultaneous addition of increasing amounts of Nab2p and Pab1p (Figure 1A, lanes 3–7), or a relative titration comprising a fixed amount (100 ng) of one of the poly(A)-binding proteins and increasing amounts of the other one (Figure 1A, lanes 8–17). Surprisingly, an apparent competition between the two proteins was observed when 50 ng of each polypeptides were used instead of 100 ng of one or the other (Figure 1A, compare lane 3 with lanes 8 and 13). However, control of the poly(A) tail length was efficiently achieved at the end of all titrations in both assays (Figure 1A, lanes 7, 12 and 17), as already observed with either Nab2p or Pab1p alone (19). These results suggested that the presence of both Nab2p and Pab1p did not lead to any significant stimulation or inhibition of polyadenylation in vitro. Also, this assay did not indicate which of the two proteins was responsible for proper termination of polyadenylation. However, it was interesting to notice that the polyadenylated species formed at protein concentrations required to achieve a full polyadenylation regulation were more reminiscent to reactions performed with Nab2p only rather than to those where Pab1p was used alone [Figure 1A, lanes 7, 12, 17; Figure 1B lane 7; Figure 6B lanes 3, 14, 30, 37, 50; Figure 7A, lane 6; and see reference (19)]. Indeed, hyperpolyadenylated species typical of polyadenylation reactions performed in the presence of Pab1p alone (Figure 1B and C ‘gel B’) could hardly be detected at the end of the titrations (Figure 1C), as previously observed (19), even when Pab1p was more abundant than Nab2p (Figure 1A, lane 12 and Figure 1C).
Figure 1.
Effect of the simultaneous presence of Nab2p and Pab1p on polyadenylation regulation in a reconstituted reaction in vitro. (A) Cleavage and polyadenylation reactions were reconstituted in vitro as described in Materials and Methods with the CYC1 radiolabeled precursor (lane 1), TAP-purified machinery [CF IA+CPF] and recombinant Nab4p/Hrp1p/CF IB. Poly(A) tail length control was assayed by addition of recombinant Pab1p in combination with recombinant Nab2p either as concomitant (lanes 3–7) or relative (lanes 8–17) titrations. (B) Cleavage and polyadenylation reactions were reconstituted as in A, but poly(A) tail length control was assayed by addition of recombinant Pab1p only (lanes 3–7). (C) Quantification of hyperpolyadenylation occurring in the indicated lanes of gels in A and B. The abundance of polyadenylated species with sizes higher than the 309 nt precursor was quantified with ImageJ and expressed as percentage of the maximum hyperpolyadenylation that takes place in the absence of any poly(A)-binding proteins lanes 2 of A and B. Gels and corresponding lane numbers are specified along with their respective value. Reaction products were analyzed on a 6% polyacrylamide-8.3 M urea denaturing gel and visualized by phosphorimaging. M, labeled MspI-digested pBR322 was used as a molecular weight marker and corresponding sizes are indicated in number of nucleotides. Upstream and downstream cleavage products are indicated by grey and white rectangles respectively. AAAA represents the poly(A) tail.
Figure 6.
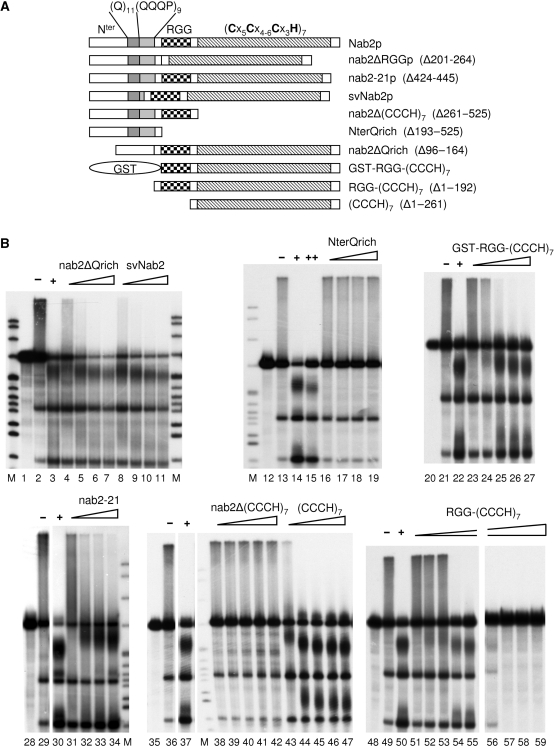
Poly(A) tail length control activity of various Nab2p deletion mutants. (A) Schematic representation of various Nab2p deletion mutants. The numbers in brackets indicate the extent of the corresponding deletion. Nter, amino terminal part of the protein. (Q)11, poly-glutamine stretch comprising residues 106 to 116. (QQQP)9, nine successive repeats of the tetrapeptide Gln-Gln-Gln-Pro comprising residues 121 to 156. RGG, arginine/glycine rich region of the protein comprising residues 201 to 264. (Cx5Cx4-6Cx3H)7, seven repeats of the indicated zinc-finger domain sub-type spanning residues 262 to 473. (B) Cleavage and polyadenylation reactions were reconstituted in vitro as described in Materials and Methods with the CYC1 precursor (lanes 1, 12, 20, 28, 35, 48), the TAP-purified 3′-end formation machinery [CF IA+CPF], recombinant CF IB/Nab4p, and either Nab2p or the indicated mutant protein as polyadenylation regulation factor. − or + indicate reactions in the absence or in the presence of 420 nM of wild-type Nab2p (500 ng in 20 µl), respectively; ++ indicates a reaction with 1.6 µM of Nab2p (2 µg in 20 µl). Triangles represent increasing concentrations of the indicated mutant protein. In the cases of nab2-21, nab2ΔQrich, svNab2, NterQrich, the concentrations used were: 84, 168, 250 and 420. For GST-RGG-(CCCH)7, nab2Δ(CCCH)7, (CCCH)7, the concentrations were: 84, 168, 250, 334 and 420 nM. For RGG-(CCCH)7, the concentrations were 26, 52, 78, 104 and 130 (lanes 51–55) and 260, 390, 520 and 650 nM (lanes 56–59). Reaction products were analyzed and visualized as in Figure 1. M, molecular weight markers were as described in Figure 1. Lanes 28–34 were on the same gel, and lanes 35–47 as well.
Figure 7.
Functional analysis of the RGG-box region of Nab2p. (A) Cleavage and polyadenylation reactions were reconstituted in vitro as described in Materials and Methods with the CYC1 precursor, the TAP-purified 3′-end formation machinery [CF IA+CPF], recombinant CF IB/Nab4p, and increasing amounts of either Nab2p (lanes 2–6) or nab2ΔRGG mutant protein (lanes 7–10) as polyadenylation regulation factors. Lane 1, unreacted precursor. (B) Polyadenylation reactions of the gel-purified polyadenylated CYC1pA were performed in vitro with TAP-purified CPF, 40 ng of recombinant CF IB/Nab4p, and increasing amounts of recombinant nab2ΔRGG mutant protein (lanes 3–5). Reactions were further supplemented with the same amount of TAP-purified CPF after 1 h (lane 6) as in Figure 4A. 3′-end accessibility was challenged by addition poly(A) polymerase to the reaction (lane 7) as in Figure 4A. Reaction products were analyzed and visualized as in Figure 1. Lane 1, unreacted precursor. M, molecular weight markers were as described in Figure 1. (C) RNA 3′-end accessibility of Nab2p/poly(A)60 or nab2ΔRGG/poly(A)60 complexes was challenged by titrations of recombinant poly(A) polymerase Pap1p. Five hundred nanograms of Nab2p or nab2ΔRGGp were pre-incubated with 3 nM of poly(A)60 for 15 min at 30°C, and polyadenylation reactions were initiated by addition of the indicated Pap1p amounts and further incubated for 1 h. RNA contents were then extracted, separated on a 8% polyacrylamide-8.3 M urea denaturing gel and visualized by phosphorimaging. Lane 1: poly(A)60 alone. M, molecular weight markers were as described in Figure 1. (D) Polyadenylation efficiency of poly(A)60 complexed to either Nab2p or nab2ΔRGGp was calculated from experiments such as in C. The abundance of polyadenylated species with sizes higher than the 60 nt precursor was quantified with ImageJ, normalized to the integrated density in each entire lane and expressed as a ratio to the abundance of polyadenylated species higher than the 60 nt precursor in the absence of Pap1p (as in C, lane 2). Values for 5, 10, 24, 42 and 960 ng of Pap1p are means from three independent experiments ± standard error. Values for 100, 300 and 500 ng of Pap1p are means from two independent experiments ± standard error.
In order to test whether Nab2p could take advantage over Pab1p in the control of the poly(A) tail length during de novo synthesis, we made use of Pab1p's ability to recruit the poly(A) nuclease (PAN) to detect any Pab1p-binding event. To actually measure the competition between Pab1p and Nab2p, we used extracts prepared from a pab1Δ strain and added in a first experiment a fixed amount of recombinant Pab1p (500 ng per lane, see Materials and Methods). Kinetics of polyadenylation were assayed on the CYC1 pre-cleaved precursor. As reported previously (16), it showed a bi-phasic reaction where the synthesis of the poly(A) tail to a physiological length occurred after 20 min and was followed by its shortening (Figure 2, lanes 1–9). This shortening was due to the PAN complex (19) and revealed the binding of Pab1p to the poly(A) tail since it is essential for PAN activity (17). We then determined the amount of endogenous Nab2p present in the pab1Δ extracts and found 57 ± 4 ng of Nab2p in 30 µg of extracts (mean of three Western blots ± standard deviation, data not shown). Therefore, we supplemented the extracts with 440 ng of recombinant Nab2p together with 500 ng of Pab1p (per 30 µg of extract) and performed similar polyadenylation kinetics. Surprisingly, the second phase of the reaction, i.e. shortening of the poly(A) tails, did not happen in the simultaneous presence of both proteins (Figure 2, lanes 14–18), compared to the Pab1p-alone conditions (Figure 2, lanes 5–9). In addition, it is worth noting that the overall mRNA degradation initiated during the second phase of the kinetics with Pab1p was absent when Nab2p was present (Figure 2, compare lanes 8, 9 to lanes 17, 18). The lack of exonuclease activity and mRNA degradation in the presence of both proteins (Figure 2, lanes 10–18) was similar to what was observed with extracts in the presence of Nab2p only (18). This absence of shortening was not due to a slower pace of the reaction since time-course experiments performed for 240 min showed the same pattern (data not shown). This suggested that the poly(A) tail length control observed in the assay reported in Figure 2 may be mainly achieved through the binding of Nab2p and not Pab1p. Alternatively, Pab1p may also be partially loaded onto the poly(A) tail together with Nab2p but the latter would prevent the activation of the Pab1p-dependent PAN while rendering the 3′ ends inaccessible to exonucleolytic degradation. In any case, these two possibilities are not mutually exclusive. However, this experiment highlighted the essential role of Nab2p, either in the presence or absence of Pab1p, in maintaining newly synthesized poly(A) tails intact.
Figure 2.
Predominant action of Nab2p versus Pab1p in poly(A) tail length control. Polyadenylation kinetics on the pre-cleaved CYC1 precursor (CYC1pre). Two 12-fold reactions were set up and kinetics were initiated by addition of radiolabeled CYC1pre. A 20 µl reaction contains: 30 µg of pab1Δ (YAS394) 3′-end-processing competent yeast cell extract and either 500 ng of recombinant Pab1p (lanes 1–9) or a combination of 500 ng of recombinant Pab1p with 440 ng of recombinant Nab2p (lanes 10–18). Twenty microliters from each reaction mixture were withdrawn at the indicated time points, and the RNA content was extracted, separated on a 6% polyacrylamide-8.3 M urea denaturing gel and visualized by autoradiography. M, molecular weight markers were as described in Figure 1.
Endonucleolytic cleavage of a mature polyadenylated CYC1 RNA is prevented by Nab2p or Pab1p
In mammals, U-rich cis-acting elements located downstream of the cleavage site enhance the overall efficiency of the reaction by allowing the binding of CstF to the pre-mRNA, thereby strongly improving cleavage (31,32). In yeast, similar sequences are sometimes found very close to the cleavage site which participate to the binding of CPF (33). However, such U-rich elements are not ubiquitously found on natural pre-mRNAs but this has no dramatic impact on their ability to be processed in vivo and in vitro (33). This suggested that a mature polyadenylated mRNA also lacking these downstream elements, as they are replaced by a poly(A) tail, might theoretically be processed again, at least in vitro. Therefore, we tested the possibility that either Nab2p or Pab1p could protect a polyadenylated CYC1 transcript (termed CYC1pA) from being re-cleaved by the mRNA 3′-end formation complex.
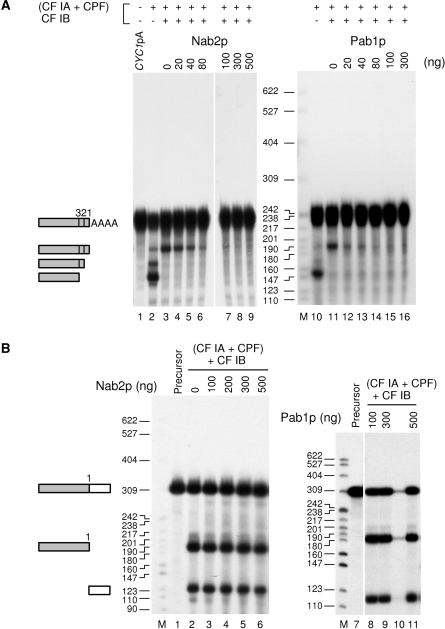
In the absence of Nab4p/Hrp1p (Figure 3A, lanes 2 and 10), CYC1pA RNA was surprisingly re-cleaved efficiently at the natural poly(A) site (numbered 1) and at two other upstream positions (numbered 2 and 3) already detailed for the CYC1 pre-mRNA (24). It was astonishing that a mature yeast mRNA could be such a good substrate for cleavage by the processing machinery in vitro in the absence of Nab4p. But more surprisingly, after addition of Nab4p/Hrp1p, the mature CYC1pA could still be cleaved. However, cleavage occurred at the major poly(A) site only (Figure 3A, lanes 3 and 11). This demonstrated that in this particular situation the essential role of Nab4p/Hrp1p in controlling cleavage is not sufficient to protect the mature mRNA from the endonucleolytic cleavage activity of the 3′-processing machinery. However, addition of increasing amounts of either Nab2p or Pab1p to the reaction abolished the cleavage of the CYC1pA RNA (Figure 3A, lanes 4–9 and 12–16). As a control, addition of recombinant poly(U)-binding protein Pub1p had no effect (data not shown). Moreover, cleavage was not prevented when Nab2p or Pab1p were added to a standard cleavage reaction where the CYC1 precursor was used as substrate (Figure 3B). Therefore, this inhibition by a poly(A)-binding protein required a polyadenylated RNA. Inhibition of cleavage of CYC1pA seemed slightly more efficient with Pab1p. Although the weak efficiency of CYC1pA cleavage indicates that this mature RNA was not a good substrate on its own (compare Figure 3A, lane 3 and Figure 3B, lane 2), these results clearly showed that a fully processed mRNA can still be cleaved by the yeast 3′-end processing machinery. In addition to Nab4p/Hrp1p, both Nab2p and Pab1p were required to prevent this phenomenon.
Figure 3.
Cleavage inhibition of mature polyadenylated CYC1pA by Pab1p or Nab2p. (A) Cleavage assays with gel-purified mature polyadenylated CYC1pA were done in an in vitro reconstituted reaction using the TAP-purified 3′-end-processing machinery [CF IA+CPF] and 40 ng of recombinant Nab4p when specified. Inhibition of cleavage was challenged by increasing amounts of either Nab2p (lanes 4–9) or Pab1p (lanes 12–16). The major cleavage site is indicated by a number 1, and the alternative cryptic cleavage sites are numbered 2 and 3, as previously described (24). Lanes 1–9 were on the same gel. Lane 1, unreacted precursor. (B) Cleavage reactions with the CYC1 precursor (lanes 1 and 7) were performed as in A, except that 40 ng of CF IB/Nab4p were included in all assays. Inhibition of cleavage was tested by adding increasing amounts of either Nab2p (lanes 2–6) or Pab1p (lanes 8–11). Lanes 7–11 were on the same gel. Lane 10 contained a spillover of lane 9 and was therefore skipped. Lane 1, unreacted precursor. Reaction products were analyzed and visualized as in Figure 1. M, molecular weight markers were as described in Figure 1.
Accessibility to adenylation of a mature CYC1pA RNA
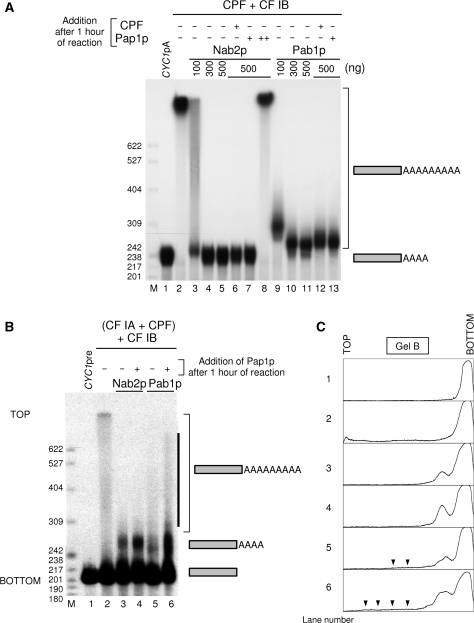
To test now whether a mature mRNA, i. e. 3′-end cleaved and polyadenylated transcript, could still be a substrate for polyadenylation as it was for cleavage, we set up an assay with the purified factors CPF and CF IB but leaving away CF IA to prevent cleavage to occur. CPF is known to bind to the poly(A) site and polyadenylate an RNA substrate on its own (13). In such a system, CYC1pA was efficiently elongated further by CPF (Figure 4A, lane 2). Addition of recombinant Nab2p completely inhibited CYC1pA adenylation (Figure 4A, lanes 3–5). Surprisingly, adding recombinant Pab1p instead had a similar effect but to a lesser extent since the transcripts ran slightly higher in the gel than the CYC1pA substrate itself, even at the highest concentrations tested (Figure 4A, lanes 9–11). Thus, Nab2p and Pab1p were both able to maintain the integrity of the fully processed mRNA against the cleavage activity of the 3′-end machinery but only Nab2p was able to fully protect the mature poly(A) tails from being further elongated.
Figure 4.
Polyadenylation of mature polyadenylated CYC1 and 3′-end accessibility assays. (A) Polyadenylation reactions of gel-purified polyadenylated CYC1pA were performed in vitro with TAP-purified CPF, 40 ng recombinant CF IB/Nab4p, and increasing amounts of recombinant Nab2p (lanes 3–5) or Pab1p (lanes 9–11). Reactions were further supplemented with the same amount of TAP-purified CPF after 1 h and the reactions were pursued for an additional hour (lanes 6 and 12). 3′-end accessibility was tested after 1 h of reaction by addition of 24 ng of yeast poly(A) polymerase Pap1p to reactions containing the highest concentrations of Nab2p (lanes 7 and 8) or Pab1p (lane 13). Reactions were further incubated for another hour. ‘++’ denotes addition of a 1.5 molar excess of poly(A) polymerase compared to Nab2p instead of 24 ng. Lane 1, unreacted precursor. (B) Polyadenylation reactions with CYC1pre RNA (lane 1, unreacted precursor) were performed with the TAP-purified 3′-end-processing machinery [CF IA+CPF] with 40 ng recombinant CF IB/Nab4p and 500 ng of either recombinant Nab2p (lanes 3 and 4) or Pab1p (lanes 5 and 6). Accessibility to further elongation was challenged after 1 h reaction as in A. The solid black bar highlights the increase of hyperpolyadenylated species upon addition of recombinant Pap1p to the Pab1p-containing reaction. (C) Densitometric analysis of the gel in B using ImageJ. Black arrows are highlighting the significant amount of hyperpolyadenylated species present in Pab1p-containing reactions in comparison with Nab2p-containing reactions. TOP and BOTTOM designate corresponding positions on the gel in B. Lane numbers of the gel in B are indicated on the left side of each corresponding densitogram. Reaction products were analyzed and visualized as in Figure 1. M, molecular weight markers were as described in Figure 1.
To challenge the robustness of this inhibition mediated by Nab2p, we supplemented the assays with either more CPF or free recombinant yeast poly(A) polymerase after 1 h of reaction and incubated them for an additional hour. In the case of Nab2p, no change could be detected in the polyadenylation state of CYC1pA (Figure 4A, lanes 6 and 7). Addition of 1.5 molar excess of poly(A) polymerase over Nab2p led to the strong polyadenylation of CYC1pA, suggesting that the 3′ ends of the poly(A) tails were not irreversibly protected by Nab2p (Figure 4A, lane 8). Unexpectedly, when Pab1p-mediated inhibition was similarly challenged, CYC1pA could slightly be adenylated further (Figure 4A, lanes 12 and 13).
In order to verify whether this elongation by adenylation could occur in vitro in standard 3′-end processing conditions, we tested the accessibility of the 3′ ends of the poly(A) tails starting with the pre-cleaved CYC1 transcript and the entire machinery. In this system, CYC1pre RNA was polyadenylated and addition of 500 ng of Nab2p or Pab1p triggered poly(A) tail length control, although hyperpolyadenylated products were visible only with Pab1p (Figure 4B and C, compare lane 2 with lanes 3 and 5). Addition of recombinant poly(A) polymerase to assays containing either Nab2p or Pab1p after 1 h of reaction stimulated polyadenylation (Figure 4B and C, compare lane 3 with 4 and lane 5 with 6). However, this additional Pap1p also increased the length and the amount of hyperpolyadenylated species in the Pab1p-containing reaction (Figure 4B and C, lanes 5, 6). These long poly(A) species have been already observed when Pab1p is used as a poly(A) tail length regulator in fully reconstituted 3′-processing assays in vitro [Figure 1 and see reference (19)]. No noticeable change could be observed upon addition of poly(A) polymerase to the Nab2p-containing reaction except the stimulation of polyadenylation (Figure 4B and C, compare lanes 3 and 4).
Therefore, although both poly(A)-binding proteins could trigger polyadenylation termination in vitro, these results strongly suggested that Nab2p can more specifically prevent accessibility to the 3′ ends of the mature poly(A) tail.
Nab2p and Pab1p can inhibit poly(A) polymerase with similar efficiencies
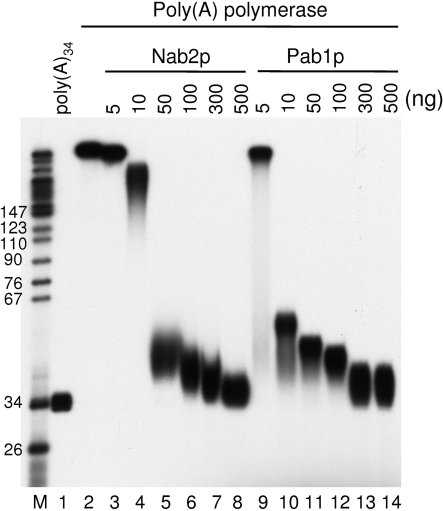
It has been previously proposed that Pab1p may inhibit poly(A) polymerase activity by coating the poly(A) (34). Given that Nab2p and Pab1p have comparable affinities for poly(A) [10.5 and 13 nM, respectively (18,35)], the Nab2p feature highlighted above was quite unexpected. No interaction has been reported between the poly(A) polymerase and either Pab1p or Nab2p that could lead to an inhibition of the enzyme and explain the strong and efficient poly(A) tail length control observed with Nab2p. However, such a difference in poly(A) polymerase inhibition could still be possible. To test this hypothesis, a poly(A)34 precursor was prepared from bulk poly(A) (see ‘Materials and methods’ section), radiolabeled at its 5′ end (Figure 5, lane 1), and incubated with recombinant yeast poly(A) polymerase (Figure 5, lane 2). The elongation by the enzyme was then tested in the presence of increasing amounts of either Nab2p (Figure 5, lanes 3–8) or Pab1p (Figure 5, lanes 9–14). Both poly(A)-binding proteins inhibited polyadenylation with comparable efficiencies. Therefore, it is unlikely that the differences between Nab2p and Pab1p in their ability to maintain the length of the poly(A) tails relies solely on Pap1p inhibition discrepancies. This particular feature of Nab2p prompted us to address more specifically the mechanisms used by Nab2p to perform its poly(A) tail length control activity.
Figure 5.
Nab2p and Pab1p inhibit poly(A) polymerase Pap1p with similar efficiencies. Polyadenylation reactions with 5′-end radiolabeled poly(A)34 precursor were done with 24 ng of recombinant poly(A) polymerase Pap1p in a final volume of 20 µl. Pap1p activity was challenged by addition of increasing amounts of either Nab2p (lanes 3–8) or Pab1p (lanes 9–14) recombinant proteins. Reaction products were analyzed on an 8% polyacrylamide-8.3 M urea denaturing gel and visualized by autoradiography. Lane 1, unreacted precursor. M, molecular weight markers were as described.
Poly(A) tail length control by Nab2p requires the zinc finger domains
In order to determine which parts of Nab2p participate in the regulation of polyadenylation, several deletions were constructed on the basis of known regions and putative domains present in the primary sequence of the protein (Figure 6A). Glutamine-rich regions are found in a wide array of proteins involved in RNA metabolism such as Rna15p, Pcf11p and hnRNP U. The N-terminal part of Nab2p comprises such a Q-rich region organized as a Q stretch followed by repeats of the QQQP tetrapeptide. The number of these repeats has been shown to vary among S. cerevisiae strains (22). We have reconstructed a short variant of Nab2p (referred to as svNab2p) harboring two QQQP repeats instead of nine (see Experimental procedures). In our reconstituted reaction, this svNab2p proved to be as effective as the full-length Nab2p with regards to the control of polyadenylation (Figure 6B, lanes 8–11). Strikingly, deletion of the entire Q-rich region had no deleterious effect on Nab2p activity either (Figure 6B, lanes 4–7). Besides, the N-terminal part of Nab2p alone—including this Q-rich region—was unable to control poly(A) tail length during de novo synthesis (Figure 6B, lanes 16–19). Interestingly, the replacement of this N-terminal Q-rich part of the protein by an irrelevant polypeptide of similar size (glutathione-S-transferase, GST) did not affect Nab2p activity (Figure 6B, lanes 23–27). These results suggested that the entire N-terminal part of the protein does not contain regions essential for the polyadenylation regulation function of Nab2p in vitro and that the variable Q-rich region does not play a role in this process either.
Deletion of part of the sixth zinc-finger down to the seventh one resulted in a cold-sensitive mutant described as nab2-21 allele (18). This mutation led to abnormally long poly(A) tails and to an mRNAs export defect (36). However, at the permissive temperature hyperpolyadenylated mRNAs could reach the cytoplasm in the nab2-21 mutant, therefore uncoupling polyadenylation termination defects from export (18). To test the ability of nab2-21 protein to regulate mRNA polyadenylation in vitro, we expressed it as a recombinant protein and assayed it in 3′-end processing reactions. As anticipated, nab2-21p showed a marked alteration to control polyadenylation (Figure 6B, lanes 31–34). This was particularly noticeable at low concentrations where nab2-21p exhibited almost no activity (Figure 6B, lane 31) compared to wild type Nab2p (see for instance Figure 7A, lane 3). Removal of all seven zinc-finger domains completely abolished poly(A) tail length control (Figure 6B, lanes 38–42). The defects observed in 3′-end processing assays with nab2-21p and nab2Δ(CCCH)7 correlated with impaired and even absent poly(A)-binding activities respectively (Supplementary Figure 2A). Therefore, the nab2-21 protein could reproduce in vitro the poly(A) tail length control defect observed in vivo in the nab2-21 mutant strain. The zinc-finger domains of Nab2p and more particularly the two last zinc fingers, are essential for the poly(A) binding and polyadenylation regulation activities of Nab2p.
Interestingly, the (CCCH)7 construct (containing the zinc-fingers region and the C-terminal part of the protein) was not sufficient to generate a specific control of polyadenylation since even the downstream fragment, which in normal conditions does not receive a poly(A) tail, appeared to be partially polyadenylated (Figure 6B, lanes 43–47). The presence of the arginine–glycine rich region in conjunction with the (CCCH)7 domain seemed to restore specificity (Figure 6B, lanes 51–55). However, this RGG-(CCCH)7 construct was less efficient than the full length Nab2p (Figure 6B, compare lanes 50 and 55) and even inhibitory at concentrations similar to those usually required for efficient polyadenylation control (Figure 6B, compare lanes 56 and 57 to Figure 7A lanes 5 and 6). Such a strong inhibition was not observed even with 1.6 µM of wild-type Nab2p under the same reaction conditions (Figure 6B, lane 15). Surprisingly, the presence of the GST polypeptide at the N-terminal end of the RGG-(CCCH)7 construct allowed the protein to terminate polyadenylation efficiently without any noticeable inhibition (Figure 6B, compare lanes 22 and 27) and to display a concentration-dependent activity similar to the wild-type Nab2p (compare Figure 6B, lanes 23–27 to Figure 7A, lanes 3–6). These results suggested that Nab2p requires an N-terminal moiety in addition to the RGG-(CCCH)7 regions to perform its function in mRNA 3′-end formation. It is tempting to hypothesize that the requirement for this N-terminal polypeptide might reflect the need for a specific size and/or shape of the protein.
The RGG-box region of Nab2p participates in the non-accessibility of the mRNA 3′ end
Arginine–glycine rich (RG-rich) regions, and more particularly RGG-box motifs, are predicted to be unstructured. They participate in protein–protein interactions and are involved in specific or non-specific RNA binding (37–42). The RGG-box region of Nab2p has previously been described as an M9-type nuclear localization sequence and therefore constitutes a docking site for Nab2p β2 importin, Kap104p (23). Removal of this part of the protein gave mixed results in terms of efficiency concerning the regulation of polyadenylation. Titration assays showed that nab2ΔRGGp was less efficient at similar concentrations than the wild-type protein which resulted in hyperpolyadenylated species, even at the highest concentrations tested (Figure 7A, compare lanes 3–6 to lanes 7–10, respectively). However, in contrast to nab2-21p or nab2Δ(CCCH)7 mutants, a significant amount of normal polyadenylated mRNA species was formed, indicating that deletion of the RGG-box only partially impaired Nab2p's competence to control poly(A) tail length. It is worth noting that nab2ΔRGGp behaved similarly to the wild-type Nab2p on a Superdex-200 gel-filtration column, suggesting that the overall shape of the protein may not be affected (data not shown).
Since nab2ΔRGGp might be impaired in its RNA-binding properties, we analyzed its ability to bind poly(A) in a classical poly(A)-Sepharose-binding assay under an ionic strength of 300 mM KCl (see Supplementary methods). As shown in Supplementary Figure 2A, nab2ΔRGGp poly(A)-binding efficiency was not significantly reduced compared to Nab2p (P > 0.05). This is consistent with earlier results showing that the Nab2p RGG-box in conjunction with Nab2p N-terminal part is not sufficient to bind poly(A) (22). We analyzed more precisely the effect of the RGG-box deletion on binding affinity to poly(A)20 in an electromobility gel shift assay (see Supplementary methods). Kdapp values determined from four independent experiments were 6.5 ± 1.2 nM for Nab2p and 11 ± 4.3 nM for nab2ΔRGGp (mean ± standard error). These values are in a range consistent with the Kdapp of 10.5 nM previously obtained by filter-binding assays (18). Nab2p and nab2ΔRGGp Kdapp were not significantly different (P > 0.05; P = 0.36). While nab2ΔRGGp binding fitted nicely with a model of a unique binding site, Nab2p-binding isotherm deviated noticeably from this pattern by displaying an apparent positive co-operativity (Supplementary Figure 2B). Therefore, even if deletion of the RGG-box had no effect on Nab2p Kdapp or its ability to bind poly(A), it clearly led to the loss of the protein-binding co-operativity.
We have shown that Pab1p incapacity to regulate polyadenylation efficiently could be at least explained by its inability to prevent accessibility to the 3′ ends of the poly(A) tails (Figure 4). We thus made the assumption that the similar defect observed with nab2ΔRGGp might be due to the loss of this particular feature of Nab2p. As in Figure 4A, we tested whether this mutant could prevent CYC1pA transcript from being further polyadenylated. As hypothesized, nab2ΔRGGp behaved exactly as Pab1p in these assays: it was unable to completely avoid extra adenylation of the mature transcript (Figure 7B, lanes 3–5) and this partial inhibition could even be more perturbed by addition of TAP-purified CPF or yeast recombinant poly(A) polymerase after 1 h of reaction (Figure 7B, lanes 6 and 7), as it was the case with Pab1p (Figure 4). This experiment showed that the RGG-box region is involved in the capacity of Nab2p to prevent access to the 3′ end of the poly(A) tail and that this property is essential for optimal polyadenylation termination.
Since there was still a possibility that inaccessibility of the 3′ ends may occur in conjunction with another protein partner of the reaction, we tested whether Nab2p or nab2ΔRGGp alone could protect the poly(A) tail from additional adenylation. Nab2p/poly(A)60 or nab2ΔRGGp/poly(A)60 complexes were pre-assembled at protein and RNA concentrations similar to those of our standard 3′-end-processing reaction and increasing concentrations of yeast recombinant Pap1p were added. Poly(A)60 was used to be consistent with the average length of the poly(A) tails in yeast. In the range of Pap1p concentrations found in our standard 3′-end-processing reactions, Nab2p alone prevented this RNA from being efficiently polyadenylated (Figure 7C, lanes 3–6), whereas nab2ΔRGGp did not inhibit polyadenylation as efficiently (Figure 7C, lanes 11–14). Densitometric analysis of multiple experiments clearly highlighted that nab2ΔRGGp/poly(A)60 complexes were 2 to 2.6-fold more efficiently adenylated than the Nab2p/poly(A)60 complexes in these conditions (Figure 7D, 5–42 ng of Pap1p). However, this difference was abolished when the poly(A) polymerase concentration was 5, 12, 20 or 40-fold higher than that found in our standard 3′-end-processing reactions (Figure 7C, lanes 7–10 and 15–18; Figure 7D, 100–960 ng of Pap1p).
These results suggested that Nab2p is necessary and sufficient to prevent the poly(A) tails from being elongated further after they have reached their physiological length in vitro. Furthermore, the RGG-box region of the protein does play an essential role in this particular feature of the protein.
Nab2p/poly(A) and Pab1p/poly(A) complexes display different topologies
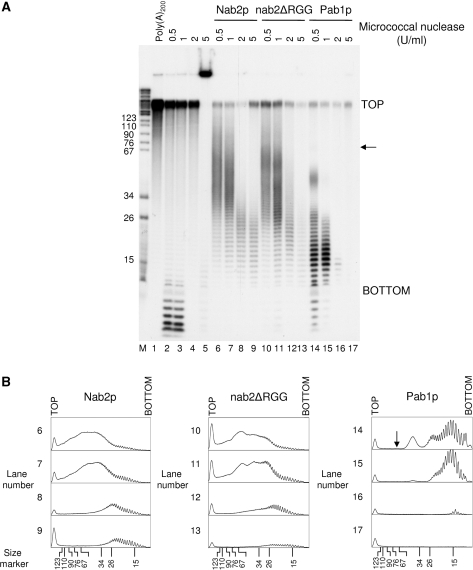
To test whether the functional differences we observed between Nab2p and Pab1p might be due to the way both proteins cover the poly(A) tail, we analyzed the topology of Nab2p/poly(A), nab2ΔRGGp/poly(A) and Pab1p/poly(A) complexes by micrococcal nuclease digestion. At low concentrations of nuclease, Pab1p protected poly(A) species of 73 (indicated by the black arrow), 46, 25 and 16 adenosine residues on average (Figure 8A, lane 14). Since Pab1p has a binding site of 12 nt and covers about 23–25 nt (43), these species are likely to represent the binding of three, two and one molecules onto the poly(A), respectively. The 16-nt species might correspond to 25-nt species further trimmed and result in the most protected polymer close to a single binding site of Pab1p. The relative abundance of each species likely reflected the easiness by which the enzyme could cleave between two Pab1p entities. This assumption was supported by lanes 15 and 16 of Figure 8A where increase of the nuclease concentrations led to the almost complete disappearance of the longest species and left single binding site species of 16 nt (lane 15) or 14 nt (lane 16) mainly. These results suggested that Pab1p binds to poly(A) as a linear deposit of contiguous monomers. It is worth noting that similar experiments have given such results for the mammalian Pab1p homologue PABPC [poly(A)-binding protein cytoplasmic] (44).
Figure 8.
Topology analysis of Nab2p/poly(A), nab2ΔRGGp/poly(A) and Pab1p/poly(A) complexes. (A) Poly(A)200 alone (lanes 1–5) or poly(A)200/protein complexes (lanes 6–17) were digested with micrococcal nuclease (NS7) at the indicated concentrations as described in Materials and Methods. RNA fragments were extracted, ethanol precipitated and subjected to electrophoresis on a 12% polyacrylamide-8.3 M urea gel. Visualization was done by autoradiography. The black arrow points to the weak signal of the 73-nt long species. (B) Densitometric analysis of the gel shown in A using ImageJ. TOP and BOTTOM indicate corresponding positions on the gel as in A. The black arrow is as in A.
At low concentrations of micrococcal nuclease, Nab2p and nab2ΔRGGp gave a digestion pattern very different from that of Pab1p. These proteins protected a population of poly(A) ranging from 37 to 67 nt with an average maximum size of 51 nt in the case of Nab2p, and from 27 to 67 nt with an average maximum size of 45 nt in the case of nab2ΔRGGp (Figure 8A, lanes 6 and 10). Besides, nab2ΔRGGp profile showed three weak maxima at 59, 41 and 29 nt among the protected population (more visible on Figure 8B, densitogram 10). Raising the nuclease concentration to 1 U/ml had globally no effect on Nab2p and nab2ΔRGGp profiles but resulted in a better resolution of the maxima observed in the nab2ΔRGGp profile (Figure 8A, lanes 7 and 11 and corresponding densitograms in Figure 8B). Further increase in micrococcal nuclease concentrations gave protected poly(A) species of 25 and 23 nt in the case of Nab2p, and 23 and 21 nt in the case of nab2ΔRGGp on average (Figure 8A, lanes 8, 9 and 12, 13). Therefore, Nab2p may cover ∼20 nt, and deletion of the RGG-box had only a weak effect on the length of a single binding site. However, it significantly altered the ability of the protein to protect efficiently long poly(A) species. The apparent resistance of the Nab2p/poly(A) and nab2ΔRGGp/poly(A) complexes to the nuclease treatment until 1 U/ml contrasted with the Pab1p profile obtained (Figure 8A, lane 15), where the nuclease could digest more efficiently and led directly to formation of single binding sites poly(A) species. These results showed that Nab2p may bind to poly(A) in a manner slightly different from the linear deposit of contiguous monomers as observed for Pab1p. The overall topology resulting from Nab2p binding to poly(A) seemed to protect the poly(A) tail more efficiently from the nuclease activity, which is consistent with the stability of the polyadenylated mRNA when Nab2p is used as a polyadenylation regulation factor compared to Pab1p (Figure 2, lanes 6–9 and 15–18).
DISCUSSION
Pre-mRNA 3′-end processing has been studied in yeast and mammals concurrently. Even if slight differences may still subsist in the overall organization of the machineries involved and in the sequence of events of the reaction, multiple strong similarities have been revealed. In this context, the regulation of nuclear mRNA poly(A) tail synthesis has been well described in higher eucaryotes: this process involves PABPN1, also termed PABP2, which binds to the growing poly(A) tail and acts as a molecular ruler that indicates the moment for termination (11). In yeast, the mechanism currently proposed for this particular step suggests that the binding of the most abundant poly(A)-binding Pab1p to the poly(A) tails would recruit the PAN complex that trim the polymer to its physiological length (17). Our recent work has complicated this model by showing that Nab2p could function as an mRNA polyadenylation regulator (18). The present analysis of the nab2-21 mutant protein confirms that this is an intrinsic property of Nab2p. We have also suggested that PAN may act at a later stage of mRNA metabolism than de novo poly(A) tail synthesis, and that the sole presence of a poly(A)-binding protein is sufficient to trigger poly(A) tail length control (19). To further reinforce this latter point, we have purified CF IA and CPF from a pab1Δ yeast strain by TAP-tagging. We could observe that the factors purified in this mutant context were fully functional (Supplementary Figure 3).
Here, the comparative study of Nab2p and Pab1p activities in vitro with either yeast cell extracts or reconstituted systems confirms our earlier proposal that Nab2p is favored compared to Pab1p when the two proteins are in competition. This result is surprising since both polypeptides display similar affinities for poly(A) and that no known interactions between Nab2p and core components of the 3′-end-processing machinery have been reported. Although a two-hybrid interaction has been detected between Nab2p and Nab4p, we have not observed any physical interaction between the two proteins in vitro using gel filtration (unpublished results). The two-hybrid result may thus reflects the relationships that Nab2p and Nab4p have with regards to their common karyopherin Kap104p. Even if one cannot rule out a yet undiscovered interaction with any of the 3′-end-processing components, an explanation brought by the present study seems to come from the way Nab2p binds to the poly(A).
Our results demonstrate the ability of Nab2p to protect a population of poly(A) species (40–70 nt) that surprisingly parallels the range of size of the average poly(A) tails added to the mRNA (such as those seen in our in vitro 3′-end-processing assays). Although deletion of the RGG-box seemed to weakly affect the protection pattern (Figure 8), performing the micrococcal digestion at 37°C instead of 30°C exacerbated the defects (Supplementary Figure 4), whereas Pab1p profile showed protection of single binding site species only. In addition, denaturing the RNA before incubation with the proteins and performing the micrococcal digestion at 37°C confirmed that Nab2p covers and efficiently protects ca. 20-nt long units (see Supplementary Figure 5, lanes 6–8, where a ∼40 nt species is detected), whereas nab2ΔRGGp could only protect single binding site species, like Pab1p. Finally, higher sensitivity to the nuclease of nab2ΔRGGp/poly(A) complex was reproducibly seen and comparable to the sensitivity of the Pab1p/poly(A) complex (compare lanes 12, 13 and 16, 17 with lanes 8, 9 in Figure 8A, Supplementary Figure 4 and Supplementary Figure 5).
One noticeable consequence of the manner this protein binds to the poly(A) is the ability of Nab2p to ‘hide’ the 3′-end of the mature polyadenylated mRNA. This feature clearly relies on its RGG-box region. Although this kind of motif has been reported to be involved in protein/protein interactions, we have been unable to detect any Nab2p/Nab2p complex formation either by gel filtration or pull-down assays with a GST-Nab2p and an untagged version of the protein (unpublished data). Surprisingly, binding of Nab2p to poly(A) was very sensitive to 2 mg/ml of heparin and resulted in a 10-fold increase of its Kdapp, whereas binding of nab2ΔRGGp was not (unpublished data). Moreover, Nab2p non-specific binding is salt-sensitive (45). These results suggest that the RGG-box region may be involved in non-specific interactions with the phosphate backbone of the RNA presumably via electrostatic contacts. These additional contacts may have a role in a higher order organization of the Nab2p/poly(A) ribonucleoprotein particle that efficiently protects a defined range of poly(A) tail lengths.
Here, a parallel must be made with the mammalian polyadenylation regulator PABPN1. First, this 33 kDa protein also leads to smeary and poorly defined poly(A) species in similar micrococcal protection experiments (44,46). Second, PABPN1 also contains an arginine/glycine rich domain, although without defined RGG-boxes, but located in its C-terminal part. This region of the protein similarly confers a non-specific RNA-binding activity highly sensitive to heparin that is essential for PABPN1 function (47), as we have observed for Nab2p. This RG-rich region is also responsible for the binding co-operativity observed with PABPN1 but this property is linked to the self-association of the protein (47). Although we did not detect any Nab2p self-interaction in the absence of RNA, the loss of an RGG-box-dependent co-operativity observed in our gel-shift experiments with the 20-nt long poly(A) and nab2ΔRGGp is intriguing, since the length of the poly(A) covered by this mutant does not seem to be strongly changed. Yet, we cannot rule out the possibility that protein/protein interactions may occur only when Nab2p is bound to the RNA. Third, it is known that binding of PABPN1 to the long mammalian poly(A) tails leads to the formation of a 20-nm wide globular RNP (12). It has been hypothesized that this structure may disrupt the tripartite [CPSF-mPAP-PABPN1] complex and trigger the release of the machinery from the mRNA. Since yeast poly(A) tails are 50–80-nt long on average, current models should take into account the fact that Pab1p and Nab2p cover about 20 nt and therefore that the yeast poly(A) tail can accommodate a maximum of three to four molecules onto it, albeit with up to 2-fold larger molecular weights than that of PABPN1. As both Pab1p and Nab2p are able to prevent cleavage of the mature mRNA but that only Nab2p can also avoid further adenylation, it is tempting to hypothesize that release of the yeast machinery from the mRNA may only occur in the presence of Nab2p. Finally, the poly(A) tails synthesized are stable and efficiently protected from degradation by nucleases in the presence of Nab2p or PABPN1 which is not observed with Pab1p [(11) and this study].
One noticeable difference between Nab2p and PABPN1 is that PABPN1 does not seem to prevent accessibility to the 3′-ends, even with 1.5 molar excess of PABPN1 over the poly(A) polymerase (11). This may be explained by the fact that PABPN1 is involved in the stimulation and processivity of the polyadenylation reaction itself and in poly(A) tail length control at the same time.
We did not formally demonstrated the importance of a defined size or shape for Nab2p to function in poly(A) tail length control. However, the presence of a globular polypeptide located in the N-terminal part of the protein seems to be critical for the proper activity of Nab2p. Indeed, all of the constructs displaying a polyadenylation control close to wild-type bared an N-terminal polypeptide ranging from ∼17 to 26 kDa [Nab2p, svNab2p, nab2ΔQrich, GST-RGG-(CCCH)7]. This extension, directly located in front of the RGG-box region, appears to be actually globular (in the case of the GST moiety) or predicted to be so. The requirement for a defined size or shape of Nab2p to control polyadenylation and the way the protein covers the poly(A) may explain the physiological length of the yeast mRNA poly(A) tails added during 3′-end processing. Thus, it will be interesting to determine the structure of Nab2p bound to a poly(A)60 molecule.
The contribution of the RGG-box in the Nab2p/poly(A) RNP organization we observed has another importance because this motif is also the docking site for Nab2p importin, Kap104p. Since their interaction is known to impair Nab2p RNA-binding activity and its role in polyadenylation (19), one can predict that binding of Kap104p in the cytoplasm may trigger important remodeling of the mRNP.
Previous reports have shown that the zinc-finger region of Nab2p is essential for cell viability (18,48). In the present study, additional arguments favor Nab2p as the yeast poly(A) tail length regulating factor and demonstrate that its poly(A)-binding properties of the protein is essential for that function. It is worth noting that in contrast, Pab1p does not require its high-affinity poly(A)-binding sites to sustain cell viability (35,43). While preparing this report, a putative Schizosaccharomyces pombe PABPN1 homologue has been identified and called Pab2 (49). Although surprisingly non-essential for cell viability, pab2 null mutants display poly(A) tail length defects very similar to those exhibited by the S. cerevisiae nab2 mutants (18). The authors proposed that S. pombe Pab2 may regulate polyadenylation in a manner more similar to what we observed with Nab2p than with Pab1p. This suggested that poly(A) tail length control in S. pombe may use a protein similar to the one found in higher eukaryotes, but the mechanism used would remain closer to that of S. cerevisiae. Therefore, poly(A) tail length control very likely relies not only on a specific poly(A)-binding protein but also on its relationships with the rest of the 3′-end processing machinery.
It is now well established that even if Pab1p has a cytoplasmic steady-state localization, it nevertheless shuttles between the nucleus and the cytoplasm just like its mammalian counterpart, but mostly via an mRNA-independent pathway (29,50). The precise role of Pab1p in the nucleus is not well defined. However, its presence seems to be required for the release of mature mRNAs from the site of transcription and may thereby represent a marker for mRNP (29). In addition, mammalian PABPC has recently been found associated with nuclear polyadenylated pre-mRNAs (51). Although we did not observe any dramatic stimulating or inhibitory effect on polyadenylation regulation in the presence of both Pab1p and Nab2p in vitro, it is conceivable that, after synthesis of the poly(A) tail under the control of Nab2p only, at least one Pab1p molecule may displace the loading of Nab2 proteins and eventually cause PAN initial trimming and mRNA export. Protection against cleavage of the newly processed mRNAs before their export to the cytoplasm would emerge as an additional and important function for Nab2p and/or Pab1p in yeast. The composition of the mRNP would then be changed into a Pab1p-only bound poly(A) tail during export and/or in the cytoplasm due to the coordinated action of Kap104p, ready to re-import Nab2p to the nucleus, and the higher concentrations of Pab1p in this cellular compartment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank John Aitchison for the generous gift of the plasmid pGEX2TK-nab2ΔRGG, Roy Parker for the pab1Δ strain YRP1130, and Maurice S. Swanson for helpful discussions. We thank Geneviève Demaison for her invaluable technical assistance. This work was supported by grants from the CNRS, the French Ministère de la Recherche Scientifique, La Fondation pour la Recherche Médicale/Fondation BNP-Paribas (to L.M.-S.). N.V. was supported by a pre-doctoral fellowship from the French Ministère de la Recherche et de la Technologie. Funding to pay the Open Access publication charges for this article was provided by La Fondation pour la Recherche Médicale/Fondation BNP-Paribas.
Conflict of interest statement. None declared.
REFERENCES
- 1.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 4.De Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerwitz Y, Kuhn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 2003;22:3705–3714. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahle E. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 1991;266:3131–3139. [PubMed] [Google Scholar]
- 9.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 11.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 12.Keller RW, Kuhn U, Aragon M, Bornikova L, Wahle E, Bear DG. The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. J. Mol. Biol. 2000;297:569–583. doi: 10.1006/jmbi.2000.3572. [DOI] [PubMed] [Google Scholar]
- 13.Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winstall E, Sadowski M, Kuhn U, Wahle E, Sachs AB. The Saccharomyces cerevisiae RNA-binding protein Rbp29 functions in cytoplasmic mRNA metabolism. J. Biol. Chem. 2000;275:21817–21826. doi: 10.1074/jbc.M002412200. [DOI] [PubMed] [Google Scholar]
- 15.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minvielle-Sebastia L, Preker PJ, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl Acad. Sci. USA. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown CE, Sachs AB. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dheur S, Nykamp KR, Viphakone N, Swanson MS, Minvielle-Sebastia L. Yeast mRNA Poly(A) tail length control can be reconstituted in vitro in the absence of Pab1p-dependent Poly(A) nuclease activity. J. Biol. Chem. 2005;280:24532–24538. doi: 10.1074/jbc.M504720200. [DOI] [PubMed] [Google Scholar]
- 20.Sachs AB, Deardorff JA. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 21.Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee DC, Aitchison JD. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J. Biol. Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- 24.Minvielle-Sebastia L, Beyer K, Krecic AM, Hector RE, Swanson MS, Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer S, Urbanke C, Wahle E. Equilibrium studies on the association of the nuclear poly(A) binding protein with poly(A) of different lengths. Biochemistry. 2002;41:6082–6089. doi: 10.1021/bi0160866. [DOI] [PubMed] [Google Scholar]
- 26.Butler JS, Sadhale PP, Platt T. RNA processing in vitro produces mature 3′ ends of a variety of Saccharomyces cerevisiae mRNAs. Mol. Cell. Biol. 1990;10:2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 28.Minvielle-Sebastia L, Preker PJ, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 29.Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preker PJ, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 31.Weiss EA, Gilmartin GM, Nevins JR. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991;10:215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 33.Dichtl B, Keller W. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 2001;20:3197–3209. doi: 10.1093/emboj/20.12.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lingner J, Radtke I, Wahle E, Keller W. Purification and characterization of poly(A) polymerase from Saccharomyces cerevisiae. J. Biol. Chem. 1991;266:8741–8746. [PubMed] [Google Scholar]
- 35.Deardorff JA, Sachs AB. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J. Mol. Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 36.Hilleren P, Parker R. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA. 2001;7:753–764. doi: 10.1017/s1355838201010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J. Biol. Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 38.McBride AE, Cook JT, Stemmler EA, Rutledge KL, McGrath KA, Rubens JA. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J. Biol. Chem. 2005;280:30888–30898. doi: 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 39.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 40.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mears WE, Rice SA. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghisolfi L, Kharrat A, Joseph G, Amalric F, Erard M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur. J. Biochem. 1992;209:541–548. doi: 10.1111/j.1432-1033.1992.tb17318.x. [DOI] [PubMed] [Google Scholar]
- 43.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baer BW, Kornberg RD. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J. Cell. Biol. 1983;96:717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly SM, Pabit SA, Kitchen CM, Guo P, Marfatia KA, Murphy TJ, Corbett AH, Berland KM. Recognition of polyadenosine RNA by zinc finger proteins. Proc. Natl Acad. Sci. USA. 2007;104:12306–12311. doi: 10.1073/pnas.0701244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baer BW, Kornberg RD. Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc. Natl Acad. Sci. USA. 1980;77:1890–1892. doi: 10.1073/pnas.77.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhn U, Nemeth A, Meyer S, Wahle E. The RNA binding domains of the nuclear poly(A)-binding protein. J. Biol. Chem. 2003;278:16916–16925. doi: 10.1074/jbc.M209886200. [DOI] [PubMed] [Google Scholar]
- 48.Marfatia KA, Crafton EB, Green DM, Corbett AH. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 2003;278:6731–6740. doi: 10.1074/jbc.M207571200. [DOI] [PubMed] [Google Scholar]
- 49.Perreault A, Lemieux C, Bachand F. Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J. Biol. Chem. 2007;282:7552–7562. doi: 10.1074/jbc.M610512200. [DOI] [PubMed] [Google Scholar]
- 50.Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 51.Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]