Abstract
Meganucleases cut long (>12 bp) unique sequences in genomes and can be used to induce targeted genome engineering by homologous recombination in the vicinity of their cleavage site. However, the use of natural meganucleases is limited by the repertoire of their target sequences, and considerable efforts have been made to engineer redesigned meganucleases cleaving chosen targets. Homodimeric meganucleases such as I-CreI have provided a scaffold, but can only be modified to recognize new quasi-palindromic DNA sequences, limiting their general applicability. Other groups have used dimer-interface redesign and peptide linkage to control heterodimerization between related meganucleases such as I-DmoI and I-CreI, but until now there has been no application of this aimed specifically at the scaffolds from existing combinatorial libraries of I-CreI. Here, we show that engineering meganucleases to form obligate heterodimers results in functional endonucleases that cut non-palindromic sequences. The protein design algorithm (FoldX v2.7) was used to design specific heterodimer interfaces between two meganuclease monomers, which were themselves engineered to recognize different DNA sequences. The new monomers favour functional heterodimer formation and prevent homodimer site recognition. This design massively increases the potential repertoire of DNA sequences that can be specifically targeted by designed I-CreI meganucleases and opens the way to safer targeted genome engineering.
INTRODUCTION
By definition, meganucleases are sequence-specific endonucleases with large (12–45 bp) cleavage sites (1), and they can be used to achieve very high levels of gene targeting efficiencies in mammalian cells and plants (2–6). Indeed, meganuclease-induced recombination is an efficient and robust method for genome engineering; the nuclease cuts the genome and a supplied exogenous DNA recombines near the break to repair or mutate the region. The major limitation until recently was the requirement for the prior introduction of a meganuclease target site in the locus of interest, but this has now been overcome by developing protein engineering approaches (7–9).
In nature, meganucleases are essentially represented by homing endonucleases (HEs), a widespread family of endonucleases including hundreds of proteins (10,11). These proteins are encoded by mobile genetic elements which propagate by a process called ‘homing’: the endonuclease cleaves a cognate allele from which the mobile element is absent, thereby stimulating a homologous recombination event that duplicates the mobile DNA into the recipient locus (12,13). Given their natural function and their exceptional cleavage properties in terms of efficacy and specificity, HEs provide ideal scaffolds to derive novel endonucleases for genome engineering. Data have accumulated over the last decade, allowing a good characterization of the LAGLIDADG family, the largest of the four HE families (10).
LAGLIDADG refers to the only sequence actually conserved throughout the family, and is found in one or (more often) two copies in the protein. Proteins with a single motif, such as I-CreI (14), form homodimers and cleave palindromic or pseudo-palindromic DNA sequences, whereas the larger, double-motif proteins, such as PI-SceI are monomers and cleave non-palindromic targets. Nine different LAGLIDADG proteins have been crystallized, showing a very striking core structure conservation that contrasts with the lack of similarity at the primary sequence level (10,11,15–23). In this core structure, two characteristic αββαββα folds, contributed by two monomers, or two domains in double LAGLIDAG proteins, are facing each other with a 2-fold symmetry. DNA binding depends on the four β-strands from each domain, folded into an anti-parallel β-sheet, and forming a saddle on the DNA helix major groove. The catalytic site is central, formed with contributions from helices of both monomers. In addition to this core structure, other domains can be found; for instance, the intein PI-SceI has a protein splicing domain, and an additional DNA-binding domain (18,24).
The extensive structural conservation within the meganuclease family has encouraged the mutagenesis and construction of chimeric and single chain HEs (25–27), which withstood extensive modifications (25–27). Seligman and co-workers (28,29) used a rational approach to substitute specific individual residues of the I-CreI αββαββα fold, and could observe substantial cleavage of novel targets. The same kind of approach was applied to I-SceI recently by another group (30). In a similar way, Gimble and co-workers (31) modified the additional DNA-binding domain of PI-SceI, and could obtain variant proteins with altered binding specificity. Recent work has shown that it is possible to obtain a large number of locally altered variants of the I-CreI meganuclease that recognize a wide variety of targets (7), and to use and assemble them by a combinatorial process, to obtain entirely redesigned mutants with chosen specificity (8,9). These variants can be used to cleave genuine chromosomal sequences and open a wide range of applications, including the correction of mutations responsible for inherited monogenic diseases (5). A limiting factor that still remains for the widespread use of the I-CreI meganuclease is the fact that the protein is a homodimer. Thus, although we have experimental evidence that mixing two meganucleases that target two different DNA sequences can result in formation of a heterodimer that recognizes a hybrid DNA sequence (7–9), this still results in a mixture of three different enzymes, including both homodimers (7).
Here, by using the protein design algorithm FoldX (32–34), we have re-designed the interaction surface of the I-CreI meganuclease to obtain an obligatory heterodimer to prefer a single DNA target sequence. Previous studies in the field have already used computational design to redesign meganuclease interfaces such that I-CreI and I-DmoI monomers can be made to dimerize or form chimeras with peptide linkage (26,35,36). The I-CreI scaffold is less amenable to such linkage because the N- and C-termini from adjacent monomers are relatively far apart (68 Å). This prompted us to consider engineering obligate heterodimers, as has been recently reported for zinc finger nucleases (37,38). As with chimeras, this approach should also reduce off-target cutting and increase the repertoire of novel sequence-specific meganucleases. Moreover, the interface mutations can immediately be applied to the many variants already selected from I-CreI combinatorial libraries (7,9). This removes one of the last hurdles on the way of using meganucleases for gene therapy and other applications.
MATERIALS AND METHODS
Generation of the KTG and QAN meganucleases
I-CreI is a dimeric HE that cleaves a 24-bp pseudo-palindromic target. Analysis of I-CreI structure bound to its natural target shows that in each monomer, eight residues establish direct interactions with seven bases (16). Residues Q44, R68, R70 contact three consecutive base pairs at position 3–5 (and −3 to −5). An exhaustive protein library versus target library approach was undertaken to engineer locally this part of the DNA-binding interface. First, the I-CreI scaffold was mutated from D75 to N to decrease likely energetic strains caused by the replacement of the basic residues R68 and R70 in the library that satisfy the hydrogen-acceptor potential of the buried D75, in the wt I-CreI structure. The D75N mutation did not affect the protein folding, but decreased the toxicity of I-CreI in over-expression experiments (data not shown). Then, positions 44, 68 and 70 were randomized and 64 palindromic targets resulting from substitutions in positions ±3, ±4 and ±5 of a palindromic target cleaved by I-CreI were generated. Screening of the 64 palindromic targets with the protein library allowed the identification of new specificities for I-CreI (7). Among these new variants mutations Q44K, R68T and R70G recognized based CCT at positions 3, 4 and 5 (KTG variant). Mutations Q44Q, R68A and R70N recognized bases GTT at positions 3, 4 and 5 (QAN variant).
FoldX design
The different heterodimers were designed using FoldX (version 2.7), an automatic protein design algorithm (32–34). As template, we used the crystal structure of meganuclease I-CreI in complex with DNA (PDB code: 1g9y.pdb). We first optimized the structure using the <RepairPDB> command of FoldX, in order to release any van der Waals clashes. We then mutated each position of interest to alanine and, using the <BuildModel> command, all models (heterodimers and homodimers alike) were generated separately three times with different seeds to allow for flexibility in surface side chains and to cover more sampling space. Finally, each model of the complex was analysed through the <AnalyseComplex> command to compute the different interaction energies, and the values were averaged over the three models.
Cloning meganuclease mutants
The two homodimerizing meganucleases KTG and QAN, based on the I-CreI meganuclease scaffold, were each mutated at up to six amino acid positions to form two compatible heterodimerizing interfaces, denoted KTG-A2 and QAN-B3. Mutations were introduced using round-the-world PCR with a Quickchange kit (Stratagene, Cat. 200518).
KTG-A2 mutations (K7R, E8R, F54W, E61R, K96R, L97F) were introduced using three complementary primer sets: (i) A1_RR_F, CAA TAC CAA ATA TAA CAG GCG GTT CCT GCT GTA CCT GGC CG, A1_RR_R, CGG CCA GGT ACA GCA GGA ACC GCC TGT TAT ATT TGG TAT TG; (ii) A1_RF_F, TCA ACT GCA GCC GTT TCT GAG ATT CAA ACA GAA ACA GGC AAA CC, A1_RF_R, GGT TTG CCT GTT TCT GTT TGA ATC TCA GAA ACG GCT GCA GTT GA; (iii) A2_WLR_F, CCA GCG CCG TTG GTG GCT GGA CAA ACT AGT GGA TAG AAT TGG CGT TGG TTA CG, A2_WLR_R, CGT AAC CAA CGC CAA TTC TAT CCA CTA GTT TGT CCA GCC ACC AAC GGC GCT GG.
QAN-B3 mutations (K7E, F54G, L58M, K96E) were introduced using three complementary primer sets: (i) B3_EE_F, CAA TAC CAA ATA TAA CGA AGA GTT CCT GCT GTA CCT GGC CG, B3_EE_R, CGG CCA GGT ACA GCA GGA ACT CTT CGT TAT ATT TGG TAT TG; (ii) B3_GME_F, CCA GCG CCG TTG GGG TCT GGA CAA AAT GGT GGA TGA AAT TGG CGT TGG TTA CG, B3_GME_R, CGT AAC CAA CGC CAA TTT CAT CCA CCA TTT TGT CCA GAC CCC AAC GGC GCT GG; (iii) B3_EL_F, TCA ACT GCA GCC GTT TCT GGA ACT GAA ACA GAA ACA GGC AAA CC, B3_EL_R, GGT TTG CCT GTT TCT GTT TCA GTT CCA GAA ACG GCT GCA GTT GA.
The first primer set was used for PCR and transformation, according to the manufacturer's instructions (Stratagene, Quikchange). Approximately 300 transformant bacterial colonies were pooled in 2 ml medium, and plasmid DNA was recovered by miniprep. This DNA was used as template for a second and then a third round of PCR with mutagenic primers. Five third-round mutants were verified by DNA sequencing.
Note that the dimer interface mutations are outside the DNA recognition region and thus the primers above are universal for any I-CreI mutant with altered specificity. Similar methods were used to make the alternative designs for the heterodimer pairs (QAN-A1, KTG-B3, QAN-B4), introducing the appropriate mutations in the oligos for mutagenic PCR (Figure 1).
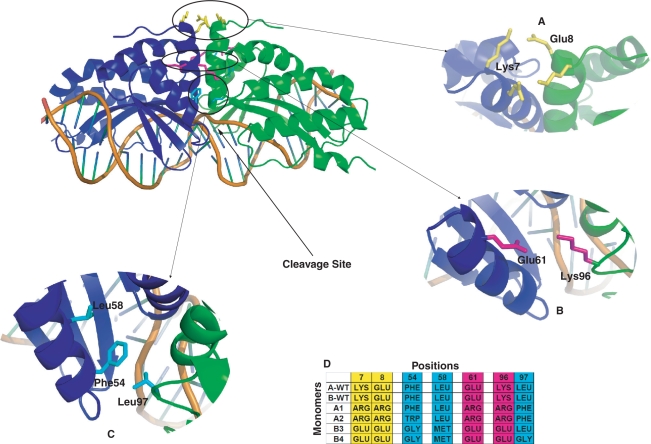
Figure 1.
Structure of the complex of meganuclease I-Cre-I (PDB:1G9Y) with its target DNA template. (A–C) Details of three modifiable interaction patches between the two monomers on the homodimer. (D) Designed heterodimeric interfaces, showing amino acid changes at each relevant position in the protein.
Production and purification of meganucleases
Fresh BL21(DE3) (Stratagene) transformants carrying the pET (Novagen) I-CreI mutants, were grown overnight in 5 ml of Luria Broth (LB plus 30 μg/ml kanamycin) at 37°C on a shaker. This pre-culture was expanded to a larger culture (1:200). At an OD600nm of 0.6–0.8, flasks were put on ice for 15 min to arrest growth. Expression was induced by adding IPTG (1 mM final) for 18 h at 16°C, and cells were harvested by centrifugation (15 min, 16 000g). Pellets were re-suspended in 30-ml ice-cold lysis buffer (50 mM Tris–HCl, 200 mM NaCl, 5 mM MgCl2, 10% Glycerol, 10 mM imidazole pH 8) containing 1 μl/μl DNAse I and the procedure was carried out at 4°C thereafter. The suspension was immediately frozen in liquid nitrogen and thawed for 16 h at 4°C on a rotating platform (60 r.p.m.). The suspension was homogenized with an Ultra Turrax T25 (Jankel & Kunkel, IKA-Labortechnik); three cycles of 1 min on ice) and then broken with an EmulsiFlex-C5 homogenizer (Avestin), each for five rounds of 500–1000 psi (pounds per square inch). The lysate was centrifuged at 150 000g for 60 min. This supernatant was cleared through a 0.45 μm filter (Millipore). A 5-ml Hi-Trap column (Amersham-Pharmacia) was loaded with two bead volumes (vol) of 250 mM NiS04, and rinsed with 3 vol of binding buffer (50 mM Tris–HCl, 300 mM NaCl, 1 mM DTT, 20% glycerol, 10 mM imidazole pH 8). The supernatant was then applied to the column and washed with washing buffer (binding buffer with 50 mM imidazole) until the A280nm returned to its basal level. Protein was eluted with elution buffer (0.3 M imidazole). The protein peak was collected and immediately applied to a dialysis membrane (MWCO = 3.5 kDa, Spectra), placed in 2 l of dialysis buffer (50 mM Tris–HCl, 200 mM NaCl, 1 mM DTT, 1 mM EDTA, 50% glycerol pH 8) at 4°C, for at least 12 h. The various stages of purification were analysed by SDS–PAGE (Supplementary Data Figure S1). The purified protein was aliquoted and snap-frozen in liquid nitrogen and stored at –80°C. We noticed that the different enzymes had slightly different apparent activities, and that enzymes containing the QAN moiety were less stable, losing activity entirely after several months of storage at −80°C. Thus, we used always an aliquot only once and within 1 month of being prepared.
Analytical centrifugation
The oligomeric state of meganucleases and mutants was investigated by monitoring sedimentation properties in centrifugation experiments; 1.04 mg of pure protein was used per sample (0.52 mg/ml of each monomer or 1.04 mg/ml of individual WT homodimers) in storage buffer (50 mM Tris–HCl, 225 mM NaCl, 1 mm EDTA, 1 mM DTT, 8% glycerol pH 8.0).
The sedimentation velocity profiles were collected by monitoring the absorbance signal at 280 nm as the samples were centrifuged in a Beckman Optima XL-A centrifuge fitted with a four-hole AN-60 rotor and double-sector aluminium centrepieces (18 6000g, 4°C). Molecular weight distributions were determined by the C(s) method implemented in the Sedfit (39) and UltraScan 7.1 software packages (http://www.ultrascan.uthscsa.edu). Buffer density and viscosity corrections were made according to data published by Laue et al. (40) as implemented in UltraScan 7.1. The partial specific volume of meganucleases and mutants was estimated from the protein sequence according to the method by Cohn and Edsall (41).
Co-expression of the designed monomers
In order to remove the His tag from the QAN-B3 monomer, it was excised from the parent plasmid pCLS1214 (pET-series) with NcoI/NotI (New England Biolabs). This fragment was then cloned into similarly cut pCDFDuet1 plasmid (Novagen). TOP10 ultracompetent cells (Invitrogen) were transformed with this mixture and selected in 50 μg/ml Streptomycin–Spectinomycin sulphate. Clones were verified by DNA sequencing.
BL21(DE3) ultracompetent cells were co-transformed with 10 ng of each plasmid (pCLS1211-KTG-A2 and pCDFDuet1-QAN-B3). The double transformants were selected by growing the transformed colonies in presence of Kanamycin and Streptomycin–Spectinomycin sulphate. The purification was performed essentially as above.
DNA digestion assays
Cleavage of the target sequences was determined as previously described (25) with modifications: co-expressed, purified enzymes were diluted to 1 µg/µl in fresh dialysis buffer (in the case of the designed monomers which were purified separately, 1.5 µg of each monomer was added, i.e. 0.5 µg/µl each). Enzymes were stored at −80°C. Reaction mixtures contained appropriate amounts of enzyme and DNA target, as indicated. DNA targets were made from purified 3.2-kb plasmid containing the appropriate target sequences (pre-linearized with XmnI) and 225 mM NaCl in a 20 µl final reaction volume. The digestion mixtures were incubated for 60 min at 37°C in a water bath and then mixed with 2.5 µl volume of Stop solution (10×), modified from Wang et al. (14) (50% Glycerol, 0.1 M EDTA, 0.5% SDS, 1 mg/ml Proteinase K, 0.25% bromophenol blue). Samples were incubated for 30 min more at 37°C, and then half of each sample was visualized on a 1% agarose gel.
For competition assays, DNA target sites of characteristic lengths were constructed by PCR from the appropriate plasmid template. These templates were incubated as above, for the times and reagent concentrations indicated in each individual experiment. The gels were scanned and quantitated for image intensity of cleavage bands using imageJ. Activity curves were determined by non-linear regression, using Kaleidagraph 4.0 for the equation: Cleavage (%) = m1 × m0/(m0 + m2), where m0 is protein concentration (in μM), and the parameters m1 and m2 represent maximum cleavage (%) and enzyme concentration for 50% cleavage (μM), respectively.
RESULTS
Protein design
To design the heterodimeric interface of I-CreI, we used the X-ray structure of the homodimer determined at 2.05 Å resolution (PDB: 1g9y), bound to its cognate DNA target sequence. The aim was to facilitate heterodimerization and at the same time to prevent the formation of homodimers, or at least make them thermodynamically unstable.
A large part of the dimerization interface of the homodimer is composed of two α-helices (Lys7 to Gly19 in both monomers), arranged in a coiled-coil. The two helices are very close to each other, packing in the centre mainly through the backbone, making them unsuitable for re-design. The amino acids below these helices (Asp20 and onward) are contacting the DNA and are thus responsible for both the activity (active site) and specificity (DNA recognition) of the endonucleases. These functions alone prevent any of these residues to be modified in the design process. This left us with few possibilities to enforce the heterodimerization. After careful examination of the structure, we identified three patches of interactions involved in the interface that could be disturbed and changed in the dimers, without impairing their binding capacity or their enzymatic activity (Figure 1).
The most obvious one of these is the region above the two helices (Figure 1A), where Lys7 and Glu8 in one monomer establish favourable electrostatic interactions with the corresponding residues in the other monomer. In order to keep this interaction in the heterodimer, and at the same time impair monomer formation, we decided to replace them with two arginines in one monomer (named monomer A hereafter) and two glutamates in the other (called monomer B). Thus, AA and BB homodimers would undergo an electrostatic repulsion whereas AB heterodimer formation would be electrostatically favourable.
The second patch was chosen with the same idea of creating small electrostatic imbalances for homodimers, relative to heterodimers, but is positioned on each side of the coiled-coil; a double cluster of charged residues is made by the Lys96 and the Glu61 of each monomer (Figure 1B). To re-enforce the electrostatic effects of the first mutation site, we decided to mutate the second site with two arginines in monomer A, and two glutamates in monomer B, thus making a charged triangle in each monomer (positive in A, negative in B).
The third region of interest is the region around the middle of the two helices involved in the interaction surface and is mainly composed of hydrophobic interactions and hydrogen bonds, making a kind of minicore. As the H-bond network is quite intricate and extends all the way to the active site, we decided to perturb only the hydrophobic patch made by residues Tyr12, Phe16, Val45, Trp53, Phe54, Leu55 and Leu58 of one monomer, with residue Leu97 of the other monomer (the latter acting like a cap closing the hydrophobic pocket; Figure 1C). We re-designed these two pockets in order to introduce strong Van der Waals’ Clashes in the homodimers without disturbing the hydrophobic interactions in the heterodimers (i.e. without creating cavities and steric clashes). For this we decided to introduce bulky residues in monomer A (respectively Phe or Trp for position 54 and Phe for position 97 and small residues in monomer B (Gly and Leu, respectively). A glycine could be introduced at position 97 to give more space to position 54 when it is mutated in tryptophan. As a result, AA homodimers develop huge steric hindrance, preventing their formation, and BB homodimers contain big cavities, making them unstable. By contrast, the minicore of AB heterodimers should be filled efficiently by these compatible amino acids. Finally, we mutated Leu58 to methionine in monomer B, to prevent any cavity formation in the heterodimer, due to the introduction of the small side chains.
We thus defined two types of monomer A, A1 and A2, depending of the nature of the amino acid at position 54, respectively Phe or Trp, and two types of corresponding monomer B, B3 and B4, the later differing by a mutation in Glycine at position 97 to accommodate with the Trp mutation of monomer A2 (Figure 1D). The different mutations were tested with FoldX to model all homodimers (A1:A1, A2:A2, B3:B3 and B4:B4) and heterodimers (A1:B3, A2:B3 and A2:B4) and to get the different interaction energies (Table 1). Of all the heterodimers, two constructions, A1:B3 and A2:B3, presented a computed interaction energy close to the wild-type homodimer (Table 1). The last construction, A2:B4, presented a significant decrease in interaction energy compared to the wild-type homodimer but was nonetheless significantly higher than the mutant homodimers. Conversely, A1:A1, A2:A2, B3:B3 and B4:B4 homodimers were all much destabilized and thus these species were expected to remain monomeric.
Table 1.
FoldX calculated interaction energies (kcal/mol) between wild-type and designed homodimers and heterodimers
| Dimers | Difference in interaction energies between mutants and wild-type (kcal/mol) |
|---|---|
| A2_B3 | 0.13 |
| A1_B3 | 0.22 |
| A2_B4 | 3.20 |
| B3_B3 | 7.39 |
| A1_A1 | 8.30 |
| A2_A2 | 8.53 |
| B4_B4 | 11.96 |
The best binding energy (A2_B3) corresponds with the best in vitro result.
Optimizing conditions for specific DNA cleavage
To verify that we were able to design a specific heterodimer correctly, we employed two meganuclease variants that recognize different DNA sequences (7). These variants both harbour an Asp75 to Asn mutation that decreases energetic strains caused by the replacement of the basic residues Arg68 and Arg70; these arginines normally satisfy the hydrogen-acceptor potential of the buried Asp75 in the I-CreI structure. We used the meganuclease denoted as ‘KTG’, which differs from the WT at positions 44, 68 and 70 and recognizes the bases CCT at positions 4, 5 and 6 of the DNA target. The other meganuclease is called ‘QAN’, differs from the WT at the same positions, and recognizes the bases GTT at positions 4, 5 and 6 of the DNA target. These two enzymes have been obtained by screening of a library of I-CreI derivatives mutated at positions 44, 68 and 70 (7). Throughout this paper, we denote the target DNA sequences by a 6-base code, with the first three bases corresponding to positions 4, 5 and 6 of the target sequence and the second three to the same positions in the complementary DNA sequence; the two triplets are separated by a slash (/). Thus, the target of the KTG enzyme is CCT/AGG that for the QAN target is GTT/AAC, and the mixed DNA target for the heterodimer KTG-QAN is denominated as GTT/AGG.
For the WT meganuclease I-CreI, it has been reported (14) that the ideal conditions for digestion of its target DNA are: 20 mM Tris–HCl (pH 8.0–9.0) with 10 mM MgCl2, and the enzyme is reportedly inhibited above 25 mM NaCl ionic strength. However, when using the KTG and QAN enzymes, we actually found that optimal (although not perfect) specificity and activity were achieved around 225 mM NaCl concentration (Supplementary Data, Figure S2). At lower ionic strengths, there was suboptimal specificity. This suggests that strong binding of only one of the monomers to the DNA is enough to allow digestion. Increasing ionic strength to 225 mM both improves the activity of the enzymes towards their targets and reduces the digestion of the mixed template. Nonetheless, further increasing NaCl concentration to 250 and 300 mM actually slightly reduced cleavage activity (data not shown). This behaviour could be explained by the ionic strength decreasing the affinity for DNA (thus preventing binding if only one monomer establishes specific interactions in the dimer). As a result of these tests, we selected the following optimal buffer for digestion of our meganuclease designs: 25 mM HEPES (pH 8), 5% Glycerol, 10 mM MgCl2 and 225 mM NaCl (see Materials and methods section).
Expression and characterization of the designed mutants
The designed mutants A1, A2, B3 and B4 were obtained by site-directed mutagenesis (Stratagene, QuikChange Kit) of the original KTG and QAN enzyme expression vectors, and the corresponding proteins expressed and purified (Supplementary Data S1). We did not construct every combination of possible variants but rather selected only QAN-A1, KTG-B3, KTG-A2, QAN-B3 and QAN-B4. These were designed to give coverage of all the designed heterodimer interactions A1:B3, A2:B3 and A2:B4, resulting in the heterodimers QAN-A1:KTG-B3, KTG-A2:QAN-B3 and KTG-A2:QAN-B4.
Whereas the wild-type KTG and QAN enzymes yield the majority of protein in the soluble fraction (data not shown), we found the opposite in the case of the designed enzymes: the majority of the expressed proteins remained in inclusion bodies in the pellet, only a small fraction could be purified, and even this was contaminated by other proteins (Supplementary Data S1). This was a first indication that our designed variants cannot homodimerize and thus become unstable and aggregate when expressed individually.
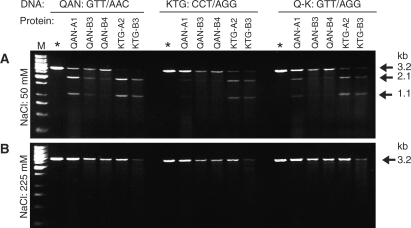
We tested the activity of the purified A1, A2, B3 and B4 enzymes on the three DNA targets (Figure 2) at low and high ionic strength (50 mM or 225 mM NaCl). At low salt, we detected general cleavage of both cognate and non-cognate targets with these mutant monomer designs. At high ionic strength, we could not detect the expected two DNA bands, although the amount of DNA decreased unspecifically in some cases, upon incubation with the enzymes (probably because of the low yield of the enzymes which resulted in a proportionally larger amount of contaminants). These results were marred by the low yield and quality of the protein obtained when the non-homodimerizing monomer designs were expressed individually; even with a large 6 l volume of bacteria yielding inadequate protein (between 0.5 and 1.5 mg/ml for designed monomers compared with 30 mg/ml for wild-type dimerizing monomers).
Figure 2.
Non-specific DNA cleavage and non-cleavage by singly expressed designed meganuclease monomer variants under different salt conditions. Approximately 3.75 µM of each purified protein was incubated with 3 nM of purified plasmid (pre-linearized with XmnI), containing either the QAN homodimer site (GTT/AAC), the KTG homodimer DNA site (CCT/AGG) or a hybrid site QAN/KTG site (Q-K: GTT/AGG). The concentration of NaCl was either (A) 50 mM or (B) 225 mM. Arrows indicate the uncut target DNA (3.2 kb) or the two bands resulting from digestion (1.1 and 2.1 kb). An asterisk (*) marks control lanes with DNA alone. One-kilobase ladders (Fermentas) are marked by M.
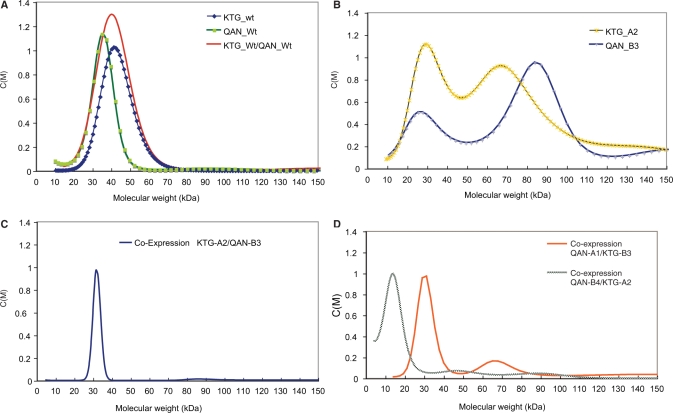
To check the oligomeric status of the purified designed enzymes we measured their size profiles by analytical ultra-centrifugation (see Figure 3 and Materials and methods section). In the case of individually expressed A1, A2, B3 and B4 proteins, we observed the appearance of the expected monomeric enzyme. However, we also saw higher molecular weight aggregates, including trimers and tetramers (Figure 3B; only KTG-A2 and QAN-B3 are shown, although similar results were obtained with the other designs). Therefore, the designed enzymes were indeed unable to homodimerize, and this may have affected their stability and aggregation properties during purification.
Figure 3.
Analytical ultra-centrifugation of the different meganucleases. (A)The wild-type monomers form homodimers of about 40 kDa (KTG-wt; QAN-wt). (B) The designed non-homodimerizing KTG-A2 and QAN-B3 form aggregates when expressed individually. (C) The co-expressed KTG-A2 and QAN-B3 form a perfect heterodimer. (D) The co-expressed QAN-B4 and KTG-A2 also form a heterodimer, to an extent, while QAN-A1 and KTG-B3 do not.
To investigate the potential for heterodimerization, we mixed equimolar quantities of the individually purified designed enzymes (QAN-A1, QAN-B3, QAN-B4, KTG-A2 and KTG-B3) in all possible combinations. In the case of the KTG-A2/QAN-B3 (the best heterodimer design), we saw the appearance of a major species corresponding to the molecular weight of the dimer, but this was not the only species formed. For the pair QAN-A1/KTG-B3 and KTG-A2/QAN-B4, we saw the appearance of new peaks of molecular mass between the monomer and dimer, and a decrease of high molecular weight aggregates (data not shown). For those combinations that should not produce a heterodimer, we did not see significant changes in the behaviour of the proteins. Overall, these results indicated that the design might have been successful but that separate expression of heterodimerizing monomers, followed by in vitro reconstitution, was not an effective strategy (probably because a large fraction of the protein was partly aggregated; see above). Thus, we decided to attempt co-expression within the bacterial cell.
Co-expression and activity of the designed mutants
The above results suggested that the heterodimer designs might have been functioning, but that the expression of the monomeric enzymes resulted in strong aggregation and thus in partly inactive enzymes. To avoid this problem, we subcloned the monomer gene expression cassettes into complementary plasmids and co-transformed into bacterial cells, such that one monomer would be expressed (with a His-tag) from the original pET-series plasmid and that the partner monomer would be expressed (without a His-tag) from a compatible pCDFDuet-I vector (Novagen). Dual antibiotic selection ensured that each cell contained both plasmids.
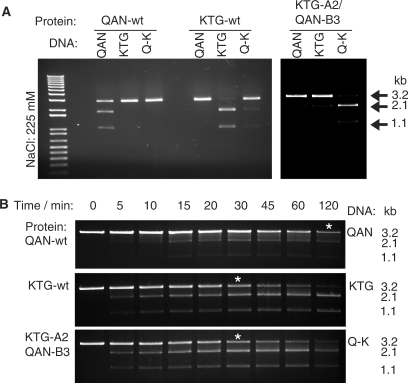
Expression analysis of the co-expressed KTG-A2/QAN-B3 proteins showed that inclusion bodies were avoided, suggesting that the previous aggregation problem had been solved. SDS–PAGE analysis of the purified enzyme subsequently revealed two bands with approximately the same amount of protein, suggesting that we were purifying the heterodimer and not homodimer (Supplementary Figure S1B). Furthermore, an analytical ultra-centrifugation of the purified proteins gave an exceptionally clean single profile at the expected molecular weight for a dimer (Figure 3C). We carried out digestion of the various DNA targets with the purified co-expressed heterodimer designs and found a clear preference for cleavage of the heterodimer DNA target (GTT/AGG), relative to the homodimeric targets (CCT/AGG and GTT/AAC) by KTG-A2/QAN-B3 (Figure 4A). Thus, we can conclude that the protein design exercise was successful and that we are able to create functional heterodimeric I-CreI enzyme variants, as long as the monomer moieties are co-expressed.
Figure 4.
Specific DNA cleavage by co-expressed wild-type (wt) and designed obligate heterodimer KTG-A2/QAN-B3 meganucleases. (A) Purified proteins were incubated with 3 nM of purified plasmid (pre-linearized with XmnI), containing either the QAN homodimer site (GTT/AAC), the KTG homodimer DNA site (CCT/AGG) or a hybrid site QAN/KTG site (Q-K: GTT/AGG). Because different constructs have different reaction optima, homodimers were used at 0.25-μM concentration (4 h, 37°C), while heterodimer was used at 0.50-μM concentration (30 min, 37°C). NaCl concentration was at 225 mM. Arrows indicate the uncut target DNA (3.2 kb) or the two bands resulting from digestion (1.1 and 2.1 kb). One-kilobase ladders (Fermentas) are marked by M. (B) The relative activities of each enzyme sample were compared in a time-course experiment against their optimal DNA sites, using 1 μM protein and 6 nM purified plasmid target. White asterisks mark the positions of the samples estimated to be closest to having 50% cleavage.
Co-expression experiments were also carried out for the other protein designs. QAN-A1/KTG-B3 proteins showed mixed results; there were indeed two bands after purification, indicating heterodimer formation. However, one band was stronger than the other and while there was specific cleavage of the heterodimer, this was at a reduced level as compared to the KTG-A2/QAN-B3 combination (data not shown). Analytical centrifugation showed formation of a dimer with a small proportion of aggregate. The third co-expression combination, KTG-A2/QAN-B4, resulted in only one band being purified and a monomer detected by analytical centrifugation (Figure 3D). Therefore, this design failed to make a heterodimer, even when co-expressed. Interestingly enough, the proportion of dimer and activity between KTG-A2/QAN-B3, QAN-A1/KTG-B3 and KTG-A2/QAN-B4 correlate perfectly well with the energies predicted by FoldX (Table 1). The best design, in terms of predicted energy in silico, forms the best heterodimer in vitro.
Although the KTG-A2/QAN-B3 heterodimer preferentially cleaved the hetero-DNA target, it could have been possible that it was less active. To rule out this, we compared the activities of the QAN, KTG and KTG-A2/QAN-B3 in a time-course (Figure 4B). This experiment shows that KTG and the heterodimer have similar activities, whereas QAN requires four times longer incubation for 50% cleavage of its substrate, using the same apparent protein concentration.
Specificity in competitive cutting assays
To measure the relative discrimination of the enzymes for homodimer and heterodimer DNA sites, we carried out competition experiments where the enzymes had access to equimolar amounts of both substrates simultaneously (Figure 5). We selected the KTG-A2/QAN-B3 heterodimer and KTG-wt for these experiments, because they exhibited similar activities against their respective targets (Figure 4B). In a time course experiment, KTG cleaved its target preferentially, although there was a slight digestion of the KTG-A2/QAN-B3 target cognate site (Figure 5A). By contrast, KTG-A2-QAN-B3 heterodimer preferentially cleaved the Q-K heterodimer DNA with little cleavage of the KTG site.
Figure 5.
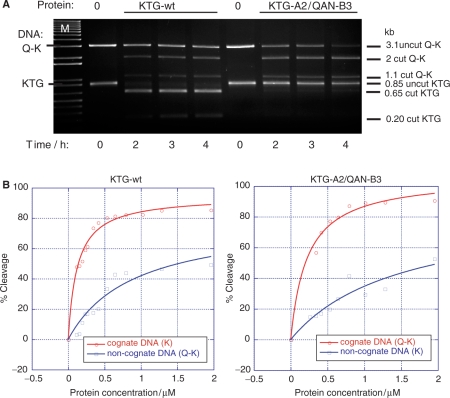
Competition assays to determine the relative specificity of KTG-wt homodimer and KTG-A2/QAN-B3 heterodimer proteins when exposed to equimolar mixtures of Q-K heterodimer DNA site (3.1-kb PCR product) and KTG homodimer DNA site (0.85-kb PCR product). Each target DNA was used at 5-nM final concentration. The different DNA targets are characteristic sizes and give specifically-sized cleavage products. (A) Time-course experiment showing the relative specific and non-specific cutting by KTG and KTG-A2/QAN-B3 enzymes. Final protein concentration = 1 μM. (B) Enzyme titration assay to determine the difference in protein concentration for 50% cleavage of cognate and non-cogate DNA target sites by each enzyme. Equimolar amounts of DNA target site PCR products (Q-K and KTG) were mixed and incubated against a dilution series of each enzyme, as indicated, for a 1 h incubation. Gels were scanned and analysed with ImageJ and Kaleidagraph.
To compare the relative cutting preferences more directly, an enzyme titration was carried out against equimolar mixtures of both DNA targets (Figure 5B). This allowed the determination of the apparent concentrations for 50% cleavage for cognate and non-cognate targets under competitive conditions: KTG-wt for cognate target = 0.1 μM; KTG-wt for non-cognate target = 1.5 μM; KTG-A2/QAN-B3 for cognate target =0.3 μM; KTG-A2/QAN-B3 for non-cognate target = 3.2 μM. Therefore, for both enzymes there was found to be an approximate 10- to 15-fold difference in enzyme concentration separating 50% cleavage of the cognate and the non-cognate targets.
In summary, these results show that although the specificities of both the parent constructs and the mutant derivatives are not absolute, we were able to design obligate heterodimer meganucleases which have similar activity to the best wt parent, and a clear cleavage preference for their heterodimer targets, whereas the original homodimers have the opposite preference for their homodimer targets.
DISCUSSION
The making of artificial endonucleases with tailored specificities has paved the way for novel applications in several fields, including gene therapy. For example, meganuclease-induced recombination can be used for the correction of mutations linked with monogenic inherited diseases such as SCID, SCA or CFTR (5). This strategy has the advantage to bypass the odds associated with current strategies of random insertion of a complementing transgene. Several reports have also shown that engineered zinc-finger nucleases can trigger efficient site-directed recombination in mammalian cultured cells, plants and insects (42–46). However, zinc finger-derived nucleases (ZFNs) have proven to be toxic in Drosophila (42,46,47) and mammalian NIHT3 cells (48–50), a genotoxic effect that is probably due to frequent off-site cleavage (45). Although HEs have shown to be less toxic (probably because of better specificity) by different groups (48–50), they can still be harmful at very high doses (51).
Off-site cleavage is severely enhanced by the formation of protein engineering by-products. Most engineered endonucleases (ZFNs and HEs) so far are heterodimers, and include two separately engineered monomers, each binding one half of the target. Heterodimer formation is obtained by co-expression of the two monomers in the same cells (9,45). This is actually associated with the formation of two unwanted homodimers (7,47), recognizing different targets, and individual homodimers can sometimes result in an extremely high level of toxicity (47). This problem is well known in the field and there have been several previous approaches to overcome it.
In the case of ZFNs, two recent reports have tackled this issue directly, with approaches that are related to the work presented here with meganucleases. ZFNs function by having two zinc finger DNA-binding domains, each fused to a FokI nuclease domain, which cuts DNA as a dimer. FokI dimerization is relatively weak and is thought to occur primarily once both DNA-binding domains have bound their target DNAs (52). Miller and co-workers (37) therefore employed a step-wise sequential rational design approach to make obligate FokI heterodimers that would function in the context of DNA-binding specificity provided by custom-designed zinc fingers. This was very successful in that not only did the zinc fingers form obligate heterodimers that cut the heterodimer DNA sites in vitro, with little or no discernible cutting of the homodimer sites, but this actually translated to a reduction in genomic DNA damage in vivo, as measured by an antibody-mediated assay for sites associated with DNA damage. In a complementary approach, Szczepek et al. (38) used computer-aided design in a similar zinc finger-FokI system to actually reduce general dimerization of FokI, thus ensuring that cleavage would tend to be at DNA sites where complementary zinc fingers bound specifically, in the correct orientation. Similarly, assays for DNA damage in vivo showed that the mutant FokI design had reduced general toxicity, while maintaining sufficient activity to drive homologous recombination.
In the case of meganucleases, previous studies have also shown that it is possible to re-design protein–protein interfaces so to gain the advantages of generating new, selective binding specificities. For example, Chevalier and colleagues (26), carried out a challenging protein computational-design approach to re-engineer new, functional fusion chimeras of I-DmoI and I-CreI endonuclease domains. This required extensive computational exploration, over 14 amino acid residue positions, and resulted in a functional chimera with eight designed point mutations. The authors concluded that it would be possible to make many new sequence-specific endonucleases by taking natural monomer domains and building such new chimeras. However, it is worth noting that in the I-DmoI–I-CreI fusion the chimera was created by connecting the two monomer domains with a short peptide linker of only 3 amino acids. This possibility was not available for making I-CreI 2-unit chimeras because the N- and C-termini of each monomer are 68 Å apart, which would require a very long linker of around 25–30 amino acids. Using a long linker could in theory result in a functional fusion of the two monomers in a single chain molecule. However, this kind of design is relatively perilous, and can result in concatemers, badly folded proteins and unstable, easily degraded linker regions. In another approach, a linker of 10 bp was used to connect two I-CreI monomers (25), but the use of this shorter sequence was only made possible by truncating the first I-CreI monomer by one-third. This deletion resulted in a shorter distance between the amino acids connected by the linker, but had also a significant impact on protein solubility (data not shown). This therefore prompted us to attempt the obligate heterodimer approach for I-CreI.
It should also be noted that there exists in the literature a history of re-engineering HE interfaces to control homodimerization itself. For example, the LAGLIDAD interface has been extensively studied by Silva and Belfort (35) who grafted residues from I-CreI helices onto I-DmoI domains. I-Dmo is a natural monomer, whereas I-CreI is a dimer, and the helix graft resulted in dimeric I-DmoI which acted as a nickase rather than a double-strand breaking nuclease. Double-strand cleavage activity could then be improved by reversion of specific helix residues. This work illustrates the difficulty sometimes encountered in re-engineering enzymes: a functional interface can yet be incompatible with full enzyme functionality. Nonetheless, this study gave insights into the relative contributions of parts of the LAGLIDAD helix, delineating the sensitivity to mutation of regions such as the C-terminal 10 residues, which contribute to the endonuclease active site. Furthermore, the authors later extended this work by re-engineering I-DmoI LAGLIDAD interfaces (using I-CreI-derived mutations) to give functional homodimer interfaces and full enzyme activity (36). They were also able to use a short 2 amino acid peptide linker to make fused dimer constructs with 3-fold higher activity over unlinked dimer. However, there was evidence that the improvement in activity was due to changes in the active site configuration rather than improved homodimerization. These studies show how interface mutations in HEs are intimately linked with cleavage activity.
In summary, the specificity problem of heterodimer meganucleases might have been approached by (i) the suppression of any dimer formation in the absence of DNA interactions (38), (ii) the design of favourable heterodimerization and unfavourable homodimerization (37) or (iii) interface re-design and direct peptide linkage to make chimeras between two monomers (25,26). The engineering of obligatory heterodimers was chosen in this study as the simplest alternative that provides functional, well-folded proteins in the context of the pure I-CreI scaffold. Using only four complementary amino acid mutations on each monomer (KTG-A2 and QAN-B3 designs), we were able to generate preferential heterodimerization and target site recognition when the monomers were co-expressed.
Hundreds of homodimeric I-CreI derivatives with locally altered specificity have been described in previous reports (7,9), and it has been shown that such proteins could be co-expressed to form heterodimers. However, the possibility to combine these proteins into obligatory heterodimers, as shown here, will dramatically improve the ability to engineer more specific reagents for genome engineering. For therapeutic applications, which require a minimal genotoxicity, this gain in specificity might simply make all the difference.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Dr Vladimir Rybin for providing assistance with analytical ultra-centrifugation. This work was funded by European Commission Framework 6 Grants (Integra, FP6-29025 and Netsensor, 012948); Cellectis S.A. Funding to pay the Open Access publication charges for this article was provided by CRG.
Conflict of interest statement. FP states an interest in the company Cellectis, SA which develops and uses meganuclease technology.
REFERENCES
- 1.Thierry A, Dujon B. Nested chromosomal fragmentation in yeast using the meganuclease I-Sce I: a new method for physical mapping of eukaryotic genomes. Nucleic Acids Res. 1992;20:5625–5631. doi: 10.1093/nar/20.21.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol. Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 6.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl Acad. Sci. USA. 1996;93:5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 8.Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 9.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevalier BS, Monnat RJ, Jr, Stoddard BL. The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat. Struct. Biol. 2001;8:312–316. doi: 10.1038/86181. [DOI] [PubMed] [Google Scholar]
- 11.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostriken R, Strathern JN, Klar AJ, Hicks JB, Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983;35:167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- 13.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Kim HH, Yuan X, Herrin DL. Purification, biochemical characterization and protein-DNA interactions of the I-CreI endonuclease produced in Escherichia coli. Nucleic Acids Res. 1997;25:3767–3776. doi: 10.1093/nar/25.19.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 16.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Jr, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 17.Spiegel PC, Chevalier B, Sussman D, Turmel M, Lemieux C, Stoddard BL. The structure of I-CeuI homing endonuclease: Evolving asymmetric DNA recognition from a symmetric protein scaffold. Structure. 2006;14:869–880. doi: 10.1016/j.str.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Moure CM, Gimble FS, Quiocho FA. Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence. Nat. Struct. Biol. 2002;9:764–770. doi: 10.1038/nsb840. [DOI] [PubMed] [Google Scholar]
- 19.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J. Mol. Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 20.Silva GH, Dalgaard JZ, Belfort M, Van Roey P. Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J. Mol. Biol. 1999;286:1123–1136. doi: 10.1006/jmbi.1998.2519. [DOI] [PubMed] [Google Scholar]
- 21.Ichiyanagi K, Ishino Y, Ariyoshi M, Komori K, Morikawa K. Crystal structure of an archaeal intein-encoded homing endonuclease PI-PfuI. J. Mol. Biol. 2000;300:889–901. doi: 10.1006/jmbi.2000.3873. [DOI] [PubMed] [Google Scholar]
- 22.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama H, Shimamura T, Imagawa T, Shirai N, Itoh T, Sako Y, Miyano M, Sakuraba H, Ohshima T, Nomura N, et al. Structure of a hyperthermophilic archaeal homing endonuclease, I-Tsp061I: contribution of cross-domain polar networks to thermostability. J. Mol. Biol. 2007;365:362–378. doi: 10.1016/j.jmb.2006.09.066. [DOI] [PubMed] [Google Scholar]
- 24.Pingoud V, Grindl W, Wende W, Thole H, Pingoud A. Structural and functional analysis of the homing endonuclease PI-sceI by limited proteolytic cleavage and molecular cloning of partial digestion products. Biochemistry. 1998;37:8233–8243. doi: 10.1021/bi980013d. [DOI] [PubMed] [Google Scholar]
- 25.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 27.Steuer S, Pingoud V, Pingoud A, Wende W. Chimeras of the homing endonuclease PI-SceI and the homologous Candida tropicalis intein: a study to explore the possibility of exchanging DNA-binding modules to obtain highly specific endonucleases with altered specificity. Chembiochem. 2004;5:206–213. doi: 10.1002/cbic.200300718. [DOI] [PubMed] [Google Scholar]
- 28.Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30:3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R, Jr, Stoddard BL, Seligman LM. Isolation and characterization of new homing endonuclease specificities at individual target site positions. J. Mol. Biol. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 30.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 31.Gimble FS, Moure CM, Posey KL. Assessing the plasticity of DNA target site recognition of the PI-SceI homing endonuclease using a bacterial two-hybrid selection system. J. Mol. Biol. 2003;334:993–1008. doi: 10.1016/j.jmb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 33.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc. Natl Acad. Sci. USA. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva GH, Belfort M. Analysis of the LAGLIDADG interface of the monomeric homing endonuclease I-DmoI. Nucleic Acids Res. 2004;32:3156–3168. doi: 10.1093/nar/gkh618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva GH, Belfort M, Wende W, Pingoud A. From monomeric to homodimeric endonucleases and back: engineering novel specificity of LAGLIDADG enzymes. J. Mol. Biol. 2006;361:744–754. doi: 10.1016/j.jmb.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 37.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 38.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 39.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 41.Cohn EJ, Edsall JT. Proteins, Amino Acids and Peptides. New York: Reinhold; 1943. [Google Scholar]
- 42.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 43.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 44.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol. Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene targeting in Drosophila with zinc-finger nucleases. Genetics. 2006;172:2391–2403. doi: 10.1534/genetics.105.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, III, Segal DJ, Weitzman MD, Cathomen T. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 49.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 50.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 51.Gouble A, Smith J, Bruneau S, Perez C, Guyot V, Cabaniols JP, Leduc S, Fiette L, Ave P, Micheau B, et al. Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J. Gene Med. 2006;8:616–622. doi: 10.1002/jgm.879. [DOI] [PubMed] [Google Scholar]
- 52.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]