Abstract
Vascular endothelial growth factor A (VEGF-A) is a potent secreted mitogen critical for physiological and pathological angiogenesis. Regulation of VEGF-A occurs at multiple levels, including transcription, mRNA stabilization, splicing, translation and differential cellular localization of various isoforms. Recent advances in our understanding of the posttranscriptional regulation of VEGF-A are comprised of the identification of stabilizing mRNA-binding proteins and the discovery of two internal ribosomal entry sites (IRES) as well as two alternative initiation codons in the 5′UTR of the VEGF-A mRNA. We have previously reported that VEGF-A translation initiation at both the AUG and CUG codons is dependent on the exon content of the coding region. In this report, we show that the expression of different VEGF-A isoforms is regulated by a small upstream open reading frame (uORF) located within an internal ribosome entry site, which is translated through a cap-independent mechanism. This uORF acts as a cis-regulatory element that regulates negatively the expression of the VEGF 121 isoform. Our data provide a framework for understanding how VEGF-A mRNAs are translated, and how the production of the VEGF 121 isoform is secured under non-hypoxic environmental conditions.
INTRODUCTION
The growth of blood vessels, a process known as angiogenesis, is essential for organ growth and repair. An imbalance in this process contributes to numerous inflammatory, ischemic, immune and malignant disorders. The vascular endothelial growth factor A (VEGF-A) is a growth and survival factor for endothelial cells, playing an essential role in numerous physiological and pathological angiogenic processes throughout embryonic development and during adulthood (1,2). In the quiescent vasculature of adult organs, VEGF is produced at basal level and protects endothelial cells from apoptosis. However, in a number of physiological situations, such as oestrus in the female reproductive organs, wound repair, adaptation to hypoxia as well as in many pathological situations like proliferative retinopathies, arthritis, psoriasis and of course cancer, VEGF level increases (3–5). VEGF acts as the key mediator of tumor angiogenesis by stimulating the growth of new blood vessels from nearby capillaries and providing tumors with access to oxygen and nutrients they need to grow and metastasis.
Both the deletion of a single VEGF allele (6,7) and modest VEGF over-expression (8) result in embryonic lethality due to improper vascularization. These experiments on transgenic mice have clearly demonstrated that the expression level of VEGF is under very tight regulatory control, at least during development. Numerous studies have been devoted to understand the regulation of expression of this factor. The importance of regulating VEGF expression is further demonstrated by comparative genomic sequence analyses, which reveal the conservation of regulatory elements throughout numerous species. These elements are summarized in Figure 1.
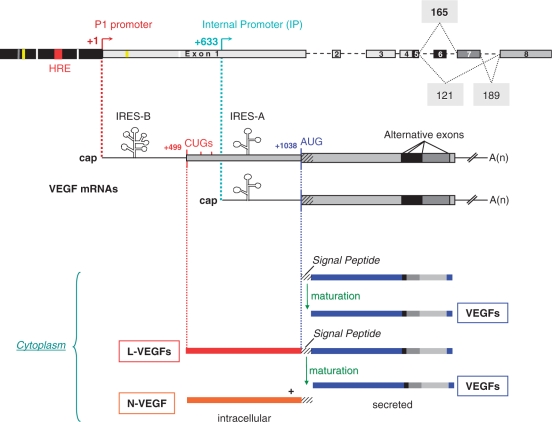
Figure 1.
VEGF gene structure. The VEGF genomic structure is schematized with 5′UTR containing: P1, TATA less promoter region including hypoxia responsive element (HRE) (13); IP, internal promoter directs the transcription of a truncated mRNA. Three coding exons are alternatively spliced and give rise to several polypeptide, the most abundant are the 121, 165 and 189 amino acids isoforms. The 1038 nucleotides upstream from the AUG contain two IRESes (A and B) and in frame CUGs translation initiation codons that direct the synthesis of L-VEGF, which cleavage generates secreted-VEGF plus the intracellular N-VEGF.
(1) The TATA-less promoter region (P1) includes consensus sites for transcription factor such as Sp1/Sp3, ER, PRE, AP2, Egf1, STAT3 and HIF1 among many others (9–13). Several studies have demonstrated that numerous stimuli activate VEGF transcription, including growth factors, hypoxia, hormones, oncogenes and tumor suppressors (9,14).
(2) A second internal promoter (IP) located 633 nucleotides downstream from the P1, directs the transcription of a truncated mRNA (15).
(3) The eight exons as well as the introns of the VEGF gene are highly conserved between human, rodent, dog and chicken. Alternative splicing gives rise to at least nine different transcript variants corresponding to nine polypeptide isoforms of 121, 145, 148, 162, 165, 165b, 183, 189 and 206 amino acids (AA) (16–22). While functions for all isoforms have not yet been fully defined, most VEGF producing cells appear to preferentially express VEGF 121, VEGF 165 and VEGF 189. These three major isoforms of VEGF have different properties concerning receptor binding and extracellular localization. VEGF 121 is fully diffusible, whereas VEGF 165 and 189 isoforms are able to bind to heparan sulfate on the cell surface and in the extracellular matrix (23). Each isoform contributes to the formation of a VEGF gradient that is essential for the process of tumor neo-vascularization. The more soluble isoform acts at distal sites to promote vascular recruitment, and the extracellular membrane-associated isoforms promote local expansion of capillary beds (24).
(4) The coding region is flanked by a long 5′-untranslated region (5′UTR). In human, this region spans 1038 nucleotides and contains three in frame alternative CUG start codons, the first one located 539 nucleotides upstream from the classical AUG initiation codon (25–27) and two internal ribosome entry sites (IRESes) (15,28,29). The first identified IRES (IRES A) is located within the 300 nucleotides upstream of the AUG start codon and IRES B, 377 nucleotides in length, is located 24 nucleotides upstream of the first CUG codon. The two IRESes control translation at the two initiation codons as demonstrated in vitro (15,28) and in vivo (30).
Translation initiated at the CUG codon directs the synthesis of L-VEGF, a precursor further cleaved to generate on one hand secreted VEGF and on the other hand N-VEGF, a 23-kDa NH2-specific fragment of 206 AA, which remains intracellular (25,27,31). Translation from the AUG codon at position 1038 only generates secreted VEGF isoforms. Interestingly, mRNA transcribed from the internal promoter lacks both IRES B and the CUG start codons.
(5) The 1881 bp-long 3′-untranslated region (3′UTR) contains multiple alternative polyadenylation signals, and a large number of AU-rich elements (AREs) (32,33). VEGF mRNA stability is regulated in response to several stimuli through the binding of stabilizing and destabilizing proteins to ARE located in his 3′UTR (34–39).
VEGF, which is regulated at every conceivable stage of its production, thus represents a paradigm for gene regulation.
We have previously reported that the initiation of translation at both AUG and CUG codons is dependent on the exon content of the coding region. In this work, we reveal a new level in the regulation of VEGF expression. We have analyzed, for the two described transcription initiation sites, the relationship between alternatively spliced mRNAs and the use of alternative translation initiation codons. We identified an upstream ORF (uORF), within the IRES A, which acts as a regulatory element essential for the control of isoform expression.
MATERIAL AND METHODS
Plasmid constructions
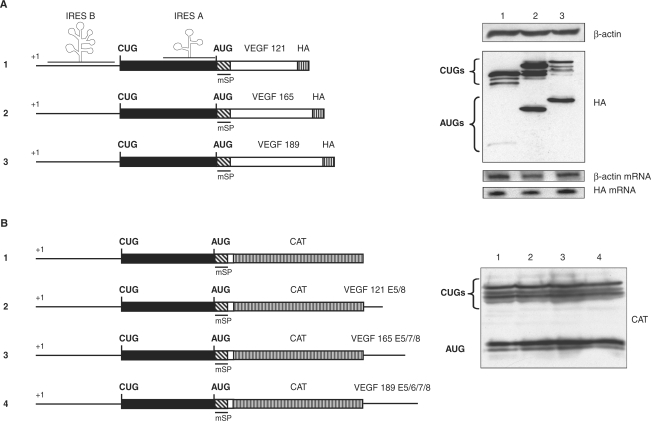
The constructs p121mSPHA, p165mSPHA and p189mSPHA (Figure 2A) containing the 5′UTR, the coding sequence of each vegf cDNA isoforms, the mutation of the signal peptide and an hemaglutinine (HA) tag at the 3′end of the vegf cDNA; the construct pminiVEGF (Figure 3B), which contains the vegf cDNA until exon 4, the mutation of the signal peptide, the genomic sequence between exon 4 and exon 8 and an HA tag, and the constructs pAUG121mSPHA, pAUG165mSPHA, pAUG189mSPHA (Figure 3A), which start 24 nucleotides upstream from the AUG codon, have already been described (40).
Figure 2.
Effect of VEGF-A alternative spliced sequences on the translation initiation. (A) Left, schematic representation of the constructs used for transfection experiments. The 5′UTR is shown as thickened line, the VEGF coding region is shown as a black box when beginning at the CUG and white box when beginning at the AUG. The mutated signal sequence is represented as hatched box. A HA tag for immunoblot detection is present at the 3′ end. The two IRESes are represented. The constructs encode the three VEGF isoforms (p121mSPHA, p165mSPHA and p189mSPHA numbered 1, 2 and 3) and give rise to the VEGF and the L-VEGF proteins. Right, constructs were transiently transfected into HeLa cells. Their expression was analyzed by western immunoblotting using an anti-HA antibody. The CUGs represent L-VEGF proteins and the AUGs represent VEGF proteins. (B) Left, schematic representation of the constructs used for transfection experiments. CAT reporter is shown as striped box in frame with VEGF coding sequence, VEGF non-coding sequences are shown as thickened line. The constructs, encoding VEGF-CAT with alternative spliced sequence of each isoform of 189, 165, 121 AA as non-coding sequence pVCmSP189, pVCmSP165 and pVCmSP121 numbered 1, 2 and 3. Right, constructs were transiently transfected into HeLa cells. Their expression was analyzed by western immunoblotting using an anti-CAT antibody. The CUGs represent L-VEGF-CAT fusion proteins and the AUGs represent VEGF-CAT fusion proteins.
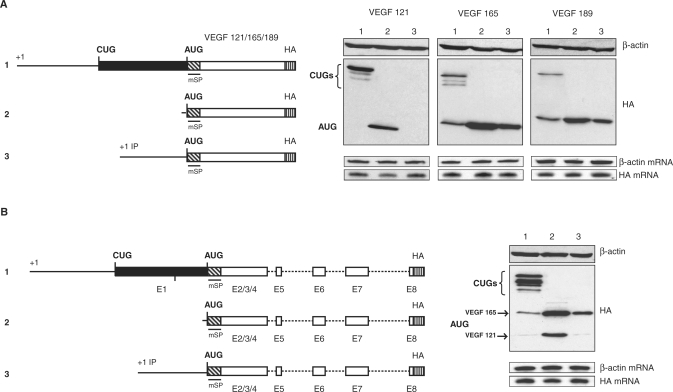
Figure 3.
Role of the VEGF-A 5′UTR on alternative translation initiation control. (A) Schematic representation of the constructs used for transfection experiments: constructs, encoding each isoform and the VEGF minigene, beginning at the main promoter (p121mSPHA, p165mSPHA, p189mSPHA and pminiVEGF, numbered 1), constructs beginning 24 nucleotides upstream from the AUG (pAUG121mSPHA, pAUG165mSPHA, pAUG189mSPHA and pAUGminiVEGF, numbered 2), and constructs beginning at the internal promoter start site +633 (pIP121mSPHA, pIP165mSPHA, pIP189mSPHA and pIPminiVEGF numbered 3). (B) Each plasmid were transiently transfected into HeLa cells. Their expression was analyzed by western immunoblotting using an anti-HA antibody. An anti-β-actin was used to control protein loading. The constructs 1 give rise to the VEGF and the L-VEGF proteins and the other constructs encode only the AUG initiated VEGF proteins. RPA analyses were performed to normalize transfection with a vector specific probe to quantify mRNA and with a β-actin probe to control RNA quantity.
The constructs p5′121, p5′165, p′5189 and pVCmSP have already been described (28). The addition of VEGF 189, 165, 121 cDNAs sequence at the 3′ extremity of the CAT cDNA were obtained by PCR. Amplification products of the p5′189, p5′165 and p5′121 plasmids with the primers Ex5 sens and VEGF 3′ reverse (Table 1) were then digested at the BglII, and EcoRI sites and cloned into the BglII and EcoRI digested pVCmSP to create the plasmids pVCmSP189, pVCmSP165 and pVCmSP121, respectively (Figure 2B).
Table 1.
Oligonucleotide sequences
| Oligonucleotides | Sequences (5′→3′) |
|---|---|
| Ex5 sens | AAGAAAGATAGAGCAAGA |
| VEGF 3′ REV | TTTGAATTCTCACCGCCTCGGCTTGTC |
| XbaI IP | ATATCTAGACGGGGCTCGC GGCGTCGCACTGAAAC |
| Ex4 REV | CTGCATTCACATTTGTTGTGCTGTAG |
| −50 AUGXbaI | GCTCTAGAGCCGGAGAGGGAGCGCGAGC |
| −100 AUGXbaI | GCTCTAGAAAGAGTAGCTCGCCGAGGCG |
| −200 AUGXbaI | GCTCTAGAGGAAGCCGGGCTCATGGACG |
| −300 AUGXbaI | GCTCTAGAGAAGTGCTAGCTCGGGCCG |
| mATGu | GGAAGCCGGGCTCTTGGACGGGTGAGG |
| mSO | CATGGACGGGGAGGCGGC GGTGTGCGCAGAC |
| RT Ex4 sens | CCAGATTATGCGGATCAAACC |
| RT 3′HA REV | TCACCGCCTCGGCTTGTCAC |
To obtain plasmid pAUGminiVEGF the XbaI/BsmI fragment of pAUG189mSPHA was inserted into XbaI/BsmI digested pminiVEGF (Figure 3B).
To obtain pIP121mSPHA, pIP165mSPHA, pIP189mSPHA and pIPminiVEGF, we digested a fragment obtained by PCR amplification of p189mSPHA with forward primer XbaI-IP and reverse primer Ex4 rev (Table 1), with XbaI/BsmI for insertion into XbaI/BsmI digested p121mSPHA, p165mSPHA p189mSPHA and pminiVEGF, respectively (Figure 3A–C).
To obtain plasmid containing serial deletion from the internal promoter, we digested plasmids p121mSPHA, p165mSPHA p189mSPHA and pminiVEGF with XbaI/BsmI, the fragment was gel purified. We digested with XbaI/BsmI PCR products amplified with reverse primer Ex4 rev and forward primer −50AUGXbaI, −100AUGXbaI, −200AUGXbaI, −300AUGXbaI. Each fragment, corresponding to each VEGF cDNA isoforms, was ligated with fragments corresponding to each deletion. We obtain the constructs: p-50AUG121mSPHA, p-100AUG121mSPHA, p-200AUG121mSPHA, p-300AUG121mSPHA, p-50AUG165mSPHA, p-100AUG165mSPHA, p-200AUG165mSPHA, p-300AUG189mSPHA, p-50AUG189mSPHA, p-100AUG189mSPHA, p-200AUG189mSPHA, p-300AUG189mSPHA, p-50AUGminiVEGF, p-100AUGminiVEGF, p-200AUGminiVEGF, p-300AUGminiVEGF (Figure 4A).
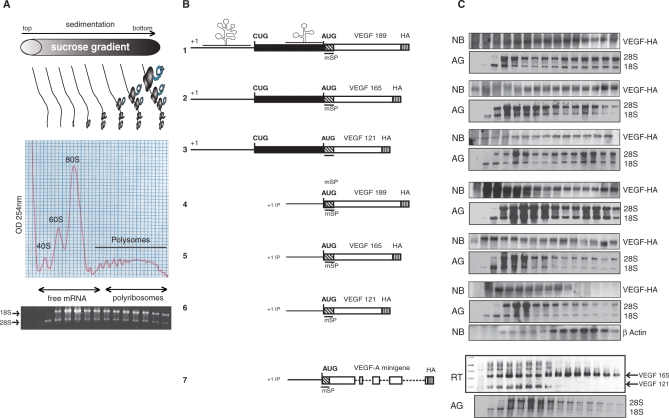
Figure 4.
Analysis of the VEGF mRNA polyribosomal distribution. (A) Schematic representation of polysomal and free mRNAs distribution after HeLa cells transfection. The diagram represents the polysome distribution profiles obtained after centrifugation of cytoplasmic lysates over sucrose gradient then fractionated from top to bottom. RNA was isolated from each fraction and visualized on agarose gel by Sybr green staining. (B) Schematic representation of the constructs used for transfection experiments: construct encoding each VEGF isoforms, beginning at the main transcription start site (p121mSPHA, p165mSPHA and p189mSPHA, numbered 1, 2 and 3) or at internal promoter start site (pIP121mSPHA, pIP165mSPHA, pIP189mSPHA numbered 4, 5 and 6) and the VEGF minigene beginning at internal promoter start site (pIPminiVEGF numbered 7). (C) From each fraction, RNA was prepared for agarose gel analysis of 18S and 28S rRNAs (AG) and for northern blot (NB) of VEGF and β-actin for lane 6. For the minigene construct (lane 7), RNA were analyzed by RT–PCR (RT) since it was difficult to resolve them by northern blot analysis, with primer that amplify fragments corresponding to each VEGF isoforms.
All the uORF mutations were performed using site-directed mutagenesis (QuickChange, Stratagene, La Jolla, CA), the primers are described in the Table 1. To introduce the mutation of the uORF's start codon, we used primer reverse and forward mATGu, with the plasmid pIP121mSPHA, pIP165mSPHA pIP189mSPHA and pIPminiVEGF, leading respectively to the constructs pIP121mORFmSPHA, pIP165mORFmSPHA, pIP18mORF9mSPHA and pIPminiVEGF (Figure 4B and C).
To obtain the construct pIPmSOmSP189HA, which contains the mutation of uORF's stop codon, we use the plasmid pIP189mSPHA with the primer mSO reverse and forward (Table 1). To obtain the other isoform pIPmSOmSP121HA pIPmSOmSP165HA and pIPmSOminiVEGF, we digeted a fragment obtained by PCR amplification of pIPmSOmSP189HA with forward primer XbaI-intP and reverse primer Ex4 rev, with XbaI/BsmI for insertion into XbaI/BsmI digested pIP121mSPHA, pIP165mSPHA and pIPminiVEGF (Figure 4B and C).
Cell culture
HeLa cell line, a human uterus carcinoma cell line of epithelial origin (obtained from ATCC n°. CCL2™), were maintained in DMEM supplemented with 10% FCS, 1% glutamine, 1% amphotericin and 0.1% gentamycin. All the cells were grown in a humidified atmosphere of 5% CO2 at 37°C.
For hypoxic conditions, HeLa cells were cultured at 37°C with 5% CO2, 94% N2 and 1% O2 in a hypoxic incubator (Binder GmbH, Tuttligen, France).
Transient transfection
Cells were seeded into 100 mm dishes 1 day prior to transfection, allowed growing to 50–70% confluency. According to the manufacturer's instructions, 8 μg of plasmid were transfected into the cells with 16 μl of the JetPEI reagent (Polyplus Transfection Illkirch, France) in NaCl. Cells lysates were prepared 24 h posttransfection for protein or mRNA quantification and analysis.
SiRNA transfection
eIF4E targeting and duplex control siRNA (Dharmacon, Lafayette, CO) were transfected in HeLa cells (5 × 105) using Interferin (Polyplus Transfection Illkirch, France) according the manufacturer's protocol. After 48 h, cells were cotransfected (3 μg of plasmids and 10 nM siRNA) with Lipofectamine 2000 (Invitrogen, Cergy-Pontoise, France), and harvested 24 h later for protein and RNA analyses.
Protein extracts and western blotting
Western blotting was performed as previously described (41). Briefly, transfected cells were scraped in phosphate-buffered saline, collected by centrifugation, resuspended in sample lysis buffer [20% (sodium dodecyl sulfate) SDS, 50% glycerol, 50 mM Tris pH7.8, Bromophenol blue] in the presence of 1% β-mercaptoethanol and 1% dithiothreitol 1 M and subjected to sonication. Total proteins were quantified by BCA assay (Interchim, Montluçon, France). The 30 μg protein samples were heated for 2 min at 95°C and separated by 12.5% polyacrylamide gel (PAGE), before being transferred onto a nitrocellulose membrane. HA-tagged VEGF proteins were detected using the HA antibody (HA.11, BabCO Eurogentec, Herstal, Belgium) (dilution 1:1000). CAT-fusion VEGF proteins were detected using the rabbit polyclonal CAT antibody prepared in the laboratory (dilution 1:10 000). eiF4E protein was detected using specific polyclonal antibody (Cell Signaling, Danvers, MA) (dilution 1:1000). The protein signal was normalized using an anti-β-actin monoclonal antibody. (AC-15, Sigma-Aldrich, Lyon, France) (dilution 1:10 000). Signals were detected using a chemiluminescence ECL kit (Amersham Pharmacia Biotech, Piscataway, NJ).
Preparation of RNA probes for ribonuclease protection assays and northern blot
To quantify cell transfection, a probe designed for anneal the 3′ end of construct was amplified from p189PSmHA using the forward primer CGGATCTTTTCCCTCTGCCAA and the T7 containing reverse primer TAATACGACTCACTATAGGGCAAACTCTAAACCAAATACTC, designed to amplify a 190-nt fragment spanning the 3′UTR of the pSCT from the HA tag to the polyadenylation signal.
To normalize RPA quantification, a β-actin probe was designed using forward primer TGTACGCCAACACAGTGCTGTC and T7 containing reverse primer TAATACGACTCACTATAGGGTAGAAGCATTTGCGGTGGACG to amplify a 250-nt fragment.
RNA probe labeled with Biotin-14-CTP (Invitrogen, Cergy-Pontoise, France) were synthesized using an in vitro transcription kit (Maxiscript; Ambion, Austin, TX). Probes of validated length was gel-purified on 5% polyacrylamide 8M urea gel, eluted in probe elution buffer (Ambion, Austin, TX) overnight at 37°C and ethanol precipitated.
To analyze mRNAs distribution into polysomes, northern blot was performed using a DNA probe amplified by PCR from p189SPmHA with forward primer −50XbaI and reverse primer Ex4 rev (Table 1). To prove mRNA integrity, northern blot was performed using a DNA probe amplified with the primers used for RPA probe synthesis. After gel purification, the fragment was labeled using Biotin ULS labeled probe (Biotin ULS Labaling Kit, Fermentas Canada, Ontario).
RNA preparation, Rnase protection assay and northern blot
Total RNA was isolated from transiently transfected cells with the Trizol reagent (Invitrogen, Cergy-Pontoise, France) and treated with 1 μl of DNA free (Ambion, Austin, TX). RPA analysis was performed using the RPA III kit (Ambion, Austin, TX) following the protocol provided by the manufacturer. The hybridization was carried out by overnight incubation at 42°C of 1ng of labeled probe and 2 μg of RNA samples. The hybridization mixture was then treated with ribonuclease A/T1 mix for 30 min at 37°C. Digested samples were ethanol precipitated and separated on a 5% polyacrylamide 8 M urea gel followed by semi-dry electroblotting onto Positively Charged Nylon Membranes (BrightStar-Plus, Ambion, Austin, TX). After transfer, membranes were UV cross-linked and the bands were visualized using secondary chemiluminescent detection (BrightStar BioDetect, Ambion, Austin, TX).
Integrity of each mRNAs was verified by northern blot as previously described (42).
Sucrose-gradient fractionation, polysome-associated RNA analysis, northern blot and RT–PCR
Sucrose-gradient fractionation was performed essentially as described. Extracts from HeLa cells were prepared by lysis at 4°C in extraction buffer (10 mM Tris–HCl, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% Nonidet-P40 and 500 U/ml RNAsin), and nuclei were removed by centrifugation (12 000g, 10 s, 4°C). The supernatant was supplemented with 20 mM dithiothreitol, 150 μg/ml cycloheximide, 665 μg/ml heparin and 1 mM phenylmethylsulfonyl fluoride and centrifuged (12 000g, 5 min, 4°C) to eliminate mitochondria. The supernatant was layered onto a 5 ml or a 10 ml linear sucrose gradient (15–40% sucrose [w/v] supplemented with 10 mM Tris–HCl, pH 7.5, 140 mM NaCl, 1.5 mM MgCl2, 10 mM dithiothreitol, 100 μg/ml cycloheximide, and 0.5 mg/ml heparin) and centrifuged in a SW50.1Ti or SW41Ti rotor (Beckman, Villepinte, France) for 2 h at 160000 g and at 4°C, without brake. Fractions of 300 μl were collected and digested with 100 μg proteinase K in 1% SDS and 10 mM EDTA (30 min, 37°C). RNAs were then recovered by phenol–chloroform–isoamyl alcohol extraction, followed by ethanol precipitation. Finally, the fractions containing the mRNA were precipitated with 2 M LiCl on ice at 4°C overnight. After centrifugation (12 000g, 15 min at 4°C), pellets were washed with 70% ethanol prestored at 20°C, air dried and resuspended in appropriate volumes of RNAse-free water.
RNAs were analyzed either by electrophoresis on denaturing 1.2% formaldehyde agarose gels and subsequent northern blotting or by cDNA amplification as previously described (43), using specific VEGF primers (RTEx4 sens, RT3′HA rev).
RESULTS
Previously, we have demonstrated that VEGF can be generated through both AUG initiated translation and cleavage of a larger L-VEGF precursor protein that is initiated from one of multiple upstream, in frame CUG codons (25). In order to detect differences between translational initiation at AUG and CUG codons for each VEGF mRNA splice variant, we constructed expression plasmids containing VEGF 121, VEGF 165 or VEGF 189 with mutated signal peptide sequences (40).
The VEGF 121 AUG shut-off requires the full-length mRNA sequence
Using this approach, we have demonstrated that translation initiation at a CUG is always efficient regardless of which splice variant is expressed, whereas initiation at the AUG depends upon which exons are present in the mRNA (40). VEGF 121 is expressed through initiation events using the CUG start codons, which only generate high molecular weight L-VEGF. In contrast, VEGF 189 and 165 splice variants lead to the production of both CUG and AUG-initiated proteins (Figure 2A).
To determine how the alternatively spliced VEGF sequence influences the initiation-codon choice, we fused the first exon of VEGF, containing the 5′UTR, in-frame AUG start codon and mutated signal peptide, with the CAT reporter gene and inserted at the 3′ end the sequence corresponding to exons 5 to 8 of VEGF 121, 165 or 189. Translation initiation still occurred at both CUG codons and the AUG codon, disregardless of the VEGF sequence fused at the 3′ end (Figure 2B). This shows that the mere presence of the VEGF 121 isoform specific primary sequence was not sufficient to reproduce the AUG blockade. We can conclude that the VEGF 5–8 alternative exons influence the choice of the initiation codon only in the context of the full length VEGF mRNA. Consequently, we hypothesized that the AUG shut-off in the VEGF 121 mRNA depends upon RNA structures that require continuity from the 5′UTR to the vicinity of the stop codon. These results highlight the role of the full VEGF mRNA sequence in this regulation and suggest that there is something specific within the VEGF 121 isoform sequence that inhibits AUG initiated translation.
Role of the 5′UTR in the VEGF 121 AUG shut-off mechanism
It was previously established that an alternative promoter is positioned within the human VEGF 5′UTR (15). The +1 transcription initiation site is located at +633 downstream from the classical start site. Furthermore, the internal promoter's insensitivity to hypoxia indicates that it is used independently of the main promoter region. Moreover, transcripts initiated from the internal promoter are of special interest because they cannot encode L-VEGF isoforms. In addition, these transcripts can be translated by either a cap-dependent or IRES-A-dependent mechanism from the AUG codon (15).
The role of the two 5′ sequences was investigated by comparing the expression of each splice variant fused to the different 5′UTRs. A deletion of most of the VEGF 5′UTR (24 remaining nucleotides) was used as a control (Figure 3, lanes 2). VEGF 165 and 189 AUG initiated form was expressed, disregardless of the length of the 5′UTR (Figure 3A), while VEGF 121 AUG initiated form was only detectable from the control construct (Figure 3A, VEGF 121, lane 2). The integrity of all mRNAs expressed after transfection was verified by northern blot (Supplementary data). Quantification of these mRNAs was determined by RNase protection assays (Figure 3). For each spliced variant, we can see that VEGF mRNAs are expressed at comparable levels, indicating that the lack of the 121 AUG initiated form (Figure 3A lanes 1 and 3) is due to a posttranscriptional regulation.
To examine this regulation in a more physiological context, minigene plasmids were constructed (see materials and methods). These minigene constructs should allow, after alternative splicing of the pre-mRNA, the expression of the different VEGF isoforms. While alternative splicing generating VEGF 189 was very inefficient in HeLa cells, results obtained with VEGF 121 and 165 parallel those obtained with the cDNA constructs (Figure 3B) and only the construct lacking a 5′UTR permitted expression of the VEGF 121 AUG initiated form (Figure 3B, lane 2). Since we have previously shown that the half-lives of the VEGF 121, 165, and 189 proteins are very similar (40), we investigated whether this regulation was exerted through a translational regulation mechanism and we examined the association of the different transcripts with polysomes. For transfections using full-length constructs (Figure 4, lane 1–3), mRNAs associated with polysomes were recovered. Identical results were obtained with constructs in which VEGF 165 and 189 transcripts began at the internal promoter (Figure 4, lanes 4, 5). The same construction with VEGF 121 clearly showed a very poor association of this mRNA to polysomes (Figure 4, lane 6). Because the northern blot is not discriminative enough to identify the different VEGF mRNA isoforms, we analyzed the VEGF mRNA distribution by RT–PCR after transfection with the IP minigene construct. Interestingly, VEGF 165 mRNA was present in fractions containing heavy polysomes, whereas the 121 encoding mRNA was found to be recovered only in the free mRNA, monosome and disome fractions (Figure 4, lane 7). These results corroborate those obtained with cDNA transfections and reinforce the previous finding showing that the AUG shut-off mechanism of VEGF 121 is independent of the splicing mechanism.
The minimal 5′ region involved in the control of the AUG usage contains a small ORF
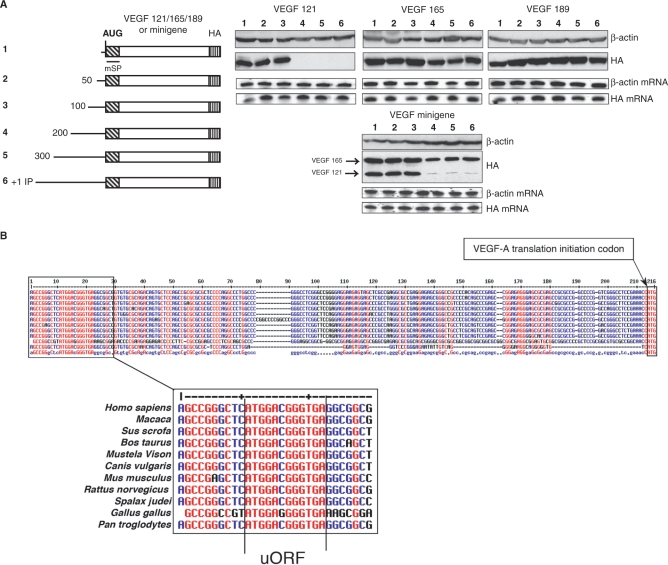
A deletion analysis was performed to locate the minimal sequence responsible for the AUG translational blockade of VEGF 121 starting with the region between position 633 (IP) and 24 nt upstream from the AUG (Figure 5A). It is clear that the 200 nt upstream from the AUG were sufficient to mediate the specific AUG blockade of VEGF 121 (Figure 5A, lanes 4) and the region between 100 and 200 was necessary for this effect (Figure 5A, lanes 3 versus 4). These results were confirmed by those obtained with the minigene constructs (Figure 5A).
Figure 5.
Identification of the minimal region involved in translation initiation control. (A) Left, schematic representation of the deletion within the 5′ leader performed in the IP constructs encoding the three VEGF isoforms and the minigene. Right, expression of AUG-initiated VEGF isoforms were analyzed by western immunoblotting using an anti-HA antibody. Anti-β-actin was used to control protein loading. RPA analyses were performed to normalize transfection with a vector specific probe to quantify mRNA from transfection and with a β-actin probe to control RNA quantity. (B) Partial alignment of the VEGF mRNA 5′ untanslated region of several species. Conserved nucleotides are shown in red. Main VEGF AUG translation initiation codons (on the right) are framed. The lower part shows a magnification of the region containing the uORF. The uORF, extremely well conserved among several species, is positioned in +1 frame relative to the VEGF ORF in human mRNA. This uORF would be initiated at uAUG (position 852) and be terminated at position 863, 175 nucleotides upstream from the VEGF AUG, and would produce a putative three amino-acid long polypeptide.
It is well documented that the 5'UTRs of vertebrate mRNAs contain a variety of features that affect the translation efficiency of the main coding sequence (44). These include the length and putative secondary structures of the 5'UTR, the sequence context of the initiation codon as well as the presence of upstream AUG codons (uAUGs). Interestingly, inspection of the human VEGF 5’UTR revealed the presence of a unique and short uORF precisely within the region between 200 and 100 nucleotides upstream of the AUG initiation codon. This uORF is highly conserved between species (Figure 5B) and begins at AUG 852, which is 186 nucleotides upstream from the main AUG (using the human sequence).
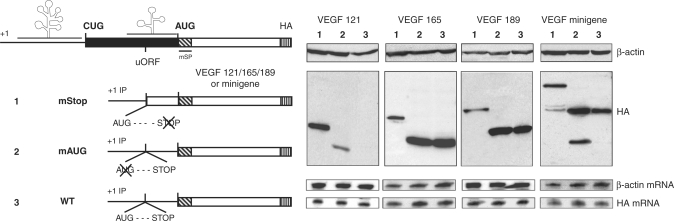
We first investigated whether this uORF was translated. Since it is impossible to visualize the encoded tripeptide, the uORF stop codon was mutated so that initiation at the uAUG would generate a 62 AA amino-terminally extended VEGF. This extended VEGF protein was always detected (Figure 6, lane 1) demonstrating that the uAUG was efficiently used whatever the splice variant. A consequence of this construction is the loss of the VEGF AUG 1038 initiated form.
Figure 6.
Role of the uORF in the translational initiation control. Left, schematic representation of construct with uORF mutation. The internal promoter beginning constructs encoding each VEGF isoform or the VEGF minigene either with wild type uORF (pIP121mSPHA, pIP165mSPHA, pIP189mSPHA and pIPminiVEGF numbered 3), a mutated uORF AUG (pIP121mORFmSPHA, pIP165mORFmSPHA, pIP18mORFmSPHA and pIPminiVEGFmORF numbered 2) or a mutated uORF stop codon, which made the uORF in frame with the VEGF-HA (pIPmSOmSP121HA, pIPmSOmSP165HA, pIPmSOmSP189HA and pIPminiVEGFmSO numbered 1). Right, expression of AUG-initiated VEGF isoforms and uORF were analyzed by western immunoblotting using an anti-HA antibody and anti-β-actin to control loading of protein. RPA analyses were performed to normalize transfection with a vector specific probe to quantify mRNA from transfection and with a β-actin probe to control RNA quantitation.
To gain an insight into the role of the uORF, we mutated the uAUG. This mutation increased AUG 1038 initiation translation of VEGF 121 isoform, but had no effect on the VEGF 165 or 189 expression level (Figure 6, lanes 2 and 3). Remarkably, results obtained with the minigene construct exactly parallel those achieved with the cDNA constructs (Figure 6, right panel).
Taken together, these data demonstrate that the uORF serves as a cis acting regulatory element for VEGF protein isoform expression.
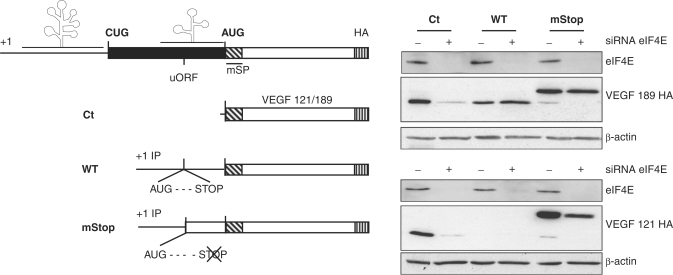
Translation of the uORF is mostly cap-independent
Since the uORF is located within IRES-A, we investigated whether the uAUG was translated by a cap- or IRES-dependent mechanism. In order to accomplish this, we performed a knockdown directed against the cap-binding protein eIF4E using a validated siRNA. As shown in Figure 7, the eIF4E knockdown inhibited most of the synthesis of the 121 or 189 control constructs (Ct) in which the IRES sequences were deleted. Remarkably, expression from the VEGF 189 AUG in a wild-type (Figure 7, WT) context was unaffected by the inhibition of cap-dependent translation indicating that the VEGF AUG is completely IRES-dependent. As expected, the VEGF 121 AUG initiated form remained undetectable (Figure 7, WT). Finally, we can see that the uAUG is principally cap-independent for both the 121 and 189 constructs (Figure 7, mStop). One can postulate that the uAUG is able to exhaust small ribosomal subunits recruited at the cap site, since modest cap-dependent initiation was detected downstream, at the VEGF AUG (Figure 7, mStop).
Figure 7.
Translation of the uORF after eIF4E knockdown. Left, schematic representation of construct with uORF mutation (described in the legend of Figure 6). Right, following eIF4E (+) or control (−) siRNA transfection, expression of AUG-initiated VEGF 121 and 189 isoforms constructs (schematized on the left) were analyzed using an anti-HA antibody and anti-β-actin to control loading of protein. Anti-eIF4E was used to verify knockdown efficiency. RPA analyses were performed to normalize transfection with a vector specific probe to quantify mRNA from transfection and with a β-actin probe to control RNA quantitation.
DISCUSSION
VEGF has been shown to be a key mediator of angiogenesis in diverse physiological and pathological processes such as embryogenesis, the female oestrus cycle, diabetic retinopathy and tumor development (1,45). The role of VEGF in developmental angiogenesis is emphasized by the finding that loss of a single VEGF allele (6,7), as well as modest VEGF overexpression (8) result in defective vascularization and early embryonic lethality.
Moreover, it has been demonstrated that a single nucleotide polymorphism in the VEGF gene (634C-G) results in IRES-B dysfunction and a 17% reduction in CUG initiated translation and expression of L-VEGF. This polymorphism was correlated with increased risk in motor neuron degeneration in amyotrophic lateral sclerosis (ALS) (46). Taken together, these findings clearly show that a small variation in the level of VEGF can induce deleterious phenotypes and underline the importance of VEGF expression regulation both during development and in the adult.
VEGF gene expression is controlled at many levels including transcription start sites (10,15), inducibility of promoters (11–13,15), mRNA stability through the binding of regulatory proteins to the 3′UTR (33–35,38), mRNA translation via IRES sequences located in the 5′UTR (15,28,29) and use of alternative initiation codons (25–27). These different control elements have been previously characterized and studied independently of each other. In this work, we have attempted to integrate several control elements by reconstituting messengers that possess the original +1 transcription start site, various 5′UTRs and the different alternatively spliced region (either as cDNAs or using the genomic sequence). By this approach, we have revealed another layer in the already complex translational control of VEGF expression: the role of a uORF.
Previously, we have demonstrated that differences in the exon content between VEGF 121 and VEGF 165/189 control AUG-initiated translation in the larger mRNA. This mechanism proceeds independently of the 3′UTR. Here, we first demonstrated that this control also involves sequences located in the 5′UTR, upstream of the AUG start codon and downstream of the internal promoter +1 start point (Figure 1). We next identified that an active uORF is located within this VEGF region, itself situated within the IRES-A responsible for this regulation.
Generally, uORFs impose a constitutive barrier to the scanning ribosome and reduce the number of ribosomes that gain access to the main AUG codon. In the case of VEGF, our data have established that the uORF is mostly translated by a cap-independent mechanism (Figure 7). Therefore, initiation at the main AUG could be either directed by the IRES or could be the consequence of reinitiation after IRES-dependent uORF translation. It has been established that reinitiation tends to be less efficient when the intercistronic distance is short or after translation of long uORFs. The size of the VEGF uORF (three codons) and the relatively long intercistronic region (175 nt) might, therefore, be predicted to have positive effects on reinitiation efficiency (47). However, our experimental data suggest that after uORF translation in the VEGF 121 mRNA, ribosomes could not reinitiate at the main AUG (Figures 2 and 3). This regulation could be explained by a modulation of uORF effects due to specific conditions involving trans-acting factors that function in concert with another portion of the messenger. This mode of action is exemplified by the case of Her-2 mRNA for which the 3′UTR can override translational repression mediated by a uORF within the 5′UTR (48). Specific proteins binding to a sequence element within the 3′UTR allow ribosome reinitiation at the main AUG after uORF translation. These results have defined a novel mechanism by which translational control of genes harboring a 5′ uORF can be modulated by elements located downstream. Because of the mechanistic resemblance between Her-2 and VEGF, one can hypothesizes that controlled reinitiation takes place during VEGF mRNA translation and that this control is inhibited in the 121 context. On the other hand, reinitiation probably only provides a minor contribution since the mutation of the uAUG in 165 or 189 mRNAs does not generate a significant change in the use of the main AUG (Figure 6).
Alternatively, uORFs translation could affect the local RNA structure. In the case of the arginine/lysine transporter (cat-1) mRNA, it has been demonstrated that induction of IRES activity requires the translation of a small uORF located within the IRES (49). The translation of the uORF unfolds an inhibitory structure in the mRNA leader creating an active IRES through RNA–RNA interactions between the 5′ end of the leader and downstream sequences (49), which leads to the stimulation of CAT-1 synthesis. Thus, uORFs have the potential to affect local RNA structure and could thereby affect mRNA translation. In the case of VEGF, one can hypothesize that uORF translation unfolds the IRES structure eliciting a conformational change. This modification only has a minor or transitory consequence for the 165 and 189 mRNAs, or when the VEGF ORF has been replaced (Figure 2B). Conversely, an inactive IRES conformation seems to be locked in the VEGF 121 context. It will be of interest to identify trans-acting factors [ITAFs, (50)] that could specifically bind the 165 and 189 mRNAs and stabilize the active structure of the IRES or, conversely, ITAFs that bind to the 121 form and lock the IRES in an inactive conformation as previously described for the FGF-2 IRES, which is silenced by p53 binding (51).
This hypothesis is also supported by the fact that the majority of VEGF 121 mRNA, initiated at the internal promoter, cosediments with free mRNAs or monosomes but not with polysomal fractions (Figure 4). In contrast, VEGF 165 and 189 mRNAs are distributed throughout the polysome's gradient, indicating their efficient translation (Figure 4). Cooperation of several mRNA segments might also explain the fact that this negative control could only be assessable in the context of the wild-type VEGF mRNA, and not when the VEGF ORF has been replaced by a reporter gene (Figure 2B).
As suggested by our experimental data, the uORF is not necessary for IRES formation but is involved in the specific AUG shut-off that exists within the VEGF 121 mRNA.
Our current inability to predict a priori conditions that alter uORF function means that uORFs, which appear to be constitutive inhibitors, may turn out to be regulated by specific sequences, trans-acting factors or conditions that have not yet been identified.
The VEGF uORF seems to act as a cis regulatory element that controls VEGF protein isoform expression. The important biological properties that distinguish the VEGF isoforms are their heparin and heparan sulfate binding affinities. In contrast to VEGF 165 and VEGF 189, VEGF 121 does not bind to the extracellular matrix and its consequent diffusibility endows it with a more potent angiogenic activity (52). Moreover, it has been shown by comparing differential VEGF expression patterns in tumors versus normal cells that there is a shift in the percentage of VEGF isoforms that are expressed, with a significant increase in VEGF 121 associated with a decrease of VEGF 165 and 189 in neoplastic cells (53).
In pathological conditions, the hypoxic responsive promoter is preferentially activated leading to the expression of a single mRNA that permits IRES-dependent translation initiation at both CUG codons and the AUG start codon (Figure 1). We have shown that VEGF 121 is mainly expressed through initiation events utilizing the CUG start codons (L-VEGF) followed by maturation of this amino-terminally extended protein (Figure 1). Hence, transcriptional activation of this VEGF promoter would lead to an increased expression of the most diffusible VEGF isoform. Conversely, since the hypoxic responsive promoter is weakly active during normoxia, the translational regulation described in this article could represent a mechanism that rigorously inhibits VEGF 121 synthesis in noninduced conditions and particularly when the internal promoter is active.
This work provides additional evidence that the very high complexity required for the control of VEGF gene expression necessitates investigation of several control elements simultaneously (such as uORF, alternatively spliced sequences, alternative start codons and IRESs). VEGF, which is regulated at every conceivable stage of gene expression, represents a paradigm for gene regulation. Regarding this complexity, we are still a long way from understanding how events both inside and outside cells work together to control and coordinate VEGF expression and activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by a grant from the Ligue Nationale Contre le Cancer as an Equipe Labellisée, from the Comité de Haute-Garonne de la Ligue Contre le Cancer and from an ACI Canceropôle. Amandine Bastide receives fellowships from the ‘Ministère de l’Education Nationale et de la Recherche’ and from the ‘Association pour la Recherche contre le Cancer (ARC)’. We thank Dr A.C. Prats and Dr Jason Scott Iacovoni for critical reading of the article. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N. The role of VEGF in the regulation of physiological and pathological angiogenesis. EXS. 2005;94:209–231. doi: 10.1007/3-7643-7311-3_15. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara N. Role of vascular endothelial growth factor in the regulation of angiogenesis. Kidney Int. 1999;56:794–814. doi: 10.1046/j.1523-1755.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- 4.Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat. Rev. Cancer. 2002;2:795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- 5.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 6.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 7.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O'Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 8.Miquerol L, Langille BL, Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127:3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- 9.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr. Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 10.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy AP, Levy NS, Iliopoulos O, Jiang C, Kaplin WG, Jr, Goldberg MA. Regulation of vascular endothelial growth factor by hypoxia and its modulation by the von Hippel-Lindau tumor suppressor gene. Kidney Int. 1997;51:575–578. doi: 10.1038/ki.1997.82. [DOI] [PubMed] [Google Scholar]
- 12.Milanini J, Vinals F, Pouyssegur J, Pages G. p42/p44 MAP kinase module plays a key role in the transcriptional regulation of the vascular endothelial growth factor gene in fibroblasts. J. Biol. Chem. 1998;273:18165–18172. doi: 10.1074/jbc.273.29.18165. [DOI] [PubMed] [Google Scholar]
- 13.Pages G, Pouyssegur J. Transcriptional regulation of the vascular endothelial growth factor gene–a concert of activating factors. Cardiovasc. Res. 2005;65:564–573. doi: 10.1016/j.cardiores.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat. Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 15.Akiri G, Nahari D, Finkelstein Y, Le SY, Elroy-Stein O, Levi BZ. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- 16.Bates DO, Cui TG, Doughty JM, Winkler M, Sugiono M, Shields JD, Peat D, Gillatt D, Harper SJ. VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res. 2002;62:4123–4131. [PubMed] [Google Scholar]
- 17.Houck KA, Ferrara N, Winer J, Cachianes G, Li B, Leung DW. The vascular endothelial growth factor family: identification of a fourth molecular species and characterization of alternative splicing of RNA. Mol. Endocrinol. 1991;5:1806–1814. doi: 10.1210/mend-5-12-1806. [DOI] [PubMed] [Google Scholar]
- 18.Jingjing L, Xue Y, Agarwal N, Roque RS. Human Muller cells express VEGF183, a novel spliced variant of vascular endothelial growth factor. Invest. Ophthalmol. Vis. Sci. 1999;40:752–759. [PubMed] [Google Scholar]
- 19.Lange T, Guttmann-Raviv N, Baruch L, Machluf M, Neufeld G. VEGF162, a new heparin-binding vascular endothelial growth factor splice form that is expressed in transformed human cells. J. Biol. Chem. 2003;278:17164–17169. doi: 10.1074/jbc.M212224200. [DOI] [PubMed] [Google Scholar]
- 20.Poltorak Z, Cohen T, Sivan R, Kandelis Y, Spira G, Vlodavsky I, Keshet E, Neufeld G. VEGF145, a secreted vascular endothelial growth factor isoform that binds to extracellular matrix. J. Biol. Chem. 1997;272:7151–7158. doi: 10.1074/jbc.272.11.7151. [DOI] [PubMed] [Google Scholar]
- 21.Tischer E, Mitchell R, Hartman T, Silva M, Gospodarowicz D, Fiddes JC, Abraham JA. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J. Biol. Chem. 1991;266:11947–11954. [PubMed] [Google Scholar]
- 22.Whittle C, Gillespie K, Harrison R, Mathieson PW, Harper SJ. Heterogeneous vascular endothelial growth factor (VEGF) isoform mRNA and receptor mRNA expression in human glomeruli, and the identification of VEGF148 mRNA, a novel truncated splice variant. Clin. Sci. 1999;97:303–312. [PubMed] [Google Scholar]
- 23.Houck KA, Leung DW, Rowland AM, Winer J, Ferrara N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J. Biol. Chem. 1992;267:26031–26037. [PubMed] [Google Scholar]
- 24.Grunstein J, Masbad JJ, Hickey R, Giordano F, Johnson RS. Isoforms of vascular endothelial growth factor act in a coordinate fashion To recruit and expand tumor vasculature. Mol. Cell Biol. 2000;20:7282–7291. doi: 10.1128/mcb.20.19.7282-7291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huez I, Bornes S, Bresson D, Creancier L, Prats H. New vascular endothelial growth factor isoform generated by internal ribosome entry site-driven CUG translation initiation. Mol. Endocrinol. 2001;15:2197–2210. doi: 10.1210/mend.15.12.0738. [DOI] [PubMed] [Google Scholar]
- 26.Meiron M, Anunu R, Scheinman EJ, Hashmueli S, Levi BZ. New isoforms of VEGF are translated from alternative initiation CUG codons located in its 5′UTR. Biochem. Biophys. Res. Commun. 2001;282:1053–1060. doi: 10.1006/bbrc.2001.4684. [DOI] [PubMed] [Google Scholar]
- 27.Tee MK, Jaffe RB. A precursor form of vascular endothelial growth factor arises by initiation from an upstream in-frame CUG codon. Biochem. J. 2001;359:219–226. doi: 10.1042/0264-6021:3590219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huez I, Creancier L, Audigier S, Gensac MC, Prats AC, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller DL, Dibbens JA, Damert A, Risau W, Vadas MA, Goodall GJ. The vascular endothelial growth factor mRNA contains an internal ribosome entry site. FEBS Lett. 1998;434:417–420. doi: 10.1016/s0014-5793(98)01025-4. [DOI] [PubMed] [Google Scholar]
- 30.Bornes S, Prado-Lourenco L, Bastide A, Zanibellato C, Iacovoni JS, Lacazette E, Prats AC, Touriol C, Prats H. Translational induction of VEGF internal ribosome entry site elements during the early response to ischemic stress. Circ. Res. 2007;100:305–308. doi: 10.1161/01.RES.0000258873.08041.c9. [DOI] [PubMed] [Google Scholar]
- 31.Rosenbaum-Dekel Y, Fuchs A, Yakirevich E, Azriel A, Mazareb S, Resnick MB, Levi BZ. Nuclear localization of long-VEGF is associated with hypoxia and tumor angiogenesis. Biochem. Biophys. Res. Commun. 2005;332:271–278. doi: 10.1016/j.bbrc.2005.04.123. [DOI] [PubMed] [Google Scholar]
- 32.Levy NS, Goldberg MA, Levy AP. Sequencing of the human vascular endothelial growth factor (VEGF) 3′ untranslated region (UTR): conservation of five hypoxia-inducible RNA-protein binding sites. Biochim. Biophys. Acta. 1997;1352:167–173. doi: 10.1016/s0167-4781(97)00052-3. [DOI] [PubMed] [Google Scholar]
- 33.Dibbens JA, Miller DL, Damert A, Risau W, Vadas MA, Goodall GJ. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol. Biol. Cell. 1999;10:907–919. doi: 10.1091/mbc.10.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claffey KP, Shih SC, Mullen A, Dziennis S, Cusick JL, Abrams KR, Lee SW, Detmar M. Identification of a human VPF/VEGF 3′ untranslated region mediating hypoxia-induced mRNA stability. Mol. Biol. Cell. 1998;9:469–481. doi: 10.1091/mbc.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 36.Onesto C, Berra E, Grepin R, Pages G. Poly(A)-binding protein-interacting protein 2, a strong regulator of vascular endothelial growth factor mRNA. J. Biol. Chem. 2004;279:34217–34226. doi: 10.1074/jbc.M400219200. [DOI] [PubMed] [Google Scholar]
- 37.Shih SC, Mullen A, Abrams K, Mukhopadhyay D, Claffey KP. Role of protein kinase C isoforms in phorbol ester-induced vascular endothelial growth factor expression in human glioblastoma cells. J. Biol. Chem. 1999;274:15407–15414. doi: 10.1074/jbc.274.22.15407. [DOI] [PubMed] [Google Scholar]
- 38.Ciais D, Cherradi N, Bailly S, Grenier E, Berra E, Pouyssegur J, Lamarre J, Feige JJ. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- 39.Coles LS, Bartley MA, Bert A, Hunter J, Polyak S, Diamond P, Vadas MA, Goodall GJ. A multi-protein complex containing cold shock domain (Y-box) and polypyrimidine tract binding proteins forms on the vascular endothelial growth factor mRNA. Potential role in mRNA stabilization. Eur. J. Biochem. 2004;271:648–660. doi: 10.1111/j.1432-1033.2003.03968.x. [DOI] [PubMed] [Google Scholar]
- 40.Bornes S, Boulard M, Hieblot C, Zanibellato C, Iacovoni JS, Prats H, Touriol C. Control of the vascular endothelial growth factor internal ribosome entry site (IRES) activity and translation initiation by alternatively spliced coding sequences. J. Biol. Chem. 2004;279:18717–18726. doi: 10.1074/jbc.M308410200. [DOI] [PubMed] [Google Scholar]
- 41.Armstrong F, Lamant L, Hieblot C, Delsol G, Touriol C. TPM3-ALK expression induces changes in cytoskeleton organisation and confers higher metastatic capacities than other ALK fusion proteins. Eur. J. Cancer. 2007;43:640–646. doi: 10.1016/j.ejca.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Touriol C, Roussigne M, Gensac MC, Prats H, Prats AC. Alternative translation initiation of human fibroblast growth factor 2 mRNA controlled by its 3′-untranslated region involves a Poly(A) switch and a translational enhancer. J. Biol. Chem. 2000;275:19361–19367. doi: 10.1074/jbc.M908431199. [DOI] [PubMed] [Google Scholar]
- 43.Armstrong F, Duplantier MM, Trempat P, Hieblot C, Lamant L, Espinos E, Racaud-Sultan C, Allouche M, Campo E, Delsol G, et al. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene. 2004;23:6071–6082. doi: 10.1038/sj.onc.1207813. [DOI] [PubMed] [Google Scholar]
- 44.Sonenberg N. mRNA translation: influence of the 5′ and 3′ untranslated regions. Curr. Opin. Genet. Dev. 1994;4:310–315. doi: 10.1016/s0959-437x(05)80059-0. [DOI] [PubMed] [Google Scholar]
- 45.Carmeliet P. Angiogenesis in health and disease. Nat. Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 46.Lambrechts D, Storkebaum E, Morimoto M, Del-Favero J, Desmet F, Marklund SL, Wyns S, Thijs V, Andersson J, van Marion I, et al. VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat. Genet. 2003;34:383–394. doi: 10.1038/ng1211. [DOI] [PubMed] [Google Scholar]
- 47.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehta A, Trotta CR, Peltz SW. Derepression of the Her-2 uORF is mediated by a novel post-transcriptional control mechanism in cancer cells. Genes Dev. 2006;20:939–953. doi: 10.1101/gad.1388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, Zhou L, Snider MD, Scheuner D, Kaufman RJ, et al. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- 50.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2007;8:1033–1035. doi: 10.1038/sj.onc.1210777. [DOI] [PubMed] [Google Scholar]
- 51.Prats AC, Prats H. Translational control of gene expression: role of IRESs and consequences for cell transformation and angiogenesis. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:367–413. doi: 10.1016/s0079-6603(02)72075-8. [DOI] [PubMed] [Google Scholar]
- 52.Zhang HT, Scott PA, Morbidelli L, Peak S, Moore J, Turley H, Harris AL, Ziche M, Bicknell R. The 121 amino acid isoform of vascular endothelial growth factor is more strongly tumorigenic than other splice variants in vivo. Br. J. Cancer. 2000;83:63–68. doi: 10.1054/bjoc.2000.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Catena R, Muniz-Medina V, Moralejo B, Javierre B, Best CJ, Emmert-Buck MR, Green JE, Baker CC, Calvo A. Increased expression of VEGF121/VEGF165-189 ratio results in a significant enhancement of human prostate tumor angiogenesis. Int. J. Cancer. 2007;120:2096–2109. doi: 10.1002/ijc.22461. [DOI] [PubMed] [Google Scholar]